Abstract
We identified a conserved sequence within the Muscle creatine kinase (MCK) promoter that is critical for high-level activity in skeletal and cardiac myocytes (MCK Promoter Element X [MPEX]). After selectively enriching for MPEX-binding factor(s) (MPEX-BFs), ICAT-based quantitative proteomics was used to identify MPEX-BF candidates, one of which was MAZ (Myc-associated zinc finger protein). MAZ transactivates the MCK promoter and binds the MPEX site in vitro, and chromatin immunoprecipitation analysis demonstrates enrichment of MAZ at the endogenous MCK promoter and other muscle gene promoters (Skeletal α-actin, Desmin, and α-Myosin heavy chain) in skeletal and cardiac myocytes. Consistent with its role in muscle gene transcription, MAZ transcripts and DNA-binding activity are upregulated during skeletal myocyte differentiation. Furthermore, MAZ was shown to bind numerous sequences (e.g., CTCCTCCC and CTCCACCC) that diverge from the GA box binding motif. Alternate motifs were identified in many muscle promoters, including Myogenin and MEF2C, and one motif was shown to be critical for Six4 promoter activity in both skeletal and cardiac myocytes. Interestingly, MAZ occupies and is able to transactivate the Six4 promoter in skeletal but not cardiac myocytes. Taken together, these findings are consistent with a previously unrecognized role for MAZ in muscle gene regulation.
The mouse Muscle creatine kinase (MCK) gene serves as a useful model for understanding muscle-specific gene transcription since its expression is restricted to terminally differentiated striated muscle, where it is one of the most abundantly expressed genes. Transcription of MCK is regulated by an upstream enhancer (−1256 to −1050), a proximal promoter (−358 to +7), and an intron 1 modulatory region (+740 to +1731). The upstream enhancer confers muscle-specific expression to reporter constructs in cell culture, transgenic mice, and virus-mediated gene transfer (1, 13, 24, 30, 31, 56, 61) and contains at least seven control elements, which were identified by footprinting, deletion/mutation analysis, and gel mobility shift assays (1, 8, 16, 39, 41). The sequences and relative positions of elements in the MCK enhancer (CArG/SRE, AP2, MEF3, A/T-rich, left and right E-boxes, and MEF2) are highly conserved between mammalian species, and many of the associated transcription factors have been identified (11, 19, 20, 27, 39, 68).
In contrast to the upstream enhancer, very little is known about the highly conserved (−358 to +7) region comprising the MCK proximal promoter. Studies with cultured cells and transgenic mice have demonstrated that the MCK promoter alone is capable of driving muscle-specific expression (30, 56) and that it is ∼40-fold more active in skeletal than in cardiac myocytes (C. L. Himeda and S. D. Hauschka, unpublished data). However, the MCK promoter also synergizes with the upstream enhancer to drive much higher expression of reporter constructs in both types of striated muscle (13, 56). The MCK promoter is known to contain a highly conserved E-box and CArG site, both of which are important for expression in skeletal and cardiac muscle (46, 56). Interestingly, in contrast to the upstream enhancer of MCK, which displays high sequence conservation only in its seven known control elements, the promoter is highly conserved throughout (∼70% identical between humans and mice), suggesting a complex mechanism of regulation and the presence of multiple control elements that have yet to be identified.
Here we describe a new control element in the MCK promoter, MCK Promoter Element X (MPEX), which is important for promoter activity in both skeletal and cardiac myocytes. The MPEX sequence is rich in guanines and cytosines, a feature characteristic of binding sites for a large number of transcription factors (AP2, MAZ, and the prodigious Sp/KLF family). After selectively enriching for MPEX-binding factors (MPEX-BFs) from skeletal myocytes, we used a quantitative proteomics strategy to identify candidates in an unbiased approach. One of the strongest of these candidates was MAZ (myc-associated zinc finger protein).
MAZ is a six-Cys2-His2 zinc finger transcription factor that recognizes the GA box motif (GGGAGGG) and GC-rich sequences. Originally identified as a factor binding the P2 promoter of c-myc (51), it is ubiquitously expressed in human and mouse tissues (58, 59) and has been implicated in a wide range of transcriptional roles. MAZ appears to be important for transcription termination between closely spaced human complement genes and at an alternative termination site within an intron of mouse IgM-D (3, 5). In an in vitro coupled transcription-polyadenylation assay, the MAZ binding site promotes pausing of RNA polymerase II and the activation of polyadenylation (73). MAZ has been implicated in the activation of a number of genes, including RAG-2, Adenovirus major late, CD4, serotonin 1a receptor, CLC-K1, PNMT, parathyroid hormone receptor, ENT-1, insulin I, and PPAR-γ1 (9, 14, 25, 37, 49, 50, 66, 70, 71, 72). In addition, MAZ has been shown to repress its own gene, as well as c-myc, eNOS, Sp4, telomerase, and eosinophil-derived neurotoxin (28, 34, 57, 60, 62, 69). Despite its abundant expression in skeletal and cardiac muscle (58, 59), the functional role of MAZ in these tissues has not been explored, and to date, no MAZ knockout mouse has been described.
We show that MAZ is able to transactivate the MCK promoter in differentiated skeletal myocyte cultures and that it binds the MPEX site in gel shift interference assays. Using chromatin immunoprecipitation (ChIP) analysis, we show that MAZ is present at the MCK promoter and other muscle gene promoters in skeletal and cardiac myocytes. In addition, we found that MAZ transcripts and DNA-binding activity are upregulated during skeletal muscle differentiation. After further characterizing the binding preferences of MAZ, we discovered that divergent MAZ motifs are highly abundant in the promoters of both regulatory and structural muscle genes, and the function of one such motif was shown to be critical for the activity of the Six4 promoter in both skeletal and cardiac myocytes. Interestingly, MAZ occupies and is able to transactivate the Six4 promoter in skeletal but not cardiac myocytes, thereby underscoring the complexities of transcriptional control in different cell types. Taken together, these data indicate important and previously unknown roles for MAZ in the regulation of muscle-specific genes.
MATERIALS AND METHODS
Plasmid constructs.
Reporter plasmids −80MCKCAT, −358MCKCAT, enh-358MCKCAT, and pUCSV2PAP have been previously described (1, 56), as has the construct containing the full-length MAZ cDNA under the control of the cytomegalovirus promoter in plasmid pCGN (50). Three tandem copies of the MCK promoter sequence (GGGCCCCTCCCTGGGGACAGCCCC) containing the wild-type MPEX sequence (underlined) flanked by SalI sites were ligated into the SalI site of −80MCKCAT to generate (MPEX)3−80MCKCAT. The Six4 promoter was PCR amplified from mouse genomic DNA using forward primer 5′-TAATATTTAGCATGCGTCCATCTCAGGGTTTCTGC-3′ (SphI site underlined) and reverse primer 5′-TAATATTTATCTAGAATAGCTGCTTTCTGCCGTTC-3′ (XbaI site underlined). The 643-bp product was digested with SphI and XbaI and then cloned into the pCAT plasmid (29) to generate −643Six4CAT. The mutations listed in Fig. 1A and Fig. 8A were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions. All constructs were sequenced to verify the integrity of the inserted DNA.
FIG. 1.
MPEX is required for full MCK enhancer-promoter activity in skeletal and cardiac myocytes. (A) Partial sequence alignment of MCK promoter (−85 to −51 for mouse) from multiple mammalian species. The MPEX sequence (solid line) and a conserved 3′ region (dashed line) are indicated. The wild-type mouse sequence (WT) used as a probe in Fig. 2A, as well as mutations within this sequence (M1, M2, and M3), tested in panels B and C and Fig. 2A, are shown. Unaltered bases are indicated as asterisks. (B and C) Analysis of MPEX mutations in skeletal (B) or cardiac (C) myocytes. MM14 skeletal myocytes or primary neonatal cardiomyocytes were transfected with constructs containing the CAT reporter under the control of either the 358-bp MCK promoter (−358MCKCAT) or the MCK enhancer linked to the promoter (enh-358MCKCAT) and the PAP reference plasmid. Activities of the wild-type constructs compared to those of constructs containing mutation M1, M2, or M3 are shown. For each panel, data are plotted as the mean value and standard deviation of the CAT/PAP ratio determined for each culture dish, and the activity of the wild-type construct is set at 100.
FIG. 8.
A divergent MAZ motif is essential for high-level activity of the Six4 promoter in skeletal and cardiac myocytes. MAZ transactivates and occupies the Six4 promoter in skeletal but not cardiac myocytes. (A) The Six4 promoter contains two conserved, divergent MAZ motifs. Partial sequence alignment of the mouse and human Six4 promoter (−156 to −94 for mouse) is shown. The wild-type sequences for mouse and human are shown, as well as mutations in the two variant MAZ motifs tested in panels B and C. (B and C) Analysis of mutations in the Six4 promoter MAZ motifs and transactivation by MAZ. MM14 skeletal myocytes or primary neonatal cardiomyocytes were transfected with constructs containing the CAT reporter under the control of the 643-bp 5′-flanking sequence of mouse Six4 and the PAP reference plasmid. Activity of the wild-type (WT) construct compared to those of constructs containing mutation M1 or M2 in MAZ1 or mutation M1 or M2 in MAZ2 is shown. For the last two bars, the wild-type reporter construct was transfected or not with 0.5 μg of MAZ expression vector (MAZ) or pCGN control vector (CTL). Data are plotted as the mean value and standard deviation of the CAT/PAP ratio determined for each culture dish, and the activity of the wild-type construct is set at 100. (D) MAZ occupies the Six4 promoter in skeletal but not cardiac myocytes. ChIP assays were performed using either MM14 skeletal myocytes or primary neonatal cardiomyocytes. Chromatin was immunoprecipitated using MAZ-specific antibodies or nonspecific rabbit IgG and then analyzed by qPCR using primers specific for the MCK promoter. Data are represented as enrichment by MAZ-specific antibodies (MAZ) relative to nonspecific IgG (CTL). Data are plotted as the mean values and standard deviations from three independent ChIP experiments (three replicate PCRs for each).
Cell culture.
