Abstract
The OTT-MAL/RBM15-MKL1 fusion protein is the result of the recurrent translocation t(1;22) in acute megakaryocytic leukemia in infants. How it contributes to the malignancy is unknown. The 3′ fusion partner, MAL/MKL1/MRTF-A, is a transcriptional coactivator of serum response factor (SRF). MAL plays a key role in regulated gene expression depending on Rho family GTPases and G-actin. Here we demonstrate that OTT-MAL is a constitutive activator of SRF and target gene expression. This requires the SRF-binding motif and the MAL-derived transactivation domain. OTT-MAL localizes to the nucleus and is not regulated by upstream signaling. OTT-MAL deregulation reflects its independence from control by G-actin, which fails to interact with OTT-MAL in coimmunoprecipitation experiments. Regulation cannot be restored by reintroduction of the entire MAL N terminus into the fusion protein. OTT-MAL also caused a delayed induction of the MAL-independent, ternary complex factor-dependent target genes c-fos and egr-1 and the mitogen-activated protein kinase/Erk pathway. With testing in heterologous tissue culture systems, however, we observed considerable antiproliferative effects of OTT-MAL. Our data suggest that the deregulated activation of MAL-dependent and -independent promoters results in tissue-specific functions of OTT-MAL.
The OTT-MAL/RBM15-MKL1 fusion protein is the product of a balanced translocation t(1;22)(p13;q13) in infant acute megakaryocytic leukemia (AMKL; FAB M7) (3, 11-13). At the time of their discovery, little was known about either gene, hence the names OTT (one twenty-two) and RBM15 (RNA-binding motif protein 15) for the 5′ sequences and MAL (megakaryocytic acute leukemia) and MKL-1 (megakaryoblastic leukemia) for the 3′ sequences. The breakpoint in the first intron of OTT and the third intron (variant translocation) or fourth intron (common translocation) of MAL leaves nearly the full-length (f.l.) coding region of both proteins intact (11-1, 15).
OTT encodes a protein containing three RNA recognition motifs (RRM) and a spen paralog and ortholog C-terminal (SPOC) domain. It belongs to the Spen family of proteins, with OTT, MINT/SHARP, and OTT3 being the three known mammalian orthologs of the Drosophila spen (split ends) and spenito genes (6-8). The RRM motifs are thought to bind to nucleic acids (16, 32), whereas the highly conserved SPOC domain, at least of SHARP, interacts with SMRT and NCoR corepressor complexes (2, 6, 25). OTT, as well as MINT/SHARP and Drosophila spen, orchestrates Notch-regulated transcription (8-10, 17, 25). Spen family members were also found in the spliceosome and are implicated in mRNA splicing and export (6, 32). Deficiency of either OTT or MINT in mice causes embryonic lethality around embryonic day 9.5 or 13.5, respectively, demonstrating that they are essential genes with nonredundant functions (9, 22).
The fusion partner MAL (synonyms, MKL1 and MRTF-A) is a potent transcriptional coactivator of the myocardin-related transcription factor (MRTF) family, which is implicated in gene expression by serum response factor (SRF) (reviewed in reference 21). MAL is required for the activation of a subset of SRF target genes which are independent of mitogen-activated protein kinase (MAPK)-regulated ternary complex factors (4, 15). A variant isoform of MAL, BSAC, was identified as a suppressor of tumor necrosis factor-induced cell death (24). All MRTF family members contain N-terminal RPEL motifs, a basic box harboring the SRF interaction motif, a glutamine-rich stretch, a SAP (SAF-A/B, acinus, PIAS) domain, a leucine zipper motif required for dimerization, and a strong C-terminal transactivation domain (11, 13, 15, 29, 31). The ubiquitously expressed MAL protein is regulated by serum and Rho family GTPases, which change the intracellular actin dynamics (15, 26). MAL responds to these changes by altered binding to G-actin via its RPEL motifs (15, 19). G-actin regulates MAL activity in three ways; it inhibits nuclear import of MAL, enhances nuclear export, and represses transcriptional activation of SRF targets (28). Treatment with the actin-binding drug latrunculin B or ectopic expression of actin and nonpolymerizable actin mutants represses MAL nuclear accumulation and activation of SRF (20, 26). Conversely, stimulation by serum or cytochalasin D, which dissociates the actin-MAL complex, potentiates SRF activation by MAL (15, 28).
The lack of material, due to associated myelofibrosis and the young age of patients, has precluded analysis of tissues or cell lines from patients expressing OTT-MAL. However, biochemical and functional analyses can readily be undertaken with nonmyeloid cells. Comparing the properties of each partner and the fusion product may help identify functional and mechanistic alterations. A previous study suggests that the fusion protein possesses an enhanced ability to activate SRF, although the molecular basis for this has remained unclear (4). Here we show that OTT-MAL constitutively activates SRF target gene expression even in the absence of stimuli. Neither positive nor negative upstream signals affect OTT-MAL activity, showing that the fusion protein is functionally deregulated. Moreover, OTT-MAL aberrantly activates ternary complex factor (TCF)-dependent gene expression. Rather than enhancing cell growth, OTT-MAL had an antiproliferative effect, suggesting complex and tissue-specific functions of OTT-MAL.
MATERIALS AND METHODS
Plasmids, cells, and reagents.
Details are available upon request. The sequence of human OTT-MAL was cloned into pcDNA3 or pEF harboring an N-terminal hemagglutinin (HA) tag or a C-terminal Flag tag. For deletions, see Fig. 2A. All constructs were verified by sequencing. OTT-123MAL was generated by cloning the amplified N-terminal murine MAL (f.l.) sequence (Leu-1 to Arg-122) into the Xba site of human OTT-MAL. OTT-MAL YY and MAL YY, harboring alanines instead of tyrosines in the SRF-binding motif LKYHQYI (31), were cloned into pCMX-HA. The SRF reporter (p3D.A-Luc) and plasmids pRL-TK, pMLV-LacZ, pSRF-VP16, pEF-C3, pEF-Flag-Actin(s), pEF-Flag-NLS-Actin(s), and pEF-MAL (f.l.) have been described previously (15, 20). NIH 3T3 and NIH 3D.A-FosHA cells (1) were cultivated in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (Gibco). Mo7e cells were grown in RPMI medium supplemented with 10% fetal calf serum (FCS) and 20% conditioned medium as a source of interleukin-3. UT7/mpl cells have been described elsewhere (18). For generation of inducible cell lines, HEK293-TR cells (Invitrogen) were transfected with HA-OTT-MAL or HA-MAL subcloned into pcDNA4-TO (Invitrogen) and selected by 2.5 μg/ml blasticidin (Calbiochem) and 500 μg/ml phleomycin (Zeocin; Invitrogen). Latrunculin B and cytochalasin D were from Calbiochem.
