Abstract
Colony-stimulating factor 1 (CSF-1) is the main growth factor controlling the development of macrophages from myeloid progenitor cells. However, CSF-1 also regulates some of the key effector functions of macrophages (e.g., phagocytosis and cytokine secretion). The endosomal SNARE protein syntaxin 7 (Stx7) regulates vesicle trafficking events involved in phagocytosis and cytokine secretion. Therefore, we investigated the ability of CSF-1 to regulate Stx7. CSF-1 upregulated Stx7 expression in primary mouse macrophages; it also upregulated expression of its SNARE partners Vti1b and VAMP8 but not Stx8. Additionally, CSF-1 induced the rapid serine phosphorylation of Stx7 and enhanced its binding to Vti1b, Stx8, and VAMP8. Bioinformatics analysis and results from experiments with kinase inhibitors suggested the CSF-1-induced phosphorylation of Stx7 was mediated by protein kinase C and Akt in response to phosphatidylinositol 3-kinase activation. Based on mutagenesis studies, CSF-1 appeared to increase the binding of Stx7 to its SNARE partners by inducing the phosphorylation of serine residues in the Habc domain and/or “linker” region of Stx7. Thus, CSF-1 is a key regulator of Stx7 expression and function in macrophages. Furthermore, the effects of CSF-1 on Stx7 may provide a mechanism for the regulation of macrophage effector functions by CSF-1.
Macrophages are a key component of the immune system, where they function as sentinels to detect pathogens (e.g., bacteria, viruses, and fungi) (18, 26). The phagocytosis and intracellular killing of pathogens by macrophages play a central role in host defense (19). Macrophages express a range of receptors (e.g., Fc, complement, and scavenger receptors) that enable them to phagocytose pathogens (1, 19, 51). The subsequent presentation of pathogen-derived peptides to T cells is important for the development of acquired immunity (15, 33). Additionally, the secretion of a range of cytokines (e.g., tumor necrosis factor [TNF], interleukin 6 [IL-6], and IL-12) and chemokines (e.g., CCL2 and CXCL8) by macrophages helps orchestrate both the innate and adaptive immune responses to infection (15, 18, 26).
The effector functions of macrophages are reliant upon the coordinated trafficking of intracellular vesicles from one compartment to another (e.g., trafficking of cytokine-containing vesicles from the endoplasmic reticulum to the plasma membrane) and fusion between different vesicles (e.g., fusion of phagosomes with lysosomes) (49). SNARE proteins are intimately involved in intracellular vesicle trafficking (22, 25, 55). They are a group of relatively small (∼15 to 40 kDa), largely membrane-associated proteins that are characterized by a conserved region of around 60 amino acids, referred to as a SNARE domain, and typically a short C-terminal stretch of hydrophobic residues that facilitates their anchoring to cellular membranes. Generally, R-SNARE proteins are found on the vesicle delivering the cargo, whereas Q-SNAREs are typically anchored to the vesicle target membrane (e.g., plasma membrane, phagosomal membrane, etc.). The docking and fusion of transport vesicles with target membranes are mediated by the direct interaction of R-SNAREs with Q-SNAREs to form trans-SNARE complexes consisting of one R-SNARE protein and two to three Q-SNARE proteins (22, 25, 55).
Although colony-stimulating factor 1 (CSF-1) is the main growth factor governing the proliferation, differentiation, and survival of macrophages (48), it also modulates several macrophage effector functions, including cytokine secretion, phagocytosis, and macropinocytosis. For example, CSF-1 primes macrophages for enhanced TNF, IL-6, and IL-12p40 secretion in response to lipopolysaccharide (LPS) (50). Conversely, pharmacologic inhibitors of the CSF-1 receptor suppress the LPS-induced production of these cytokines both in vitro (24) and in vivo (13). CSF-1 has also been reported to enhance the phagocytosis of bacteria, fungi, and parasites by macrophages (11, 17, 45, 47). Given the role of SNARE proteins in regulating secretory and endocytic pathways, CSF-1 could potentially modulate the immune functions of macrophages by governing the expression, localization, and/or activity of SNARE proteins. The Qa-SNARE protein syntaxin 7 (Stx7), which interacts with Q-SNAREs Vti1b and Stx8 and the R-SNARE vesicle-associated membrane protein 8 (VAMP8), regulates late endosome fusion (3, 43). Stx7 has also been implicated in phagocytosis (12) and TNF secretion (37, 38) by macrophages. Therefore, we investigated the effects of CSF-1 on Stx7 in macrophages. The findings presented indicate that CSF-1 regulates Stx7 expression and function and suggest that CSF-1 may indeed modulate the effector functions of macrophages, at least in part, via its ability to regulate Stx7.
MATERIALS AND METHODS
Reagents.
Cell culture medium and supplements, fetal calf serum, SuperScript III reverse transcriptase, precast sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and anti-V5 antibodies were from Invitrogen. Pfu DNA polymerase and restriction enzymes were supplied by Promega. Precast Criterion gels and isoelectric focusing strips (pH 3 to 10) were obtained from Bio-Rad. The Stx7 and 18S rRNA real-time PCR probe sets were obtained from ABI. The affinity-purified rabbit polyclonal anti-Stx7 and anti-VAMP8 antibodies were as described previously (34), while the mouse monoclonal anti-Stx8 and anti-Vti1b antibodies were purchased from BD Biosciences. Anti-phospho-Erk1/2, anti-phospho-Akt, and anti-Akt antibodies were from Cell Signaling Technology. The anti-Erk2 and anti-CSF-1 receptor (C-20) antibodies were provided by Santa Cruz Biotechnology Inc. Protein G-Sepharose, enhanced chemiluminescence reagents, and [32P]orthophosphate (10 mCi/ml) were provided by GE Healthcare, while Complete protease inhibitors were obtained from Roche Biochemicals. The protein kinase C (PKC) inhibitors GF109203X and Gö6983, Akt inhibitors Akt VIII and Akt X, and phosphatidylinositol (PI) 3-kinase inhibitors wortmannin and LY294002 were from Calbiochem.
Mouse bone marrow-derived macrophages.
The use of mice in this study was approved by the Melbourne Health Animal Ethics Committee. Bone marrow-derived macrophages were obtained by culturing bone marrow cells from 6- to 8-week-old, female C57BL/6 mice in Dulbecco's modified Eagle's medium supplemented with 5,000 U/ml recombinant CSF-1, 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM GlutaMax-1 for 6 to 7 days at 37°C in a humidified atmosphere of 5% CO2 (27).
Real-time PCR analysis of gene expression.
Total RNA was isolated with an RNeasy Mini kit (Qiagen) and then reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR was performed using an ABI PRISM 7900HT sequence detection system and predeveloped TaqMan probe/primer combinations for mouse Stx7 and 18S rRNA from ABI. Threshold cycle numbers were transformed using the ΔΔCT and relative value method as described by the manufacturer.
Cell lysis.
Macrophages were lysed directly in tissue culture dishes with NP-40 lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10% glycerol, 1 mM sodium orthovanadate, 0.1 mM sodium molybdate, 10 mM NaF, 10 mM β-glycerophosphate, and Complete protease inhibitors) for 30 to 60 min on ice. The lysates were clarified by centrifugation at 13,000 × g for 10 min at 4°C and the protein concentrations of the supernatants then measured with a Bio-Rad protein assay kit. The cell lysates were either used immediately or stored at −70°C.
SDS-PAGE, Western blotting, and immunoprecipitation.
One-dimensional SDS-PAGE was performed according to standard procedures using 10% and 14% SDS-PAGE gels. For two-dimensional SDS-PAGE, isoelectric focusing (IEF) strips (pH 3 to 10; linear, 11 cm) were passively rehydrated in 8 M urea, 0.5% Triton X-100, 0.5% Pharmalytes (Bio-Rad), and 10 mM dithiothreitol (DTT) overnight at room temperature. Cell lysates were first concentrated using Centricon microconcentrators (Millipore Corp.), and then aliquots of the concentrated lysates (containing 400 μg of protein) were diluted with IEF sample solution (8 M urea, 50 mM DTT, 0.2% Pharmalytes, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate and 0.002% bromophenol blue). The samples were absorbed into IEF strips and IEF performed for 30,000 V·h at 20°C using a Protean IEF cell (Bio-Rad). The focused IEF strips were stored at 20°C overnight prior to resolution in the second dimension. The IEF strips were equilibrated in 0.5 M Tris (pH 8.8), 6 M urea, 2% SDS, 20% glycerol, and 2% DTT for 15 min before being loaded onto 10% Criterion precast gels, which were subjected to electrophoresis for 2 h at 100 V. The separated proteins were then transferred to polyvinylidene difluoride (PVDF) membranes and the membranes Western blotted with anti-Stx7 antibodies. For immunoprecipitation assays, aliquots of cell lysate (containing 500 to 1,000 μg protein) were precleared with protein G-Sepharose beads for 1 h at 4°C with mixing. The mixtures were then centrifuged at 13,000 × g for 5 min at 4°C and the cleared supernatants retained. One to two micrograms of the appropriate antibody was added to the supernatants and the samples incubated for 4 h at 4°C with mixing. Immune complexes were captured by the addition of protein G-Sepharose and incubating for an additional 1 h at 4°C with mixing. Following four washes with lysis buffer, the immunoprecipitates were subjected to Western blotting.