Mouse MM14 skeletal myoblasts were grown on 100-mm collagen-coated tissue culture dishes in proliferation medium (Ham's F-10C supplemented with 15% horse serum and 2 ng/ml basic fibroblast growth factor) as described previously (10, 44). For transient transfections, log-phase cultures at ∼4 × 105 cells/dish were induced to differentiate by rinsing cultures twice with saline G and then switching to differentiation medium (Ham's F-10C supplemented with 1.5% horse serum and 6 μg/ml insulin). Cells were maintained for 48 h in differentiation medium prior to harvesting. For myocyte nuclear extracts and chromatin immunoprecipitations, 100-mm dishes were plated with ∼1 × 105 log-phase cells/dish, grown to near confluence (∼4 × 106 cells/dish), and then allowed to differentiate in proliferation medium without additional fibroblast growth factor for 4 to 6 days prior to harvesting. All cultures contained >90% terminally differentiated myocytes as assessed by immunostaining of a parallel culture with the myosin-specific antibody MF-20. This procedure produced ∼7 × 106 differentiated myonuclei per 100-mm dish. For myoblast nuclear extracts, cells were grown in proliferation medium as described above and harvested at ∼5 × 105 cells/dish. Ventricular cardiomyocytes from 1- to 3-day-old Sprague-Dawley rats were isolated and cultured as described elsewhere (45). These cultures contained >90% cardiomyocytes as assessed by immunohistochemistry with the MF-20 antibody.
Preparation of nuclear extracts.
Crude nuclear extracts from cultured cells were prepared as previously described (12) using a cocktail of several protease inhibitors (P8340; Sigma). Total protein in the extracts was quantitated by the Bradford method (6).
Coupling of DNA to magnetic beads.
Streptavidin-linked magnetic beads (M-280; Dynal) were coupled to biotinylated, double-stranded oligonucleotides according to the Dynal protocol at a concentration of 20 pmol DNA/mg beads. Oligonucleotides contained either the −91 to −70 sequence from the mouse MCK promoter (MPEX sequence shown in bold) linked to the −53 to −38 sequence (MPEX*; TCTCAGGGGCCCCTCCCTGGGG/AGTCACACCCTGTAGG) (−91 to −70/−53 to −38) or the −91 to −54 sequence containing the M1 and M3 mutations (mutated bases underlined) (MPEX-mt*; TCTCAGGGGCACATACATTGTGCCCGACACGCATGGCT) (see Results for rationale of sequence selection). The efficiency of coupling was verified by agarose gel electrophoresis.
MPEX-BF enrichment.
All enrichment steps were performed at 4°C. Crude nuclear extracts from ∼1.3 × 109 MM14 myocytes (∼62 mg of total protein) were pooled, and equal amounts (∼31 mg of each) were incubated for 90 min on a rotating platform with 26 mg of either MPEX* or MPEX-mt* beads plus a 100-fold molar excess of poly(dI-dC). MPEX* beads were washed with buffer [10 mM Tris-Cl (pH 7.4), 0.1 M NaCl, 0.1 mM EDTA, 0.5 mM Tris (2-carboxyethyl)phosphine (TCEP)] containing a 10-fold molar excess of MPEX-mt* oligonucleotide competitor, and MPEX-mt* beads were washed with buffer containing a 10-fold molar excess of MPEX* competitor. Beads were washed once as described above and then three times in buffer containing a 10-fold molar excess of poly(dI-dC) for 5 min each. Bound proteins were eluted from the beads for 10 min with wash buffer adjusted to 1 M ammonium bicarbonate (AMBIC).
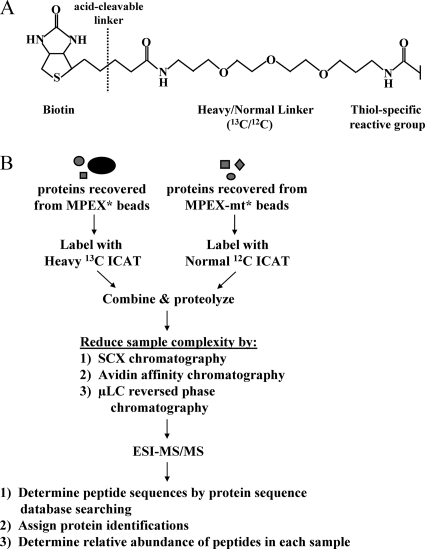
ICAT labeling and preparation of peptides for microcapillary reversed-phase liquid chromatography-tandem mass spectrometry (μLC-MS/MS).
Proteins eluted from MPEX* and MPEX-mt* beads were concentrated under vacuum and then denatured by adding sodium dodecyl sulfate (SDS) to 0.15% and heating for 8 min at 100°C. Proteins were reduced with 4 mM TCEP for 30 min at 37°C. Isotopically heavy and normal cleavable isotope-coded affinity tag (ICAT) reagents (Applied Biosystems, Inc.) were added to MPEX* and MPEX-mt* samples, respectively, according to the manufacturer's directions, and samples were incubated for 2 h at 37°C. Labeling reactions were quenched by adding β-mercaptoethanol to 10 mM and incubating for 20 min at 37°C. Samples were combined and digested for 150 min with 3 μg endoproteinase Lys-C (Boehringer-Mannheim), which cleaves on the carboxy-terminal side of lysine residues. The SDS concentration was reduced to 0.02% by adding 20 mM Tris-HCl (pH 8.3) and 1 mM EDTA, and the sample was digested overnight at 37°C with 15 μg trypsin (sequencing grade modified; 1:20 [wt/wt]; Promega), which cleaves on the carboxy-terminal side of arginine and lysine residues. The sample was diluted to 6 ml with buffer A (5 mM KH2PO4 [pH 3], 25% acetonitrile), and the pH was adjusted to 3 with 10% trifluoroacetic acid. Peptides were purified on a strong cation exchange (SCX) column and eluted in 1 ml with 5 mM KH2PO4 (pH 3)-25% CH3CN-360 mM KCl. Eluted peptides were evaporated to ∼500 μl, diluted threefold with 2× phosphate-buffered saline (PBS), and neutralized with 1 M AMBIC. ICAT-labeled peptides were purified from unlabeled peptides by passing over a monomeric avidin cartridge (ABI) and washing with 2× PBS (pH 7.2), 1× PBS (pH 7.2), and 20% methanol-50 mM AMBIC. Peptides were eluted in 30% acetonitrile-0.4% trifluoroacetic acid and dried under reduced pressure. Dried peptides were resuspended in cleaving reagent (Applied Biosystems, Inc.) and incubated for 2 h at 37°C to cleave the biotin moiety from the labeled peptides. Cleaved peptides were dried under reduced pressure, resuspended in buffer A (described above), then fractionated by passing over a Partisphere SCX cartridge, washing with buffer A, and eluting in six stepwise fractions at 30, 60, 90, 120, 250, and 500 mM ammonium acetate.
μLC-MS/MS and data analysis.
SCX fractions were pressure loaded onto 10-cm by 75-μm fused silica microcapillary reversed-phase columns (5-μm Magic C18 beads; Michrom Bioresources) equilibrated with 10% methanol, 0.4% acetic acid, and 0.005% heptafluorobutyric acid. Peptides were resolved by running 80-min gradients of 10 to 40% buffer C (100% methanol, 0.4% acetic acid, 0.005% heptafluorobutyric acid) at 0.3 μl/min and analyzed by μLC-MS/MS using an LTQ ion trap mass spectrometer (ThermoFinnigan) as previously described (22). Cysteine-containing tryptic peptides were identified by searching MS/MS spectra against a mouse protein sequence database (International Protein Index; downloaded on 11 October 11 2005) using SEQUEST as described elsewhere (15). Data were quantified and analyzed essentially as previously described (23) using the XPRESS and INTERACT computer programs, respectively. The algorithms PeptideProphet (36) and ProteinProphet (43) were used to determine the probability that peptide and protein assignments were correct. A probability cutoff value of 0.9 was used for each analysis.
Gel mobility shift assays.
Gel shift and gel shift interference assays were carried out as previously described (16) and with the following modifications. Double-stranded oligonucleotide probes were end labeled with 32P and purified on a 12% polyacrylamide gel. Briefly, 1 to 2 μg of nuclear extract was mixed with 20 μg of bovine serum albumin in 10 mM Tris-Cl (pH 7.4), 0.1 M NaCl, 0.1 mM EGTA, 0.5 mM dithiothreitol, 0.3 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride, and 4% glycerol. Mouse monoclonal antibodies to human MAZ produced in vivo in mouse ascites fluid (3) were purchased from the Cell Culture Hybridoma Facility at Stony Brook University (MAZ hybridoma 133). Control mouse ascites fluid (M-8273) was purchased from Sigma. Incubations with antisera or unlabeled oligonucleotide competitors were carried out at room temperature for 15 min prior to the addition of probe. The mixtures were incubated with 15 pg of probe/μl and 36 ng of poly(dI-dC)/μl for 15 min at room temperature and then loaded onto 4% polyacrylamide gels. Electrophoresis was carried out in 0.5× Tris-borate-EDTA buffer at 200 V for 1.5 h at 4°C. Gels were dried and analyzed using a PhosphorImager (Molecular Dynamics). The sequences of the double-stranded mouse MCK oligonucleotides used for probes or competitors are shown in Fig. 1A. For the competitors used in Fig. 7A, the forward sequences are as follows: human MCK, 5′-AGAACTCCTCCCTGGGGACAACCCCTCCCAGC-3′; cat MCK, 5′-AGCTCCCTTCCCCGGGGGGCAGCCCCTCCCAG-3′; dog MCK, 5′-AGCTCCCTTCCCTGGGGGCAGCCCCTCCCAGC-3′; bovine MCK, 5′-AGCCCGCTCCCAAGGGGCAGCCCTTCCCAGCC-3′; MAZ-Des, 5′-CTGCAGCTGTCAGGGGAGGGGCGCCGGGGGGT-3′ (−63 to −69); MAZ-Skact, 5′-TTGGAGCCAGTTGGGAGGGGCAGACAGCTGGG-3′ (−238 to −269).
FIG. 7.
MAZ recognizes sequences that diverge from the established motif. (A to C) Labeled MPEX probe was mixed with 2 μg of MM14 skeletal myocyte nuclear extracts and analyzed via gel shift assays. (A) MPEX species variants compete for MAZ binding. Oligonucleotides containing the MPEX sequences from the mouse, human, cat, dog, and bovine MCK promoters, as well as those containing MAZ motifs from the Desmin and Skeletal α-actin promoters (MAZ-Des and MAZ-Skact, respectively), were tested as competitors for MAZ binding. (B) MAZ recognizes MPEX sequences containing changes in the conserved bases of the MAZ motif. Oligonucleotides containing the mouse MPEX sequence with changes (underlined) in the conserved bases of the MAZ motif were tested as competitors for MAZ binding. (C) Divergent motifs from MAZ target genes compete for MAZ binding. Oligonucleotides containing the mouse MPEX sequence with changes (underlined) in or flanking the established MAZ motif, as well as an oligonucleotide containing the GAGA box from the rat Insulin I promoter (MAZ-rIns1), were tested as competitors for MAZ binding. For panels A to C, the effect of a competitor containing the mouse MPEX site with the M1-M3 mutation (M1-M3), which does not compete for MAZ binding, is shown for comparison. The arrowhead indicates free probe. (D) Summary of MAZ recognition motifs. These include the established sequence (GA box), which is also recognized by the Sp/KLF family, and variant motifs (from A to C) which were found to compete well for MAZ binding. Although the established sequence does not include it, we found that an additional C (present in the MCK promoter MPEX sequence and many of the MAZ target genes) is required for optimal binding (in panel C, compare lane 2 with lanes 3 to 5).