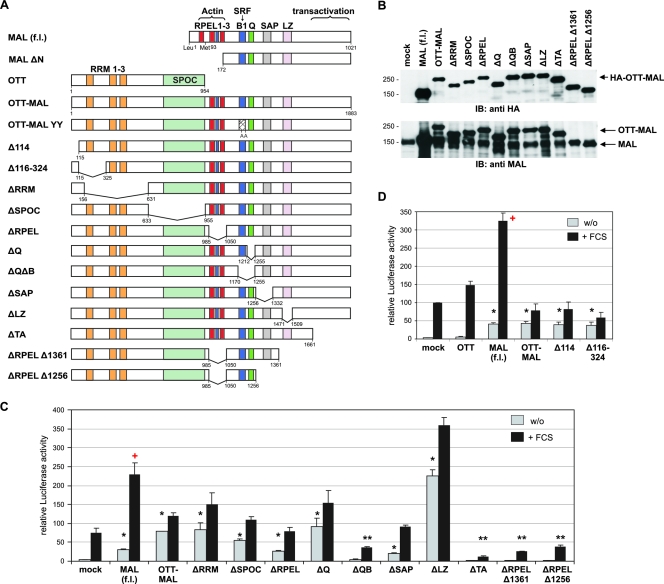
FIG. 2.
OTT-MAL activates SRF through the MAL transactivation domain. (A) Schematic drawing of the OTT-MAL fusion protein and the mutant constructs used in this study. LZ, leucine zipper. (B) Expression levels of MAL and OTT-MAL in transiently transfected NIH 3T3 cells. Total lysates were immunoblotted (IB) with anti-HA (upper panel) or anti-MAL (lower panel) antibodies. The values on the left are molecular sizes in kilodaltons. (C) Deletions in OTT-MAL covering the C-terminal domain block the SRF luciferase reporter activation in NIH 3T3 cells transiently transfected with 50 ng of the indicated constructs. (D) N-terminal deletions in OTT-MAL do not abrogate activation or restore synergistic regulation by serum. SRF luciferase reporter activity was analyzed following serum starvation overnight and normalized to Renilla luciferase, as before. Error bars indicate the SEM (n = 3). Single asterisks indicate significant activation (P < 0.05), double asterisks indicate significant repression (P < 0.05), and the plus sign indicates synergy with FCS (P < 0.02, according to an unpaired Student t test). w/o, without FCS.
Reporter assays, immunoprecipitations, and Western blotting.
Transfections of NIH 3T3 cells were carried out with Lipofectamine (Invitrogen) according to the manufacturer's protocols, as described previously (20). For luciferase assays, 35,000 cells/1-cm-diameter dish (12-well plate) were transfected with 15 ng p3DA-Luc, 40 ng pRL-TK, and 50 ng pMLV-LacZ together with the indicated amounts of plasmids in a total of 500 ng DNA. For UT7 and Mo7e cells, 5 ×106 to 8 ×106 cells were electroporated at 250 V with 2.5 to 10 μg reporter and 5 to 10 μg of OTT-MAL expression plasmids. Luciferase activity was measured with a dual-luciferase assay kit (Promega) and normalized to either pRL-TK luciferase (after 1 day) or pMLV-LacZ activity (after 2 days), as indicated. Figures show percentages of induction compared to SRF-VP16 (80 ng) or n-fold induction compared to the unstimulated control. Error bars usually indicate the standard error of the mean (SEM) of three independent experiments. Statistical analysis was done by unpaired Student t test. Statements of synergistic effects upon simultaneous stimulation were statistically tested as described previously (30).
Immunoprecipitation of actin-MAL complexes was done as described previously (19). HEK293 cells, 4 × 106/10-cm-diameter dish, were transfected with 3 μg of pEF-Flag-actin constructs by using Lipofectamine. The following day, cells were further cultivated with 0.5% FCS and 1 μg/ml doxycycline for 24 h. Flag-tagged actins were precipitated with M2-agarose (Sigma), and proteins were blotted with anti-HA antibody-peroxidase conjugate (3F10; Roche) or anti-Flag antibody-peroxidase conjugate (M2; Sigma). For visualizing proteins in radioimmunoprecipitation assay lysates, anti-phospho-Erk (1:1,000; Cell Signaling), anti-pan-Erk (1:1,000; Transduction Laboratories), antitubulin (1:10,000; Sigma), and rabbit anti-MAL (1:500) antibodies were used subsequent to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting according to standard protocols.
Immunofluorescence microscopy.
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and blocked with 10% FCS-1% gelatin-0.05% Triton X-100 in phosphate-buffered saline. Staining conditions were as follows: anti-Flag antibody (rabbit; Sigma-Aldrich), 1:100; rhodamine-phalloidin (Molecular Probes), 1:50; anti-HA antibody (mouse; Babco), 1:500; Alexa Fluor 488- or Alexa Fluor 546-conjugated anti-mouse antibody (immunoglobulin G [heavy and light chains]; Invitrogen), 1:1,000; tetramethyl rhodamine isocyanate- or fluorescein isothiocyanate-conjugated anti-rabbit antibody (Dako Cytomation), 1:40. Microscopy was performed with a Zeiss Axioplan 2 and a 63×, numerical aperture 1.4 oil immersion objective fitted with appropriate filters (Chroma). Pictures were taken with a cooled monochrome SPOT RT charge-coupled device camera (Diagnostic Instruments) with MetaVue software (Universal Imaging), and images were processed with Photoshop (Adobe Systems). Growth curves were determined in triplicate with a cytometer (Beckman-Coulter).
Quantitative real-time (RT) PCR.
RNA preparation (Qiagen) and first-strand cDNA synthesis (ABgene) were done according to the manufacturers' protocols. For cDNA synthesis, 1 μg of RNA and anchored oligo(dT) primers were used. For cDNA quantitation, 1/40 of the reverse transcription reaction mixture was mixed with gene-specific primers (0.5 μM), MgCl2 (3 mM), and LightCycler FastStart DNA Master Sybr green I mix (1.5 μl; Roche) to a total volume of 15.5 μl. The primers used for quantitative RT-PCR are as follows: alas1, CTGCAAAGATCTGACCCCTC (forward) and CCTCATCCACGAAGGTGATT (reverse); acta2, CGGTGCTGTCTCTCTATGCC (forward) and AGCAGTAGTAACGAAGGAATAGCCA (reverse); c-fos, TAGCCTCTCTTACTACCACTCACC (forward) and GAATGAAGTTGGCACTGGAG (reverse); egr-1, ACCTGACCGCAGAGTCTTTTC (forward) and GCCAGTATAGGTGATGGGGG (reverse). The PCR was carried out on a LightCycler instrument (Roche) according to the manufacturer's instructions. Calculation was done by the ΔΔCt method.
RESULTS
OTT-MAL constitutively activates SRF.
