Abstract
Background:
Programmed DNA-reorganization and DNA-elimination events take place frequently during cellular differentiation. An extreme form of such processes, involving DNA reorganization, DNA elimination and DNA fragmentation, is found during macronuclear differentiation in hypotrichous ciliates. Ciliated protozoa can therefore serve as a model system to analyze the molecular basis of these processes during cellular differentiation in eukaryotic cells.
Results:
Using a biological approach to identify cis-acting sequences involved in DNA fragmentation, we show that in the hypotrichous ciliate Stylonychia lemnae sequences required for specific DNA processing are localized in the 3'- and the 5'-subtelomeric regions of the macronuclear precursor sequence. They can be present at various positions in the two subtelomeric regions, and an interaction between the two regions seems to occur. Sequence comparison revealed a consensus inverted repeat in both subtelomeric regions that is almost identical to the putative Euplotes chromosome breakage sequence (E-Cbs), also identified by sequence comparison. When this sequence was mutagenized, a processed product could no longer be detected, demonstrating that the sequence plays a crucial role in DNA processing. By injecting a construct into the developing macronucleus, which exclusively contains the subtelomeric regions of the Stylonychia αl-tubulin gene, we show that subtelomeric regions are not only required but are also sufficient for DNA processing in Stylonychia.
Conclusions:
Our results indicate that an inverted repeat with the core sequence 5'-TGAA present in both subtelomeric regions acts as a Cbs in Stylonychia. The results allow us to propose a mechanistic model for DNA processing in this ciliate.
Background
Programmed DNA-reorganization and DNA-elimination processes are frequently observed in differentiating eukaryotic cells. Examples include the mating type switch in yeast [1], the antigen variation in trypanosomes and other parasitic flagellates [2,3], the specific DNA elimination observed during embryogensis of nematodes, Cyclops and Sciara [4,5,6], and the processing of mammalian immunoglobulin and T-cell receptor genes [7,8,9]. However, these processes are most extreme in hypotrichous ciliates, which therefore serve as model systems to study the molecular basis of programmed DNA reorganization, DNA elimination and specific DNA fragmentation during cellular differentiation [10].
One characteristic of ciliated protozoa is the occurrence of two morphologically and functionally different nuclei in one cell: the generative micronucleus and the somatic macronucleus, which is responsible for transcription during vegetative growth. After sexual reproduction, a new macronucleus is formed as a derivative of the micronucleus while the old macronucleus degenerates. The stages of macronuclear development in hypotrichous ciliates, such as Oxytricha, Euplotes or Stylonychia, are summarized in Figure 1 [11,12]. First, a DNA-synthesis phase leads to the formation of polytene chromosomes. During this developmental stage, transposon-like elements, as well as short internal eliminated sequences (IES), are excised in the form of DNA circles [13,14,15,16]. The polytene chromosomes then disintegrate and the DNA to be eliminated is enclosed into vesicles where DNA degradation takes place; depending on the organism up to 95% of the DNA is eliminated. The remaining DNA is specifically fragmented into small DNA molecules and telomeric sequences are added de novo. In a second DNA-synthesis phase, each molecule is amplified to a specific copy number, resulting in the somatic macronucleus containing millions of individual DNA molecules, each carrying one gene and all control sequences required for replication and expression (for reviews see [10,15,17]).
Figure 1.

Macronuclear development in S. lemnae (adapted from [11,12]). The arrow indicates the time point of injection. DNA from injected cells was prepared either about 6 hours after injection or from vegetative cells; the arrowheads show these two time points.