For the first four oligonucleotides, the sequences that align with the MAZ site in the mouse MCK promoter are underlined. For the last two oligonucleotides, the MAZ sequences are underlined. Competitors used in Fig. 7B and C are based on the mouse MCK sequence in Fig. 1A, with changes to the MAZ motif indicated within the figure. The sequence of MAZ-rIns1 is 5′-GGGGGCAGAGAGGAGGTGCTTTGGACTATAAA-3′ (37).
Transient transfections.
Proliferating MM14 cells and primary neonatal rat cardiomyocytes were transfected using a standard calcium phosphate technique (1) with 8 μg of chloramphenicol acetyltransferase (CAT) reporter plasmid and 2 μg of the placental alkaline phosphatase (PAP) reference plasmid (pUCSV2PAP). Four hours later, cells were glycerol shocked; MM14 cells were switched to differentiation medium (as described above), and cardiomyocytes were switched to serum-free medium (Dulbecco's modified Eagle's medium/M199 [4:1] supplemented with 6 μg/ml insulin and amphotericin B [Fungizone]). Cells were harvested 48 h after glycerol shock and analyzed for CAT and PAP activity (1). For each construct tested, at least two plasmid preparations were used. The results of independent transfections are shown as the average of at least two transfections of four plates each.
ChIP assays.
ChIP assays were performed from cultured MM14 skeletal myocytes or primary neonatal rat cardiomyocytes using the Fast ChIP method (42), with some modifications. Cells were fixed in 1% formaldehyde in Ham's F-10C for 10 min and subjected to Dounce homogenization 10× prior to sonication. Cells were sonicated for 10 rounds of 15-s pulses at 100% power output on a Model 100 sonic dismembrator (Fisher Scientific) to shear the DNA to a ladder of ∼200 to 800 bp, and the efficiency of shearing was verified by agarose gel electrophoresis. Chromatin was immunoprecipitated using three different MAZ-specific antibodies: mouse monoclonal antibodies (described above) or rabbit polyclonal antibody QC11541 or QC3520 (generous gifts of Aviva Systems Biology) or equivalent amounts of control mouse ascites fluid (M-8273; Sigma) or nonspecific rabbit immunoglobulin G (IgG) (sc-2027; Santa Cruz Biotechnology, Inc.). Quantitative PCR was performed using forward and reverse primers (250 nM) and the 2× SensiMix DNA kit (Quantace Ltd.). Reaction conditions were 40 cycles of 94°C for 15 s, 51.8°C for 30 s, and 72°C for 30 s. Mouse primer sequences are as follows: MCK promoter, F (5′-CGCCAGCTAGACTCAGCACT-3′) (−238 to −219) and R (5′-GAGGAGCCTACAGGGTGTGA-3′) (−32 to −51); Myosin heavy chain (MyHC) IIa promoter, F (5′-TCTCTCCCATGTTCCTCTAGTGT-3′) (−783 to −761) and R (5′-AGTTGCATGCCTTCAACAAT-3′) (−485 to −504); α-MyHC promoter, F (5′-TCTCCTCTATCTGCCCATCG-3′) (−276 to −257) and R (5′-TGCCTAAATTTGGAGTCCTCTG-3′) (−58 to −79); Skeletal α-actin promoter, F (5′-GTGAGCCTTGGAGCCAGTT-3′) (−276 to −258) and R (5′-GTCCCCTTGCACAGGTTTT-3′) (−10 to −28); and Desmin promoter, F (5′-ATAAGGCAGGCATCCAAATG-3′) (−278 to −259) and R (5′-TTTGTAGCCCTCCTGACATC-3′) (−31 to −50). PCR products were analyzed on a 1.5% agarose gel to verify the correct size of the product and the specificity of primer annealing.
Quantitative reverse transcriptase PCR (qRT-PCR) assays.
MM14 skeletal myoblasts were allowed to differentiate for 0, 12, 24, 48, and 72 h. RNA was extracted using the Qiagen RNeasy kit, according to the manufacturer's instructions, and parallel plates for each time point were fixed and stained for MyHC expression. DNase I (Promega) was added to 1 μg of total RNA for 15 min at room temperature and then inactivated with 2.5 mM EDTA and heating to 65°C for 10 min. DNase-treated RNA was primed with oligo(dT) (20 ng/μl) at 90°C for 2 min and then reverse transcribed using deoxynucleoside triphosphates (1 mM), RNasin (Promega), and Moloney murine leukemia virus reverse transcriptase (Promega) for 1 h at 42°C followed by 5 min at 95°C. Quantitative PCR was carried out using 40 ng cDNA, forward and reverse primers (300 nM each), and Sybr green PCR master mix (Applied Biosystems). Reaction conditions were 40 cycles of 94°C for 15 s, 53.5 to 55°C (depending on primer Tm) for 30 s, and 72°C for 30 s. Primers used were specific to mouse MAZ (F, 5′-AGGACCGCATGAGTTACCAC-3′; R, 5′-GCTGCCTCACATTTCTCACA-3′) or 18S rRNA (F, 5′-CGCCGCTAGAGGTGAAATTCT-3′; R, 5′-CGAACCTCCGACTTTCGTTCT-3′). Each PCR was performed in triplicate on RNA from four separate plates per time point. PCR products were analyzed on a 1.5% agarose gel to verify the correct size of the product and the specificity of primer annealing.
RESULTS
The MPEX sequence is required for full MCK promoter activity in skeletal and cardiac myocytes.
To identify new control elements within the MCK promoter, we undertook a mutagenic analysis of highly conserved sequences between −358 and +7. This analysis revealed a GC-rich sequence ∼40 bp upstream of the TATA box, which we named MPEX (Fig. 1A). In transient transfections of cultured skeletal myocytes, mutations M1 and M2 in MPEX resulted in ∼90%- and ∼70%-reduced activity of a reporter construct containing the MCK promoter, whereas mutation M3 just downstream of MPEX had little effect (Fig. 1B). Typically, mutations in the MCK promoter are less deleterious in the presence of the powerful MCK enhancer (1); however, the M1 and M2 mutations in MPEX still reduced activity of a reporter construct containing both the enhancer and promoter by ∼60% and ∼35% (Fig. 1B).
When we performed similar experiments with neonatal rat cardiomyocytes, M1 and M2 reduced activity of the MCK promoter construct by ∼60%, whereas M3 had no effect (Fig. 1C). Interestingly, in the context of the MCK enhancer and promoter, M1 and M2 were even more deleterious, reducing activity of each construct by ∼80% (Fig. 1C). The enhanced effect of M1 and M2 in the presence of the MCK enhancer in cardiomyocytes is in contrast to the slightly dampened effect seen in skeletal myocytes. We speculate that this may be due to synergy of the factor(s) binding MPEX with enhancer-binding factors that are present in cardiomyocytes but not in skeletal myocytes. Although further experiments will be needed to address this possibility, these results indicate that the MPEX sequence is important for MCK promoter activity in both skeletal and cardiac myocytes.
MPEX binds a specific factor(s) in skeletal myocyte nuclear extracts.
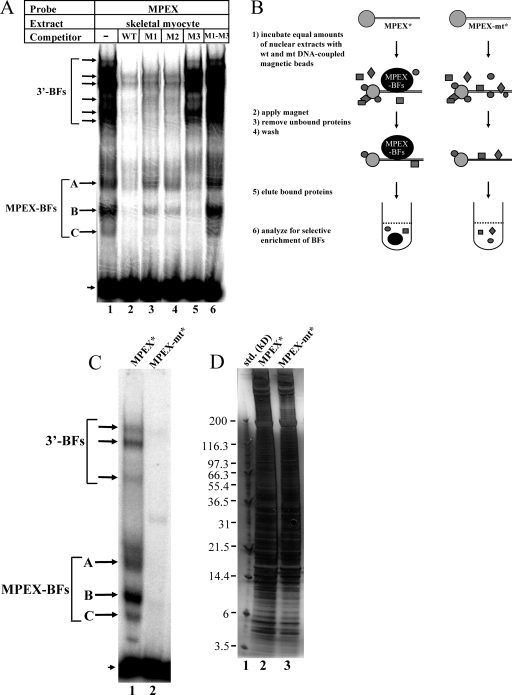
Gel shift assays were performed to determine whether MPEX is recognized by specific nuclear factors. The sequences of probe and competitor oligonucleotides are shown in Fig. 1A. Two different types of binding to the wild-type probe were observed using skeletal myocyte nuclear extracts (Fig. 2A). A multitude of low-mobility shifted complexes were competed away by unlabeled wild-type oligonucleotide and by oligonucleotides containing mutations in the MPEX sequence (M1 and M2) but not by those containing a mutation in the 3′-flanking sequence (M3). This is consistent with the detection of multiple factors (3′-BFs) that specifically recognize the sequence downstream of MPEX (Fig. 2A). In contrast, three high-mobility complexes were competed away by wild-type and M3 oligonucleotides but not competed as well by M1 and M2 oligonucleotides (Fig. 2A), consistent with the detection of one or more factors that specifically recognize MPEX (MPEX-BFs). Interestingly, an oligonucleotide containing mutations in both MPEX and the 3′-flanking sequence (M1-M3) was completely unable to compete for MPEX-specific binding (Fig. 2A). Since a portion of the 3′-flanking sequence is very similar to the MPEX sequence (Fig. 1A, dotted line), it is reasonable to postulate that MPEX-BFs preferentially bind MPEX but are also capable of recognizing the downstream sequence. Thus, when both MPEX and the downstream sequence are mutated (M1-M3), there is no competition for binding.
FIG. 2.