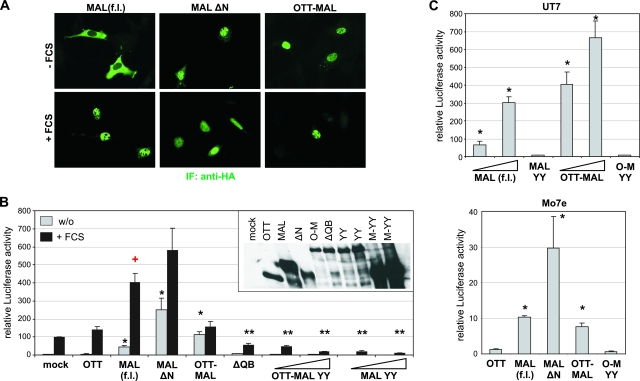
We have previously shown that MAL (synonyms, MRTF-A and MKL-1) acts as a serum-regulated transcriptional coactivator for SRF and accumulates in the nucleus upon signaling (15). We therefore investigated the localization and activity of the fusion protein OTT-MAL (synonym, RBM15-MKL1) resulting from the common (1, 22) translocation in human AML M7. When expressed in NIH 3T3 fibroblasts, OTT-MAL was found almost exclusively in the nucleus (Fig. 1A). Constitutive nuclear accumulation was also shown by MAL ΔN, which lacks the regulatory RPEL domain. In contrast, MAL (f.l.) was predominantly cytoplasmic in serum-starved cells. This demonstrated that regulation of nuclear shuttling is severely disrupted in OTT-MAL compared to MAL (f.l.).
FIG. 1.
OTT-MAL is constitutively activated. (A) Intracellular localization of OTT-MAL in comparison with MAL (f.l.) and MAL ΔN. NIH 3T3 cells transfected with HA-tagged MAL or OTT-MAL were stimulated for 1 h with 15% serum or left untreated and subjected to staining with anti-HA antibodies (green). IF, immunofluorescence. (B) OTT-MAL is transcriptionally active and requires binding to SRF. NIH 3T3 cells were transfected with SRF reporter 3D.A-Luc, pRL-tk, and the indicated constructs. Fifty or 150 ng of OTT-MAL YY and MAL YY plasmid was transfected, in which the two tyrosine residues required for SRF binding were mutated to alanine (31). Following serum starvation overnight, cells were stimulated with 15% serum for 7 h (as indicated) prior to lysis. Shown is the mean relative luciferase activity, normalized to Renilla luciferase, of three independent experiments. The inset shows the expression levels of the transfected constructs obtained by blotting with anti-HA antibody. Only MAL (f.l.) shows synergistic activation together with serum stimulation (P < 0.01; indicated by the red plus sign). w/o, without FCS. (C) SRF activation by OTT-MAL in UT7 and Mo7e human megakaryocytic cells. Cells were transfected by electroporation with the indicated constructs. Shown is the mean relative luciferase activity, normalized to Renilla luciferase, after 1 day (Mo7e) or LacZ activity after 2 days (UT7). Error bars indicate the SEM of three independent experiments. Single asterisks indicate significant activation (P < 0.05), and double asterisks indicate significant repression (P < 0.05), according to an unpaired Student t test.
We then tested how OTT-MAL affects SRF activity. OTT-MAL activated an SRF-dependent luciferase reporter construct comparably to serum stimulation (Fig. 1B). In contrast, expression of the OTT sequences alone had no effect. Unlike MAL (f.l.), however, OTT-MAL showed only slight additive but not synergistic SRF activation when used for costimulation with serum (P < 0.05; for a statistical test of synergism versus additivity, see reference 30). This suggests that OTT-MAL activity is not potentiated by serum-induced signaling.
The N-terminal RPEL domain was previously shown to mediate nucleocytoplasmic shuttling and regulation (15, 28). While the OTT-MAL rearrangement retains two of the three RPEL motifs, its behavior is similar to that of MAL ΔN, which lacks all RPEL motifs; MAL ΔN also activates SRF in a nonsynergistic manner with serum (Fig. 1B). However, OTT-MAL activated the reporter in starved cells somewhat less effectively than MAL ΔN, possibly reflecting its lower expression level (Fig. 1B, inset). In contrast, the basal activity of MAL (f.l.) which is overexpressed severalfold is considerably lower.
To investigate whether OTT-MAL requires binding to SRF, we utilized OTT-MAL ΔQB, which harbors a deletion including the SRF-binding domain. ΔQB did not activate SRF in the reporter assay (Fig. 1B). To gain more detailed insights into the role of SRF binding of OTT-MAL, we mutated the tyrosines in the SRF-binding motif LKYHQYI (31) to alanines. Mutation of the tyrosine residues in either MAL or OTT-MAL abrogated SRF activation and caused dominant negative effects on serum stimulation. This demonstrates that OTT-MAL activates SRF in a manner which depends on the known SRF interaction surface.
Whereas we mainly analyzed OTT-MAL in the best-understood signaling context of fibroblasts to allow comparison with MAL, we also tested megakaryocytic cells, in which the OTT-MAL translocation is linked to AML. Following transfection of the UT7 and Mo7e cell lines by electroporation, we observed an SRF reporter activation by MAL and OTT-MAL comparable to that in fibroblasts (Fig. 1C). Although the upstream regulation of MAL in hematopoietic cells is unknown, activation by OTT-MAL again required the SRF-binding domain. This result suggests that no fundamental differences exist in megakaryocytic cells regarding SRF activation by OTT-MAL.
OTT-MAL requires the MAL activation domain.
To analyze which domains are essential for OTT-MAL activity, we tested the deletion mutant constructs depicted in Fig. 2A. Protein expression levels are shown in Fig. 2B. A complete loss of constitutive SRF activation was found in mutant constructs lacking the C terminus, demonstrating that the MAL-derived transcriptional activation domain is required for SRF induction (Fig. 2C). Moreover, these constructs significantly repressed SRF activation by serum stimulation. A similar repression was observed for ΔQB, as before. This suggests that loss of either SRF binding or transactivation results in a dominant negative protein which sequesters endogenous MAL and SRF in inactive complexes, respectively.
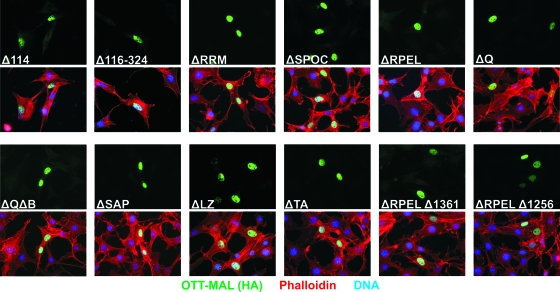
Vice versa, conserved regions of OTT, such as the RRM and the SPOC domain, were dispensable for constitutive transactivation (Fig. 2C). Similar results were obtained with HeLa cells (T. Mercher, D. Bluteau, and O. A. Bernard, unpublished data). Further deletions at the N terminus, covering two predicted N-terminal nuclear localization signals (NLSs) or the first RRM, also failed to abolish reporter activation (Fig. 2D). This indicates that the constitutive activity of OTT-MAL cannot be assigned to a specific single region of the OTT protein sequence. Furthermore, none of the OTT-MAL constructs showed synergistic SRF activation (P < 0.05) when used for costimulation with serum, in contrast to MAL (f.l.) (Fig. 2C and D). When analyzed for intracellular localization, all OTT-MAL mutant proteins were found almost exclusively in the nucleus, even in the absence of serum (Fig. 3). This demonstrates that proper regulation of transcriptional activity and nuclear shuttling cannot be restored by deleting a single region. Together, our data suggest that either multiple domains or steric hindrance is responsible for the deregulated activity of OTT-MAL.