The molecular mechanisms of these processes are still not well understood, but in the holotrichous ciliate Tetrahymena, and lately in the hypotrichous ciliate Euplotes, conserved cis-acting sequences have been identified that are involved in directing the process of specific fragmentation. In Tetrahymena thermophila, a 15 base pair (bp) sequence, the chromosome breakage sequence (Cbs), is located in the eliminated sequences flanking the macronuclear precursor [18,19,20]. Sequence comparisons in Euplotes crassus have identified a 5 bp sequence, the proposed E-Cbs, that either resides inside the macronuclear precursor at position 18, or is located in the flanking micronuclear-specific DNA 12 bp from the macronuclear precursor [21,22]. A model for chromosome fragmentation in E. crassus was proposed on the basis of the positions of the E-Cbs and the finding of overlapping sequences, involving a 6 bp staggered cut on both sides of the macronuclear precursor [22,23]. To date, no consensus sequence has been found at defined positions near the fragmentation sites in Stylonychia lemnae [24]. We therefore decided to identify cis-acting sequences involved in DNA fragmentation and telomere addition by injecting modified macronuclear precursor sequences into the developing macronuclei. Recently, we demonstrated that injection of such a construct (pCE5) into the developing macronucleus resulted in correct fragmentation and de novo telomere addition [25]. This construct contained two macronuclear precursor sequences homologous to a 1.1 kb and a 1.3 kb macronuclear DNA molecule ([26]; GenBank accession numbers X72955 and X72956). They are separated by an 11 bp spacer and are flanked by micronuclear-specific sequences. To distinguish between the injected precursor sequence and the endogenous macronuclear DNA molecule, the 1.3 kb precursor sequence was modified by inserting a 500 bp polylinker sequence. Moreover, we showed that neither sequences of the neighboring 1.1 kb macronuclear precursor sequence nor flanking micronuclear-specific sequences are required for specific fragmentation and telomere addition. Deletion of 70 bp of the 3' end of the 1.3 kb precursor sequence resulted in no detectable processing, however, indicating that a sequence located in this subtelomeric region is indeed required for fragmentation and/or telomere addition. In contrast, deletion of 69 bp of the 5' end still led to a processed product. Surprisingly, this processed product contained the subtelomeric sequences that were deleted in the construct, suggesting the presence of a so far uncharacterized proofreading mechanism during macronuclear development. Only after deletion of 520 bp of the 5' end was a processed product no longer observed [27].
Here, we show that both subtelomeric regions are required for correct DNA fragmentation but, at least in the case of the 1.3 kb precursor sequence, the distance of cis-acting sequences from the fragmentation sites are different in the 3' and 5' region. In addition, we show that the subtelomeric regions of the α1-tubulin gene are sufficient for correct DNA processing. Sequence analysis of all these regions revealed the presence of an inverted repeat with a sequence almost identical to the core E-Cbs described in Euplotes crassus.
Results
All deletion constructs used in this study are based on the construct pCE5 (Figure 2a [25]). As shown previously by PCR and Southern analysis [25,27], the concentration of the modified 1.3 kb macronuclear DNA molecule derived from the injected construct was always very low compared with the copy number of the endogenous 1.3 kb DNA molecule [27]. This may be due to a very tight copy number control mechanism occurring during macrouclear development, which allows only a very limited amplification of the injected material, independent of the amount injected. All results presented in this study are therefore based on PCR analysis as described previously [14,25,27].
Figure 2.
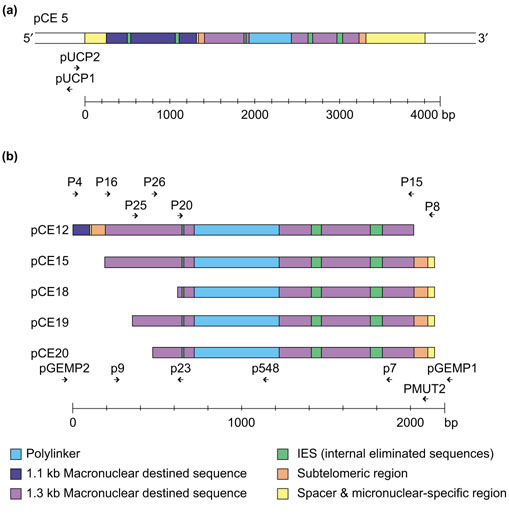
The constructs used for injection into the macronuclear anlagen. (a) Vector pCE5 contains two macronuclear precursor sequences homologous to a 1.1 and a 1.3 kb macronuclear DNA molecule. The 1.1 kb homologous precursor sequence contains 2 IES, and the 1.3 kb homologous sequence contains 3 IES. The two precursor sequences are separated by an 11 bp spacer and are flanked by micronuclear-specific sequences on both sides [26]. A 500 bp polylinker was inserted to modify the 1.3 kb homologous macronuclear precursor sequences [25]. (b) pCE12-20: deletion constructs derived from pCE5. Primers used for construction of the deletion vectors and for PCR analysis are shown above and below the maps. The sequences of the primers are listed in Table 1.