Detection and selective enrichment of MPEX-BFs from skeletal myocytes. (A) MPEX binds specific factors in skeletal myocytes. Labeled MPEX probe was mixed with 2 μg of MM14 skeletal myocyte nuclear extracts and analyzed via gel shift assays. Oligonucleotides containing the wild-type MPEX sequence (WT) or a mutation in the downstream flanking sequence (M3) competed for MPEX-specific complex formation (MPEX-BFs, arrows corresponding to A, B, and C), whereas oligonucleotides containing mutations in the MPEX sequence (M1 and M2) competed less effectively (sequences shown in Fig. 1A). Specific binding to the downstream flanking sequence of MPEX was also observed (3′-BFs). These complexes were competed away by oligonucleotides containing mutations in the MPEX sequence (M1 and M2) but not in the downstream sequence (M3). The arrowhead indicates free probe. (B) Selective enrichment of MPEX-BFs from skeletal myocytes. Equal amounts of crude MM14 skeletal myocyte nuclear extracts were incubated with beads coupled to oligonucleotides containing either the wild-type (wt) MPEX sequence (MPEX*) or the MPEX sequence containing the M1 and M3 mutations (MPEX-mt*). Gray shapes indicate other nuclear proteins. (C) Gel shift assay showing binding to the MPEX probe of proteins eluted from MPEX* and MPEX-mt* beads. Both MPEX-BFs (arrows corresponding to A, B, and C) and 3′-BFs were recovered from MPEX* beads but not MPEX-mt* beads. The arrowhead indicates free probe. (D) Silver-stained SDS-polyacrylamide gel showing the distribution of total protein recovered from MPEX* and MPEX-mt* beads. For panels C and D, ∼0.1% of total eluted protein was loaded in each lane.
MPEX-specific complexes were observed in gel shift assays using nuclear extracts from various cell types (e.g., skeletal and cardiac muscle, liver, and cultured fibroblasts) (C. L. Himeda and S. D. Hauschka, unpublished data); however, these complexes did not migrate to the same relative positions, suggesting that different proteins, isoforms, or cofactors may be associated with this sequence in different cell types.
Selective enrichment of MPEX-BFs and quantitative proteomic identification of candidates.
The MPEX sequence is highly GC rich and contains putative binding sites for many different transcription factors, including AP2, MAZ, and the Sp/KLF family, which includes 26 known members. While we could have tested candidates individually, this approach suffers from two major drawbacks: (i) it depends on the availability of specific antisera for each candidate, and many are not available, and (ii) it excludes any unknown factors or factors with poorly characterized binding motifs. For these reasons, we decided to take an unbiased proteomic approach to identify one or more of the factors that bind MPEX. MPEX-BFs were partially purified from skeletal myocytes using a selective enrichment strategy similar to one we employed in a previous study (27). This strategy is outlined and described in Fig. 2B.
First, streptavidin-linked magnetic beads were coupled to biotinylated oligonucleotides containing either a wild-type or mutant MPEX site. In designing the wild-type oligonucleotide, we wanted to abolish or reduce binding to the sequence immediately downstream of MPEX (Fig. 1A; 3′-BFs in Fig. 2A) while retaining some flanking sequence. Therefore, we designed the “wild-type” oligonucleotide (MPEX*) to contain the −91 to −70 sequence from the mouse MCK promoter, which contains the MPEX site, linked to the −53 to −38 sequence. Deletion of the sequence immediately downstream of MPEX (−69 to −54) did substantially reduce binding of 3′-BFs (compare Fig. 2A and C). We confirmed by gel shift assays that the deletion had no negative effect on MPEX-BF binding and did not create any specific binding site (C. L. Himeda and S. D. Hauschka, unpublished data).
In designing the mutant oligonucleotide (MPEX-mt*), it was important to use a sequence that had no affinity for MPEX-BFs; therefore, this oligonucleotide contains both the M1 and M3 mutations within the −91 to −54 sequence. As shown in Fig. 2A, the presence of both mutations within an oligonucleotide is necessary to completely prevent MPEX-specific binding. Both MPEX* and MPEX-mt* oligonucleotides are 38 bp long to minimize differences in nonspecific binding between the two constructs.
Both sets of beads were incubated with equal amounts of skeletal myocyte nuclear extracts and then washed, and the bound proteins were eluted. Eluted proteins were analyzed by gel shift assays for recovery of MPEX-BFs and by SDS-polyacrylamide gel electrophoresis followed by silver-staining for total protein composition (Fig. 2C and D). In pilot studies, approximately 80% of MPEX-specific binding was recovered, while overall enrichment was ∼160-fold (C. L. Himeda and S. D. Hauschka, unpublished data). As expected, MPEX-BFs were recovered from MPEX* beads but not MPEX-mt* beads (Fig. 2C). However, as seen in our previous study (27), staining for total protein revealed a large number of copurifying proteins in each sample, and no unique bands in the eluent from wild-type versus mutant beads were discernible (Fig. 2D).
Identification of MAZ as an MPEX-BF candidate by quantitative proteomics.
The use of ICAT reagents (Fig. 3A) to identify and quantify proteins from complex mixtures has been described elsewhere (22, 52). ICAT protocols have now become well established and have been applied both in specific factor identification and in global protein profiling (63). We used an ICAT-based quantitative proteomic strategy to identify proteins that are specifically enriched in samples purified with MPEX* beads compared to those purified with MPEX-mt* beads. This strategy is outlined in Fig. 3B, and a detailed description is provided in the figure legend. Overall, 464 unique, cysteine-containing peptide sequences with a PeptideProphet probability of >0.9 were detected, representing 183 unique proteins with a ProteinProphet probability of >0.9 (36, 43). The vast majority of these peptides (∼87%) were present at approximately equal ratios in MPEX* samples versus MPEX-mt* samples. Only 13 proteins showed >2-fold enrichment in MPEX* samples versus MPEX-mt* samples. Of the nine known transcription factors that were enriched, MAZ appeared to be the most promising candidate based on the number of peptides identified and the levels of their enrichment. Five unique peptides were detected for MAZ, ranging from ∼6- to ∼70-fold enrichment (Table 1). Other enriched factors will be described separately.
FIG. 3.
Quantitative proteomic identification of MPEX-BF candidates. (A) Structure of the ICAT tag. (B) Quantitative proteomic strategy for identifying MPEX-BFs. Protein samples recovered from MPEX* and MPEX-mt* beads were labeled with isotopically heavy (13C) or normal (12C) forms of the ICAT reagent, respectively. The labeled protein samples were combined and proteolyzed with endoproteinase Lys-C and trypsin. Sample complexity was then reduced by sequential chromatography. Peptides were fractionated by SCX chromatography, after which ICAT-labeled, cysteine-containing peptides were isolated from each of six fractions by avidin affinity chromatography. Peptides in each fraction were further resolved and analyzed by μLC-electrospray ionization (ESI)-MS/MS. Peptide sequences were determined by searching MS/MS spectra against a mouse protein database using the search algorithm SEQUEST. The relative abundance of peptides recovered from MPEX* versus MPEX-mt* beads was determined from the ratio of the heavy and normal signal intensities for each peptide pair. Heavy/normal ratios of at least 2:1 indicate selective enrichment of the corresponding candidate protein within the nuclear factor complex bound to oligonucleotides containing the wild-type MPEX sequence.
TABLE 1.
Peptides corresponding to MAZ in MPEX* versus MPEX-mt* DNA affinity-purified samples
| Peptidea | Heavy/normal ratiob | Charge state |
|---|---|---|
| R.AHTVRHEEKVPCHVCGK | 68.4:1 | +3 |
| R.HEEKVPCHVCGK | 21.0:1 | +3 |
| K.LSHSDEKPYQCPVCQQR | 18.6:1 | +2 |
| 50.4:1 | ||
| >6.20:1c | +3 | |
| R.QVHSTERPFKCEK | 20.0:1 | +2 |
| R.QVHSTERPFKCEKCEAAFATK | 19.1:1 | +3 |
| R.QVHSTERPFKCEKCEAAFATKDR | >5.70:1c | +3 |
| R.SHDGAVHKPYNCSHCGK | 11.0:1 | +2 |
| 30.8:1 | +3 | |
| K.VHSQGPHHVCELCNK | 15.8:1 | +2 |
Peptides were analyzed by μLC-ESI-MS/MS, and protein identifications were assigned using the SEQUEST algorithm to search a mouse protein sequence database. ICAT-labeled cysteine residues are indicated in bold, and dots refer to sites of tryptic cleavage. PeptideProphet probability scores for each peptide were >0.9.
The relative abundance of each peptide in heavy (isolated from MPEX* beads) versus normal (isolated from MPEX-mt* beads) ICAT-labeled samples was calculated using the XPRESS software program and is expressed as a ratio.
In cases where a signal is detected for a heavy-ICAT-labeled peptide but no signal is detected for the corresponding normal-ICAT-labeled peptide, each ratio is at least the value reported.
MAZ transactivates the minimal MCK promoter.
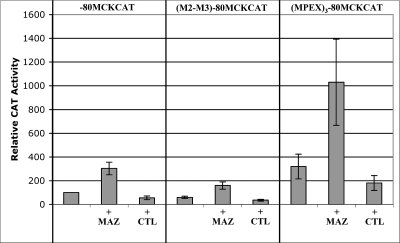
To determine whether MAZ is able to transactivate the MCK promoter, we performed cotransfection experiments using the minimal promoter sequence (−80 to +7) which is only known to contain the MPEX site and the TATA box. In addition to a reporter construct containing the minimal MCK promoter (−80MCKCAT), we also tested the M2-M3 mutation (Fig. 1A) in −80 ([M2-M3]−80MCKCAT), as well as three multimerized copies of the MPEX sequence upstream of −80 ([MPEX]3−80MCKCAT). Each of these constructs was transfected into cultured skeletal myocytes with or without a MAZ expression plasmid or the empty pCGN vector.
MAZ transactivated the minimal MCK promoter by ∼3-fold, whereas the control vector had no effect (Fig. 4). Interestingly, MAZ also transactivated (M2-M3)−80MCKCAT, although to a lesser extent (Fig. 4), suggesting that under these particular transactivation conditions, part of the MAZ-mediated response is independent of the MPEX site. Since MAZ is a ubiquitous factor, this could be an indirect effect of MAZ-mediated upregulation of other genes (perhaps members of the general transcription machinery). When three additional copies of MPEX were present ([MPEX]3−80MCKCAT), the construct was ∼3-fold more active than the minimal MCK promoter construct, and overexpression of MAZ resulted in an additional threefold increase in activity of the multimerized construct (Fig. 4). Taken together, these results indicate that MAZ is able to transactivate the MCK promoter.
FIG. 4.
MAZ transactivates the minimal MCK promoter. MM14 skeletal myocytes were transfected with constructs containing the CAT reporter under the control of the 80-bp MCK minimal promoter (−80MCKCAT), the MCK minimal promoter containing the M2-M3 mutation ([M2-M3]−80MCKCAT; sequence in Fig. 1A), or three copies of the wild-type MPEX site upstream of the MCK minimal promoter ([MPEX]3−80MCKCAT). Each reporter construct was transfected with or without 0.5 μg of MAZ expression vector (MAZ) or pCGN control vector (CTL). Data are plotted as the mean value and standard deviation of relative CAT activity, with activity of −80MCKCAT set at 100.
MAZ binds the MPEX sequence and is enriched at the MCK promoter and other muscle gene promoters.