FIG. 3.
Intracellular localization of OTT-MAL deletions in the absence of serum. NIH 3T3 cells were transfected with the HA-tagged OTT-MAL deletion mutant constructs indicated, and serum-starved cells were stained with anti-HA antibodies and phalloidin on the following day. Shown are immunofluorescence micrographs for OTT-MAL (green, upper panels) and merged images (lower panels). Nuclei were stained blue with Hoechst 33258.
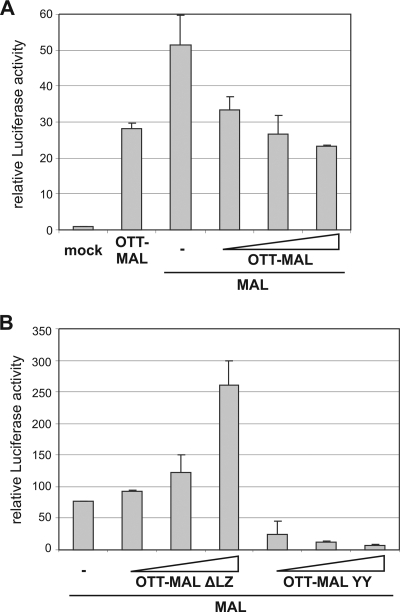
Surprisingly, deletion of the leucine zipper motif of OTT-MAL, which facilitates dimerization in MAL (15), resulted in enhanced activation of the reporter (Fig. 2C). This does not merely reflect increased protein amounts, since the expression of ΔLZ is comparable to that of wild-type OTT-MAL and endogenous MAL (Fig. 2B). Rather, it suggested that the leucine zipper limits the transactivating potential of OTT-MAL, possibly by dimerization with regulated, endogenous MAL. To functionally test the potential heterodimerization, we transfected MAL together with OTT-MAL into HeLa cells. Increasing amounts of OTT-MAL reduced the MAL-mediated reporter activation (Fig. 4A). Deletion of the leucine zipper in OTT-MAL abrogated this negative interference with MAL and restored a dose-dependent additive activation of the SRF reporter (Fig. 4B). In contrast, the SRF binding-deficient mutant OTT-MAL YY effectively prevented MAL from activating SRF (Fig. 4B), similar to the dominant negative effects on serum stimulation noted above. Together, these results suggest that OTT-MAL forms heterodimers with MAL via the leucine zipper and that such complexes have reduced transcriptional activity.
FIG. 4.
OTT-MAL interferes with MAL via the leucine zipper dimerization domain. (A) HeLa cells were cotransfected with the SRF reporter, MAL (0.4 μg), and various amounts (0.4, 0.6, and 1 μg) of OTT-MAL. (B) Cotransfection of HeLa cells with MAL (0.2 μg) and various amounts of OTT-MAL ΔLZ or OTT-MAL YY. Luciferase was measured as before. Shown is the mean luciferase activity of duplicates (error bars indicate the half range).
Independence of OTT-MAL from upstream signals.
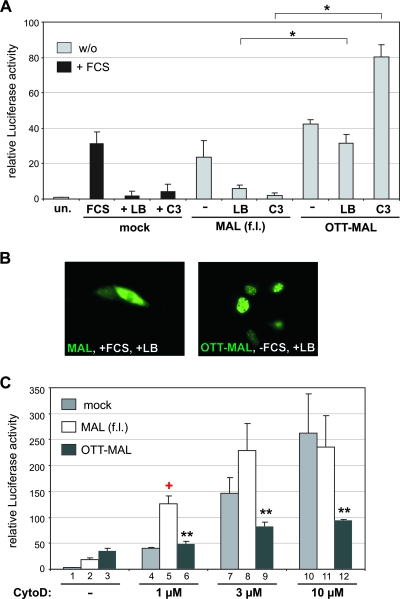
MAL is regulated by Rho family GTPases and G-actin (15). Different actin-binding drugs can regulate MAL either positively and negatively: cytochalasin D induces SRF activity by dissociating MAL from its inhibitory interaction with G-actin, whereas latrunculin B blocks this dissociation, thereby inhibiting SRF (15, 28). We thus tested the dependence of OTT-MAL on Rho-actin signaling. Whereas both serum-stimulated and MAL-mediated SRF activities were efficiently inhibited by latrunculin B treatment or C3 transferase expression, OTT-MAL-induced luciferase activity was not affected; indeed, SRF activity increased upon blockade of RhoA (Fig. 5A). In addition, the nuclear localization of OTT-MAL did not change in the presence of latrunculin B, which readily results in nuclear export of MAL, even in the presence of serum (Fig. 5B). Together, these results show that OTT-MAL activity is independent of Rho-actin signaling.
FIG. 5.
OTT-MAL cannot be regulated by actin drugs or RhoA, in contrast to MAL. (A) No inhibition of OTT-MAL by either latrunculin B (LB; 0.3 μM) or C3 transferase (20 ng). Transiently expressing cells were serum starved for 40 h. Treatment with latrunculin B was for 8.5 h, 30 min prior to serum stimulation of mock-transfected control cells for 8 h. SRF luciferase reporter activity was normalized to LacZ and is shown as n-fold activation compared to unstimulated control cells (un.). Error bars, SEM (n = 4). (B) Latrunculin B inhibits nuclear accumulation of MAL but not of OTT-MAL. Transfected cells were pretreated with latrunculin B 30 min prior to FCS stimulation, when indicated, and stained with anti-HA antibodies to visualize the MAL and OTT-MAL proteins. (C) No synergism between OTT-MAL and cytochalasin D. Cells were transfected with the SRF reporter and a vector control (mock), MAL, or OTT-MAL. Following serum starvation overnight, cells were treated with 1, 3, or 10 μM cytochalasin D (CytoD) or left untreated (−). As controls, cells were transfected with SRF-VP16 (set at 100). Cells were lysed after 7 h, and luciferase activities were determined. Shown are mean relative luciferase activities, normalized to Renilla, with error bars indicating SEM (n = 3). Single asterisks indicate significant activation (P < 0.01), double asterisks indicate significant repression (P < 0.05), and the red plus sign indicates synergism of MAL and cytochalasin D (P < 0.02).
