Abstract
Actin is a key regulator of RNA polymerase (pol) II transcription. In complex with specific hnRNPs, it has been proposed that actin functions to recruit pol II coactivators during the elongation of nascent transcripts. Here, we show by affinity chromatography, protein-protein interaction assays, and biochemical fractionation of nuclear extracts that the histone acetyltransferase (HAT) PCAF associates with actin and hnRNP U. PCAF and the nuclear actin-associated HAT activity detected in the DNase I-bound protein fraction could be released by disruption of the actin-hnRNP U complex. In addition, actin, hnRNP U, and PCAF were found to be associated with the Ser2/5- and Ser2-phosphorylated pol II carboxy-terminal domain construct. Chromatin and RNA immunoprecipitation assays demonstrated that actin, hnRNP U, and PCAF are present at the promoters and coding regions of constitutively expressed pol II genes and that they are associated with ribonucleoprotein complexes. Finally, disruption of the actin-hnRNP U interaction repressed bromouridine triphosphate incorporation in living cells, suggesting that actin and hnRNP U cooperate with PCAF in the regulation of pol II transcription elongation.
Eukaryotic gene transcription requires dynamic alterations of chromatin structure that are mediated by chromatin remodeling complexes and histone modifying enzymes (23, 28).
For transcription competence, histone acetylation allows the switch between repressive and permissive chromatin structures through direct effects on nucleosome stability and through establishment of binding sites for regulatory proteins. Considerable progress in understanding the role of histone acetylation came from the discovery that the Saccharomyces cerevisiae transcription coactivator GCN5 and, more recently, other yeast and metazoan transcription cofactors are histone acetyltransferases (HATs). Mammalian homologs of the yeast cofactor GCN5 include PCAF and GCN5L (2, 27, 32, 33, 35). PCAF and GCN5L are encoded by distinct genes, and their expressions are differential and complementary in various tissues (33, 35). Moreover, GCN5L is essential for mouse development, whereas PCAF is dispensable (33, 34). Human GCN5L and PCAF form parts of distinct multiprotein HAT complexes, namely the PCAF complex (17), the TFTC complex (1), and the STAGA complex (12). While these mammalian HAT complexes are still incompletely characterized, they have related but not identical subunit compositions.
PCAF/GCN5, together with the p300/CREB-binding protein, is among the best-studied transcriptional coactivators (11). PCAF has been proposed to facilitate long-distance transcriptional enhancement by direct association with enhancer sequences (7). PCAF is also known to acetylate free histones or nucleosomes, primarily on lysine 14 of histone H3 (28), and its requirement as coactivator or HAT has been demonstrated for myogenesis and nuclear receptor-mediated and growth factor-signaled activation (28). However, it is presently unclear whether PCAF is also involved in the maintenance of efficient transcription elongation.
Recent reports have shown that in gene transcription, nuclear actin plays a key role as a component of chromatin remodeling complexes, pre-messenger ribonucleoprotein (pre-mRNP), and messenger RNP (mRNP) particles associated with all three eukaryotic RNA polymerases (pol) (18, 22). Studies performed with the 40S RNP fraction isolated from rat liver extracts have shown that actin associates with a specific subset of hnRNPs (20). Among them, hnRNP U was found to interact directly with actin through a specific and conserved actin-binding site located in the hnRNP U C terminus (U-C) (8). Given that disruption of the actin-hnRNP U interaction abolished pol II transcription in living cells, it was proposed that the actin-hnRNP U complex associated with the elongating pol II facilitates efficient elongation of mRNA transcripts, presumably through recruitment of transcriptional coactivators (8, 16, 22). In support of this attractive model, actin bound to the Chironomus tentans hnRNP hrp65-2 was recently described as a molecular platform for recruitment of the HAT P2D10 on active genes (21, 26).
Here, we show evidence that in human cells, the HAT PCAF is associated with actin, hnRNP U, and elongating pol II and that disruption of the actin-hnRNP U interaction represses pol II transcription, presumably via a chromatin-based mechanism. We speculate that PCAF recruitment to active genes for productive pol II transcription requires the specific actin-hnRNP U interaction.
MATERIALS AND METHODS
Antibodies.
The rabbit polyclonal peptide-specific antibody to hnRNP U (CED17) was raised using the peptide CEDYKQRTQKKAEVEGK as antigen. The same peptide antigen was conjugated to Sulfolink resin (Pierce) and also used for affinity purification of the rabbit serum. The goat anti-rabbit antibody used for CED17 visualization after microinjections was obtained from Invitrogen. Mouse monoclonal antibodies to pol II (clone 8WG16), to the Ser2-phosphorylated pol II carboxy-terminal domain (CTD) (clone H5), to the TATA box binding protein (TBP; clone 1TBP18), and to hnRNP U (clone 3G6), as well as the rabbit polyclonal antibody to histone H3 and acetylated H3K9, were purchased from AbCam. Mouse monoclonal antibodies against PCAF (clone E-8) and actin (clone AC-74) were from Santa Cruz Biotechnology and Sigma, respectively. The monoclonal antibody to bromouridine triphosphate (BrUTP) and polyclonal antibody to green fluorescent protein (GFP) were from Roche and Clontech, respectively. The S-protein-horseradish-peroxide conjugate was purchased from Novagen. Goat anti-mouse Alexa 568 and donkey anti-rabbit Alexa 488 antibodies were purchased from Invitrogen. The polyclonal antibody to CBP20 was a gift from Neus Visa (Stockholm University, Sweden). The monoclonal antiactin antibody (clone 1C7) was a gift from Cora-Ann Schoenenberger (Biocentrum, University of Basel, Switzerland). The goat polyclonal anti-TFIIIC220 and the mouse monoclonal anti-ASF/SF2 antibodies were gifts from Neus Visa (Stockholm University, Sweden) and Adrian Krainer (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), respectively.
Cloning, expression, and purification of hnRNP and CTD constructs.
The hnRNP U N-terminal (U-N; amino acids 1 to 250), the hnRNP U middle domain (U-M; amino acids 251 to 550), the hnRNP U actin-binding C-terminal (U-C; amino acids 551 to 825), and the pol II CTD constructs were cloned from human cDNA, expressed in Escherichia coli, and purified by affinity chromatography. The forward primer 5′-CGCGGATCCATGAGTTCCTCGCCTGTTAATG and the reverse primer 5′-CCCAAGCTTTCATTCAACAGGTGGCTGAGGAGA were used for cloning the hnRNP U-N construct; the forward primer 5′-CATGCCATGGCAGATCGTCTCAGTGCTTCTTCC and the reverse primer 5′-CCGCTCGAGTCACTGCCACCATCATCTTATC were used for cloning the hnRNP U-M construct; the forward primer 5′-GGAATTCCGGAAGCAAATGGCAGATACTG and the reverse primer 5′-CCGCTCGAGTCAATAATATCCTTGGTGATAATG were used for cloning the hnRNP U-C construct; and the forward primer 5′-CGGGATCCCAAGGTGGTGCCATGTCTCCC and the reverse primer 5′-CCGGAATTCAGTTCTCCTCGTCACTGTC were used for the pol II CTD construct. After intermediate TA cloning into the pGEMTeasy plasmid (Promega), the hnRNP U and pol II CTD constructs were cloned into the pET30a(+) plasmid vector containing a His tag and an S tag for bacterial expression (Novagen). All four plasmid constructs were verified by restriction mapping and DNA sequencing.
The hnRNPU plasmid constructs were expressed in E. coli strain BL21(λDE3)/pLysE (Novagen) in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h. The proteins were purified from the corresponding bacterial lysates by elution with an imidazole gradient (0.015 to 0.3 M) from a Co-BD Talon resin, using the manufacturer's instructions (BD Biosciences). The final yield of purified hnRNP U constructs was 200 ng/μl. The pol II CTD construct was also expressed in E. coli BL21(λDE3)/pLysE (Novagen) in the presence of 1 mM IPTG, but induction was performed at 18°C for 16 h. The recombinant pol II CTD construct was purified from bacterial lysates by affinity purification on protein S-agarose according to the instruction manual (Novagen). The yield of pure CTD coupled to S-protein agarose was estimated to be 100 ng/μl of bed volume.