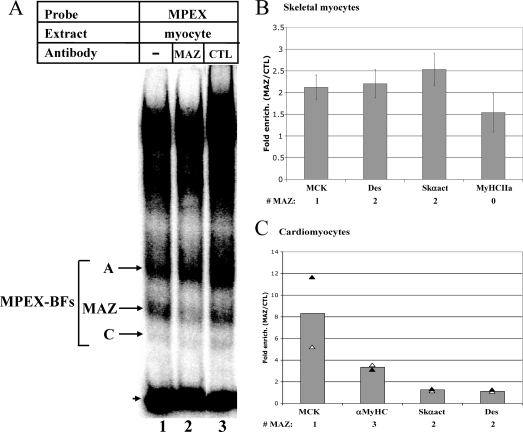
The MAZ binding motif (GGGAGGG or CCCTCCC) is present within the MPEX sequence in the mouse MCK promoter. To confirm the ability of MAZ to bind MPEX, gel shift interference assays were performed using MAZ-specific antibodies. Antibodies to MAZ significantly reduced formation of the MPEX-specific complex “B” (Fig. 5A, lane 2), whereas nonspecific antibodies had no effect (lane 3). Since the MAZ antibodies used in this experiment are a mixture of 12 different monoclonal antibodies (3), presumably one or more of these antibodies recognizes the DNA-binding domain of MAZ, precluding its binding to the probe.
FIG. 5.
MAZ binds the MCK promoter and other muscle gene promoters in skeletal and cardiac myocytes. (A) MAZ binds the MPEX sequence. Labeled MPEX probe was mixed with 2 μg of MM14 skeletal myocyte nuclear extracts and analyzed via gel shift interference assay. MAZ-specific antibodies reduced formation of the indicated complex (MAZ), whereas nonimmune antibodies (CTL) had no effect. The arrowhead indicates free probe. (B) MAZ occupies muscle gene promoters in skeletal myocytes. ChIP assays were performed using MM14 skeletal myocytes and three different MAZ-specific antibodies or nonspecific rabbit IgG/mouse ascites. Immunoprecipitated chromatin was analyzed by qPCR using primers specific to the promoters of MCK (MCK), Desmin (Des), Skeletal α-actin (Skαact), and Myosin heavy chain IIa (MyHCIIa). (C) MAZ occupies muscle gene promoters in cardiomyocytes. ChIP assays were performed as for panel B, except that neonatal cardiomyocytes were used and primers specific for the α-Myosin heavy chain promoter (αMyHC) were used in place of primers to MyHC IIa. For panels B and C, data are represented as n-fold enrichment of the indicated promoter region by MAZ-specific antibodies (MAZ) relative to nonspecific antibodies (CTL). Each bar represents the average for three independent ChIP experiments (for panel B) or two independent ChIP experiments (for panel C), with three replicate PCRs per experiment. The number of MAZ binding motifs in the region flanked by the PCR primers is indicated (# MAZ).
To determine whether MAZ binds the MCK promoter in vivo, ChIP assays were performed using chromatin from skeletal and cardiac myocytes. Immunoprecipitation of chromatin from skeletal myocytes using three different MAZ antibodies yielded ∼2-fold enrichment of the MCK promoter over that obtained with nonspecific IgG or control ascites (Fig. 5B). When we assessed other muscle genes for MAZ occupancy, we found that the Desmin and Skeletal α-actin promoters, each of which contains two MAZ motifs, were enriched to an extent similar to that of the MCK promoter whereas the Myosin heavy chain (MyHC) IIa promoter, which contains no MAZ motifs, displayed lower enrichment (Fig. 5B). The slight apparent enrichment of MAZ at the MyHC IIa promoter is likely due to a number of GC-rich regions, which might act as cryptic MAZ-binding sites (see Fig. 7).
We performed similar experiments with neonatal rat cardiomyocytes using two of the three MAZ antibodies (the third was not available in a sufficient quantity for additional ChIP assays). In this cell type, we found that antibodies to MAZ yielded ∼8-fold enrichment of the MCK promoter over that obtained with nonspecific IgG (Fig. 5C). We also saw ∼3-fold enrichment of the α-MyHC promoter, which contains three MAZ motifs and is specifically active in cardiomyocytes, whereas the Skeletal α-actin promoter showed little enrichment in these cells (Fig. 5C). Surprisingly, the Desmin promoter, which drives minimal expression of this gene in all muscle types (40), was also not enriched, suggesting that the presence of a MAZ motif in an active promoter does not necessarily correlate with its occupancy by MAZ. Nonetheless, the enrichment of several muscle-specific genes in both skeletal and cardiac myocytes suggests a novel role for MAZ as a regulator of muscle gene transcription.
MAZ transcripts are upregulated during skeletal myocyte differentiation.
To examine the temporal regulation of MAZ during muscle differentiation, we performed qRT-PCR on mRNA isolated from skeletal myoblasts and myocytes at various stages of differentiation. MAZ transcripts increased twofold by 12 h of differentiation and sixfold by 72 h (Fig. 6A). Taken together with the occupancy of MAZ at muscle-specific gene promoters (Fig. 5B), the severalfold upregulation of MAZ expression concomitant with skeletal muscle differentiation suggests that activation of MAZ transcription is important for the expression of muscle structural genes.
FIG. 6.
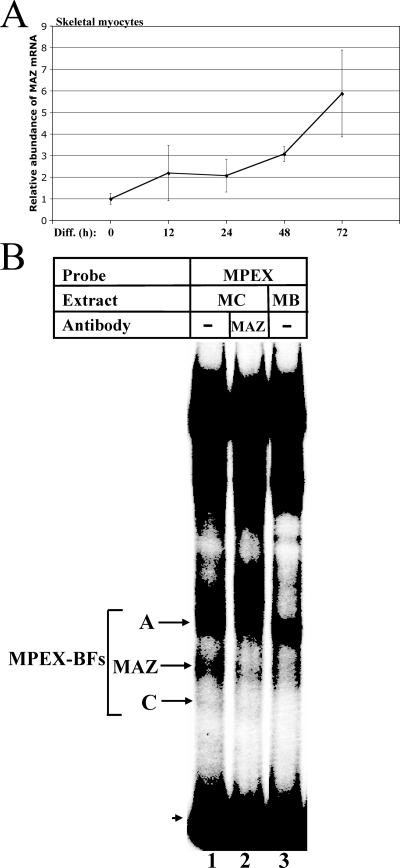
MAZ transcripts and DNA-binding activity are upregulated during skeletal myocyte differentiation. (A) MAZ transcripts increase during skeletal myocyte differentiation. MM14 skeletal myoblasts were allowed to differentiate for 0, 12, 24, 48, and 72 h and then harvested, and RNA was isolated. qRT-PCR was performed using primers specific to MAZ or 18S rRNA. Data are represented as the n-fold change in mRNA abundance for MAZ relative to 18S rRNA. Student's t test P values are 0.001 for 0 h versus 48 h and 0.05 for 0 h versus 72 h. (B) MAZ DNA-binding activity increases during skeletal myocyte differentiation. Labeled MPEX probe was mixed with 2 μg of either MM14 skeletal myocyte (MC) or myoblast (MB) nuclear extracts and analyzed via gel shift assay. The MAZ-specific band (reduced by MAZ-specific antibodies; lane 2) is indicated. The arrowhead indicates free probe.
MAZ DNA-binding activity is upregulated during skeletal myocyte differentiation.
To determine whether functional MAZ protein, in addition to MAZ transcripts, is upregulated during muscle differentiation, we compared the levels of MAZ binding to the MPEX probe in gel shift assays (Fig. 6B). The band corresponding to MAZ in skeletal myocyte nuclear extracts (Fig. 6B, lane 1) is greatly diminished in extracts from undifferentiated skeletal myoblasts (lane 3), confirming that levels of functional MAZ protein (i.e., factor with the ability to bind DNA) are upregulated in differentiated myocytes.
MAZ recognizes sequences that diverge from the established binding motif.
The MAZ binding site (GGGAGGG or CCCTCCC) is present within the MPEX sequence in the mouse MCK promoter, but surprisingly, this sequence is not fully conserved among other mammalian species (Fig. 1A). To test the ability of MAZ to bind these variant sequences, we compared them as competitors in gel shift assays. As expected, the canonical MAZ motifs within the Desmin and Skeletal α-actin promoters each competed as well as the mouse MPEX sequence for MAZ binding (Fig. 7A). This is consistent with the results of our ChIP analysis, which demonstrated MAZ occupancy within these muscle gene promoters (Fig. 5B). Interestingly, the human, cat, dog, and bovine sequences were also able to compete for MAZ binding (Fig. 7A), although the cat and bovine sequences did not compete as effectively as the mouse sequence. Furthermore, oligonucleotides containing mutations at each of the conserved bases in the MAZ motif of MPEX also competed as well as the mouse sequence (Fig. 7B). These data indicate that the DNA motif recognized by MAZ may be substantially broader than previously thought.
To better determine the spectrum of sequences that MAZ recognizes, we compared MAZ binding sites from known MAZ target genes and designed MPEX oligonucleotides to incorporate any changes from the established site (CCCTCCC). These sequences were then tested as competitors in gel shift assays (Fig. 7C). Each of these divergent sequences competed well for MAZ binding to the mouse MPEX probe (Fig. 7C, lanes 3 to 13) compared to the M1-M3 MPEX mutant sequence, which displayed no competition for binding (lane 15). Interestingly, the GAGA box from the rat Insulin 1 promoter (AGAGAGGAGGTG), which has been shown to bind hamster MAZ (37), did not compete well for MAZ binding in our assay (Fig. 7C, lane 14). Although we have not attempted a comprehensive analysis of MAZ binding (which would require a binding site selection approach), our studies have uncovered at least nine divergent motifs which MAZ appears to recognize as well as the established motif (summarized in Fig. 7D).
Divergent MAZ motifs are abundant in striated muscle genes.
Of the nine variant MAZ motifs that were identified by gel shift competition studies (Fig. 7), two have also been shown to bind members of the Sp/KLF family (Fig. 7D) (33). We therefore searched for the presence of the remaining seven sequences in the regulatory regions of other muscle genes (Table 2). Divergent MAZ motifs are present in a wide array of both skeletal and cardiac muscle genes, including those encoding structural proteins (e.g., Skeletal α-actin and β-MyHC) and key transcription factors (e.g., SRF, Myogenin, MEF2C, and Six4). The sequence CTCCTCCC, which corresponds to the divergent MAZ motif in human MCK MPEX, appears more often in these muscle genes (14 instances) than CCCTTCCC (7 instances), CTCCACCC (5 instances), CTCCGCCC (4 instances), CCCCTACC (3 instances), CCCCTCCA (3 instances), or CCCCTCAC (1 instance). In many of these gene promoters, multiple divergent motifs are present. While it is possible that Sp/KLF factors may also be capable of binding to some of these sequences, the identification of these variant MAZ-binding motifs in a wide range of muscle genes is consistent with an extensive role for MAZ in muscle gene regulation.