Next we used increasing amounts of cytochalasin D to activate SRF in cells transfected with MAL, OTT-MAL, or a vector control. At low levels of cytochalasin D, MAL and the drug synergistically activated the SRF reporter (P < 0.02) (Fig. 5C). In contrast, OTT-MAL does not show synergistic activation with cytochalasin D; only a slight increase, depending on the amounts of cytochalasin D, is observable. Comparing the dose-response curves of OTT-MAL-, MAL-, and mock-transfected cells actually showed that OTT-MAL inhibited the ability of higher concentrations of cytochalasin D to activate the reporter, in contrast to MAL (Fig. 5C, compare bars 10 and 12). This indicates that OTT-MAL activity is not positively regulated by cytochalasin D-induced changes of G-actin, in contrast to MAL. Together, the results suggest that the constitutive activity of OTT-MAL is caused by uncoupling from upstream signaling pathways.
Deregulation of OTT-MAL.
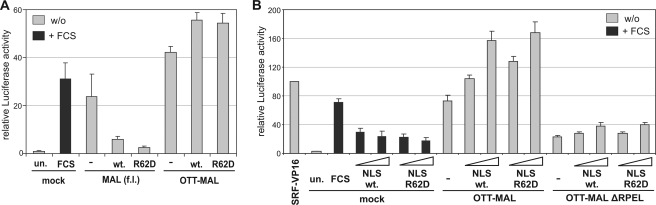
We previously showed that ectopic expression of wild-type actin and nonpolymerizable point mutant actin R62D inhibits MAL and SRF (15, 20). To test whether OTT-MAL is inhibited by actin, SRF reporter activity was determined in cells cotransfected with actin constructs. OTT-MAL-mediated SRF activation was not affected by either wild-type actin or actin R62D, in contrast to MAL (Fig. 6A). Since these actins are mostly cytoplasmic, whereas OTT-MAL appears to be entirely nuclear, we also tested actins which are fused to an NLS. These NLS-actins readily accumulate in the nucleus and colocalize there with MAL but still efficiently inhibit SRF activity (15, 20). However, OTT-MAL-induced SRF activity was not inhibited (Fig. 6B). Rather, an elevation of the SRF reporter was observed when increased levels of NLS-actins were expressed, reminiscent of the effect seen when C3 is coexpressed. No difference was found with OTT-MAL ΔRPEL, excluding the possibility that actin is directly enhancing OTT-MAL activity by binding to the RPEL motifs. Together, these results suggest that OTT-MAL is not regulated by G-actin.
FIG. 6.
Ectopic expression of actin constructs is unable to regulate OTT-MAL. (A) Cells were cotransfected with MAL or OTT-MAL and either wild-type actin or nonpolymerizable actin R62D. SRF luciferase reporter activity was normalized to LacZ and is shown as n-fold activation compared to unstimulated control cells (un.). (B) Effects of actin constructs fused to an NLS on SRF activity. Increasing amounts (100 and 300 ng) of either wild-type NLS-actin (wt.) or NLS-actin R62D were cotransfected with vector control (mock), MAL, OTT-MAL, or OTT-MALΔRPEL into NIH 3T3 cells. One day after transfection, SRF-luciferase activity was determined and normalized to pRL-tk. Shown is the average of three independent experiments, with error bars indicating the SEM. w/o, without FCS.
OTT-MAL fails to bind actin.
Critical for the activation of MAL is its dissociation from G-actin, rather than its nuclear accumulation per se (28). We therefore tested whether deregulation of OTT-MAL reflects a decreased interaction with actin. To analyze this, stable HEK293 cell lines harboring a doxycycline-inducible OTT-MAL expression plasmid were generated. Following induction, coimmunoprecipitation failed to reveal any protein complexes between OTT-MAL and either actin R62D, NLS-actin R62D, or wild-type NLS-actin. (Fig. 7A). However, coexpressed MAL was readily detectable in Flag-tagged R62D mutant actin immunoprecipitates, demonstrating that such complexes readily form in these clonal HEK293 cells. This indicates that OTT-MAL cannot bind to actin, despite the presence of two intact RPEL motifs which suffice in MAL for actin binding.
FIG. 7.
OTT-MAL fails to interact with actin. (A) HEK293 cells harboring a doxycycline-inducible OTT-MAL expression vector were transiently transfected with Flag-tagged actin R62D, NLS-actin R62D, wild-type (wt.) NLS-actin, or actin S14C. As a control, cells were cotransfected with HA-MAL and Flag-tagged actin R62D (lane 1). Following induction of HA-OTT-MAL expression by doxycycline (1 μM, 24 h), cell lysates were subjected to coimmunoprecipitation (IP) with anti-Flag-agarose. Precipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted (IB) with antibodies against HA (MAL and OTT-MAL) and Flag (actins). Expression was controlled by immunoblotting of the total lysates (lower panels). The values on the left of the upper panel are molecular sizes in kilodaltons. (B) Reconstitution of the entire N-terminal RPEL domain in OTT-MAL does not restore regulation by serum or actin. The graph shows the created OTT-123MAL construct harboring the entire region N terminal to the second RPEL motif in MAL (f.l.) (compare to Fig. 1A). NIH 3T3 cells transfected with OTT-123MAL and either the vector control or actin R62D were stained with anti-HA antibody (green, top panel) and phalloidin or anti-Flag antibody (red, merged images in the lower panel). Nuclei were stained blue with Hoechst 33258.
To test whether close proximity to OTT-derived sequences hampers binding of and regulation by actin, we engineered an OTT-123MAL construct harboring the entire MAL N terminus containing all three RPEL motifs. However, OTT-123MAL was still nuclear, even when actin R62D was coexpressed (Fig. 7B). Moreover, OTT-123MAL constitutively activated SRF and was not regulated by increasing amounts of latrunculin B, C3 transferase, wild-type actin, actin R62D, or NLS-actin R62D (data not shown). Similarly, the deletions of OTT-derived sequences in ΔRRM and ΔSPOC did not restore proper regulation of OTT-MAL, which was, in contrast, evident for MAL and in control cells (not shown). This suggested that deregulated OTT-MAL activity is not caused by a single domain in OTT nor by a lack of N-terminal MAL sequences.
Induction of gene expression.
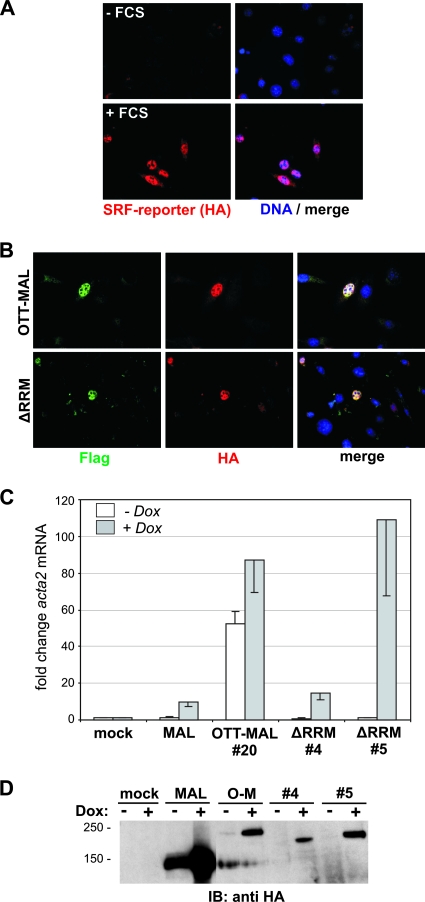
The RRM and the SPOC domain of OTT were suggested to have functions in splicing and chromatin remodeling via corepressor recruitment, respectively (6, 22, 32). Since the luciferase reporter plasmid is not integrated into the genome and does not require splicing, we sought to analyze the regulation of integrated and endogenous genes. For this, NIH 3T3 cells harboring a stainable, integrated FosHA fusion gene driven by the same SRF-controlled promoter as in the luciferase reporter (1) were used. Both OTT-MAL and the ΔRRM deletion induced the expression of the reporter protein in transfected cells in the absence of serum (Fig. 8B). In contrast, control cells required serum stimulation for induction of this stainable SRF reporter protein (Fig. 8A). This suggests that, regardless of its RRM motifs, OTT-MAL is sufficient to activate the expression of integrated, intron-containing target genes, although we cannot exclude the possibility of reduced splicing or export efficiencies on other genes.