Metabolic labeling and phosphoamino acid analysis.
Macrophages were metabolically labeled with [32P]orthophosphate by incubating the cells in phosphate-free Dulbecco's modified Eagle's medium supplemented with [32P]orthophosphate (4 mCi/6-cm dish) for 6 h. Stx7 was immunoprecipitated from lysates of labeled macrophages, subjected to SDS-PAGE, and then transferred to a PVDF membrane. Following exposure to X-ray film, Stx7 bands were excised and the pieces of membrane washed extensively with water prior to being hydrolyzed in 6 M HCl for 90 min at 100°C. The phosphoamino acid contents of the hydrolysates were determined by one-dimensional high voltage electrophoresis on cellulose-coated thin-layer chromatography plates as previously described (10).
Expression vectors and mutagenesis.
A mammalian expression vector encoding an N-terminal V5-tagged form of mouse Stx7 (i.e., pEF-V5-Stx7) was created by PCR using the plasmid pcDNA-HA-Stx7 (34) as the template and the primers F1 (5′-ACG CGT TCT TAC ACT CCG GGG ATT GGT GGG GAC TCT-3′) and R1 (5′-ACG CGT TCA GCC TTT CAG TCC CCA TAC GAT GAG ACA-3′). The PCR product generated was digested with MluI and cloned into pEF-BOS-V5. The expression vector pEF-V5-Stx7S/A, in which Ser-125, Ser-126, and Ser-129 in Stx7 are replaced by alanine residues, was created by overlapping PCR using primers F1 and R-mutagenic 1 (5′-GG AAA ACC ACC CGC TAC CCT GGC GGC GGC TCG CAC TCG AGC-3′) and F-mutagenic 1 (5′-GCT CGA GTG CGA GCC GCC GCC AGG GTA GCG GGT GGT TTT CC-3′) and R1. The expression vector pEF-V5-Stx7S/E, in which Ser-125, Ser-126, and Ser-129 in Stx7 are replaced by glutamic acid residues, was created by overlapping PCR using primers F1 and R-mutagenic 2 (5′-AGG AAA ACC ACC CTC TAC CCT CTC TTC GGC TCG CAC TCG AGC-3′) and F-mutagenic 2 (5′-GCT CGA GTG CGA GCC GAA GAG AGG GTA GAG GGT GGT TTT CCT-3′) and R1. The inserts from pEF-V5-Stx7, pEF-Stx7S/A, and pEF-V5-Stx7S/E were subsequently excised with XbaI and cloned into the retroviral vector pMX-pie (40).
Retroviral transduction of bone marrow cells.
Replication-defective, ecotropic retroviruses expressing V5-tagged Stx7 (or mutants thereof) were generated by transfecting the BOSC23 packaging cell line (41) with the appropriate pMX-pie-based V5-Stx7 construct. Retroviral supernatants were harvested 48 h later, filtered through a 0.45-μm syringe filter, and supplemented with 10 mM HEPES (pH 7.1) and 8 μg/ml Polybrene. The viral supernatants were then used to transduce mouse bone marrow cells by spin infection (57). C57BL/6 bone marrow cells, which had been cultured for 2 days in the presence of CSF-1, were seeded in 24-well tissue culture plates (2 × 106 cells per well) and the plates centrifuged at 500 × g for 5 min at 30 to 37°C. The growth medium was removed and replaced with 2 ml of retroviral supernatant. The plates were then centrifuged at 1,500 × g for 90 min at 30 to 37°C, after which the viral supernatants were replaced with fresh growth medium containing CSF-1. A second round of spin infection was performed 24 h later. The cells were then cultured in the presence of CSF-1 until a homogenous population of bone marrow-derived macrophages was obtained (typically 4 to 5 days).
RESULTS
CSF-1 upregulates Stx7 expression in macrophages.
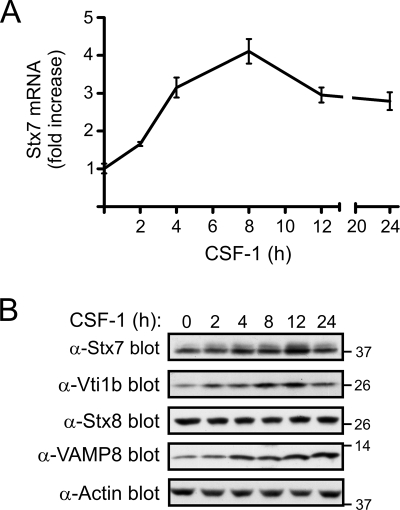
Given that CSF-1 can enhance phagocytosis, pinocytosis, and cytokine secretion by macrophages (11, 13, 24, 30, 39, 44, 50), we examined the effect of CSF-1 on the expression levels of Stx7, a Qa-SNARE protein that has been reported to play a role in these processes (9, 12, 38). Real-time PCR analysis revealed that Stx7 mRNA levels were upregulated as early as 2 h following the stimulation of mouse bone marrow-derived macrophages with CSF-1 and maximally induced (∼3- to 4-fold increase) by around 8 h (Fig. 1A). Stx7 mRNA levels declined slightly thereafter but remained above prestimulus levels for at least a further 16 h (Fig. 1A). In order to confirm that the CSF-1-induced upregulation of Stx7 gene expression resulted in a corresponding increase in the protein levels of Stx7, lysates of macrophages that had been stimulated with CSF-1 for up to 24 h were subjected to Western blotting with an anti-Stx7 antibody. CSF-1 induced an approximately twofold increase in Stx7 protein levels by 12 h poststimulation (Fig. 1B). Stx7 is known to form an endosomal SNARE complex with three other SNARE proteins, namely, Vti1b (a Qb-SNARE), Stx8 (a Qc-SNARE), and the R-SNARE VAMP8, to mediate membrane fusion between late endosomes (3, 43). Therefore, the ability of CSF-1 to regulate the expression of Stx8, Vti1b, and VAMP8 was also investigated. While Stx8 expression was unaffected by CSF-1, an increase in Vti1b expression was detected as early as 2 h post-CSF-1 stimulation (Fig. 1B). Maximal upregulation of Vti1b expression (∼2.5- to 3-fold) occurred by around 8 to 12 h poststimulation (Fig. 1B). VAMP8 protein expression was also upregulated approximately threefold by CSF-1 and remained elevated for at least 24 h (Fig. 1B).
FIG. 1.
Regulation of Stx7 expression by CSF-1. (A) Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being stimulated with CSF-1 for the time indicated. Total RNA was then extracted and reverse transcribed into cDNA, which was then subjected to quantitative real-time PCR. 18S rRNA was used as the internal control. Stx7 mRNA levels are relative to its expression in the absence of CSF-1, which was given an arbitrary value of 1.0. Stx7 mRNA levels (mean ± standard error) at each time point were measured in triplicate, and the data are representative of three experiments. (B) Macrophages were cultured as for panel A and cell lysates subsequently subjected to Western blotting with the indicated antibodies. The membrane was also probed with an antiactin antibody to assess loading. The positions of molecular mass markers (in kDa) are indicated on the right. α-Stx7, anti-Stx7; other antibodies are similarly indicated.