TABLE 2.
Divergent MAZ motifs are abundant in striated muscle genes
| Gene | Species | Divergent MAZ motifsa | Positions |
|---|---|---|---|
| Serum Response Factor | Mouse | GGGAAGGG | −6 to +2 |
| CTCCTCCC | −208 to −201 | ||
| CTCCACCC | −568 to −561 | ||
| CCCCTCCA | −571 to −564 | ||
| Skeletal α-actin | Mouse | GGGTGGAG | +64 to +71 |
| GGGAAGGG | −83 to −76 | ||
| CTCCTCCC | −495 to −488 | ||
| CTCCTCCC | −680 to −673 | ||
| β-MyHC | Mouse | CCCTTCCC | −480 to −473 |
| CTCCTCCC | −816 to −809 | ||
| CTCCTCCC | −860 to −853 | ||
| Muscle creatine kinase | Human | CTCCTCCC | −82 to −75 |
| Mouse | CCCCTCAC | −1564 to −1557 | |
| CTCCGCCC | −1585 to −1578 | ||
| Myosin light chain 2V | Mouse | CTCCTCCC | −433 to −426 |
| CCCCTCCA | −2451 to −2444 | ||
| CCCCTACC | −2276 to −2269 | ||
| Six5 | Mouse | GGGCGGAG | −286 to −279 |
| GGGAAGGG | −308 to −301 | ||
| CTCCTCCC | −357 to −350 | ||
| Vimentin | Mouse | CTCCACCC | −535 to −528 |
| GGTAGGGG | −925 to −918 | ||
| CTCCTCCC | −2055 to −2048 | ||
| MEF2C | Mouse | CTCCTCCC | −113 to −106 |
| GGGAGGAG | −1200 to −1193 | ||
| M1 muscarinic AChR | Rat | CTCCTCCC | +326 to + 333 |
| CCCCTCCA | −722 to −715 | ||
| Myogenin | Mouse | GGGAAGGG | −153 to −146 |
| CCCTTCCC | −432 to −425 | ||
| Neuronal AChR β4 | Rat | CTCCACCC | −57 to −50 |
| CCCTTCCC | −63 to −56 | ||
| Six4 | Mouse | CTCCTCCC | −105 to −98 |
| GGGTGGAG | −152 to −145 | ||
| CaChR α-1 | Mouse | CCCCTACC | −190 to −183 |
| MyoD (PRR) | Mouse | CTCCGCCC | −114 to −107 |
| Six1 | Human | CTCCTCCC | −497 to −490 |
| Telokin | Mouse | GGGCGGAG | −1329 to −1322 |
Sequences correspond to divergent MAZ motifs, shown in Fig. 7D, which have not been shown to bind Sp/KLF factors.
A divergent MAZ motif is essential for high-level activity of the Six4 promoter in skeletal and cardiac myocytes.
Although it contains no conserved perfect matches to the established MAZ binding site, the mouse Six4 promoter contains two divergent MAZ motifs that are fully conserved between mice and humans (MAZ1 and MAZ2) (Fig. 8A). To determine the functional importance of these sequences, we cloned the 643 bp upstream of the mouse Six4 transcription start site (48) and made two individual mutations in each divergent MAZ motif (Fig. 8A). Whereas mutations in the upstream motif, MAZ1, had little effect on promoter activity, either mutation in MAZ2 decreased activity of the Six4 promoter by ∼5-fold in skeletal myocytes (Fig. 8B) and by ∼7- to 8-fold in neonatal cardiomyocytes (Fig. 8C), suggesting that this divergent MAZ sequence is biologically relevant. Interestingly, the sequence of MAZ2 (CTCCTCCC) is somewhat overrepresented among the genes in Table 2, suggesting that this sequence may represent an important and hitherto-overlooked cis element for MAZ regulation of muscle genes.
MAZ transactivates and is enriched at the Six4 promoter in skeletal but not cardiac myocytes.
To verify the ability of MAZ to transactivate the Six4 promoter, we performed cotransfection experiments with skeletal and cardiac myocytes using the wild-type Six4 promoter construct and either the MAZ expression vector or the control vector. Interestingly, we found that overexpressed MAZ transactivated the Six4 promoter ∼7-fold in skeletal myocytes but had no effect in cardiomyocytes (Fig. 8B and C). Consistent with this, ChIP analysis demonstrated ∼2-fold enrichment of MAZ at the Six4 promoter in skeletal myocytes but no enrichment in cardiomyocytes (Fig. 8D), pointing to different modes of Six4 transcriptional regulation in skeletal versus cardiac muscle.
DISCUSSION
These studies have described the identification and characterization of a previously unknown control element (MPEX) in the MCK proximal promoter. Using selective enrichment followed by quantitative proteomics, we identified MAZ as one of the factors binding the MPEX sequence. Using ChIP studies, we confirmed in vivo binding of MAZ to the MCK promoter, as well as other skeletal and cardiac muscle genes. Consistent with its enrichment at the promoters of MCK, Skeletal α-actin, and Desmin, we found that MAZ transcripts and DNA-binding activity are upregulated over the course of skeletal muscle differentiation. This is, to our knowledge, the first specific demonstration of muscle gene regulation by the MAZ transcription factor.
As in our previous identification of a nonconsensus Six4 control element in the MCK enhancer (27), quantitative proteomics proved to be a powerful strategy for identifying low-abundance binding factors in a complex mixture. Out of 183 total proteins, only 9 were identified as MPEX-BF candidates. The presence of multiple MPEX-specific bands in our gel shift assays suggested that more than one factor might bind this sequence, and indeed, we were able to confirm binding of several additional candidates (data will be reported elsewhere). MAZ appeared to be one of the strongest candidates, with five unique peptides enriched in MPEX* versus MPEX-mt* DNA affinity-purified samples. Six of eleven predicted cysteine-containing tryptic peptides in mouse MAZ (55%) were identified. It is likely that the other peptides were not detected due to duty cycle limitations of the mass spectrometer (limited time over which the instrument can take measurements).
MAZ is surprisingly diverse in its functional roles, being implicated in polyadenylation and transcriptional termination, activation, and repression. Furthermore, since MAZ is only one of a plethora of factors that recognize GC-rich sequences, transcriptional regulation of its target promoters is likely to be complex and highly dynamic. Sp1 and MAZ, for instance, have been shown to compete for binding to the same or overlapping sites (25, 49, 50, 57), which are often present more than once within a given promoter. Interestingly, phosphorylation by casein kinase II has been shown to enhance DNA binding of MAZ (64), whereas it inhibits DNA binding of Sp1 (2). In the course of our studies, we discovered that the low-mobility complexes binding the sequence downstream of MPEX (3′-BFs) contain Sp1. However, mutations in this downstream region have little effect on MCK promoter activity in skeletal or cardiac myocytes, at least under the conditions tested.
Based on the high similarity of their binding sites, it is likely that MAZ, Sp1, and related factors regulate their target genes by mechanisms that depend on relative binding-site affinity, as well as the local abundance and modification state of each factor and its respective cofactors. An example of this type of complexity is seen in the promoter of RUSH/SMARCA3, a progesterone target gene, which contains a GC-rich element that is bound by Egr-1, Sp1, Sp3, MAZ, MZF1, and c-Rel in gel shift assays using nuclear extracts from progesterone-treated rabbits (26). The relatively low enrichment of MAZ on its target muscle gene promoters in our ChIP studies is consistent with such a scenario, where multiple factors are all capable of interacting with a single element and thus might compete for occupancy of the site. An additional complexity is that many transcription factors (including MAZ and Sp1) have dual functions in the activation and repression of their target genes. One mechanism by which such a factor might preferentially recruit either coactivators or corepressors is dependent on the exact sequence of its binding site (core and flanking sequences). Pit-1, for instance, adopts different structural conformations upon binding to slightly different sequences, which allows it to recruit either positive or negative coregulators (55). Thus, correct regulation of MAZ target genes may depend on subtle differences in DNA binding specificity for multiple, slightly variant control elements.
To better characterize the DNA-binding requirements of MAZ, we tested a number of MPEX sequence variants (from the MCK promoters of other species and the MAZ motifs of other genes) as competitors in gel shift assays. Since the mouse MPEX sequence contains the established MAZ motif, the effectiveness of each competitor was compared to that of the mouse sequence; all of these sequences competed, although with different efficiencies. After expanding the MAZ motif to include the divergent sequences that competed particularly well and excluding sequences that are also known to bind members of the Sp/KLF family, we searched for the presence of these motifs in striated muscle genes. Divergent MAZ motifs are abundant in both skeletal and cardiac muscle genes, and interestingly, the sequence corresponding to the variant MAZ motif in human MCK MPEX (CTCCTCCC) appeared to be overrepresented. When we tested the functionality of the two divergent MAZ motifs in the Six4 promoter, we found that only mutations in the overrepresented 3′ MAZ motif were deleterious. Although this sequence was found to be critical for Six4 promoter activity in both skeletal and cardiac myocytes, we found that MAZ was able to transactivate the Six4 promoter only in differentiated skeletal muscle cells and indeed was enriched at the endogenous Six4 promoter only in skeletal myocytes. Thus, it appears that MAZ binds a divergent motif to facilitate full transactivation of the Six4 promoter in skeletal muscle, but another factor (perhaps one of the other MPEX-BF candidates identified in our proteomic strategy) performs this function in cardiac muscle. Since the 3′ MAZ motif in the Six4 promoter (CTCCTCCC) does not match the consensus binding site for any known transcription factor, we suspect it was overlooked as an important control element within the Six4 promoter, although other putative elements flanking it (MEF3, c-Rel, and CdxA) were readily identified (48). It is also interesting to note that this variant MAZ motif could not have been predicted based on an alignment of known MAZ binding sites, since the 3′-most C in the established motif appears to be practically invariant (present in the Desmin, Skeletal α-actin, RAG-2, Ad major late, CLC-K1, PNMT, Serotonin 1a receptor, c-myc, and CD4 genes).