The failure to detect a processed product from injected deletion constructs can be due to either the deletion of cis-acting sequences required for DNA fragmentation and/or telomere addition, or the deletion of sequences required for subsequent amplification and replication of the DNA molecule. We have shown that a sequence located within the first 70 bp of the 3'-subtelomeric region of the 1.3 kb macronuclear precursor sequence is required for DNA fragmentation. After deletion of this region, fragmentation does not occur at either the 3' or the 5' end [27]. This implied that either the 3'-subtelomeric sequence is the only Cbs present or it has to interact specifically with other sequences located in the 5'-subtelomeric region. We therefore tested construct pCE18, in which 520 bp of the 5'-subtelomeric region were deleted for these two possibilities: failure to become fragmented versus deletion of sequences required for amplification and replication. pCE18 and control constructs pCE5 and pCE12, in which 70 bp of the 3'-subtelomeric sequences were deleted, were each injected into the macronuclear anlagen of 20-25 exconjugants in the polytene chromosome stage. The injected cells were allowed to continue macronuclear development for another 6-10 h until fragmentation was completed [27]. DNA was prepared and a PCR analysis was performed using primer combinations in which either both primers were derived from the modified macronuclear molecule (P9/P548; P20/P548), or only one primer came from the modified macronuclear molecule while the other primer hybridized to pUC19 sequences in the case of pCE5 (pUCP1/P20; pUCP2/P548) or to pGEM sequences in the case of pCE12 and pCE18 (pGEMP1/P20; pGEMP2/P548) and to micronuclear-specific sequences (pUCP2/P8; pGEMP2/P8; P20/P8) flanking the insert. As a control, all primer combinations were tested using the different constructs as templates (Figure 3c,d). In pCE5-injected cells, a PCR product was obtained only when both primers were derived from the modified 1.3 kb macronuclear molecule (Figure 3b, lane 5), whereas no products were obtained if one primer was derived from the flanking vector DNA (lanes 1-3) or from the 3'-flanking micronuclear specific sequences (lane 4). These results indicate that, at this stage of macronuclear development, pCE5 was fragmented completely. After injection of pCE12 and pCE18, however, all primer combinations led to amplified products (Figure 3a, lanes 1-3; Figure 3b, lanes 6-10), showing that some unfragmented plasmid remains in the cells, suggesting that chromosome breakage is reduced or does not occur at all. Sequences required for this process must therefore have been deleted. In uninjected cells no primer combination, with the exception of P20/P8, led to PCR products. Sometimes, this primer combination resulted in a PCR product amplified from micronuclear sequences. The signal obtained was always very weak and 500 bp smaller than that derived from the polylinker-containing processed construct. From these results we concluded that sequences required for fragmentation of the 1.3 kb macronuclear molecule are located not only in the first 70 bp of the 3'-subtelomeric region, but also in a region between 69 bp and 520 bp downstream of the 5' end of the 1.3 kb precursor molecule. Similar to the observation made with the 3'-deletion construct pCE12, no fragmentation was observed in pCE18, either at the 5' or at the 3' end, strongly suggesting that an interaction between sequences located in both subtelomeric regions is required for DNA fragmentation.
Figure 3.

The effect of 3' and 5' deletions in the 1.3 kb macronuclear precursor sequence on DNA fragmentation. pCE18 and, as controls, pCE5 and pCE12 [27] were injected into the macronuclear anlagen in the polytene chromosome stage of 20-30 exconjugant cells. DNA was isolated from these cells 6-10 h later and a PCR analysis was performed. The PCR products were separated on a 1% agarose gel. M, molecular weight marker (1 kb ladder, Gibco BRL). (a) PCR products from cells injected with pCE12 using the primer combinations P9/P548 (lane1), pGEMP1/P20 (lane 2) and pGEMP2/P23 (lane 3). (b) PCR products from cells injected with pCE5 (lanes 1-5) using the primer combinations pUCP1/P20 (lane 1); pUCP2/P8 (lane 2); pUCP2P548 (lane 3); P20/P8 (lane 4) and P20/P548 (lane 5). Lanes 6-10: PCR products from cells injected with pCE18 using the primer combinations pGEMP1/P20 (lane 6); pGEMP2/P8 (lane 7); pGEMP2/P548 (lane 8); P20/P8 (lane 9) and P20/P548 (lane 10). (c,d) Control PCR using pCE12 (C), pCE5 and pCE18 (d) construct DNA as templates. PCR reactions in (c) are identical to those shown in (a), and PCR reactions in (d) are identical to those shown in (b).