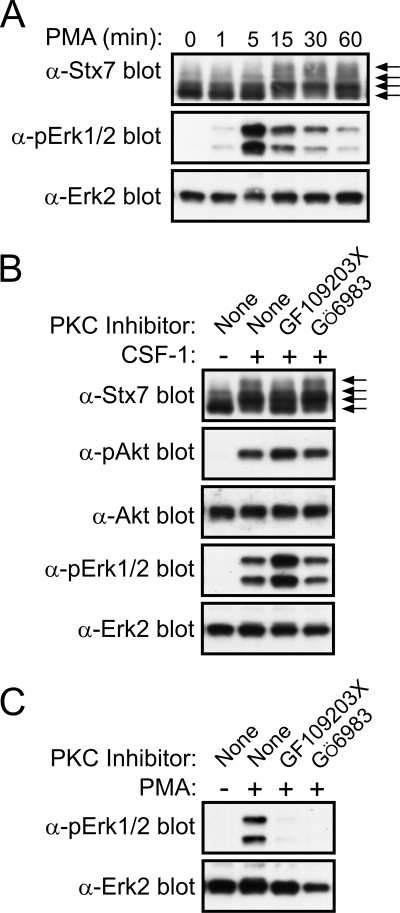
CSF-1 induces serine phosphorylation of Stx7.
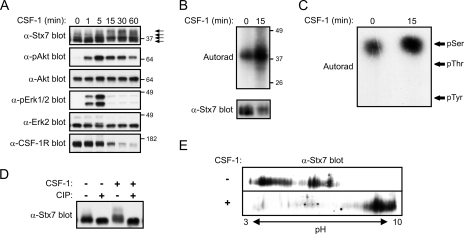
CSF-1 stimulation of macrophages also appeared to cause a slight reduction in the electrophoretic mobility of Stx7 (Fig. 1B). To examine further this apparent effect of CSF-1 on Stx7, CSF-1-deprived macrophages were subjected to acute stimulation with CSF-1 (i.e., for up to 60 min); lysates of the cells were then analyzed by Western blotting. A decrease in the electrophoretic mobility of Stx7 was detected as early as 5 min post-CSF-1 stimulation, with the change in mobility evident for at least 60 min (Fig. 2A). When SDS-PAGE conditions were optimal, it was possible to detect at least three to four isoforms of Stx7 in lysates of CSF-1-stimulated macrophages (Fig. 2A). Akt and Erk1/2 activation preceded the CSF-1-induced reduction in the electrophoretic mobility of Stx7, while degradation of the CSF-1 receptor temporally correlated with the effects of CSF-1 on Stx7 (Fig. 2A). The changes in the electrophoretic properties of Stx7 suggested that Stx7 may have become phosphorylated. Antiphosphotyrosine Western blotting of anti-Stx7 immunoprecipitates failed to reveal detectable CSF-1-induced tyrosine phosphorylation of Stx7 (data not shown), thus suggesting that Stx7 may have become phosphorylated on serine and/or threonine residues in response to CSF-1. This idea was tested by metabolically labeling macrophages with [32P]orthophosphate prior to stimulating them with CSF-1 and immunoprecipitating Stx7. Analysis of the anti-Stx7 immunoprecipitates revealed that although Stx7 was phosphorylated in the absence of CSF-1, CSF-1 stimulation led to a marked increase in the phosphorylation of Stx7 (Fig. 2B). Phosphoamino acid analysis revealed that Stx7 from both unstimulated and CSF-1-stimulated macrophages was phosphorylated exclusively on serine residues (Fig. 2C). To establish if phosphorylation of Stx7 accounted for the appearance of the different isoforms detected by Western blotting, lysates of macrophages that had been stimulated with CSF-1 or had been left unstimulated were treated with calf intestinal alkaline phosphatase (CIP). As shown in Fig. 2D, the more slowly migrating isoforms of Stx7 largely disappeared when the cell lysates were treated with CIP. The effects of CSF-1 on the electrophoretic properties of Stx7 were also examined by two-dimensional SDS-PAGE. Stx7 from macrophages that had been deprived of CSF-1 exhibited a range of pI values but could generally be divided into two major populations (Fig. 2E); however, only a single population of Stx7, which had a higher pI, was detected following CSF-1 stimulation (Fig. 2E). Phosphorylated proteins are more acidic than their nonphosphorylated counterparts, and thus, the increase in pI was unexpected. Furthermore, treating the lysates with CIP prior to two-dimensional SDS-PAGE did not abrogate the CSF-1-induced increase in pI (data not shown). Therefore, the different Stx7 “isoforms” detected by two-dimensional SDS-PAGE may have represented different Stx7 complexes rather than differentially phosphorylated Stx7. Supporting this contention is the fact that urea, which was used during the isoelectric-focusing step to promote protein denaturation, has previously been used to stabilize Stx7-Vti1b-Stx8 complexes in order to allow their analysis by ion-exchange chromatography (2).
FIG. 2.
CSF-1-induced phosphorylation of Stx7 in macrophages. (A) Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being stimulated with CSF-1 for the time indicated. The cells were then lysed and the lysates subsequently subjected to Western blotting with the indicated antibodies. The positions of molecular mass markers (in kDa) and the different Stx7 isoforms (arrows) are indicated on the right. α-Stx7, anti-Stx7; other antibodies are similarly indicated. (B and C) Macrophages that had been metabolically labeled with [32P]orthophosphate were stimulated with CSF-1 for 15 min and then lysed. (B) Stx7 was immunoprecipitated from the lysates using anti-Stx7 antibodies and the immunoprecipitates subsequently subjected to autoradiography (upper panel) or Western blotting with an anti-Stx7 antibody (lower panel). (C) Stx7 that had been immunoprecipitated from the [32P]orthophosphate-labeled macrophages in panel B was excised from the PVDF membrane and subjected to phosphoamino acid analysis. The positions of phosphoamino acid standards (pSer, phosphoserine; pThr, phosphothreonine; pTyr, phosphotyrosine) are indicated on the right. (D) Macrophages were deprived of CSF-1 for 16 h before being stimulated with CSF-1 for 15 min and then lysed in the absence of phosphatase inhibitors. Aliquots of the lysates were incubated with CIP or in reaction buffer alone for 30 min at 37°C. The lysates were then subjected to Western blotting with an anti-Stx7 antibody. (E) Lysates of macrophages that had been left unstimulated or which had been stimulated with CSF-1 for 15 min were subjected to two-dimensional SDS-PAGE analysis followed by Western blotting with an anti-Stx7 antibody.
CSF-1 stimulation enhances the binding of Stx7 to its partner SNARE proteins.
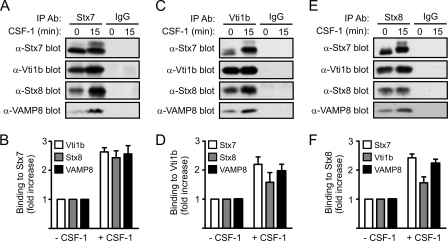
Given that Stx7 regulates vesicles trafficking in concert with other SNARE proteins (e.g., Vti1b, Stx8, and VAMP8) (3, 34, 43), the effects of CSF-1 on the interaction of Stx7 with other endosomal SNARE proteins were examined. Vti1b, Stx8, and VAMP8 coimmunoprecipitated with Stx7 from CSF-1-deprived macrophages; however, acute CSF-1 stimulation resulted in a two- to threefold increase in their coimmunoprecipitation with Stx7 (Fig. 3A and B). The coimmunoprecipitation of Vti1b, Stx8, and VAMP8 with Stx7 was specific since they were not detected in immunoprecipitates using an irrelevant control antibody (Fig. 3A). Reciprocal coimmunoprecipitation experiments with anti-Vti1b (Fig. 3C and D) and anti-Stx8 (Fig. 3E and F) antibodies likewise demonstrated increased complex formation between Stx7, Vti1b, Stx8, and VAMP8 in response to CSF-1 stimulation.
FIG. 3.
Effects of CSF-1 on Stx7 SNARE complexes in macrophages. Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being stimulated with CSF-1 for 15 min. Following cell lysis, Stx7 (A and B), Vti1b (C and D), and Stx8 (E and F) were immunoprecipitated from the lysates using anti-Stx7, anti-Vti1b, and anti-Stx8 antibodies (α-Stx7, α-Vti1b, and α-Stx8), respectively. Lysates were also immunoprecipitated with irrelevant control antibodies. (A, C, and E). The immunoprecipitates were subsequently subjected to Western blotting with the indicated antibodies. (B, D, and F) Quantified data are presented as the increase in SNARE protein binding following CSF-1 stimulation relative to that in unstimulated macrophages. Data represent the means (± standard errors) of at least three experiments.
Bioinformatics analysis of potential phosphorylation sites in Stx7.