Adding to the complexity of MAZ-mediated gene regulation, several MAZ-related factors (MAZR, MAZi, and THZif-1) have been described. MAZR (seven-Cys2-His2 zinc finger protein with 72% sequence similarity in zinc fingers 2 to 6) is only loosely related to MAZ, although it binds a similar G-rich sequence ([G/C]GGGGGGGG[A/C]C) (38). Like MAZ, MAZR is widely expressed, although it is most abundant in thymus, fetal liver, and bone marrow (38). MAZi (99.3% amino acid identity to human MAZ) was discovered in human pancreatic islet cells and shown to recognize both single- and double-stranded DNA (65). While THZif-1 has 98.2% amino acid similarity to MAZ, it contains a variant second zinc finger (only 33.3% similar to MAZ) and is capable of binding single-stranded, double-stranded, and triple-helical DNA (35, 54). Interestingly, MAZR and MAZi appear to activate, whereas THZif-1 represses, expression of c-myc. It is not currently known whether the latter two factors are the products of alternative splicing or encoded by separate genes. In our studies, we found that MAZ from mouse skeletal myocyte nuclear extracts was incapable of recognizing the insulin I GAGA box, which had been previously reported to bind MAZ from a hamster insulinoma cell line (37). This apparent discrepancy in MAZ binding motifs also hints at the possibility of species- or cell type-specific MAZ isoforms or related factors.
To the best of our knowledge, only two proteins have been reported to interact with MAZ: DCC, a type I membrane protein and putative tumor suppressor, and FAC1, a transcription factor. DCC has been shown to associate with MAZ during neuronal differentiation of P19 cells, which correlates with a loss of expression of c-myc, a MAZ target gene (67). FAC1 has been shown to repress MAZ-mediated activation of the simian virus 40 promoter and to colocalize with MAZ to plaque-like structures in the brains of Alzheimer's patients (32). While DCC and FAC1 are expressed in heart tissue (17, 32, 53), a role for these factors in regulating MAZ-mediated gene expression in cardiac muscle has not been addressed. The identification of coregulators of MAZ in both skeletal and cardiac muscle will be a critical step toward understanding how MAZ mediates its transcriptional effects in these tissues.
In skeletal and cardiac myogenesis, key transcription factors and their cofactors regulate the genes required for the specification, formation, and maintenance of distinct anatomical muscles. In cardiac myogenesis, these key factors include Nkx2.5, GATA, HAND, MEF2, Tbx20, and myocardin (7). In skeletal myogenesis, these factors include the myogenic regulatory factor family and MEF2, which play critical roles in muscle development and differentiation (4). In addition to these restricted factors, a host of more ubiquitous transcription factors is thought to help modulate or sustain robust levels of gene expression in muscle cells. These factors include Sp1, AP2, TEF-1, SRF, the Six proteins, and MAZ. The Six proteins, in particular, have emerged as key regulators of myogenesis, with Six1/4 required for Myf5 expression during early development (18) and Six1 implicated in fiber-type switching in skeletal muscle (21).
In our studies, we found that MAZ transcripts are upregulated ∼6-fold over the course of skeletal myocyte differentiation. In contrast, we found that Myogenin transcripts are upregulated ∼50-fold over the same time course. This striking difference may reflect the different roles of myogenin versus MAZ—the former as a global activator of genes required for muscle differentiation and the latter as one of many factors that contribute to the fine-tuning of gene expression patterns. These different roles are also reflected in the relative effects of mutations in control elements that bind MAZ versus myogenin. For instance, mutations in the MCK promoter MPEX site, which binds MAZ, can result in about a twofold decrease in activity in the presence of the enhancer, whereas mutations in the MCK enhancer right E-box, which binds myogenin (47), are far more deleterious (∼16-fold decrease) (46). However, mutation of the MCK promoter MPEX site causes a greater loss of transcriptional activity than mutation of a highly conserved MCK promoter E-box element that also presumably binds myogenin and/or MyoD in skeletal muscle cells (46) and a loss of activity equivalent to that with mutations in the MCK enhancer CArG, Trex, A/T-rich, left E-box, and MEF2 control elements (1, 16, 45, 46). Our discovery of multiple variant MAZ motifs in the promoters of such key muscle regulators as Myogenin, MEF2C, and Six4, as well as confirmation of a functional MAZ control element in the Six4 promoter, are consistent with a role for MAZ in the development and maintenance of the muscle phenotype, in part through the activation of key muscle transcription factors. Understanding the mechanisms by which these factors mediate their effects and the spatiotemporal dynamics of their interactions remains a significant challenge for future studies of skeletal and cardiac myogenesis.
Acknowledgments
We thank J. Angello, J. Buskin, D. Helterline, Q. Nguyen, P. Tai, and R. Welikson for technical assistance and/or critical discussions. We are grateful to J. Klimek and D. Martin at the Institute for Systems Biology proteomics facility for help with mass spectrometry.
This work was supported by NIH-RO1-AR18860 (to S.D.H.), NIH-T32-HL007312, Experimental Pathology of Cardiovascular Disease (to C.L.H.), contract no. N01-HV-28179 from the National Heart, Lung, and Blood Institute (to S.D.H. and C.L.H.), and grant no. P50GMO76547 from the National Institute of General Medical Sciences (to J.A.R.).
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Amacher, S. L., J. N. Buskin, and S. D. Hauschka. 1993. Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol. Cell. Biol. 132753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 27213489-13495. [DOI] [PubMed] [Google Scholar]
- 3.Ashfield, R., A. J. Patel, S. A. Bossone, H. Brown, R. D. Campbell, K. B. Marcu, and N. J. Proudfoot. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 135656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkes, C. A., and S. J. Tapscott. 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell. Dev. Biol. 16585-595. [DOI] [PubMed] [Google Scholar]
- 5.Bossone, S. A., C. Asselin, A. J. Patel, and K. B. Marcu. 1992. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. 897452-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brand, T. 2003. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2581-19. [DOI] [PubMed] [Google Scholar]
- 8.Buskin, J. N., and S. D. Hauschka. 1989. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol. Cell. Biol. 92627-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, D. S., M. Handa, H. Young, A. S. Gordon, I. Diamond, and R. O. Messing. 2000. Genomic organization and expression of the mouse equilibrative, nitrobenzylthioinosine-sensitive nucleoside transporter 1 (ENT1) gene. Biochem. Biophys. Res. Commun. 277200-208. [DOI] [PubMed] [Google Scholar]
- 10.Clegg, C. H., T. A. Linkhart, B. B. Olwin, and S. D. Hauschka. 1987. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 105949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cserjesi, P., B. Lilly, C. Hinkley, M. Perry, and E. N. Olson. 1994. Homeodomain protein MHox and MADS protein myocyte enhancer-binding factor-2 converge on a common element in the muscle creatine kinase enhancer. J. Biol. Chem. 26916740-16745. [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donoviel, D. B., M. A. Shield, J. N. Buskin, H. S. Haugen, C. H. Clegg, and S. D. Hauschka. 1996. Analysis of muscle creatine kinase gene regulatory elements in skeletal and cardiac muscles of transgenic mice. Mol. Cell. Biol. 161649-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, D. D., A. Stupakoff, S. M. Hedrick, K. B. Marcu, and G. Siu. 1995. A Myc-associated zinc finger protein binding site is one of four important functional regions in the CD4 promoter. Mol. Cell. Biol. 153179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem spectral data of peptides with amino acid sequences in protein databases. J. Am. Soc. Mass Spectrom. 5976-989. [DOI] [PubMed] [Google Scholar]
- 16.Fabre-Suver, C., and S. D. Hauschka. 1996. A novel site in the muscle creatine kinase enhancer is required for expression in skeletal but not cardiac muscle. J. Biol. Chem. 2714646-4652. [DOI] [PubMed] [Google Scholar]
- 17.Fricke, C., and C. B. Chien. 2005. Cloning of full-length zebrafish dcc and expression analysis during embryonic and early larval development. Dev. Dyn. 234732-739. [DOI] [PubMed] [Google Scholar]
- 18.Giordani, J., L. Bajard, J. Demignon, P. Daubas, M. Buckingham, and P. Maire. 2007. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc. Natl. Acad. Sci. USA 10411310-11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossett, L. A., D. J. Kelvin, E. A. Sternberg, and E. N. Olson. 1989. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 95022-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grayson, J., R. Bassel-Duby, and R. S. Williams. 1998. Collaborative interactions between MEF-2 and Sp1 in muscle-specific gene regulation. J. Cell. Biochem. 70366-375. [PubMed] [Google Scholar]
- 21.Grifone, R., C. Laclef, F. Spitz, S. Lopez, J. Demignon, J. E. Guidotti, K. Kawakami, P. X. Xu, R. Kelly, B. J. Petrof, D. Daegelen, J. P. Concordet, and P. Maire. 2004. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 246253-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gygi, S. P., B. Rist, S. A. Gerber, F. Turecek, M. H. Gelb, and R. Aebersold. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17994-999. [DOI] [PubMed] [Google Scholar]
- 23.Han, D. K., J. Eng, H. Zhou, and R. Aebersold. 2001. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser, M. A., A. Robinson, D. Hartigan-O'Connor, D. A. Williams-Gregory, J. N. Buskin, S. Apone, C. J. Kirk, S. Hardy, S. D. Hauschka, and J. S. Chamberlain. 2000. Analysis of muscle creatine kinase regulatory elements in recombinant adenoviral vectors. Mol. Ther. 216-25. [DOI] [PubMed] [Google Scholar]
- 25.Her, S., R. Claycomb, T. C. Tai, and D. L. Wong. 2003. Regulation of the rat phenylethanolamine N-methyltransferase gene by transcription factors Sp1 and MAZ. Mol. Pharmacol. 641180-1188. [DOI] [PubMed] [Google Scholar]