Protein-protein interaction assays.
DNase I affinity chromatography and immunoprecipitations were performed with HeLa cell nuclear extracts, essentially as previously described (8). Immunoprecipitations were carried out using antibodies to actin, PCAF, pol II, and hnRNP U, as well as with unrelated antibodies, e.g., a polyclonal anti-GFP antibody or a nonspecific rabbit immunoglobulin G (IgG; Dako). In all cases, nuclear extracts were precleared with unconjugated protein A/G-Sepharose beads prior to incubation with the specific antibodies for immunoprecipitation. The term “mock,” used hereafter, refers to immunoprecipitations performed with nuclear extracts incubated with Sepharose beads without any of the indicated antibodies.
For pull-down experiments with the recombinant hnRNP U-N, hnRNP U-M, and hnRNP U-C constructs, the purified proteins were coupled to protein S-agarose beads as described above, and the beads were incubated with HeLa nuclear extracts. After beads were incubated for 5 h at 4°C, they were washed three times with 20 volumes of 1× phosphate-buffered saline (PBS), 0.2% NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF). Where indicated, pull-down experiments were performed after preincubation of HeLa nuclear extracts, with increasing amounts of CED17 (0 ng/μl, 7.5 ng/μl, 15 ng/μl, and 30 ng/μl) to disrupt the actin-hnRNP U interaction or with a control polyclonal rabbit anti-mouse antibody (Dako), added to a final concentration of 30 ng/μl. In all cases, to analyze bound proteins, the precipitated fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting. Detection was performed by using the chemiluminescence method according to the instruction manual (GE Healthcare).
Purification of nuclear actin complexes.
Nuclear actin-containing complexes were purified from nuclear extracts obtained from HeLa cells by size exclusion chromatography on a Superose 6HR column. Where indicated, nuclear extracts were incubated with the CED17 antibody and with nonspecific polyclonal rabbit IgGs to a final concentration of 30 ng/μl for 2 h at 4°C prior to fractionation on a Superose 6HR column.
HAT activity assay.
DNase I-bound protein fractions were subjected to HAT activity assay, carried out essentially as previously described (26). Briefly, after fractions underwent DNase I affinity chromatography, beads were resuspended in a 200-μl HAT reaction buffer containing 50 mM Tris HCl (pH 8.0), 10% glycerol, 1 mM PMSF, 1 mM dithiothreitol (DTT), 50 mM sodium butyrate (Fluka), 250 μCi [3H]acetyl coenzyme A ([3H]acetyl-CoA; GE Healthcare), and 21 ng of core histones (from Ann-Kristin Östlund Farrants, Stockholm University, Sweden). For controls, additional HAT reactions were performed with 50 μl of empty DNase I beads, with 50 μl of nuclear extracts (300 to 450 μg/ml) and 50 μl of H2O added to the reaction mixture. The reaction mixtures were incubated at 30°C for 45 min with continuous agitation. The assay was terminated by chilling the aliquots on ice. Each aliquot was spotted on phosphocellulose P81 filter paper (Upstate), from which unincorporated [3H]acetyl-CoA was eliminated by washing twice with 50 mM NaHCO3 (pH 5.3) for 5 min at room temperature. Finally, filter papers were rinsed with acetone, dried at room temperature, and transferred to scintillation tubes containing 8 ml of Emulsifier Safe (Packard). Incorporated radioactivity was estimated using a model 1600TR Packard TriCarb liquid scintillation analyzer according to the manufacturer's instructions. All HAT activity assays were repeated five times. To interpret the results from the HAT assays, we subtracted the HAT background values, measured on the core histone preparation, from the values obtained from each DNase I pull-down experiment. In addition, all readouts obtained in the presence of increasing amounts of CED17 were normalized relative to the readout obtained from the DNase I pull-down experiment performed without antibody. In all cases, the results obtained proved to be statistically relevant, as revealed by analysis of variance and t test, using Statistica software, version 7.2006 (StatSoft, Inc.).
Kinase assays for CTD phosphorylation. (i) CDK7/cycH/Mat1 Ser5 phosphorylation assay.
One-hundred microliters of protein S-agarose-bound CTD (10 μg) was taken up in 8 mM morpholinepropanesulfonic acid (MOPS)-NaOH (pH 7), 250 μM EDTA, 15 mM ATP (or 7.5 μCi [γ-32P]ATP to determine the efficiency of incorporation [data not shown]), 100 μg/ml bovine serum albumin (BSA), 10 mM Mg-acetate, 0.5% glycerol, 0.5 mM DTT, 1 mM PMSF, 10 mM β-glycerophosphate, 300 ng of CDK7/cycH/MAT1 complex (Upstate). The reaction was performed for 1 h at 30°C with continuous agitation according to the manufacturer's instruction manual (Upstate Biotechnology).
(ii) CDK9/cycT Ser2 phosphorylation assay.
Fifty microliters of protein S-agarose-bound CTD was taken up in 60 mM HEPES-NaOH (pH 7.5), 3 mM MgCl2, 3 mM MnCl2, 3 μM NaVO4, 10 mM β-glycerophosphate, 7.5 mM ATP (or 7.5 μCi [γ-32P]ATP to determine efficiency of incorporation [data not shown]), 100 μg/ml BSA, 0.5% glycerol, 0.5 mM DTT, 1 mM PMSF, and 200 ng of CDK9/cycT complex (ProQuinase). The reaction was carried out for 1 h at 30°C with continuous agitation according to the instruction manual provided by the manufacturer (ProQuinase).
(iii) Kinase assay for double CTD phosphorylation of Ser2 and Ser5 residues.
Fifty microliters of Ser5-phosphorylated CTD (see description above) bound to protein S-agarose was taken up under the CDK9/cycT buffer conditions described above. The reaction was allowed for 1 h at 30°C with continuous agitation.
In all of the above-described cases, phosphorylated CTD that was still bound to protein S-agarose beads was sequentially washed with 20 volumes of 1× PBS containing 1 mM PMSF; 20 volumes of 1× PBS containing 1 mM PMSF, 0.2% NP-40, 0.2% deoxycholate; and 20 volumes of 1× PBS containing 1 mM PMSF and 0.5 M NaCl. Beads were stored at 4°C in 1× PBS, 1 mM PMSF, and 0.02% NaN3.
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed as described by Takahashi et al. (29). The cross-linked chromatin was obtained from HeLa cells, untreated or treated with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA; final concentration of 0.3 μM, 2 h, 37°C) or 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Sigma), incubated for 1 h or 2 h at 37°C (final concentration 75 μM). Cross-linked chromatin was immunoprecipitated with antibodies to hnRNP U (both 3G6 and CED17), actin, pol II (the 8WG16 or H5 antibody), PCAF, TBP, H3, acetylated H3K9, and with a nonspecific rabbit IgG (AbCam). DNA-protein complexes were analyzed by PCR with specific primers for promoter and coding regions of the β-tubulin, S19, and GAPDH genes, as well as the 5S and 5.8S genes. (Complete nucleotide sequence data for each primer used in the ChIP assays are available upon request.) About 1/10 of the precipitated DNA was used for each PCR. PCR products were separated by electrophoresis on 2% agarose gels and visualized with ethidium bromide. All ChIP experiments were performed in triplicate.
Chromatin RNA immunoprecipitation (RIP) assays.