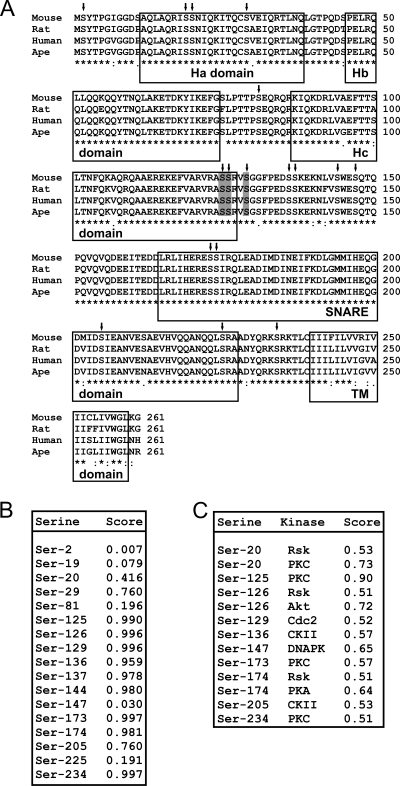
In order to identify the serine residue(s) in Stx7 that underwent CSF-1-induced phosphorylation, the amino acid sequence of Stx7 was first subjected to bioinformatics analysis. ClustalW-based sequence alignment (53) of mouse Stx7 (UniProtKB/Swiss-Prot accession number 070439) with those of the rat (accession number 070257), human (accession number 015400), and ape (accession number Q5R602) Stx7 proteins identified 17 serine residues that were evolutionarily conserved across all 4 species (Fig. 4A). Of note were the seven conserved serine residues (Ser-125, Ser-126, Ser-129, Ser-136, Ser-137, Ser-144, and Ser-147) at the C-terminal end of the Hc domain and in the “linker” region connecting the Hc domain and the SNARE domain, as well as the four conserved serine residues (Ser-173, Ser-174, Ser-205, and Ser-225) in the SNARE domain (Fig. 4A). Submission of the mouse Stx7 amino acid sequence to the NetPhos server (www.cbs.dtu.dk/services/NetPhos/) (7) identified a number of putative serine phosphorylation sites. Among these sites, Ser-125, Ser-126, Ser-129, Ser-173, and Ser-234 received the highest scores (Fig. 4B). The NetPhosK 1.0 server (www.cbs.dtu.dk/services/NetPhosK/), which predicts phosphorylation sites in proteins for specific protein kinases (8), identified Ser-20 and Ser-125 as likely sites of phosphorylation by PKC (Fig. 4C). Similarly, Ser-126 was identified as a potential phosphorylation site for Akt (Fig. 4C).
FIG. 4.
Predicted serine phosphorylation sites in Stx7. (A) The amino acid sequences of mouse, rat, human, and ape Stx7 were aligned using the ClustalW algorithm. Serine residues that are conserved across all four species are indicated with inverted arrows. Ser-125, Ser-126, and Ser-129 are shaded in gray. The Ha, Hb, Hc, SNARE, and transmembrane (TM) domains are boxed. (B) The amino acid sequence of mouse Stx7 was analyzed using the NetPhos 2.0 software program. Predicted serine phosphorylation sites in Stx7 and their corresponding probability scores are shown. (C) The amino acid sequence of mouse Stx7 was analyzed using the NetPhosK 1.0 software program. Predicted serine phosphorylation sites in Stx7, the protein kinase predicted to phosphorylate the site, and their corresponding probability scores (>0.50) are shown.
Activation of PKC triggers phosphorylation of Stx7.
Given the above findings and the fact that CSF-1 has been reported to trigger the activation of PKC in macrophages (23, 56), we tested whether treatment of macrophages with the PKC activating agent phorbol myristate acetate (PMA) resulted in the phosphorylation of Stx7. Figure 5A shows that PMA induced a rapid decrease in the electrophoretic mobility of Stx7, suggesting that Stx7 had indeed undergone phosphorylation upon PKC activation. PMA stimulation did not result in Akt activation (data not shown), but it did lead to the rapid and transient activation of Erk1/2 (Fig. 5A). To determine whether CSF-1-induced phosphorylation of Stx7 involved PKC, the effects of the PKC inhibitors GF109203X (54) and Gö6983 (20) on the electrophoretic mobility of Stx7 were examined. GF109203X partially blocked the CSF-1-induced change in Stx7 mobility, whereas Gö6983 was without effect (Fig. 5B). The efficacy of the PKC inhibitors was confirmed by demonstrating their capacity to block PMA-induced Erk1/2 activation (Fig. 5C). The ability of GF109203X to partially suppress the CSF-1-induced phosphorylation of Stx7 was not a consequence of the inhibitor impairing Akt activation by CSF-1 (Fig. 5B). These observations suggested that in addition to PKC, another serine/threonine kinase(s) also contributed to the CSF-1-induced phosphorylation of Stx7.
FIG. 5.
Phosphorylation of Stx7 by PKC in macrophages. (A) Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being stimulated with 100 nM PMA for the time indicated. The cells were then lysed and the lysates subsequently subjected to Western blotting with the indicated antibodies. The different Stx7 isoforms are indicated by arrows on the right. α-Stx7, -pErk1/2, and -Erk2 represent anti-Stx7, -pErk1/2, and -Erk2. (B) Macrophages were deprived of CSF-1 for 16 h before being treated with 0.1% dimethyl sulfoxide, 5 μM GF109203X, or 1 μM Gö6983 for 30 min. The macrophages were stimulated with CSF-1 for 15 min and then lysed. The lysates were subsequently subjected to Western blotting with the indicated antibodies (shown as in panel A). (C) Macrophages were deprived of CSF-1 for 16 h before being treated with 0.1% dimethyl sulfoxide, 5 μM GF109203X, or 1 μM Gö6983 for 30 min. The macrophages were stimulated with PMA for 15 min and then lysed. The lysates were subsequently subjected to Western blotting with the indicated antibodies.
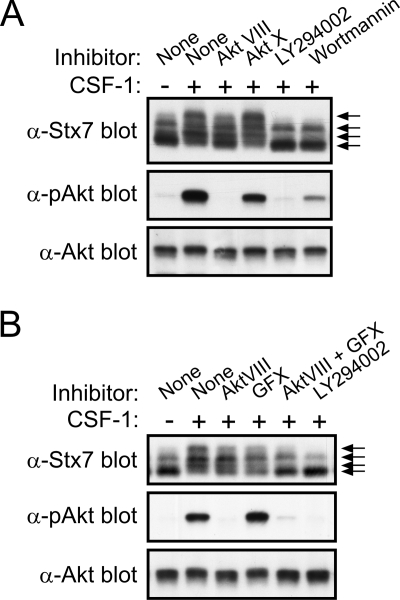
Regulation of Stx7 phosphorylation by Akt and PI 3-kinase.
Bioinformatics analysis of Stx7 also indicated that Akt, which is activated in response to CSF-1 (28), may also phosphorylate Stx7, particularly at Ser-126 (Fig. 4C). In order to establish if Akt activity was required for the CSF-1-induced phosphorylation of Stx7, macrophages were treated with the Akt inhibitors Akt VIII (6) and Akt X (52) prior to their stimulation with CSF-1. As shown in Fig. 6A, Akt inhibitor VIII partially suppressed the CSF-1-induced phosphorylation of Stx7 while completely blocking Akt activation. In contrast, Akt inhibitor X had no apparent affect on Stx7 phosphorylation and only partially blocked the activation of Akt in response to CSF-1 (Fig. 6A). Given that the activation of Akt is dependent on PI 3-kinase activity, the effects of the PI 3-kinase inhibitors LY294002 (60) and wortmannin (5) on the CSF-1-induced phosphorylation of Stx7 were also examined. Both PI 3-kinase inhibitors markedly suppressed Stx7 phosphorylation (Fig. 6A). Inhibitors of p70S6 kinase, MEK, p38 MAP kinase, and Src family kinases had no effect on the CSF-1-induced phosphorylation of Stx7 (data not shown). Because the activation of PI 3-kinase can lead to the activation of various PKC isoforms (32) and to the activation of Akt, the combined effects of the PKC inhibitor GF109203X and Akt inhibitor VIII on Stx7 phosphorylation were assessed. When used in combination, the two inhibitors reduced the CSF-1-induced phosphorylation of Stx7 to an extent that was largely comparable to that achieved with the PI 3-kinase inhibitor LY294002 (Fig. 6B). Thus, the CSF-1-induced phosphorylation of Stx7 is most likely mediated via both PKC and Akt in response to the activation of PI 3-kinase.
FIG. 6.