- 26.Hewetson, A., and B. S. Chilton. 2008. Progesterone-dependent DNA looping between RUSH/SMARCA3 and Egr-1 mediates repression by c-Rel. Mol. Endocrinol. 22813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himeda, C. L., J. S. Ranish, J. C. Angello, P. Maire, R. Aebersold, and S. D. Hauschka. 2004. Quantitative proteomic identification of Six4 as the Trex-binding factor in the muscle creatine kinase enhancer. Mol. Cell. Biol. 242132-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izzo, M. W., G. D. Strachan, M. C. Stubbs, and D. J. Hall. 1999. Transcriptional repression from the c-myc P2 promoter by the zinc finger protein ZF87/MAZ. J. Biol. Chem. 274:19498-19506. [DOI] [PubMed] [Google Scholar]
- 29.Jaynes, J. B., J. S. Chamberlain, J. N. Buskin, J. E. Johnson, and S. D. Hauschka. 1986. Transcriptional regulation of the muscle creatine kinase gene and regulated expression in transfected mouse myoblasts. Mol. Cell. Biol. 62855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaynes, J. B., J. E. Johnson, J. N. Buskin, C. L. Gartside, and S. D. Hauschka. 1988. The muscle creatine kinase gene is regulated by multiple upstream elements, including a muscle-specific enhancer. Mol. Cell. Biol. 862-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, J. E., B. J. Wold, and S. D. Hauschka. 1989. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol. Cell. Biol. 93393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan-Sciutto, K. L., J. M. Dragich, J. Caltagarone, D. J. Hall, and R. Bowser. 2000. Fetal Alz-50 clone 1 (FAC1) protein interacts with the Myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry 393206-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Krüppel-like transcription factors. Genome Biol. 4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karantzoulis-Fegaras, F., H. Antoniou, S. L. Lai, G. Kulkarni, C. D'Abreo, G. K. Wong, T. L. Miller, Y. Chan, J. Atkins, Y. Wang, and P. A. Marsden. 1999. Characterization of the human endothelial nitric-oxide synthase promoter. J. Biol. Chem. 2743076-3093. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki, H., M. Machida, M. Komatsu, H. O. Li, T. Murata, H. Tsutsui, A. Fujita, M. Matsumura, Y. Kobayashi, K. Taira, and K. K. Yokoyama. 1996. Specific regulation of gene expression by antisense nucleic acids: a summary of methodologies and associated problems. Artif. Organs 20836-848. [DOI] [PubMed] [Google Scholar]
- 36.Keller, A., A. I. Nesvizhskii, E. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 745383-5392. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy, G. C., and W. J. Rutter. 1992. Pur-1, a zinc-finger protein that binds to purine-rich sequences, transactivates an insulin promoter in heterologous cells. Proc. Natl. Acad. Sci. 8911498-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, A., H. Yamagiwa, H. Hoshino, A. Muto, K. Sato, M. Morita, N. Hayashi, M. Yamamoto, and K. Igarashi. 2000. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 201733-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lassar, A. B., J. N. Buskin, D. Lockshon, R. L. Davis, S. Apone, S. D. Hauschka, and H. Weintraub. 1989. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 58823-831. [DOI] [PubMed] [Google Scholar]
- 40.Li, H., and Y. Capetanaki. 1993. Regulation of the mouse desmin gene: transactivated by MyoD, myogenin, MRF4 and Myf5. Nucleic Acids Res. 21335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246780-786. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1179-185. [DOI] [PubMed] [Google Scholar]
- 43.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 754646-4658. [DOI] [PubMed] [Google Scholar]
- 44.Neville, C., N. Rosenthal, M. McGrew, N. Bogdanova, and S. Hauschka. 1997. Skeletal muscle cultures. Methods Cell Biol. 5285-116. [PubMed] [Google Scholar]
- 45.Nguyen, Q. G., J. N. Buskin, C. L. Himeda, C. Fabre-Suver, and S. D. Hauschka. 2003. Transgenic and tissue culture analyses of the muscle creatine kinase enhancer Trex control element in skeletal and cardiac muscle indicate differences in gene expression between muscle types. Transgenic Res. 12337-349. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen, Q. G., J. N. Buskin, C. L. Himeda, M. A. Shield, and S. D. Hauschka. 2003. Differences in the function of three conserved E-boxes of the muscle creatine kinase gene in cultured myocytes and in transgenic mouse skeletal and cardiac muscle. J. Biol. Chem. 27846494-46505. [DOI] [PubMed] [Google Scholar]
- 47.Ohkawa, Y., C. G. Marfella, and A. N. Imbalzano. 2006. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 25490-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozaki, H., K. Yamada, M. Kobayashi, S. Asakawa, S. Minoshima, N. Shimizu, M. Kajitani, and K. Kawakami. 1999. Structure and chromosome mapping of the human SIX4 and murine Six4 genes. Cytogenet. Cell Genet. 87108-112. [DOI] [PubMed] [Google Scholar]
- 49.Parks, C. L., and T. Shenk. 1997. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J. Virol. 719600-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks, C. L., and T. Shenk. 1996. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J. Biol. Chem. 2714417-4430. [DOI] [PubMed] [Google Scholar]
- 51.Pyrc, J. J., K. H. Moberg, and D. J. Hall. 1992. Isolation of a novel cDNA encoding a zinc-finger protein that binds to two sites within the c-myc promoter. Biochemistry 314102-4110. [DOI] [PubMed] [Google Scholar]
- 52.Ranish, J. A., E. C. Yi, D. M. Leslie, S. O. Purvine, D. R. Goodlett, J. Eng, and R. Aebersold. 2003. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33349-355. [DOI] [PubMed] [Google Scholar]
- 53.Reale, M. A., G. Hu, A. I. Zafar, R. H. Getzenberg, S. M. Levine, and E. R. Fearon. 1994. Expression and alternative splicing of the deleted in colorectal cancer (DCC) gene in normal and malignant tissues. Cancer Res. 544493-4501. [PubMed] [Google Scholar]
- 54.Sakatsume, O., H. Tsutsui, Y. Wang, H. Gao, X. Tang, T. Yamauchi, T. Murata, K. Itakura, and K. K. Yokoyama. 1996. Binding of THZif-1, a MAZ-like zinc finger protein to the nuclease-hypersensitive element in the promoter region of the c-MYC protooncogene. J. Biol. Chem. 27131322-31333. [DOI] [PubMed] [Google Scholar]
- 55.Scully, K. M., E. M. Jacobson, K. Jepsen, V. Lunyak, H. Viadiu, C. Carrière, D. W. Rose, F. Hooshmand, A. K. Aggarwal, and M. G. Rosenfeld. 2000. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 2901127-1131. [DOI] [PubMed] [Google Scholar]
- 56.Shield, M. A., H. S. Haugen, C. H. Clegg, and S. D. Hauschka. 1996. E-box sites and a proximal regulatory region of the muscle creatine kinase gene differentially regulate expression in diverse skeletal muscles and cardiac muscle of transgenic mice. Mol. Cell. Biol. 165058-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, J., M. Mangold, G. Suske, C. Geltinger, I. Kanazawa, K. Sun, and K. K. Yokoyama. 2001. Characterization and promoter analysis of the mouse gene for transcription factor Sp4. Gene 26419-27. [DOI] [PubMed] [Google Scholar]
- 58.Song, J., H. Murakami, H. Tsutsui, X. Tang, M. Matsumura, K. Itakura, I. Kanazawa, K. Sun, and K. K. Yokoyama. 1998. Genomic organization and expression of a human gene for Myc-associated zinc finger protein (MAZ). J. Biol. Chem. 27320603-20614. [DOI] [PubMed] [Google Scholar]
- 59.Song, J., H. Murakami, H. Tsutsui, H. Ugai, C. Geltinger, T. Murata, M. Matsumura, K. Itakura, I. Kanazawa, K. Sun, and K. K. Yokoyama. 1999. Structural organization and expression of the mouse gene for Pur-1, a highly conserved homolog of the human MAZ gene. Eur. J. Biochem. 259676-683. [DOI] [PubMed] [Google Scholar]
- 60.Song, J., H. Ugai, H. Nakata-Tsutsui, S. Kishikawa, E. Suzuki, T. Murata, and K. K. Yokoyama. 2003. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements. Int. J. Mol. Med. 11547-553. [PubMed] [Google Scholar]
- 61.Sternberg, E. A., G. Spizz, W. M. Perry, D. Vizard, T. Weil, and E. N. Olson. 1988. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol. Cell. Biol. 82896-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su, J. M., X. M. Lai, K. H. Lan, C. P. Li, Y. Chao, S. H. Yen, F. Y. Chang, S. D. Lee, and W. P. Lee. 2007. X protein of hepatitis B virus functions as a transcriptional corepressor on the human telomerase promoter. Hepatology 46402-413. [DOI] [PubMed] [Google Scholar]
- 63.Tao, W. A., and R. Aebersold. 2003. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr. Opin. Biotechnol. 14110-118. [DOI] [PubMed] [Google Scholar]
- 64.Tsutsui, H., C. Geltinger, T. Murata, K. Itakura, T. Wada, H. Handa, and K. K. Yokoyama. 1999. The DNA-binding and transcriptional activities of MAZ, a myc-associated zinc finger protein, are regulated by casein kinase II. Biochem. Biophys. Res. Commun. 262198-205. [DOI] [PubMed] [Google Scholar]
- 65.Tsutsui, H., O. Sakatsume, K. Itakura, and K. K. Yokoyama. 1996. Members of the MAZ family: a novel cDNA clone for MAZ from human pancreatic islet cells. Biochem. Biophys. Res. Commun. 226801-809. [DOI] [PubMed] [Google Scholar]
- 66.Uchida, S., S. Sasaki, and F. Marumo. 2001. Isolation of a novel zinc finger repressor that regulates the kidney-specific CLC-K1 promoter. Kidney Int. 60416-421. [DOI] [PubMed] [Google Scholar]
- 67.Ugai, H., H. O. Li, M. Komatsu, H. Tsutsui, J. Song, T. Shiga, E. Fearon, T. Murata, and K. K. Yokoyama. 2001. Interaction of Myc-associated zinc finger protein with DCC, the product of a tumor-suppressor gene, during the neural differentiation of P19 EC cells. Biochem. Biophys. Res. Commun. 2861087-1097. [DOI] [PubMed] [Google Scholar]
- 68.Vincent, C. K., A. Gualberto, C. V. Patel, and K. Walsh. 1993. Different regulatory sequences control creatine kinase-M gene expression in directly injected skeletal and cardiac muscle. Mol. Cell. Biol. 131264-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, H. Y., H. T. Chang, T. W. Pai, C. I. Wu, Y. H. Lee, Y. H. Chang, H. L. Tai, C. Y. Tang, W. Y. Chou, and M. D. Chang. 2007. Transcriptional regulation of human eosinophil RNases by an evolutionary-conserved sequence motif in primate genome. BMC Mol. Biol. 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, X., R. C. Southard, C. D. Allred, D. R. Talbert, M. E. Wilson, and M. W. Kilgore. 28 September 2007, posting date. MAZ drives tumor-specific expression of PPAR gamma 1 in breast cancer cells. Breast Cancer Res. Treat. doi: 10.1007/s10549-007-9765-7. [DOI] [PMC free article] [PubMed]
- 71.Williams, L. J., and A. B. Abou-Samra. 2000. The transcription factors SP1 and MAZ regulate expression of the parathyroid hormone/parathyroid hormone-related peptide receptor gene. J. Mol. Endocrinol. 25309-319. [DOI] [PubMed] [Google Scholar]
- 72.Wu, C. X., W. P. Zhao, H. Kishi, J. Dokan, Z. X. Jin, X. C. Wei, K. K. Yokoyama, and A. Muraguchi. 2004. Activation of mouse RAG-2 promoter by Myc-associated zinc finger protein. Biochem. Biophys. Res. Commun. 3171096-1102. [DOI] [PubMed] [Google Scholar]
- 73.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3593-600. [DOI] [PubMed] [Google Scholar]