FIG. 8.
Induction of integrated and endogenous target genes by OTT-MAL. NIH 3T3 cells harboring an integrated 3D.A-FosHA reporter gene whose expression requires proper splicing were immunostained with anti-HA antibodies (red) and Hoechst 33258 (blue). As controls, cells were either left untreated (A, top) or serum stimulated for 1 h (A, bottom). In panel B, cells were transiently transfected with either Flag-tagged OTT-MAL or OTT-MALΔRRM, kept under serum-starved conditions, and subsequently coimmunostained with anti-Flag antibodies (green), anti-HA antibodies (for detection of reporter gene expression; red), and Hoechst 33258 (blue). (C) Induction of smooth muscle α-actin by MAL, OTT-MAL, and OTT-MALΔRRM. Expression in stably transfected HEK293 clonal cells was induced by doxycycline (Dox; 1 μg/ml, 24 h), and the mRNA was isolated and subjected to quantitative RT-PCR with primers specific for acta2 and alas1. Shown is the average induction of acta2 compared to mock-transfected control cells after normalization to alas1 (error bars indicate the half range). The expression levels of the MAL, OTT-MAL, and OTT-MALΔRRM proteins in the various clonal lines are visualized in panel D by immunoblotting (IB) against HA. The values on the left are molecular sizes in kilodaltons.
We then examined the expression of an endogenous SRF target gene. HEK293 cell lines with doxycycline-inducible expression of MAL, OTT-MAL, or OTT-MAL ΔRRM were analyzed for induction of the mRNA of the known SRF target smooth muscle α-actin (acta2). Upon expression of MAL or OTT-MAL ΔRRM, acta2 mRNA was considerably induced compared to cells not treated with doxycycline and mock-transfected control cells (Fig. 8C). The cells expressing OTT-MAL showed elevated levels of acta2 already in the absence of doxycycline, indicating functional leakiness; this was nevertheless further enhanced following doxycycline addition. Induction of acta2 by OTT-MAL ΔRRM varied between clonal lines, which correlated with the expression level of the fusion protein (Fig. 8D). MAL protein expression reached higher levels than OTT-MAL, although acta2 induction was weaker; this is likely explained by its regulatability by G-actin. Together, the results indicate that the presence of the OTT-MAL fusion protein constitutively activates SRF target genes in cells, which are usually tightly controlled by Rho-actin-MAL signaling.
Aberrant induction of TCF targets.
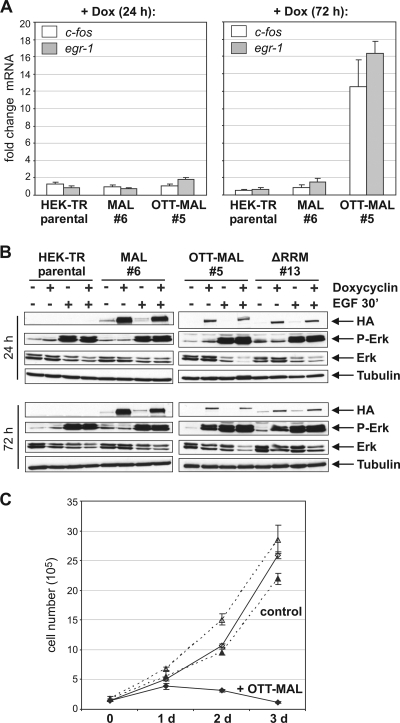
It was previously shown that OTT-MAL activates SRF promoters which are independent of MAL (4). We thus analyzed whether OTT-MAL elevated the endogenous levels of the c-fos and egr-1 mRNAs, which are ternary complex factor-dependent SRF targets. At 24 h after induction of OTT-MAL expression, no increase in c-fos or egr-1 mRNA was observed (Fig. 9A). After 72 h, however, OTT-MAL, but not MAL, resulted in elevated c-fos and egr-1 levels. This result demonstrates that OTT-MAL aberrantly activates the expression of immediate-early genes.
FIG. 9.
OTT-MAL induces delayed c-fos and egr-1 gene expression and Erk phosphorylation but inhibits cell growth. (A) Activation of c-fos and egr-1 transcription by MAL and OTT-MAL. Expression in stably transfected HEK293 clonal cells was induced by doxycycline (Dox; 1 μg/ml) for 24 h (left) or 72 h (right), and the mRNA was isolated and subjected to quantitative RT-PCR with primers specific for c-fos, egr-1, and alas1. Shown is the average induction upon doxycycline addition, normalized to alas1. Error bars, SEM (n = 3). (B) Activity of MAPK/Erk upon expression of MAL, OTT-MAL, and OTT-MALΔRRM for 24 h (top) or 72 h (bottom). As a control, Erk was activated by epidermal growth factor (EGF) treatment (25 ng/ml) for 30 min. Immunoblotting of cell lysates was performed with anti-phospho-Erk, anti-pan-Erk, antitubulin, and anti-HA (MAL/OTT-MAL) antibodies. (C) Growth curve of stably transfected HEK293 cells following induction of OTT-MAL expression (⧫) by doxycycline (1 μg/ml) for 3 days. Triangles, control HEK cell line; open symbols, without induction by doxycycline.
The Drosophila OTT ortholog spen was genetically identified as a positive regulator of the Ras pathway (5). This raised the possibility that OTT-MAL triggers ternary complex factor-dependent transcription via activation of the MAPK/Erk pathway. Expression of OTT-MAL, as well as the ΔRRM mutant protein, resulted in increased Erk phosphorylation after 72 h, but not after 24 h (Fig. 9B). This is consistent with the delayed induction of c-fos and egr-1, compared to acta2. Together, these results suggest that c-fos and egr-1 expression by OTT-MAL is a secondary consequence of increased MAPK/Erk signaling, rather than direct activation of their ternary complex factor-dependent promoters.