The RIP assays were performed according to the method described by Gilbert et al. (4), with the following modifications. Cells (107) were fixed for 10 min with 1% formaldehyde, and a chromatin fraction was prepared as described above in the presence of 24 U/ml RNAguard as RNase inhibitor (GE Healthcare). The chromatin was fragmented by sonication and precleared with recombinant protein G-Sepharose (Zymed) for 1 h at 4°C. Precleared samples were incubated overnight at 4°C with antibodies to actin (AC74), pol II (8WG16), PCAF (E8), and CBP20 and with an antibody against GFP (Clontech), used as nonspecific IgG. The antibodies were subsequently precipitated with protein G-Sepharose. Precipitated protein-nucleic acid complexes were washed six times, alternating 20 volumes of RIPA buffer (1× PBS, 1 mM PMSF, 0.2% NP-40, 0.1% deoxycholate, 0.1% SDS, 24 U/ml RNase inhibitor) and RIPA buffer supplemented with 1 M NaCl. After the final wash, the beads were resuspended in 1× PBS, 10 mM DTT, 1% SDS, and 1 mM PMSF and incubated at 70°C to reverse cross-linked RNA-protein complexes. After precipitation, nucleic acids were extracted using TRI reagent (Sigma) according to the manufacturer's instructions. Nucleic acid pellets were dissolved in 15 μl of 1× DNase I reaction buffer and incubated with 1 U of amplification-grade DNase I as described by the manufacturer (Invitrogen). The RNA samples were reverse transcribed into cDNA by Superscript II (Invitrogen), using oligo(dT) primers (Roche) and random N6 primers (Roche). The cDNA samples were then analyzed with primers specific to the H2B promoter (forward primer [positions −349 to −331] 5′-AGA CCG GCC CTA GAA GAG and reverse primer [−23 to −5] 5′-AGC AGG GTA TGA CAA GGC), to H2B mRNA (forward primer [+170 to +190] 5′-ACAAGGTTCTGAAGCAGGTC and reverse primer [+399 to +419] 5′-TAGCGCTGGTGTACTTGGTG), to S19 mRNA (forward primer 5′-ACGCGAGCTGCTTCCACAG and reverse primer 5′-AGCTGCCACCTGTCCGGC), and to tRNATyr (forward primer 5′-CCTTCGATAGCTCAGCTGGTAGAGCGGAGG and reverse primer 5′-CGGAATTGAACCAGCGACCTAAGGATGTCC) as control for the specificity of the assay. All RIP experiments were performed in duplicate.
RNase treatment.
RNase treatment of HeLa cells was performed as described previously (3). RNase-treated and -nontreated cells were then fixed and analyzed by immunofluorescence with antibodies to actin (1C7), hnRNP U (CED17), PCAF, and H4 as described previously (3). Images were taken using an epifluorescence microscope (model DMRXA; Leica). Data acquisition was performed with a 100× objective (1.3 numerical aperture) and a digital camera (model C4742-95; Hamamatsu).
Microinjections.
Nuclei (200) of 50% confluent HeLa cells, DRB treated (2 h at 37°C, final concentration of 75 μM) and nontreated, were injected with CED17 antibody (7 mg/ml) or a nonspecific rabbit IgG at the same final concentration. Microinjections were performed in HEPES-buffered (20 mM) Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (Invitrogen) in the presence of DRB (Sigma) or TSA (as described above) or with both, using an Eppendorf FemtoJet in combination with an Eppendorf InjectMan application (Eppendorf). After injections, cells were kept at 37°C in the presence of DRB and TSA for 30 min (see Fig. 7). DRB and TSA were subsequently removed by exchanging the medium (DMEM supplemented with 10% fetal bovine serum). Cells were further incubated for 1 h at 37°C and then subjected to in vivo run-on transcription assays.
FIG. 7.
Actin, hnRNP U, and PCAF are associated with constitutively expressed genes, as determined by ChIP analysis. Lysates of cross-linked cells treated or untreated with the HDAC inhibitor TSA (+TSA, −TSA, respectively) were incubated with the indicated antibodies, and coprecipitated DNA was subjected to PCR using primers that amplify promoter and coding regions of the β-tubulin, S19, and GAPDH genes, as well as the 5S and 5.8S genes. Lanes 8 and 17, PCR amplification of 0.5% input chromatin.
Run-on transcription assay.
BrUTP incorporation was performed using the transfecting reagent FuGENE 6 according to the manufacturer's protocol (Roche). Briefly, HeLa cells were incubated with a BrUTP/FuGENE 6 mixture for 15 min at 4°C and then at 37°C for another 15-min period to allow BrUTP incorporation. The reaction was stopped by replacing the cell culture medium with ice-cold PBS. Cells were immediately fixed with 3.7% formaldehyde, and the incorporated BrUTP was monitored by immunofluorescence with a mouse monoclonal anti-BrUTP antibody, revealed with an Alexa 568-conjugated goat anti-mouse antibody (Invitrogen). The injected CED17 antibody was detected with an Alexa 488-conjugated donkey anti-rabbit antibody (Invitrogen). Coverslips were mounted on glass slides in Mowiol (Calbiochem) mounting medium. Cells displaying BrUTP incorporation relative to those that did not display BrUTP incorporation were manually counted by visualization with fluorescence microscopy. Immunostaining and confocal microscopy were carried out with cells grown on coverslips, as described previously (3). Images were taken with an LSM 510 model laser scanning microscope (Zeiss).
RESULTS
The HAT PCAF interacts with actin.
To identify nuclear actin binding proteins, HeLa nuclear extracts were subjected to DNase I affinity chromatography. This method is based on the use of DNase I conjugated to CNBr-activated Sepharose and has been proven to be highly selective for coprecipitation and affinity purification of actin from nuclear extracts (8, 37). Among the DNase I-bound proteins that we analyzed on immunoblots, we confirmed the presence of actin, hnRNP U, and pol II (Fig. 1A, cf. lanes 1, 2, and 3) (8). In addition, one of the other bands revealed by Coomassie staining of the gel corresponded to the HAT PCAF on the basis of its reactivity with a PCAF-specific monoclonal antibody (Fig. 1A, lane 3). Remarkably, the HAT CBP could not be detected in the soluble nuclear extracts subjected to DNase I affinity chromatography (not shown). CBP was revealed only on immunoblots of nuclear proteins extracted under high-salt conditions, which are not compatible with nuclear actin precipitation using DNase I beads. The immunoblots shown in Fig. 1A also show that the HAT TFIIIC220 (9, 26) and the splicing factor ASF/SF2 could not be found among the actin-associated proteins. Finally, actin, hnRNP U, PCAF, and pol II could not be coprecipitated from nuclear extracts by using unconjugated Sepharose beads, supporting the specificity of DNase I affinity chromatography (Fig. 1A, lane 2). Consistently, a mouse monoclonal anti-PCAF antibody specifically coprecipitated actin, hnRNP U, PCAF, and pol II from nuclear extracts (Fig. 1B). We next cloned and expressed the recombinant hnRNP U subdomains U-N, U-M, and U-C to determine whether PCAF associated with the C-terminal domain of hnRNP U containing the actin-binding site (8). The hnRNP U constructs were cloned with an S tag for binding to protein S-agarose, and affinity chromatography was performed (Fig. 1C). For this experiment, the hnRNP U constructs were coupled to the beads and incubated with HeLa nuclear extracts. Bound proteins were separated by SDS-PAGE and analyzed on immunoblots. Figure 1 shows that the hnRNP U-C beads coprecipitated endogenous actin, pol II, and PCAF, as well as full-length hnRNP U (Fig. 1D, lane 4), but none of the proteins was coprecipitated with the hnRNP U-N or hnRNP U-M beads (Fig. 1D, cf. lanes 2 and 3). In agreement with the above-described results, actin, hnRNP U, PCAF, and pol II were specifically coprecipitated from HeLa cell nuclear extracts also with an anti-β-actin antibody (Fig. 1E, lane 4), whereas none of the proteins was coprecipitated with unrelated control antibodies (Fig. 1E, lane 3). We conclude that actin, hnRNP U, PCAF, and pol II are associated and support the view that they may be part of the same complex.
FIG. 1.
PCAF is associated with actin and hnRNP U. (A) DNase I affinity chromatography reveals the association of PCAF with nuclear actin. (B) Coimmunoprecipitation of actin, hnRNP U, and pol II with anti-PCAF antibodies from HeLa nuclear extracts. The mock experiment corresponds to nuclear extracts incubated only with protein A/G-Sepharose. Control immunoprecipitation (IP) was a polyclonal anti-GFP antibody. (C) Schematic representation of the hnRNP U constructs used in this study. The C-terminal actin-binding motif is highlighted. The amino acid residues R and K required for actin binding are in boldface. (D) Actin, PCAF, and pol II are specifically precipitated from nuclear extracts with the hnRNP U-C construct (lane 4) but not with the hnRNP U-N (lane 2) or the hnRNP U-M (lane 3) construct. (E) A complex containing actin, hnRNP U, PCAF, and pol II is coprecipitated from nuclear extracts with an anti-β-actin antibody. Numbers at left indicate molecular mass in kDa.