Effects of Akt and PI 3-kinase inhibitors on CSF-1-induced phosphorylation of Stx7 in macrophages. (A) Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being treated with 0.1% dimethyl sulfoxide, 10 μM Akt-VIII, 5 μM Akt-X, 10 μM LY294002, or 100 nM wortmannin for 30 min. The macrophages were stimulated with CSF-1 for 15 min and then lysed. The lysates were subsequently subjected to Western blotting with the indicated antibodies (anti-Stx7, anti-pAkt, and anti-Akt). The different Stx7 isoforms are indicated by arrows on the right. (B) Macrophages were deprived of CSF-1 for 16 h before being treated with 0.1% dimethyl sulfoxide, 10 μM Akt VIII, 5 μM GF109203X (GFX), 10 μM Akt VIII and 5 μM GF109203X together, or 10 μM LY294002 for 30 min. The macrophages were stimulated with CSF-1 for 15 min and then lysed. The lysates were subsequently subjected to Western blotting with the indicated antibodies.
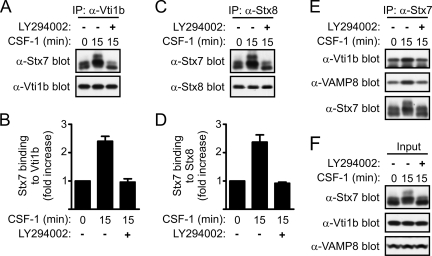
Regulation of the interaction of Stx7 with other SNARE proteins by PI 3-kinase.
PI 3-kinase-dependent phosphorylation of Stx7 by PKC and Akt may represent an important mechanism to regulate the assembly of endosomal SNARE complexes. Therefore, the effect of the PI 3-kinase inhibitor LY294002 on the binding of Stx7 to Vti1b, Stx8, and VAMP8 was examined. The ability of CSF-1 to enhance the binding of Stx7 to Vti1b was blocked when the macrophages were pretreated with LY294002 (Fig. 7A and B). The enhanced interaction between Stx7, Vti1b, Stx8, and VAMP8 in response to CSF-1 and the sensitivity of this response to LY294002 were confirmed by coimmunoprecipitation assays using antibodies against Stx8 and Stx7 (Fig. 7C to E). Western blotting of the cell lysates revealed that LY294002 had not affected the expression level of Stx7, Vti1b, or VAMP8 (Fig. 7F). Together, these findings suggest that the PI 3-kinase-dependent phosphorylation of Stx7 positively regulates its binding to partner SNARE proteins. However, we cannot exclude the possibility that the inhibitory effects of the PI 3-kinase inhibitor on SNARE complex assembly could also have been due to the impaired trafficking of the SNARE proteins to late endosomes/lysosomes.
FIG. 7.
Effects of PI 3-kinase inhibition on Stx7 SNARE complexes in macrophages. Mouse bone marrow-derived macrophages were deprived of CSF-1 for 16 h before being treated with 0.1% dimethyl sulfoxide or 10 μM LY294002 for 30 min. The macrophages were stimulated with CSF-1 for 15 min and then lysed. Vti1b (A and B), Stx8 (C and D), and Stx7 (E) were subsequently immunoprecipitated (IP) from the cell lysates with anti-Vti1b (α-Vti1b), anti-Stx8 (α-Stx8), and anti-Stx7 (α-Stx7) antibodies, respectively, and then Western blotted with the indicated antibodies. (B and D) Quantified data are presented as the increase in SNARE protein binding following CSF-1 stimulation relative to that in unstimulated macrophages. Data represent the means (± standard errors) of three experiments. (F) Western blotting of cell lysates with the indicated antibodies.
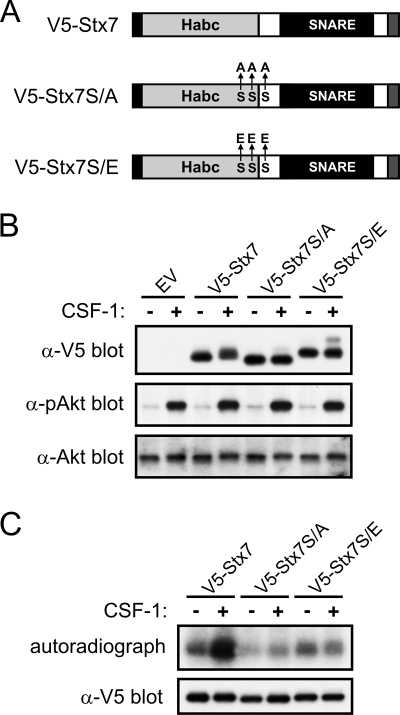
Mutation of Ser-125, Ser-126, and Ser-129 abrogates CSF-1-induced phosphorylation of Stx7.
It has recently been reported that Ser-125, Ser-126, and Ser-129 in Stx7, which we identified as potential phosphorylation sites for PKC and Akt (Fig. 4C), are phosphorylated in vivo (59). To establish if these serine residues represent the major CSF-1-induced phosphorylation sites in Stx7, mutant forms of Stx7, in which Ser-125, Ser-126, and Ser-129 were replaced with alanine or the phosphomimetic, glutamic acid (see Fig. 8A), were expressed in macrophages via retroviral transduction. Mutation of Ser-125, Ser-126, and Ser-129 to alanine largely abolished the ability of CSF-1 to induce the appearance of higher-molecular-mass isoforms of Stx7 (Fig. 8B). Furthermore, the Stx7 serine-to-alanine mutant (V5-Stx7S/A) appeared to exhibit a slightly faster electrophoretic mobility than V5-Stx7, even in the absence of CSF-1 (Fig. 8B). When the same serine residues were mutated to glutamic acid (V5-Stx7S/E), Stx7 exhibited a reduced electrophoretic mobility (Fig. 8B). This reduction in mobility was not a consequence of increased basal levels of activated Akt in macrophages expressing V5-Stx7S/E (Fig. 8B). Likewise, the inability of CSF-1 to induce a decrease in the mobility of V5-Stx7S/A was not due to a failure of CSF-1 to activate Akt (Fig. 8B). The contribution of Ser-125, Ser-126, and Ser-129 to Stx7 total phosphorylation was assessed by metabolically labeling retrovirally transduced macrophages with [32P]orthophosphate. As was the case for endogenous Stx7 (Fig. 2B), V5-Stx7 was phosphorylated in the absence of CSF-1 but underwent further phosphorylation upon CSF-1 stimulation (Fig. 8C). V5-Stx7S/A exhibited a lower level of basal phosphorylation than did V5-Stx7, and stimulation with CSF-1 did not result in any increase in its phosphorylation (Fig. 8C). The extent of V5-Stx7S/E phosphorylation, both in the absence and presence of CSF-1, was comparable to that of V5-Stx7 from unstimulated macrophages (Fig. 8C). Thus, Ser-125, Ser-126, and/or Ser-129 represent the major CSF-1-induced phosphorylation sites in Stx7.
FIG. 8.
Effects of mutating Ser-125, Ser-126, and Ser-129 on the CSF-1-induced phosphorylation of Stx7. (A) Schematic representation of V5-tagged wild-type Stx7 (V5-Stx7) and V5-Stx7 in which Ser-125, Ser-126, and Ser-129 have been replaced with alanine (V5-Stx7S/A) or glutamic acid (V5-Stx7S/E). The C-terminal transmembrane anchor domain is represented by a gray filled box. (B and C) Mouse bone marrow cells were transduced with retroviruses expressing V5-Stx7, V5-Stx7S/A, or V5-Stx7S/E or were transduced with a virus containing the empty vector (EV). The cells were cultured in the presence of CSF-1 until they differentiated into adherent macrophages. (B) The macrophages were then deprived of CSF-1 for 16 h before being stimulated with CSF-1 for 15 min. The cells were lysed and the lysates subsequently subjected to Western blotting with the indicated antibodies (anti-V5, anti-pAkt, and anti-Akt). (C) Retrovirally transduced macrophages were metabolically labeled with [32P]orthophosphate before being stimulated with CSF-1 for 15 min. The cells were then lysed and the V5-tagged Stx7 proteins immunoprecipitated using anti-V5 antibodies. The immunoprecipitates were subsequently subjected to autoradiography (upper panel) and Western blotting with an anti-V5 antibody (lower panel).
Ser-125, Ser-126, and/or Ser-129 regulate the CSF-1-dependent binding of Stx7 to other SNARE proteins.