In the course of these experiments, a certain toxicity of OTT-MAL expression became apparent. Therefore, the growth of HEK293 cells inducibly expressing OTT-MAL was analyzed. Induction of OTT-MAL expression by doxycycline sharply reduced cell proliferation, and cell numbers dropped after 2 to 3 days following induction (Fig. 9C). Thus, OTT-MAL negatively affects cell growth, despite the induction of c-fos, egr-1, and the MAPK/Erk pathway.
DISCUSSION
We show here that the oncogenic fusion protein OTT-MAL/RBM15-MKL1 activates MAL- and SRF-dependent gene expression independently of Rho-actin signaling. This constitutive activation is dependent on binding to SRF and the potent MAL-derived transcriptional activation domain. The activity of OTT-MAL is resistant to both negative and positive control by upstream signaling. We show that OTT-MAL fails to be regulated at any of the three actin-dependent levels which were previously shown for MAL (28). The failure to coprecipitate OTT-MAL-G-actin complexes suggests that defective actin binding underlies the constitutive activity of OTT-MAL.
OTT-MAL localizes to the nucleus. When we analyzed OTT alone, we also found the protein in the nucleus (data not shown), raising the possibility that the regulated nucleocytoplasmic shuttling of MAL usually observed is overridden by OTT sequences. Consistent with this, several potential NLSs were recently identified in murine OTT/RBM15, resulting in nuclear accumulation of a green fluorescent protein-OTT fusion protein (10). None of the deletions analyzed here are depleted of all putative OTT-derived NLSs, since they are stretched out over most of the OTT protein. In this respect, OTT-MAL is effectively comparable to NLS-MAL—but unlike OTT-MAL, nuclear NLS-MAL still responds to signaling via the Rho-actin pathway, which potentiates its transcriptional activity (28).
Previous work has established that, mechanistically, the critical regulatory step in the activation of target gene transcription is not nuclear accumulation but (at least partial) dissociation of the actin-MAL complex (28). Even when NLS-actin relocalizes MAL to the nucleus, it represses SRF-mediated gene expression (15, 20). Similarly, MAL accumulates in the nucleus when CRM1-dependent export is blocked by leptomycin B but SRF targets are not activated unless the actin-MAL complex is dissociated, e.g., by cytochalasin D (20, 28). In contrast, OTT-MAL constitutively activates transcription and its effect cannot be potentiated. Thus, we suggest that the constitutive activity of OTT-MAL at SRF target genes is not solely because it is always nuclear but because it escapes regulation by G-actin, similar to MAL ΔN (15). The failure to delineate this deregulation to a specific region of the OTT protein suggests that there are either multiple activating domains or, perhaps, steric blockage of G-actin binding by bulky N-terminal extensions. In both cases, the loss of binding to G-actin appears to be the critical deregulatory event when MAL is fused to OTT in t(1,22).
Consistent with this idea, transcriptional activation by OTT-MAL ΔRPEL, which lacks the potential actin-binding motifs, is not enhanced compared to the intact OTT-MAL protein. In fact, we observed a slightly reduced SRF activation by OTT-MAL ΔRPEL (Fig. 2C). It therefore remains possible that this mutant is partially defective in SRF binding. It also remains possible that the fused OTT sequences antagonize transcriptional activation by the MAL C terminus.
MAL dimerizes via the leucine zipper when forming a transcriptionally active complex with SRF and DNA (15). Whereas we consider the variations in transcriptional activation by the various mutant constructs to mainly reflect their expression level, OTT-MAL ΔLZ shows a clearly enhanced effect on SRF. Since we showed functional interaction between MAL and OTT-MAL, it is likely that much of the poorly expressed OTT-MAL protein normally forms heterodimers with endogenous MAL. Binding to MAL may even inhibit OTT-MAL, which would be relieved in ΔLZ. In line with this hypothesis, we observed slightly enhanced activities when endogenous MAL was inhibited either by C3 or ectopic expression of actins. Such conditions result in a reduced level of nuclear MAL, since Rho and actin regulate MAL import and export rates (28). We speculate that signals from activated RhoA or actin limit the activating potential of OTT-MAL via leucine zipper-dependent dimerization with endogenous MAL. However, we cannot exclude the possibility that OTT-MAL also forms homodimers with detrimental effects; maybe the fusion to OTT renders the complex too big or leads to steric hindrance.
Our study supports previous findings (4) that OTT-MAL also enhances the transcription of MAL-independent SRF target genes such as c-fos and egr-1. One possible explanation for their induction is that OTT-MAL lost its target gene specificity; however, this is inconsistent with the observed delay of MAL-independent targets. In addition, there is a similarly delayed activation of MAPK/Erk signaling by OTT-MAL (Fig. 7), and genetic studies with Drosophila have identified the OTT ortholog spen as a positive regulator of the Ras pathway (5). Thus, we favor the hypothesis that OTT-MAL indirectly activates Ras-MAPK-TCF signaling by secreted or cellular factors, ultimately resulting in the expression of immediate-early genes c-fos and egr-1.
We demonstrate here an antiproliferative effect of OTT-MAL in adherent cells, however. This finding is somewhat counterintuitive, since an “oncoprotein” would be expected to promote growth. Why does OTT-MAL not induce growth? Firstly, the inability to mimic OTT-MAL's oncogenic function in tissue culture may be caused by cell-type-specific effects or the microenvironment for cells harboring the fusion protein is lost. Consistent with such non-cell-autonomous effects, we failed to see colony formation by bone marrow cells when OTT-MAL was introduced ex vivo (Robert Slany and G. Posern, unpublished data). Secondly, another MRTF family member and SRF coactivator, the muscle-specific myocardin protein, has antiproliferative activity and was recently identified as a suppressor of malignant growth (14). It remains possible that MAL exhibits similar functions and that the deregulated activation of MAL target genes by overexpression of OTT-MAL contributes to the observed antiproliferative effect. Finally, one functional MAL allele (as well as one OTT allele) is lost in the AML M7 leukemias. This situation is not mimicked in our experiments, and we cannot exclude the possibility that such loss of alleles affects leukemogenesis.
It appears likely that functions mediated by OTT-derived sequences also affect cells harboring the fusion protein. It was previously shown that OTT, as well as the paralogs SHARP and MINT, binds to RBPJ and either activates or represses Notch-regulated gene expression, depending on the cell context (9, 10, 17). Activated Notch signaling has also been implicated in both hematopoietic stem cell immortalization and cancer (23, 27). Stimulation of Notch-regulated genes by high levels of OTT alone was recently suggested to inhibit the differentiation of myeloid precursor cells (10). Conversely, conditional knockout of OTT in adult mice caused defects in hematopoiesis, affecting both lymphoid and myeloid lineages (22). This study, however, implicated OTT as a negative regulator of megakaryocyte proliferation and suggested that OTT-MAL possibly harbors a dominant negative effect over endogenous OTT function, thereby contributing to AMKL (22). Together, we speculate that the combination of OTT dysregulation and activation of MAL-dependent and TCF-dependent target genes elicits the pathogenic effect of OTT-MAL in AMKL. Further studies will have to delineate the individual and cell type-specific contributions.