Disruption of the actin-hnRNP U complex reduces histone acetylation levels.
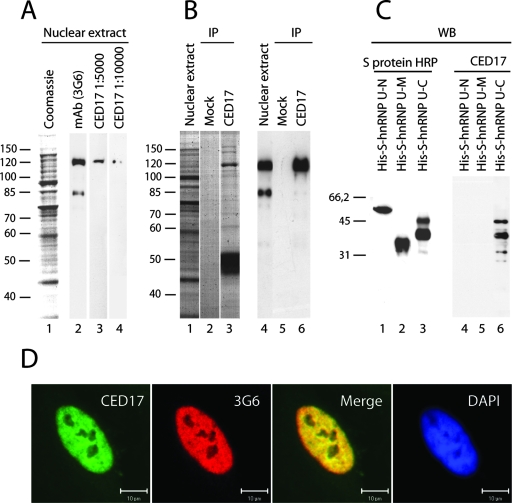
We next determined whether an intact actin-hnRNP U interaction is required for PCAF association with the nuclear actin complex. To address this question, we made a novel peptide-specific polyclonal antibody, CED17, raised against a short 15-amino-acid-long peptide encompassing the actin-binding site on the hnRNP U C terminus (8). After affinity purification using the same antigen, CED17 was found to be monoreactive for hnRNP U and to efficiently immunoprecipitate endogenous hnRNP U from HeLa nuclear extracts, and it cross-reacted with the bacterially expressed hnRNP U-C construct but not with the hnRNP U-N or hnRNP U-M construct, as revealed on immunoblots (Fig. 2A to C). In addition, CED17 displayed colocalization with the monoclonal anti-hnRNP U antibody 3G6 (Fig. 2D).
FIG. 2.
Characterization of the anti-hnRNP U antibody CED17. (A) Western blotting analysis of nuclear extracts was prepared from HeLa cells (lanes 3 and 4) and compared with the anti-hnRNP U monoclonal antibody 3G6 (lane 2). (B) Nuclear extracts were subjected to immunoprecipitations with CED17. Precipitated proteins were resolved by SDS-PAGE (lanes 1 to 3) and analyzed on immunoblots with the 3G6 antibody (cf. lanes 4 to 6). (C) CED17 specifically recognized the recombinantly expressed and purified hnRNP U-C construct as revealed on immunoblots (lane 6). HRP, horseradish peroxidase. (D) CED17 exhibits nuclear staining and colocalizes with the the monoclonal anti-hnRNP U 3G6 antibody in HeLa cells (scale bar, 10 μm). Numbers at left indicate molecular mass in kDa.
In view of its specificity, we next determined whether CED17 could disrupt the actin-hnRNP U interaction by direct competition for the actin-binding site on hnRNP U. We incubated nuclear extracts without and with increasing amounts of affinity purified CED17 and applied DNase I affinity chromatography. The DNase I-bound proteins were then resolved by SDS-PAGE and analyzed on immunoblots. The experiment shown in Fig. 3 demonstrates that incubation of nuclear extracts with increasing amounts of CED17 did not affect actin binding to the DNase I beads but it specifically released hnRNP U (Fig. 3A, cf. lanes 2 and 5). Pol II and PCAF were also released from the DNase I beads concomitantly with hnRNP U (Fig. 3A, cf. lanes 2 and 5). However, precipitations of hnRNP U, PCAF, and pol II were not affected by competition for a nonspecific rabbit IgG (Fig. 3A, lane 6). In addition, the relative levels of actin, PCAF, and pol II coprecipitated from nuclear extracts preincubated with the CED17 antibody using hnRNP U-C beads were significantly reduced (Fig. 3B, cf. lanes 3 and 4). Finally, the CED17 antibody failed to precipitate actin from nuclear extracts and coprecipitated lower amounts of both PCAF and pol II (Fig. 3C, lane 3).
FIG. 3.
Disruption of the actin-hnRNP U complex releases PCAF. (A) DNase I affinity chromatography performed with nuclear extracts preincubated with increasing amounts of CED17 antibody (+Ab) or a control antibody. Lanes 1, nuclear extract; 2, no competing antibody; 3 to 5, decreasing amounts of competing antibody; 6, control antibody. (B) Pull-down experiment using the hnRNP U-C construct bound to protein S-agarose beads. Nuclear extracts were preincubated with CED17 or a control antibody and subjected to pull-down experiments with hnRNP U-C beads. Bound proteins were resolved by SDS-PAGE and analyzed on immunoblots. Lanes 1, nuclear extract; 2, mock experiment where unconjugated beads are incubated with nuclear extracts; 3, no competing antibody; 4, preincubation with CED17; 5, preincubation with control antibody. (C) Immunoprecipitations using CED17. After nuclear extracts were precleared with protein A/G-Sepharose, they were subjected to immunoprecipitation with CED17. Bound proteins were resolved by SDS-PAGE and revealed on immunoblots. Lanes 1, nuclear extract; 2, mock experiment where protein A/G-Sepharose beads are incubated with nuclear extracts for preclearing; 3, CED17 immunoprecipitated proteins.
Next, we fractionated soluble HeLa nuclear extracts by gel filtration chromatography on a Superose 6HR column and monitored copurification of proteins on immunoblots. Actin, hnRNP U, PCAF, and pol II were found to coelute in fractions containing protein assemblies with apparent molecular masses of 2 to 3 MDa, which, significantly, did not contain histone H3 (Fig. 4A). Since actin, hnRNP U, PCAF, and pol II were found to coelute under native conditions (Fig. 4A), HeLa nuclear extracts were preincubated with the CED17 antibody or nonspecific rabbit IgGs and subsequently subjected to gel filtration chromatography on a Superose 6HR colum. As shown in Fig. 4B, after preincubation with CED17, the signals for pol II and actin were preferentially detected toward lower-molecular-weight fractions, at apparent molecular masses of about 700 kDa in comparison to their elution profiles obtained in the absence of CED17 (Fig. 4A). Under the same conditions, hnRNP U and PCAF elutions from the Superose 6HR column were also altered, with new signals detected at molecular masses less than 670 kDa (Fig. 4B). In all cases, the elution profiles were only marginally altered upon preincubation with nonspecific IgGs (Fig. 4C). Therefore, we conclude that under native conditions, CED17 specifically affected the stability of the actin complex presumably by releasing some of its core components. Overall, these results are consistent with the competition experiments (Fig. 3). They indicate that actin, PCAF, hnRNP U, and pol II are associated together as part of the same complex under native conditions and suggest that the actin-hnRNP U interaction is needed for its integrity.
FIG. 4.
Actin, hnRNP U, PCAF, and pol II are part of the same native complex. (A) Resolution of actin containing nuclear complexes by gel filtration. Fractions obtained from a Superose 6HR column were analyzed on Western blots. The position of the 670-kDa marker (thyroglobulin) is indicated. Void, void volume. (B and C) Resolution of actin-containing nuclear complexes with a Superose 6HR column following preincubation of HeLa nuclear extracts with 30 ng/μl CED17 or nonspecific polyclonal rabbit IgGs, respectively.
If PCAF is part of the same complex with actin, it is likely that the subset of nuclear actin-associated proteins precipitated with DNase I beads exhibits inherent HAT activity. To test this possibility, we set up an in vitro HAT activity assay of the DNase I-bound fraction obtained after applying DNase I affinity chromatography to HeLa nuclear extracts, after extracts were preincubated with increasing amounts of CED17. The DNase I-bound fraction was first washed and then supplemented with purified core histones and [3H]acetyl-CoA, and the reaction was allowed for 45 min at 30°C. The incorporated radioactivity was measured by liquid scintillation. Figure 3 shows that concomitantly with PCAF release from the DNase I-bound fraction, we could detect specific reduction in HAT activity in up to five independent experiments (Fig. 5 and data not shown). We suggest that the decrease in HAT activity due to disruption of the actin-hnRNP U interaction by competition with CED17 is caused primarily by PCAF release from the actin complex.
FIG. 5.