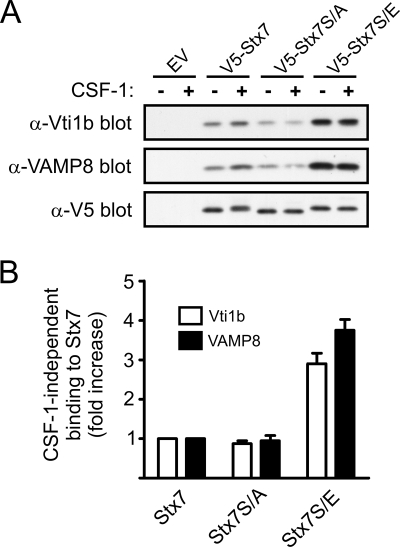
The importance of the phosphorylation of Ser-125, Ser-126, and/or Ser-129 for the binding of Stx7 to partner SNARE proteins was evaluated by immunoprecipitating the mutant forms of Stx7 from retrovirally transduced macrophages. Mutation of Ser-125, Ser-126, and Ser-129 to glutamic acid resulted in a three- to fourfold increase in the CSF-1-independent binding of Vti1b and VAMP8 to Stx7 (Fig. 9A and B). In contrast, no increase in CSF-1-independent binding of Vti1b and VAMP8 occurred when Ser-125, Ser-126, and Ser-129 were mutated to alanine (Fig. 9A and B). Furthermore, CSF-1 failed to enhance the binding of Vti1b and VAMP8 to V5-Stx7S/A (Fig. 9A). Although V5-Stx7S/E exhibited a higher level of CSF-1-independent Vti1b and VAMP8 binding than V5-Stx7, the binding of Vti1b and VAMP8 to V5-Stx7S/E was not enhanced further by CSF-1 (Fig. 9A). On the basis of these findings, it is suggested that the PI 3-kinase-dependent phosphorylation of Stx7 on Ser-125, Ser-126, and/or Ser-129 is a key regulatory event in the binding of Stx7 to its SNARE partners in macrophages.
FIG. 9.
Effects of mutating Ser-125, Ser-126, and Ser-129 in Stx7 on the assembly of Stx7 SNARE complexes. Mouse bone marrow cells were transduced with retroviruses expressing V5-Stx7, V5-Stx7S/A, or V5-Stx7S/E or were transduced with a virus containing the empty vector (EV). The cells were then cultured in the presence of CSF-1 until they differentiated into adherent macrophages. The macrophages were deprived of CSF-1 for 16 h before being stimulated with CSF-1 for 15 min. (A) The cells were then lysed and the V5-tagged Stx7 proteins immunoprecipitated using anti-V5 (α-V5) antibodies. The immunoprecipitates were subsequently subjected to Western blotting with the indicated antibodies (anti-Vti1b [α-Vti1b], anti-VAMP8 [α-VAMP8], and anti-V5). (B) Quantified data are presented as the increase in the CSF-1-independent binding of Vti1b and VAMP8 to V5-Stx7S/A and V5-Stx7S/E relative to their binding to V5-Stx7. Data represent the means (± standard errors) of three independent experiments.
DISCUSSION
Previous studies, including our own, have shown that Stx7 forms an endosomal SNARE complex with Vti1b, Stx8, and VAMP8 to mediate late endosome/lysosome fusion (3, 34, 43). In macrophages, Stx7 has been detected on mature phagosomes (12); it has also been shown to be a component of a novel SNARE complex that mediates the trafficking of the key inflammatory cytokine TNF to the plasma membrane (38). Given that CSF-1 regulates phagocytosis, pinocytosis, and cytokine secretion in macrophages (11, 13, 24, 30, 44, 50), we propose that the regulation of Stx7 expression and function by CSF-1 contributes to its regulatory effects on at least some of these effector functions.
Stx7 expression was upregulated, at both the mRNA and protein levels, by CSF-1 in mouse bone marrow-derived macrophages (Fig. 1). Vti1b and VAMP8 expression was also upregulated; however, the expression levels of Stx8 appear to be unaffected by CSF-1. LPS has been reported to increase Vti1b, Stx6, and VAMP3 expression in macrophages; moreover, their LPS-induced upregulation was necessary for efficient TNF secretion (37, 38). Gamma interferon (IFN-γ), which heightens the inflammatory response of macrophages to pathogens (e.g., increases cytokine secretion), also upregulates the expression of Vti1b, Stx6, and VAMP3 (37, 38). Whether the effects of CSF-1, LPS, and IFN-γ on SNARE protein expression are identical but perhaps additive, or just overlapping, is still to be established. Nonetheless, our findings, together with those of Stow and colleagues (37, 38), suggest that the regulatory effects of growth factors (e.g., CSF-1) and cytokines (e.g., IFN-γ) on macrophages are likely to be mediated, at least in part, via their ability to regulate the expression of the SNARE proteins that are required for specific effector functions (e.g., cytokine secretion).
Little is known about how the binding of Stx7 to its partner SNARE proteins is regulated. However, the data we have presented here suggest that phosphorylation of Stx7 is a key regulatory event in the assembly of endosomal SNARE complexes. Bioinformatics analysis identified Ser-125, Ser-126, and Ser-129 as likely sites for phosphorylation. In support of this contention, a recent mass spectrometry-based analysis of phosphorylation sites in proteins from mouse liver identified Ser-125, Ser-126, and Ser-129 as in vivo phosphorylation sites in Stx7 (59). Our analysis predicted that the phosphorylation of these sites was likely to be mediated by PKC and Akt. While pharmacologic inhibitors of PKC and Akt individually suppressed the CSF-1-induced phosphorylation of Stx7, the extent of suppression was not as great as that achieved with a PI 3-kinase inhibitor (e.g., LY294002). However, when the PKC and Akt inhibitors were used in combination, they were as effective as the PI 3-kinase inhibitor. Based on these findings, we propose that PKC and Akt are largely responsible for mediating the CSF-1-induced phosphorylation of Stx7 in response to PI 3-kinase activation, potentially via the PI 3-kinase target, phosphoinositide-dependent kinase 1 (32).
The CSF-1-induced phosphorylation of Ser-125, Ser-126, and/or Ser-129, which are located at the end of the Habc domain and at the start of the “linker” connecting the Habc and SNARE domains in Stx7, appears to positively regulate the binding of Stx7 to its SNARE partners (e.g., Vti1b and VAMP8). Notably, though, the phosphorylation of Ser-125, Ser-126, and/or Ser-129 is not obligatory for the binding of Stx7 to its partner SNARE proteins, since a Stx7 mutant (i.e., Stx7S/A), in which these amino acids had been replaced with alanine residues, could still bind Vti1b and VAMP8. This suggests that the phosphorylation of Ser-125, Ser-126, and/or Ser-129 in Stx7 regulates the rate of complex assembly between Stx7 and its partner SNARE proteins and/or the stability of such SNARE complexes.
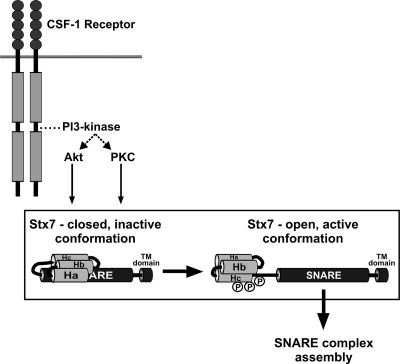
The Habc domain of Stx7 has been shown to fold back and interact intramolecularly with the SNARE domain; SNARE complex assembly is retarded in this “closed” conformation (2). However, deletion of the Habc domain releases Stx7 from this inhibitory constraint and leads to accelerated SNARE complex assembly (2). The Habc domain of the Qa-SNARE protein Stx1A also interacts with the SNARE domain to inhibit SNARE complex assembly (14, 16, 35). While an intramolecular interaction between the Habc and SNARE domains of Stx7 is likely to contribute to the regulation of endosomal SNARE complex assembly, how the switching of Stx7 from a “closed” to an “open” conformation is regulated in vivo has not been established. Our results suggest that the PI 3-kinase-dependent phosphorylation of Stx7 by PKC and Akt regulates the conformational state of Stx7. We propose that phosphorylation of Ser-125, Ser-126, and/or Ser-129 in response to PI 3-kinase activation induces a conformational change in Stx7 such that it adopts an “open” conformation in which its SNARE domain is accessible to bind other SNARE proteins (Fig. 10), thereby facilitating the assembly of Stx7 SNARE complexes.
FIG. 10.