Acknowledgments
This work was funded by the department of Axel Ullrich at the MPI of Biochemistry, by Cancer Research UK, and by DFG grant PO 1032/1 (to G.P.).
We thank Robert Slany for infection and culture of murine bone marrow cells, Johannes Schilling for his excellent work with OTT-MAL, and Henrik Daub for critical reading of the manuscript.
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Alberts, A. S., O. Geneste, and R. Treisman. 1998. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell 92475-487. [DOI] [PubMed] [Google Scholar]
- 2.Ariyoshi, M., and J. W. Schwabe. 2003. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 171909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, A., C. Civin, N. Schneider, G. Dahl, A. Pappo, P. Bowman, A. Emami, S. Gross, C. Alvarado, and C. Phillips. 1991. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a Pediatric Oncology Group Study. Blood 78748-752. [PubMed] [Google Scholar]
- 4.Cen, B., A. Selvaraj, R. C. Burgess, J. K. Hitzler, Z. Ma, S. W. Morris, and R. Prywes. 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 236597-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, F., and I. Rebay. 2000. split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr. Biol. 10943-946. [DOI] [PubMed] [Google Scholar]
- 6.Hiriart, E., H. Gruffat, M. Buisson, I. Mikaelian, S. Keppler, P. Meresse, T. Mercher, O. A. Bernard, A. Sergeant, and E. Manet. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 28036935-36945. [DOI] [PubMed] [Google Scholar]
- 7.Jemc, J., and I. Rebay. 2006. Characterization of the split ends-like gene spenito reveals functional antagonism between SPOC family members during Drosophila eye development. Genetics 173279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuang, B., S. C.-Y. Wu, Y.-A. Shin, L. Luo, and P. Kolodziej. 2000. split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 1271517-1529. [DOI] [PubMed] [Google Scholar]
- 9.Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun, T. Furukawa, Y. Taniguchi, H. Kurooka, Y. Hamada, S. Toyokuni, and T. Honjo. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18301-312. [DOI] [PubMed] [Google Scholar]
- 10.Ma, X., M. J. Renda, L. Wang, E.-C. Cheng, C. Niu, S. W. Morris, A. S. Chi, and D. S. Krause. 2007. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell. Biol. 273056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, Z., S. W. Morris, V. Valentine, M. Li, J.-A. Herbrick, X. Cui, D. Bouman, Y. Li, P. K. Mehta, D. Nizetic, Y. Kaneko, G. C. Chan, L. C. Chan, J. Squire, S. W. Scherer, and J. K. Hitzler. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28220-221. [DOI] [PubMed] [Google Scholar]
- 12.Mercher, T., M. Busson-Le Coniat, F. N. Khac, P. Ballerini, M. Mauchauffe, H. Bui, B. Pellegrino, I. Radford, F. Valensi, F. Mugneret, N. Dastugue, O. A. Bernard, and R. Berger. 2002. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes Chromosomes Cancer 3322-28. [DOI] [PubMed] [Google Scholar]
- 13.Mercher, T., M. B. Coniat, R. Monni, M. Mauchauffe, F. N. Khac, L. Gressin, F. Mugneret, T. Leblanc, N. Dastugue, R. Berger, and O. A. Bernard. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 985776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milyavsky, M., I. Shats, A. Cholostoy, R. Brosh, Y. Buganim, L. Weisz, I. Kogan, M. Cohen, M. Shatz, S. Madar, E. Kalo, N. Goldfinger, J. Yuan, S. Ron, K. MacKenzie, A. Eden, and V. Rotter. 2007. Inactivation of myocardin and p16 during malignant transformation contributes to a differentiation defect. Cancer Cell 11133-146. [DOI] [PubMed] [Google Scholar]
- 15.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113329-342. [DOI] [PubMed] [Google Scholar]
- 16.Newberry, E. P., T. Latifi, and D. A. Towler. 1999. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 3810678-10690. [DOI] [PubMed] [Google Scholar]
- 17.Oswald, F., U. Kostezka, K. Astrahantseff, S. Bourteele, K. Dillinger, U. Zechner, L. Ludwig, M. Wilda, H. Hameister, W. Knochel, S. Liptay, and R. M. Schmid. 2002. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J. 215417-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porteu, F., M. C. Rouyez, L. Cocault, L. Benit, M. Charon, F. Picard, S. Gisselbrecht, M. Souyri, and I. Dusanter-Fourt. 1996. Functional regions of the mouse thrombopoietin receptor cytoplasmic domain: evidence for a critical region which is involved in differentiation and can be complemented by erythropoietin. Mol. Cell. Biol. 162473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posern, G., F. Miralles, S. Guettler, and R. Treisman. 2004. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 233973-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posern, G., A. Sotiropoulos, and R. Treisman. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 134167-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posern, G., and R. Treisman. 2006. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16588-596. [DOI] [PubMed] [Google Scholar]
- 22.Raffel, G. D., T. Mercher, H. Shigematsu, I. R. Williams, D. E. Cullen, K. Akashi, O. A. Bernard, and D. G. Gilliland. 2007. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl. Acad. Sci. USA 1046001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy, M., W. S. Pear, and J. C. Aster. 2007. The multifaceted role of Notch in cancer. Curr. Opin. Genet. Dev. 1752-59. [DOI] [PubMed] [Google Scholar]
- 24.Sasazuki, T., T. Sawada, S. Sakon, T. Kitamura, T. Kishi, T. Okazaki, M. Katano, M. Tanaka, M. Watanabe, H. Yagita, K. Okumura, and H. Nakano. 2002. Identification of a novel transcriptional activator, BSAC, by a functional cloning to inhibit tumor necrosis factor-induced cell death. J. Biol. Chem. 27728853-28860. [DOI] [PubMed] [Google Scholar]
- 25.Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich, C. C. Tsai, M. Hon, and R. M. Evans. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 151140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98159-169. [DOI] [PubMed] [Google Scholar]
- 27.Varnum-Finney, B., L. Xu, C. Brashem-Stein, C. Nourigat, D. Flowers, S. Bakkour, W. S. Pear, and I. D. Bernstein. 2000. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 61278-1281. [DOI] [PubMed] [Google Scholar]
- 28.Vartiainen, M. K., S. Guettler, B. Larijani, and R. Treisman. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 3161749-1752. [DOI] [PubMed] [Google Scholar]
- 29.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105851-862. [DOI] [PubMed] [Google Scholar]
- 30.Wideman, L., J. Y. Weltman, J. T. Patrie, C. Y. Bowers, N. Shah, S. Story, A. Weltman, and J. D. Veldhuis. 2000. Synergy of l-arginine and growth hormone (GH)-releasing peptide-2 on GH release: influence of gender. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279R1455-R1466. [DOI] [PubMed] [Google Scholar]
- 31.Zaromytidou, A. I., F. Miralles, and R. Treisman. 2006. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol. Cell. Biol. 264134-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419182-185. [DOI] [PubMed] [Google Scholar]