Disruption of the actin-hnRNP U interaction represses HAT activity. Nuclear extracts were subjected to DNase I affinity chromatography, and bound proteins were assayed for any HAT activity by using purified core histones as substrate. DNase I pull-down experiment was performed after preincubation of nuclear extracts with increasing amounts of CED17 or a control nonspecific rabbit IgG. Measurements were performed by liquid scintillation. Counts are normalized relative to the bound protein fraction obtained in the absence of the competing CED17 antibody. Experiments were successfully reproduced up to five times. Bars depict standard deviations.
PCAF, hnRNP U, and actin associate with the pol II phosphoCTD and active genes.
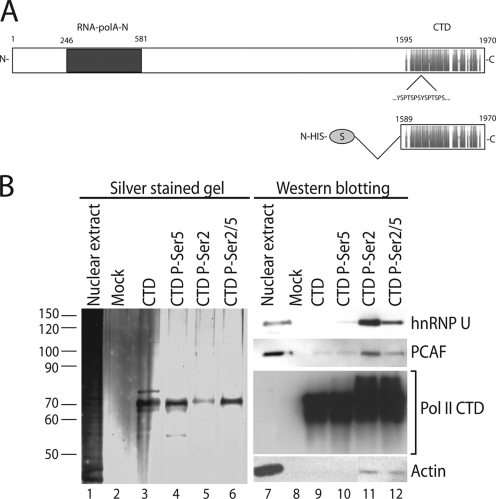
We next determined whether actin, hnRNP U, and PCAF are associated with active pol II genes, using a combination of protein-protein interaction and ChIP assays. To determine whether actin, hnRNP U, and PCAF are coupled to the pol II CTD, we cloned and expressed an S-tagged version of the human pol II CTD construct containing all conserved heptad repeats, which are normally phosphorylated on Ser2 and Ser5 during ongoing transcription (14, 25) (Fig. 6A). Purified recombinant CTD was conjugated to protein S-agarose beads. The CTD was phosphorylated by Ser2, Ser5, or both (Ser2/5) simultaneously, using recombinant cdk7 and cdk9 complexes, and the phosphorylation levels were monitored with radioactive ATP as the substrate (data not shown). Phosphorylated and nonphosphorylated CTD beads were incubated with nuclear extracts from HeLa cells, and the bound proteins were resolved by SDS-PAGE, revealed by silver staining, and analyzed on immunoblots. Figure 6B shows that actin, hnRNP U, and PCAF were coprecipitated only by the CTD constructs phosphorylated by Ser2 or Ser2/5 (Fig. 6B, cf. lanes 5 and 6 and 11 and 12). Remarkably, none of the proteins under investigation was coprecipitated with unphosphorylated CTD or with the Ser5-phosphorylated CTD (Fig. 6B, cf. lanes 3 and 4 and 9 and 10).
FIG. 6.
Actin, hnRNP U, and PCAF are associated with specific phosphorylation states of the pol II CTD. (A) Schematic representation of the recombinant S-tagged CTD construct used in this study. (B) Recombinantly expressed CTD was bound to protein S-agarose and phosphorylated with cdk7, cdk9, or with both to obtain Ser5-specifc, Ser2-specific, and Ser2/5-specific CTD phosphorylation (P) states, respectively. The beads were incubated with nuclear extracts, and bound proteins were resolved by SDS-PAGE (lanes 1 to 6) and analyzed on immunoblots (lanes 7 to 12). The mock experiment was performed by incubating unconjugated protein S-agarose beads with nuclear extracts. Numbers at left indicate molecular mass in kDa.
If actin, hnRNP U, and PCAF are simultaneously associated with the Ser2/5- and Ser2-phosphorylated states of the CTD which normally correlate with the elongating pol II, it is possible that they are also located at the gene coding region. To determine whether and how actin, hnRNP U, and PCAF associate with active genes, we performed ChIP experiments with three constitutively expressed pol II genes, the β-tubulin, S19, and GAPDH genes. For these studies, we used two different antibodies against hnRNP U, the monoclonal 3G6 antibody raised against the entire protein, and our CED17 antibody specifically raised against the actin-binding site on hnRNP U. Figure 7 shows that antibodies to pol II, PCAF, and β-actin precipitated gene promoters and coding regions (Fig. 7, cf. lanes 1, 3, and 4). In the case of hnRNP U, all gene promoters were specifically coprecipitated with the CED17 antibody but not with the 3G6 antibody (Fig. 7, cf. lanes 2 and 9). The opposite held true for the coding region of each gene analyzed, indicating that different hnRNP U epitopes are available at the promoter and coding region. As expected, an anti-TBP antibody precipitated only the gene promoters (Fig. 7, lane 5). To determine whether the association of actin, hnRNP U, PCAF, and pol II with the constitutively expressed genes is dependent on HAT activity, we subjected HeLa cells to treatment with the HDAC inhibitor TSA and performed ChIP experiments with the same antibodies. Consistent with a general decrease in the rate of histone deacetylation, TSA treatment revealed considerable quenching of the signals at both promoters and coding regions for PCAF and hnRNP U, whereas actin and pol II were marginally affected (Fig. 7). In the presence of TSA, the anti-TBP antibody did not precipitate the promoter and coding regions (Fig. 7, lane 14). As expected, in all cases, promoters and coding regions were precipitated with an anti-histone H3 antibody (Fig. 7, cf. lane 6 and 15). Independently of the TSA treatment, a control nonspecific antibody did not precipitate promoters and coding regions. Moreover, except for the anti-H3 antibody, none of the antibodies used in the ChIP experiments precipitated the 5S and 5.8S genes, altogether supporting the specificity of the assays (Fig. 7). We conclude that actin, hnRNP U, and PCAF are located at the promoters and coding regions of constitutively expressed genes. Next, we quantified the ChIP results obtained in the presence and absence of TSA treatment with respect to the steady-state levels of promoter and coding regions precipitated with the anti-H3 antibody (Fig. 8A and B). Our results indicate that pol II, hnRNP U, PCAF, and actin are present at both the promoters and the coding regions of constitutively expressed genes under normal conditions, in the absence of TSA. However, similar levels of occupancy by pol II, hnRNP U, PCAF, and actin are displayed only along the gene (Fig. 7, 8A, and 9A). Gene and promoter occupancies are altered in the presence of TSA, and these alterations are especially evident in the case of hnRNP U and PCAF (Fig. 7, 8B, and 9B). In either case, hnRNP U association with the promoter can be revealed only with the CED17 antibody, which is known to compete with actin for hnRNP U binding (Fig. 3 and 4). These results, together with data averaging from all three genes analyzed by ChIP in the presence and absence of TSA, support the idea that actin and hnRNP U are not closely associated with each other at the gene promoter (Fig. 9). We conclude that actin, hnRNP U, and PCAF are likely to become associated as part of the same complex at the gene coding region, preferentially interacting with the Ser2- and Ser2/5-phosphorylated CTD.
FIG. 8.
Relative occupancies at individual promoters and coding regions of actin, hnRNP U, and PCAF. The bar diagrams show the relative amounts of gene promoters and coding regions precipitated with antibodies against actin, hnRNP U (3G6), PCAF, β-actin, TBP, histone H3, and hnRNP U (CED17), as well as with a nonspecific rabbit IgG used as control, calculated in the absence (A) and presence (B) of TSA. Values were obtained by densitometric calculations of PCR products resolved by agarose gel electrophoresis (and revealed by ethidium bromide staining) in three independent experiments. Error bars represent standard deviations.
FIG. 9.
Average occupancies at promoter and coding regions in the absence (A) and presence (B) of TSA. Bar diagrams represent average values calculated from the analysis of all promoters and coding regions of the β-tubulin, S19, and GAPDH genes coprecipitated in three independent ChIP experiments using the indicated antibodies (two-tailed Student's t test, 0.01 < ** < 0.05). The values are normalized relative to the signal values obtained in ChIP experiments using an anti-histone H3 antibody. The levels of occupancy were calculated for each individual gene promoter and coding region in the presence and absence of TSA treatment, respectively (data not shown).
PCAF, hnRNP U, and actin associate with RNA along coding region of active genes.