A model for the regulation of the binding of Stx7 to its partner SNARE proteins by CSF-1. In the absence of CSF-1, Stx7 adopts predominantly a “closed” conformation by virtue of the Habc domain intramolecularly interacting with the SNARE domain, thereby suppressing the binding of Stx7 to its partner SNARE proteins. Upon activation of the CSF-1 receptor, Akt and PKC become activated, via PI 3-kinase, and phosphorylate Ser-125, Ser-126, and/or Ser-129 in the Hc domain and/or “linker” region of Stx7. This induces a conformational change in Stx7 that results in Stx7 adopting an “open” conformation with an enhanced capacity to bind to its SNARE partners (e.g., Vti1b, Stx8, and VAMP8).
Like Stx7, the Qb-SNARE Vti1b also contains a Habc domain; however, unlike Stx7 the Habc and SNARE domains of Vti1b do not appear to interact intramolecularly to suppress SNARE complex assembly (2). Consequently, phosphorylation of Stx7, in particular phosphorylation of Ser-125, Ser-126, and/or Ser-129, is likely to be a major determinant of the assembly of Stx7-Vti1b-Stx8 Q-SNARE complexes. Stx7 also forms a SNARE complex with Vti1b and the Qc-SNARE Stx6 to facilitate the fusion of Golgi-derived vesicles with recycling endosomes during the intracellular trafficking of TNF in macrophages (38). How Stx7 discriminates between different partner SNARE proteins (e.g., Stx6 versus Stx8) has not been elucidated. While the subcellular localization of its partner SNARE proteins, as well as the nature of the cargo to be transported by the vesicle, is likely to influence which SNARE proteins Stx7 binds, phosphorylation of Stx7 could potentially regulate the differential binding of SNARE proteins to Stx7. Our mutagenesis and metabolic labeling experiments suggested that Ser-125, Ser-126, and Ser-129 represent the major CSF-1-induced phosphorylation sites in Stx7. However, bioinformatics analysis also identified Ser-173 and Ser-234 as possible phosphorylation sites for PKC. Ser-173 is located near the start of the SNARE motif, while Ser-234 resides between the SNARE domain and the hydrophobic C-terminal membrane anchoring domain. It will clearly be important to establish if Ser-173 and Ser-234 are bona fide phosphorylation sites and if their phosphorylation status differentially regulates the binding of SNARE proteins, such as Stx6 and Stx8, to Stx7.
Stx7 also binds to non-SNARE proteins, such as class C vacuolar protein sorting (Vps) proteins (e.g., Vps33, a Sec1-like protein) (29). Class C Vps proteins control the fusion of late endosomes with lysosomes by regulating SNARE protein pairing (42, 46). Studies with yeast revealed that the class C Vps complex binds to unpaired Vam3p (a yeast syntaxin homolog) but not to Vam3p that is bound to its SNARE partners (46). It is worth noting that the Sec1 protein Munc18 interacts with and stabilizes the “closed” conformation of Stx1A (14). By analogy, the phosphorylation state, and hence conformation, of Stx7 may regulate its interaction with the Sec1-like protein Vps33.
PI 3-kinase activity is required for a range of vesicular trafficking processes in macrophages (4, 36, 58). The findings presented here on the regulation of Stx7 and the assembly of endosomal SNARE complexes by PI 3-kinase may help to explain, at least in part, the importance of PI 3-kinase activity for endocytic processes in macrophages. Moreover, the effects of CSF-1 on Stx7 may provide a mechanism for the regulation of macrophage effector functions, such as cytokine secretion, phagocytosis, and macropinocytosis, by CSF-1. Our findings may also be of relevance to the regulation of CSF-1 signaling. Endocytosis and lysosomal degradation of activated CSF-1 receptor is important for controlling CSF-1 signaling and macrophage proliferation (31). There is mounting evidence that some receptors (e.g., epidermal growth factor receptor) continue to signal during their transit through the endocytic pathway (21). The ability of CSF-1 to regulate the expression and function of the SNARE proteins that control late endosome/lysosome fusion may therefore represent a novel negative-feedback mechanism to regulate the duration and magnitude of CSF-1 signaling.
Acknowledgments
This work was supported by the Cooperative Research Centre for Chronic Inflammatory Diseases and a grant from the National Health and Medical Research Council of Australia.
We thank Heung-Chin Cheng (Department of Biochemistry and Molecular Biology, Bio21 Institute, The University of Melbourne) for assistance with phosphoamino acid analysis.
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Aderem, A., and D. M. Underhill. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17593-623. [DOI] [PubMed] [Google Scholar]
- 2.Antonin, W., I. Dulubova, D. Arac, S. Pabst, J. Plitzner, J. Rizo, and R. Jahn. 2002. The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J. Biol. Chem. 27736449-36456. [DOI] [PubMed] [Google Scholar]
- 3.Antonin, W., C. Holroyd, D. Fasshauer, S. Pabst, G. F. Von Mollard, and R. Jahn. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 196453-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki, N., M. T. Johnson, and J. A. Swanson. 1996. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 1351249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcaro, A., and M. P. Wymann. 1993. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 296297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, S. F., D. Defeo-Jones, S. Fu, P. J. Hancock, K. M. Haskell, R. E. Jones, J. A. Kahana, A. M. Kral, K. Leander, L. L. Lee, J. Malinowski, E. M. McAvoy, D. D. Nahas, R. G. Robinson, and H. E. Huber. 2005. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 385399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 2941351-1362. [DOI] [PubMed] [Google Scholar]
- 8.Blom, N., T. Sicheritz-Ponten, R. Gupta, S. Gammeltoft, and S. Brunak. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 41633-1649. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanovic, A., N. Bennett, S. Kieffer, M. Louwagie, T. Morio, J. Garin, M. Satre, and F. Bruckert. 2002. Syntaxin 7, syntaxin 8, Vti1 and VAMP7 (vesicle-associated membrane protein 7) form an active SNARE complex for early macropinocytic compartment fusion in Dictyostelium discoideum. Biochem. J. 36829-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201110-149. [DOI] [PubMed] [Google Scholar]
- 11.Cheers, C., M. Hill, A. M. Haigh, and E. R. Stanley. 1989. Stimulation of macrophage phagocytic but not bactericidal activity by colony-stimulating factor 1. Infect. Immun. 571512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, R. F., A. D. Schreiber, S. Grinstein, and W. S. Trimble. 2002. Syntaxins 13 and 7 function at distinct steps during phagocytosis. J. Immunol. 1693250-3256. [DOI] [PubMed] [Google Scholar]
- 13.Conway, J. G., B. McDonald, J. Parham, B. Keith, D. W. Rusnak, E. Shaw, M. Jansen, P. Lin, A. Payne, R. M. Crosby, J. H. Johnson, L. Frick, M. H. Lin, S. Depee, S. Tadepalli, B. Votta, I. James, K. Fuller, T. J. Chambers, F. C. Kull, S. D. Chamberlain, and J. T. Hutchins. 2005. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc. Natl. Acad. Sci. USA 10216078-16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulubova, I., S. Sugita, S. Hill, M. Hosaka, I. Fernandez, T. C. Sudhof, and J. Rizo. 1999. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 184372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon, D. T., and R. M. Locksley. 1996. The instructive role of innate immunity in the acquired immune response. Science 27250-53. [DOI] [PubMed] [Google Scholar]
- 16.Fiebig, K. M., L. M. Rice, E. Pollock, and A. T. Brunger. 1999. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 6117-123. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez, C. E., C. A. Lyman, S. Lee, C. Del Guercio, E. Roilides, J. Bacher, A. Gehrt, E. Feuerstein, M. Tsokos, and T. J. Walsh. 2001. Recombinant human macrophage colony-stimulating factor augments pulmonary host defences against Aspergillus fumigatus. Cytokine 1587-95. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, S. 1998. The role of the macrophage in immune regulation. Res. Immunol. 149685-688. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg, S., and S. Grinstein. 2002. Phagocytosis and innate immunity. Curr. Opin. Immunol. 14136-145. [DOI] [PubMed] [Google Scholar]