To characterize this region further, we constructed two more deletion vectors (pCE19 and pCE20; Figure 2b). In pCE19, 230 bp are deleted from the 5'-subtelomeric region of the 1.3 kb precursor molecule, whereas 350 bp are missing in pCE20. Again, DNA of these constructs was injected into cells of the polytene chromosome stage and the injected cells were allowed to finish macronuclear development. Macronuclear DNA of 30-40 vegetative cells derived from single injected cells was analyzed using PCR for the presence of modified macronuclear sequences from the injected construct. By using one primer hybridizing to the 1.3 kb DNA molecule and a second primer homologous to the inserted polylinker sequence, only PCR products are obtained from macronuclear DNA molecules processed from the injected construct. As shown in Figure 4, this was only the case with macronuclear DNA of pCE19 injected cells (Figure 4a,b; lane 5). No PCR product was detectable using macronuclear DNA either of uninjected cells as a control (Figure 4a,b; lane 3) or of cells after injection of pCE20 (Figure 4a,b; lanes 7). These results suggest that, in addition to sequences located in a 70 bp region at the 3' end, sequences located between 230 bp and 350 bp downstream of the 5' end are necessary for processing of this precursor sequence.
Figure 4.

Functional analysis of the 5' end of the 1.3 kb macronuclear precursor sequence. Different deletion constructs were injected into the developing macronucleus and the cells were allowed to finish macronuclear development. The presence of the injected constructs in the macronuclear DNA and their processing was analyzed by PCR. All constructs were injected into the polytene chromosome stage of the macronuclear anlagen; injection conditions were as described earlier [27]. A PCR analysis was performed using DNA of 30-40 vegetatively growing cells as template. Primers used were P9 and P7, both derived from endogenous 1.3 kb macronuclear sequences, and P548, derived from the inserted polylinker. (a) PCR products from uninjected and injected cells were separated on a 1% agarose gel. As a control a PCR using vector pCE5 as template was performed. M, molecular weight marker (1 kb ladder, Gibco BRL). Lane 1, PCR product from pCE5 vector DNA using the primer combination P9/P548. Lanes 2-3, PCR products from uninjected cells using the primer combinations P9/P7 (lane 2) and P9/P548 (lane 3). Lanes 4-5, PCR products from cells injected with pCE19 using the primer combinations P9/P7 (lane 4) and P9/P548 (lane 5). Lanes 6-7, PCR products from cells injected with pCE20 using the primer combinations P9/P7 (lane 6) and P9/P548 (lane 7). (b) The gel was hybridized with a DIG-labeled probe of the polylinker inserted into the 1.3 kb macronuclear precursor sequence. While in the case of pCE19 ((a) lane 5) no PCR product using the primer combination P9/P548 is visible in the ethidium bromide stained gel, a clear signal is observed after hybridization ((b) lane 5).
It is still noteworthy that whenever processing was obtained from a 5'-deletion construct the processed product again contained the previously deleted sequences, suggesting the presence of a correction mechanism during macronuclear development as postulated before [27]. To rule out that these results are due to PCR artefacts by template switching, we performed control experiments in which various amounts of construct DNA were mixed with total cellular DNA from uninjected cells. Under our experimental conditions, the ratio of injected construct DNA to total cellular DNA is below 10-6. In our control experiments, however, we obtained non-specific PCR amplification only by increasing the ratio of construct DNA to cellular DNA to at least 1:10 and at DNA concentrations above 50 μg/ml, as described previously [27]. These results therefore make a PCR artefact by template switching very unlikely, although homologous recombination events can not be completely ruled out with these control elements.