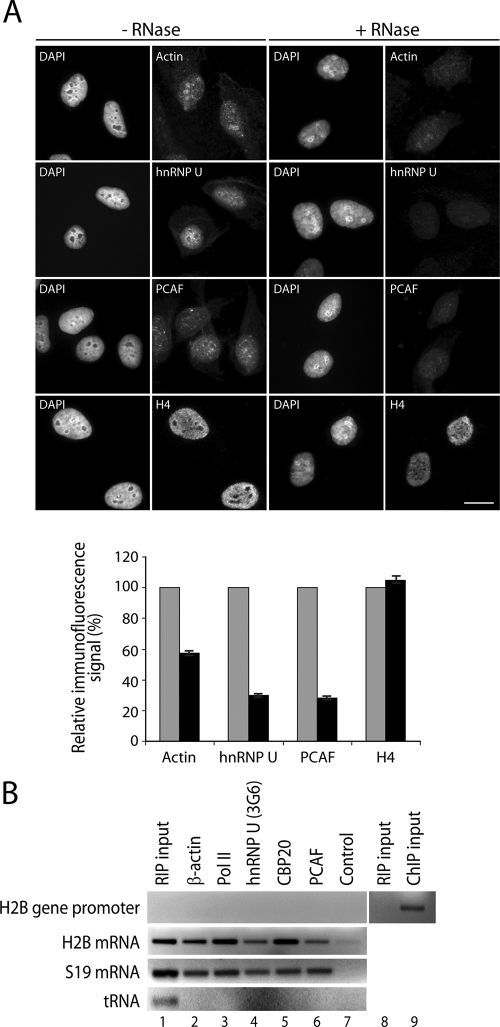
If actin, hnRNP U, and PCAF associate with elongating pol II and are present along the coding region of active genes, they may also be coupled to RNA transcripts presumably as part of RNP complexes. To start addressing this question, we performed in situ RNase treatment. HeLa cells were mildly permeabilized with Triton X-100 and then incubated with RNase A (3). After treatment, cells were fixed and studied by indirect immunofluorescence and wide-field microscopy. To analyze the distribution of actin, we used a monoclonal antibody against the actin dimer (1C7), which is known to preferentially recognize the nuclear fraction of cellular actin (24). The nuclear distributions of hnRNP U and PCAF were analyzed with CED17 and with a monoclonal anti-PCAF antibody, respectively. As expected for RNA-associated proteins, after RNase treatment was performed, we found that the bulk staining of actin, hnRNP U, and PCAF was considerably decreased from the nucleoplasm (Fig. 10A). Unbiased statistical analysis of the RNase-treated and -nontreated cells showed a 40% to 70% reduction in the overall amounts of cellular actin, hnRNP U, and PCAF (Fig. 10A). On the contrary, the overall levels of histone H4, normally associated with chromatin, were not affected by the in situ treatment with RNase A (Fig. 10A). Under the same permeabilization conditions but in the absence of RNase treatment, the distributions of actin, hnRNP U, PCAF, and H4 were not affected (Fig. 10A), altogether suggesting that in the cell nucleus, a considerable fraction of actin, hnRNP U, and PCAF is coupled to RNP complexes.
FIG. 10.
Actin, hnRNP U, and PCAF are associated with nascent ribonucleoprotein complexes. (A) The intranuclear distributions of actin, hnRNP U, and PCAF are sensitive to RNase treatment. HeLa cells were permeabilized with Triton X-100 (0.1%, 10 min, room temperature) and treated with RNase A (1 mg/ml, 10 min, room temperature) prior to fixation. After treatment, cells were immunostained with antibodies to actin, hnRNP U, PCAF, fibrillarin, and histone H4. The protein distributions were analyzed by wide-field microscopy. Quantification was performed by measurements taken from selected regions of interest corresponding to different subnuclear domains. Three different regions were selected randomly for each nucleus on the 4′,6′-diamidino-2-phenylindole (DAPI)-stained images, excluding nucleoli. The three selected regions were used to measure the Alexa 488 or Alexa 568 signals derived from the fluorochrome-conjugated secondary antibodies. The signal was quantified using ImageJ software. For each region, the mean gray value was registered after background normalization. The number of regions measured for actin, hnRNPU, PCAF, and H4 was between 89 and 67. The mean gray values were averaged and expressed as percentages on histograms. The average of the mean gray values measured in control cells was determined to be 100% of signal, and the average of the mean gray values measured after RNase treatment was expressed proportionately. Error bars represent standard deviations. (B) RIP analysis. Lysates of cross-linked cells were incubated with the indicated antibodies, and coprecipitated RNA was subjected to reverse transcription-PCR (RT-PCR) using primers that amplify the H2B gene promoter, H2B mRNA, S19 mRNA, and tRNATyr. Lane 1 shows PCR amplification of 1.5% input RNA. Lanes 8 and 9, RIP and ChIP inputs were analyzed by PCR with H2B promoter-specific primers.
To investigate the potential association of actin, hnRNP U, and PCAF with RNA transcripts still coupled to chromatin, we next performed chromatin RIP experiments with formaldehyde-cross-linked HeLa cells (4). After extraction, chromatin was fragmented by sonication and incubated with antibodies to actin (AC74), hnRNP U (3G6), PCAF, and pol II. At the end of the procedure, the RNA was extracted and reverse transcribed. A cDNA aliquot obtained by reverse transcription with oligo(dT) primers was analyzed by PCR with primers specific for the H2B promoter, H2B mRNA, and S19 mRNA. cDNA obtained by reverse transcription with random N6 oligo primers was analyzed with primers specific for tRNATyr. Figure 10 shows that antibodies to actin (AC74), pol II (8WG16), hnRNP U (3G6), and PCAF, as well as a polyclonal anti-CBP20 antibody used as a positive control, immunoprecipitated S19 and H2B mRNA. None of the antibodies used in the study precipitated a tRNA species (Fig. 10B) or the H2B gene promoter (Fig. 10B). Moreover, the H2B promoter-specific primers gave a signal only when ChIP input was used as the template in the PCR. In contrast, no signal was detected when RIP inputs were used, indicating that no DNA template contaminated the RIP assays (Fig. 10B). Finally, a nonspecific IgG did not precipitate any of the RNA species analyzed, supporting the specificity of the RIP assay (Fig. 10B). We conclude that actin, hnRNP U, and PCAF are associated with native RNP complexes along coding regions of active genes in living cells.
Actin-hnRNP U interaction assists transcription elongation via a HAT-dependent mechanism.
If actin, hnRNP U, and PCAF are part of the same complex with phosphorylated CTD, they are found along the coding region of active genes, and they are part of RNP complexes, they may cooperate in pol II transcription activation and maintenance. To start proving this possibility, we injected CED17 into HeLa cell nuclei to monitor pol II transcription under conditions that disrupt the actin-hnRNP U complex. Before injections, the elongating pol II was stalled by treatment with DRB (reviewed in reference 25), which is known to reversibly repress transcription by direct CDK inhibition. Following antibody injection, DRB was removed, and the cells were incubated with CED17 for up to 1 h. We next performed BrUTP incorporation in live cells. The levels of BrUTP incorporated in elongating RNA transcripts were monitored by indirect immunofluorescence with a monoclonal anti-BrUTP antibody and confocal microscopy. In the experiment shown in Fig. 9, more than 75% of the cells injected with CED17 exhibited significant downregulation of BrUTP incorporation (Fig. 11A, arrows) in comparison with noninjected cells. BrUTP incorporation was not affected when CED17 was injected in the cytoplasm (Fig. 11A, arrowheads) or after nuclear microinjections of a nonspecific antibody (Fig. 11A), supporting the specificity of the assay. Finally, CED17 injection did not alter the nuclear distributions of actin, hnRNP U, and PCAF, as revealed by immunofluorescence and confocal microscopy (not shown). Statistical evaluation of the relative coefficient of BrUTP incorporation (defined as the ratio between cells that did not incorporate BrUTP versus those that did incorporate BrUTP after CED17 injection) showed 2.5- to 3-fold transcription inhibition after CED17 nuclear injections. Remarkably, this effect was not encountered in control cells not treated with DRB that, instead, displayed normal levels of BrUTP incorporation due to efficient pol II transcription (Fig. 11B). Moreover, the inhibitory effect of CED17 was counteracted by treatment of cells with TSA, independently of the presence of DRB (Fig. 11B). These results together with evidence that the inhibitory effect of CED17 on global transcription is counteracted by TSA treatment suggest that disruption of the actin-hnRNP U interaction inhibits pol II transcription through a mechanism that is likely to involve PCAF.