- 20.Gschwendt, M., S. Dieterich, J. Rennecke, W. Kittstein, H. J. Mueller, and F. J. Johannes. 1996. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase C isoenzymes. FEBS Lett. 39277-80. [DOI] [PubMed] [Google Scholar]
- 21.Hoeller, D., S. Volarevic, and I. Dikic. 2005. Compartmentalization of growth factor receptor signalling. Curr. Opin. Cell Biol. 17107-111. [DOI] [PubMed] [Google Scholar]
- 22.Hong, W. 2005. SNAREs and traffic. Biochim. Biophys. Acta 1744493-517. [PubMed] [Google Scholar]
- 23.Imamura, K., A. Dianoux, T. Nakamura, and D. Kufe. 1990. Colony-stimulating factor 1 activates protein kinase C in human monocytes. EMBO J. 92423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irvine, K. M., C. J. Burns, A. F. Wilks, S. Su, D. A. Hume, and M. J. Sweet. 2006. A CSF-1 receptor kinase inhibitor targets effector functions and inhibits pro-inflammatory cytokine production from murine macrophage populations. FASEB J. 201921-1923. [DOI] [PubMed] [Google Scholar]
- 25.Jahn, R., and R. H. Scheller. 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7631-643. [DOI] [PubMed] [Google Scholar]
- 26.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20197-216. [DOI] [PubMed] [Google Scholar]
- 27.Jaworowski, A., N. J. Wilson, E. Christy, R. Byrne, and J. A. Hamilton. 1999. Roles of the mitogen-activated protein kinase family in macrophage responses to colony stimulating factor-1 addition and withdrawal. J. Biol. Chem. 27415127-15133. [DOI] [PubMed] [Google Scholar]
- 28.Kelley, T. W., M. M. Graham, A. I. Doseff, R. W. Pomerantz, S. M. Lau, M. C. Ostrowski, T. F. Franke, and C. B. Marsh. 1999. Macrophage colony-stimulating factor promotes cell survival through Akt/protein kinase B. J. Biol. Chem. 27426393-26398. [DOI] [PubMed] [Google Scholar]
- 29.Kim, B. Y., H. Kramer, A. Yamamoto, E. Kominami, S. Kohsaka, and C. Akazawa. 2001. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 27629393-29402. [DOI] [PubMed] [Google Scholar]
- 30.Knight, K. R., G. Vairo, and J. A. Hamilton. 1992. Regulation of pinocytosis in murine macrophages by colony-stimulating factors and other agents. J. Leukoc. Biol. 51350-359. [DOI] [PubMed] [Google Scholar]
- 31.Lee, P. S., Y. Wang, M. G. Dominguez, Y. G. Yeung, M. A. Murphy, D. D. Bowtell, and E. R. Stanley. 1999. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 183616-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Good, J. A., W. H. Ziegler, D. B. Parekh, D. R. Alessi, P. Cohen, and P. J. Parker. 1998. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 2812042-2045. [DOI] [PubMed] [Google Scholar]
- 33.Medzhitov, R., and C. A. Janeway, Jr. 1998. Innate immune recognition and control of adaptive immune responses. Semin. Immunol. 10351-353. [DOI] [PubMed] [Google Scholar]
- 34.Mullock, B. M., C. W. Smith, G. Ihrke, N. A. Bright, M. Lindsay, E. J. Parkinson, D. A. Brooks, R. G. Parton, D. E. James, J. P. Luzio, and R. C. Piper. 2000. Syntaxin 7 is localized to late endosome compartments, associates with Vamp 8, and is required for late endosome-lysosome fusion. Mol. Biol. Cell 113137-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munson, M., X. Chen, A. E. Cocina, S. M. Schultz, and F. M. Hughson. 2000. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat. Struct. Biol. 7894-902. [DOI] [PubMed] [Google Scholar]
- 36.Murray, J., L. Wilson, and S. Kellie. 2000. Phosphatidylinositol-3′ kinase-dependent vesicle formation in macrophages in response to macrophage colony stimulating factor. J. Cell Sci. 113337-348. [DOI] [PubMed] [Google Scholar]
- 37.Murray, R. Z., J. G. Kay, D. G. Sangermani, and J. L. Stow. 2005. A role for the phagosome in cytokine secretion. Science 3101492-1495. [DOI] [PubMed] [Google Scholar]
- 38.Murray, R. Z., F. G. Wylie, T. Khromykh, D. A. Hume, and J. L. Stow. 2005. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis factor-alpha. J. Biol. Chem. 28010478-10483. [DOI] [PubMed] [Google Scholar]
- 39.Norbury, C. C., L. J. Hewlett, A. R. Prescott, N. Shastri, and C. Watts. 1995. Class I MHC presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity 3783-791. [DOI] [PubMed] [Google Scholar]
- 40.Onishi, M., S. Kinoshita, Y. Morikawa, A. Shibuya, J. Phillips, L. L. Lanier, D. M. Gorman, G. P. Nolan, A. Miyajima, and T. Kitamura. 1996. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 24324-329. [PubMed] [Google Scholar]
- 41.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 908392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poupon, V., A. Stewart, S. R. Gray, R. C. Piper, and J. P. Luzio. 2003. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell 144015-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pryor, P. R., B. M. Mullock, N. A. Bright, M. R. Lindsay, S. R. Gray, S. C. Richardson, A. Stewart, D. E. James, R. C. Piper, and J. P. Luzio. 2004. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5590-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Racoosin, E. L., and J. A. Swanson. 1989. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J. Exp. Med. 1701635-1648.2681516 [Google Scholar]
- 45.Roilides, E., C. A. Lyman, T. Sein, R. Petraitiene, and T. J. Walsh. 2003. Macrophage colony-stimulating factor enhances phagocytosis and oxidative burst of mononuclear phagocytes against Penicillium marneffei conidia. FEMS. Immunol. Med. Microbiol. 3619-26. [DOI] [PubMed] [Google Scholar]
- 46.Sato, T. K., P. Rehling, M. R. Peterson, and S. D. Emr. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6661-671. [DOI] [PubMed] [Google Scholar]
- 47.Schonlau, F., C. Schlesiger, J. Ehrchen, S. Grabbe, C. Sorg, and C. Sunderkotter. 2003. Monocyte and macrophage functions in M-CSF-deficient op/op mice during experimental leishmaniasis. J. Leukoc. Biol. 73564-573. [DOI] [PubMed] [Google Scholar]
- 48.Stanley, E. R., K. L. Berg, D. B. Einstein, P. S. Lee, F. J. Pixley, Y. Wang, and Y. G. Yeung. 1997. Biology and action of colony-stimulating factor-1. Mol. Reprod. Dev. 464-10. [DOI] [PubMed] [Google Scholar]
- 49.Stow, J. L., A. P. Manderson, and R. Z. Murray. 2006. SNAREing immunity: the role of SNAREs in the immune system. Nat. Rev. Immunol. 6919-929. [DOI] [PubMed] [Google Scholar]
- 50.Sweet, M. J., C. C. Campbell, D. P. Sester, D. Xu, R. C. McDonald, K. J. Stacey, D. A. Hume, and F. Y. Liew. 2002. Colony-stimulating factor-1 suppresses responses to CpG DNA and expression of toll-like receptor 9 but enhances responses to lipopolysaccharide in murine macrophages. J. Immunol. 168392-399. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, P. R., L. Martinez-Pomares, M. Stacey, H. H. Lin, G. D. Brown, and S. Gordon. 2005. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 23901-944. [DOI] [PubMed] [Google Scholar]
- 52.Thimmaiah, K. N., J. B. Easton, G. S. Germain, C. L. Morton, S. Kamath, J. K. Buolamwini, and P. J. Houghton. 2005. Identification of N10-substituted phenoxazines as potent and specific inhibitors of Akt signaling. J. Biol. Chem. 28031924-31935. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 26615771-15781. [PubMed] [Google Scholar]
- 55.Ungar, D., and F. M. Hughson. 2003. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19493-517. [DOI] [PubMed] [Google Scholar]
- 56.Valledor, A. F., J. Xaus, L. Marques, and A. Celada. 1999. Macrophage colony-stimulating factor induces the expression of mitogen-activated protein kinase phosphatase-1 through a protein kinase C-dependent pathway. J. Immunol. 1632452-2462. [PubMed] [Google Scholar]
- 57.Van Parijs, L., Y. Refaeli, J. D. Lord, B. H. Nelson, A. K. Abbas, and D. Baltimore. 1999. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity 11281-288. [DOI] [PubMed] [Google Scholar]
- 58.Vieira, O. V., R. J. Botelho, L. Rameh, S. M. Brachmann, T. Matsuo, H. W. Davidson, A. Schreiber, J. M. Backer, L. C. Cantley, and S. Grinstein. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 15519-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villen, J., S. A. Beausoleil, S. A. Gerber, and S. P. Gygi. 2007. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA 1041488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlahos, C. J., W. F. Matter, K. Y. Hui, and R. F. Brown. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 2695241-5248. [PubMed] [Google Scholar]