We examined the 70 bp of the 3'-subtelomeric region and the 5'-subtelomeric regions from position 230-350 for the presence of palindromic sequences, direct and inverted repeats, and found a six-base inverted repeat of the sequence 5'-TTGAAA at position 267-272 in the 5'-subtelomeric region and at position 20-25 in the 3'-subtelomeric region. This sequence is almost identical to the core of the E-Cbs, 5'-TTGAA, described in Euplotes [21] and can be found only once in each subtelomeric region. In order to demonstrate that this sequence is indeed required for specific DNA processing, we mutagenized the 5'-TTGAAA present in the 3'-subtelomeric region into 5'-TTCAGA. Thirty exconjugant cells were injected with this construct, and the same number of cells was injected with pCE5 as a control. In 23 cells injected with pCE5, a processed product could be obtained using the primer combination P9/P548. This number corresponds to the normal success rate of our injection procedure [25,27]. However, a processed product could not be detected in any of the 30 cells injected with the mutagenized construct pCE5mut (Figure 5). The failure to detect such a product cannot be explained by a variation in the success rate of injection but rather provides significant evidence that this inverted repeat acts as a Cbs.
Figure 5.

The effect of mutagenizing the St-Cbs on DNA-processing. Cells were injected with either pCE5 (lane 1) or pCE5mut (lane 2). Cells were allowed to finish macronuclear development and a PCR reaction was performed using the primer combination P9/P548 for specific detection of the modified precursor sequence. M, molecular weight marker (1 kb ladder, Gibco BRL).
Our experiments using the 1.3 kb macronuclear precursor sequence showed that subtelomeric regions are required and are probably sufficient to direct DNA fragmentation and telomere addition. To test whether this is also true for other DNA molecules, a vector was constructed that contains only 195 bp of the leader- and 240 bp of the trailer sequences but no telomeric sequences of the Stylonychia α1-tubulin macronuclear molecule [28]. The α1-tubulin open reading frame (ORF) was replaced by the 717 bp ORF of the enhanced green fluorescent protein (GFP) [29] (Figure 6a). This construct was injected into the macronuclear anlage in the early polytene chromosome stage. Again, the injected cells were allowed to finish macronuclear development and a PCR analysis with DNA of 30-40 vegetative cells was performed. To detect the processed macronuclear molecules derived from the injected construct, a primer combination was used in which one primer (P27) hybridized to the 5'-α1-tubulin leader sequences while the second primer (GFPP2) was derived from GFP sequences (Figure 6a). As a PCR product could be obtained from cells injected with the α1-GFPtel- construct (Figure 6b,c; lanes 3), sequences required for fragmentation of the α1-tubulin macronuclear molecule must be located in the subtelomeric regions of the precursor sequences. No amplification was observed with primer GFPP2 and one primer from the flanking pUC vector sequences as primer combination using either total cellular DNA isolated from exconjugant cells 6-10 h after injection of α1-GFPtel- or from DNA isolated from injected vegetative cells as template, demonstrating that fragmentation had occurred (data not shown). These subtelomeric sequences therefore seem to be sufficient for correct processing of the macronuclear molecule. As in the case with other injected constructs, the copy number of the processed construct was very low compared with endogenous macronuclear tubulin genes, suggesting that not only sequences required for processing but also for regulating amplification are contained in the subtelomeric regions. The copy number of this product was obviously too low to allow detection of GFP expression using fluorescence microscopy.
Figure 6.

Injection of α1-GFPtel- into the developing macronucleus of the polytene chromosome stage. The presence of the injected construct in the macronuclear DNA of injected cells and its processing was analyzed by PCR. After injection, the cells were allowed to finish macronuclear development, then a PCR analysis was performed using DNA of 30-40 vegetatively growing cells as templates. Primers used were P27, derived from the endogenous α1-tubulin macronuclear molecule, and pGFPP2, derived from GFP sequences. (a) Schematic diagram of the α1-GFPtel- construct. (b) PCR products from the construct, from uninjected and injected cells were separated on an 1% agarose gel. M, molecular weight marker (1 kb ladder, Gibco, BRL). Lane 1, PCR product from construct DNA; lane 2, PCR product from uninjected cells; lane 3, PCR product from cells injected with α1-GFPtel-. The gel was hybridized with a DIG-labeled probe of the α1-GFPtel- vector DNA.
We analyzed whether the inverted repeats found in the subtelomeric region of the 1.3 kb DNA molecule are also present in the subtelomeric regions of the α1-tubulin macronuclear DNA molecule. In both subtelomeric regions, the core inverted sequence 5'-TGAA could be identified and was found to be 5'-TTGAA at position 62-66 in the 5'-subtelomeric region and 5'-TGAAA at position 45-49 in the 3'-subtelomeric region. Again, these repeats were only found once in the subtelomeric regions of this DNA molecule.