FIG. 11.
In vivo disruption of the actin-hnRNP U interaction represses pol II transcription in a HAT-dependent manner. (A) The CED17 antibody or nonspecific polyclonal rabbit IgGs were injected into nuclei or cytoplasm (as control) of HeLa cells. After cells were injected, DRB was washed away and cells were incubated for 1 h at 37°C. Cells were then subjected to BrUTP incorporation to measure the effect of the CED17 antibody on pol II transcription. The injected antibody was visualized with an Alexa 488-conjugated goat anti-rabbit antibody. The incorporated BrUTP was visualized with a monoclonal anti-BrUTP antibody. Panel A shows examples of injected and noninjected HeLa cells after treatment with DRB. Arrows indicate CED17 or IgG nuclear injections, whereas arrowheads indicate cytoplasmic injections of CED17 (scale bar, 10 μm). (B) The bar diagram shows the coefficient of BrUTP incorporation (the ratio of cells which did not incorporate BrUTP relative to those that incorporated BrUTP following CED17 injection) after cells were counted directly by microscopy. Where indicated, experiments were performed in the presence of DRB (DRB treated), TSA (TSA treated), or DRB and TSA simultaneously (DRB-TSA treated). Error bars represent standard deviations.
To further support this idea, we monitored the association of actin, hnRNP U, PCAF, and pol II with the coding region of the constitutively expressed S19 gene after stalling the elongating pol II with DRB in time course experiments. At each time point, HeLa cells were subjected to ChIP analysis using antibodies against actin, hnRNP U (3G6), PCAF, H3, and acetylated H3K9, as well as antibodies to inactive pol II (8WG16) and to the Ser2-phosphorylated pol II CTD (H5) construct. After 2 h, DRB incubation, antibodies to actin, hnRNP U, PCAF, and phosphorylated CTD did not coprecipitate the coding region of the S19 gene (Fig. 12). Under the same conditions, while immunoprecipitation levels with antibodies to H3 were not affected by inhibition of CTD phosphorylation, we observed a quenching of the signals obtained with the antibody to acetylated H3K9 (Fig. 12A). Statistical evaluations of the ChIP data with respect to the overall levels of pol II (8WG16 antibody) showed a progressive release of actin, hnRNP U, PCAF, and Ser2-phosphorylated CTD from the gene coding region (Fig. 12B). Quantification of the ChIP signals obtained with the acetylated H3K9 antibody with respect to the signals from the total H3 antibody revealed a considerable 40% reduction in H3K9 acetylation levels along the gene coding region (Fig. 12C). We conclude that the association of actin, hnRNP U, PCAF, and pol II with the coding region and H3K9 acetylation levels are sensitive to the phosphorylation state of the CTD.
FIG. 12.
Association of actin, hnRNP U, and PCAF with the gene coding region is dependent on the phosphorylated pol II CTD. (A) Lysates of cross-linked HeLa cells treated or untreated with the CDK inhibitor DRB at different time points were incubated with the indicated antibodies and coprecipitated DNA was subjected to PCR using primers that amplify coding region of the S19 gene. Lane 7, PCR amplification of 0.5% input chromatin. (B) Effect of DRB on the association of pol II CTD, hnRNP U, PCAF, and actin with active genes: densitometric quantification of relative signals obtained in the above-described ChIP experiments in the absence and presence of DRB treatment. The bar diagrams represent average values calculated from analysis of the coding region of the S19 gene coprecipitated in three independent ChIP experiments using the indicated antibodies. The values for hnRNP U, PCAF, β-actin, and phosphorylated (P)-Ser2 pol II CTD (H5 antibody) are normalized against the signals obtained using the pol II antibody 8WG16. (C) Densitometric quantification of the relative signals obtained in the ChIP experiments with the antibody against acetylated H3K9 in the absence and presence of DRB treatment. Average values from analysis of the S19 gene coding region coprecipitated in three independent ChIP experiments are normalized against the anti-histone H3 antibody.
Overall, these results also support the view that the actin-hnRNP complex cooperates with PCAF and phosphorylated pol II CTD to activate and maintain pol II transcription through a HAT-dependent mechanism.
DISCUSSION
In this study, we show evidence that in higher eukaryotes, HAT activity accompanies nuclear actin during its regulatory function in the transcription of protein-encoding genes. Consistent with this view, actin was specifically coprecipitated with hnRNP U and with the HAT PCAF, using a combination of different affinity chromatography methods. Actin, hnRNP U, and PCAF were also found to interact with the hyperphosphorylated pol II CTD. Consistent with the view that they are closely associated with elongating polymerase, ChIP analysis revealed their association with both promoter and coding regions of pol II genes. RNase treatment performed on living cells demonstrated that the nuclear distributions of actin, hnRNP U, and PCAF are sensitive to RNA. Consistent with the presence of actin and hnRNP U in pre-mRNP/mRNP complexes (6, 19, 20), RIP assays showed the association of actin, hnRNP U, and PCAF with RNA transcripts, which are still associated with chromatin. Therefore, we suggest that in mammalian cells, actin, hnRNP U, and PCAF are also likely to be integrated into nascent RNP complexes. Finally, we designed a novel peptide-specific antibody, CED17, against the actin binding motif on hnRNP U (8). CED17 specifically disrupted the actin-hnRNP U interaction with subsequent PCAF release from the actin-containing complex in affinity chromatography experiments and under native conditions. Remarkably, CED17-mediated disruption of the actin-hnRNP U complex resulted in a considerable decrease in the total HAT activity associated with the subset of proteins precipitated with nuclear actin. We suggest that the HAT activity associated with the actin complex is mediated primarily by PCAF. Based on these results, we speculate that the integrity of the actin-hnRNP U complex may be important for the association of PCAF. These mechanisms are likely to occur in vivo, since disruption of the actin-hnRNP U complex resulted in a transcriptionally repressed cellular state.
Previous reports emphasized the importance of PCAF association with promoter or enhancer regions to function as transcription coactivator, independently from pol II (7). We propose that the role of PCAF as a transcription coactivator is not restricted to promoter or enhancer regions. Given the association with the coding region of active genes and the association with the hyperphosphorylated form (Ser2/5) of the pol II CTD, we suggest that PCAF is required for transcription elongation in close association with pol II. PCAF could perform its function as part of large chromatin acetylating transcription coactivator complexes such as STAGA, which is already known to associate with components of the pre-mRNA splicing machinery (13). This idea underscores the requirement for ongoing histone modifications for efficient transcription elongation (10). In either case, we suggest that the potential involvement of PCAF in elongation is likely to require an intact actin-hnRNP U complex.
A role for actin in transcription initiation has been suggested for all three eukaryotic RNA polymerases (5, 22). Since actin is a component of growing RNP complexes (19) and specifically associates with the phosphorylated pol II (8), actin is likely to have a key role also in transcription elongation. Our present observations are consistent with this view and actually suggest that actin and hnRNP U specifically cooperate for productive elongation and may not be in contact in the initial transcriptional phases. As partly suggested by the CTD pull down experiments, the hyperphosphorylated CTD may have an important role in loading actin and hnRNP U onto the elongating pol II machinery and growing RNP complexes. We propose that once actin is recruited via CTD to nascent RNPs, hnRNP U probably functions as an adaptor protein to facilitate PCAF recruitment of active genes and to facilitate transcription elongation through a HAT-dependent mechanism. In support of this view, we found that the nuclear distributions of actin, hnRNP U, and PCAF are sensitive to RNase treatment and that all three proteins are closely associated with nascent RNPs. Based on this finding, we propose that the cooperative regulatory function of actin, hnRNP U, and PCAF on pol II transcription elongation is likely to be dependent on the presence of nascent RNA transcripts.
Consistent with recent findings of the involvement of N-WASP and the ARP2/3 complex in later phases of pol II transcription (31, 36), actin-mediated pol II transcriptional control may be sensitive to the different polymerization states of actin (30). Actin filaments that can be identified by phalloidin staining in the cytoplasm have not yet been revealed in the cell nucleus (reviewed in reference 22). Moreover, actin in complex with hnRNP U, PCAF, and pol II can be coprecipitated by DNase I affinity chromatography, which, notoriously, has a high affinity for monomeric actin (23). We have recently proposed that the transcriptionally competent actin may be present in a monomeric or oligomeric form which is different from the canonical actin filaments (16 and references therein). In agreement with this hypothesis, our present results show that the nuclear actin fraction which is sensitive to RNase treatment was detected with a monoclonal antibody specific for the actin dimer (24). It is possible that the polymerization states of actin involved in the initiation or elongation phases are different (16).
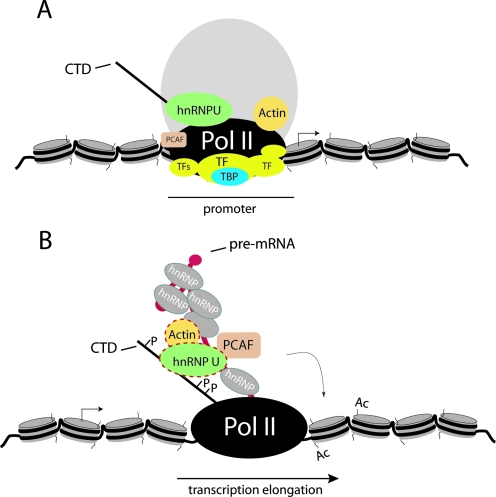
In either case, our results support a general role for actin in pol II transcription. Actin is likely to function as an allosteric regulator mediating specific protein-protein interactions for productive mRNA synthesis. The molecular mechanisms that facilitate pol II processivity by establishing and maintaining a transcriptionally active chromatin state may require the specific interaction between actin and hnRNP U to facilitate PCAF-mediated HAT activity (Fig. 13). Among the transient histone modifications coordinated by the phosphorylated pol II CTD and required for efficient pol II transcription elongation, hypoacetylated histone levels are found toward the 3′ end of active genes (10, 15). We speculate that PCAF recruitment to elongating pol II for passage of the polymerase through hypoacetylated nucleosome barriers may be facilitated by actin and hnRNP U.
FIG. 13.
Model for actin-hnRNP U-mediated control of pol II transcription elongation. (A) Actin and hnRNP U may not be in contact during transcription initiation (TF, transcription factors). (B) During transcription, actin may be recruited to the elongating transcription machinery via the hyperphosphorylated CTD and then to the nascent RNP, where actin in complex with the hnRNP U could facilitate recruitment of PCAF to the active gene to enhance the processivity of the elongating pol II.
Acknowledgments
We thank Chandrasekhar S. Raju for help with the microinjections.
This work was supported by grants from the Swedish Research Council (Vetenskapsrådet) to P.P. A.O. and A.K. were supported by a predoctoral fellowship from the Swedish Institute and by fellowships from FEBS and Boehringer Ingelheim, respectively. E.L. is supported through postdoctoral research fellowships from Fondation pour la Recherche Médicale (FRM) and Wenner-Gren Foundation (Sweden).
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 27418285-18289. [DOI] [PubMed] [Google Scholar]
- 2.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fomproix, N., and P. Percipalle. 2004. An actin-myosin complex on actively transcribing genes. Exp. Cell Res. 294140-148. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert, C., A. Kristjuhan, G. S. Winkler, and J. Q. Svejstrup. 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14:457-464. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann, W. A., L. Stojiljkovic, B. Fuchsova, G. M. Vargas, E. Mavrommatis, V. Philimonenko, K. Kysela, J. A. Goodrich, J. L. Lessard, T. J. Hope, P. Hozak, and P. de Lanerolle. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6:1094-1101. [DOI] [PubMed] [Google Scholar]
- 6.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 112655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krumm, A., L. Madisen, X.-J. Yang, R. Goodman, Y. Nakatani, and M. Groudine. 1998. Long distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl. Acad. Sci. USA 9513501-13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kukalev, A., Y. Nord, C. Palmberg, T. Bergman, and P. Percipalle. 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12238-244. [DOI] [PubMed] [Google Scholar]
- 9.Kundu, T. K., Z. Wang, and R. G. Roeder. 1999. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 191605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 11.Marmorstein, R. 2001. Structure of histone acetyl transferases. J. Mol. Biol. 311433-444. [DOI] [PubMed] [Google Scholar]
- 12.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 27323781-23785. [DOI] [PubMed] [Google Scholar]
- 13.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 191401-1415. [DOI] [PubMed] [Google Scholar]
- 15.Mellor, J. 2006. Dynamic nucleosome and gene transcription. Trends Genet. 22320-329. [DOI] [PubMed] [Google Scholar]
- 16.Obrdlik, A., A. Kukalev, and P. Percipalle. 2007. The function of actin in gene transcription. Histol. Histopathol. 221051-1055. [DOI] [PubMed] [Google Scholar]
- 17.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 9435-44. [DOI] [PubMed] [Google Scholar]
- 18.Olave, I. A., S. L. Reck-Peterson, and G. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodelling. Annu. Rev. Biochem. 71755-781. [DOI] [PubMed] [Google Scholar]
- 19.Percipalle, P., J. Zhao, B. Pope, A. Weeds, U. Lindberg, and B. Daneholt. 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 153229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Percipalle, P., A. Jonsson, D. Naschchekin, C. Karlsson, T. Bergman, and B. Daneholt. 2002. Nuclear actin is associated with a specific subset of hnRNP A/B type proteins. Nucleic Acids Res. 301725-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Percipalle, P., N. Fomproix, K. Kylberg, F. Miralles, B. Björkroth, B. Daneholt, and N. Visa. 2003. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1006475-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Percipalle, P., and N. Visa. 2006. Molecular functions of nuclear actin. J. Cell Biol. 172967-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Percipalle, P., and A.-K. Östlund Farrants. 2006. Chromatin remodelling and transcription: Be-WICHed by nuclear myosin 1. Curr. Opin. Cell Biol. 18267-274. [DOI] [PubMed] [Google Scholar]
- 24.Schoenenberger, C. A., S. Buchmeier, M. Boerris, R. Sutterlin, U. Aebi, and B. M. Jockusch. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J. Struct. Biol. 152157-168. [DOI] [PubMed] [Google Scholar]
- 25.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and the long of it. Genes Dev. 182437-2468. [DOI] [PubMed] [Google Scholar]
- 26.Sjölinder, M., P. Björk, E. Söderberg, N. Sabri, A.-K. Östlund Farrants, and N. Visa. 2005. The growing pre-mRNA recruits actin and chromatin modifying factors to transcriptionally active genes. Genes Dev. 191871-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, E. R., J. M. Belote, R. L. Schiltz, X.-J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 262948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterner, D. E., and S. L. Berger. 2000. Acetylation and transcription-related factors. Microbiol. Mol. Biol. Rev. 64435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14804-816. [PMC free article] [PubMed] [Google Scholar]
- 30.Vieu, E., and N. Hernandez. 2006. Actin's latest act: polymerizing to facilitate transcription? Nat. Cell Biol. 8650-651. [DOI] [PubMed] [Google Scholar]
- 31.Wu, X., Y. Yoo, N. N. Okuhama, P. W. Tucker, G. Liu, and J. L. Guan. 2006. Regulation of RNA polymerase II-dependent transcription by N-WASP and its nuclear binding partners. Nat. Cell Biol. 8756-763. [DOI] [PubMed] [Google Scholar]
- 32.Xu, W., D. G. Edmondson, and S. Y. Roth. 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 185659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26229-232. [DOI] [PubMed] [Google Scholar]
- 34.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 9711303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, X.-J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382319-324. [DOI] [PubMed] [Google Scholar]
- 36.Yoo, Y., X. Wu, and J. L. Guan. 2007. A novel role of the actin nucleating ARP2/3 complex in the regulation of RNA polymerase II dependent transcription. J. Biol. Chem. 2827616-7623. [DOI] [PubMed] [Google Scholar]
- 37.Zechel, K. 1980. Isolation of polymerization competent cytoplasmic actin by affinity chromatography on immobilized DNase I using formamide as eluant. Eur. J. Biochem. 110343-348. [DOI] [PubMed] [Google Scholar]