Discussion
Here we describe an experimental approach to characterize cis-acting DNA sequences involved in DNA processing during macronuclear development in Stylonychia lemnae. By sequence comparison, a putative fragmentation sequence (E-Cbs) has previously been located at defined positions either in the subtelomeric regions of macronuclear DNA molecules or in flanking micronuclear-specific sequences of Euplotes crassus [23]. In contrast, by sequence analysis of a large number of Stylonychia macronuclear DNA molecules no such sequences could be detected at defined positons in the subtelomeric regions of these molecules [24]. We therefore decided to use a biological approach to identify such sequences by injecting various constructs carrying a modified macronuclear precursor sequence into the macronuclear anlagen [25,27].
For the 1.3 kb macronuclear precursor sequence, we had already demonstrated that no micronuclear specific sequences are required for correct fragmentation and we localized a Cbs within the first 70 bp of the 3'-subtelomeric region. Deletion of the same number of base pairs from the 5' end still led to a processed product, however. Only after deletion of more than 500 bp from the 5' end could no product be detected. These results suggested that either the 3' sequence is the only Cbs required for fragmentation at both ends or that a second Cbs resides further downstream in the 5'-subtelomeric region. In fact, we now show that the 500 bp 5'-deletion construct was not fragmented, and the failure to detect a processed product from this construct was not just due to deletion of sequences required for amplification and replication later during macronuclear development. To characterize this putative Cbs in the 5'-subtelomeric region, we analyzed further deletion constructs for their ability to become processed. From these results we localize a 5'-Cbs to a position between 230 and 350. Interestingly, sequences absent from deletion constructs were restored in the processed product. Control experiments from an earlier study [27], and those made within this study, make a PCR artefact by template switching very unlikely. Homologous recombination does not seem sufficient to produce the products with the restored sequences because they were not obtained from all deletion constructs, but only from those containing the putative Cbs. Perhaps some as yet uncharacterized proofreading mechanism occurs during macronuclear development, possibly using transcripts from the old macronucleus as templates [27].
All experiments described so far were performed with only one macronuclear precursor sequence. To determine whether subtelomeric regions are not only required but are also sufficient for DNA processing in Stylonychia, a vector was constructed carrying only the subtelomeric regions of the DNA molecule encoding the α1-tubulin gene; the tubulin ORF was replaced by enhanced GFP. Correct processing of this construct could be demonstrated by PCR analysis, showing that in this case subtelomeric regions are required and indeed are sufficient for correct fragmentation and telomere addition. No or only weak GFP-expression could be detected by fluorescence microscopy. This again may be explained by the low copy number of the macronuclear molecule derived from this vector as indicated by the PCR analysis (Figure 6a,b; lane3) and would imply that sequences required for copy number control also reside within the subtelomeric regions.
A sequence comparison of the subtelomeric regions of the 1.3 kb precursor sequence containing the putative Cbs revealed the presence of a six-base inverted repeat resembling the core of the E-Cbs described in Euplotes crassus [21]. A subsequent search showed that a similar sequence is also present in the subtelomeric regions of the α1-tubulin macronuclear DNA-molecule and in the subtelomeric region of many other (over 80%) macronuclear DNA molecules from Stylonychia lemnae (data not shown). As in the case of the 1.3 kb macronuclear molecule, this sequence could only be found once in each subtelomeric region. However, in contrast to Euplotes it never occurred at the same position in both subtelomeric regions. Although it was always found within the first 60 bp of the 3' end, the position at the 5' end was more variable and in all cases examined it is localized further downstream than at the 3' end. To analyze whether this sequence is indeed involved in DNA processing we mutagenized this sequence in the 3'-subtelomeric region of the 1.3 kb precursor sequence. The fact that a processed product was never observed after injection of this construct is a strong indication that this sequence is required for specific DNA processing during macronuclear development; we therefore now call it St-Cbs.
The occurrence of Cbs at different locations in the two subtelomeric regions could be explained by the presence of several nucleases, each with different binding and cutting characteristics, a possibility that seems very unlikely. Moreover, we showed that already only one deletion, either in the 3'- or 5'-subtelomeric region, resulted in the lack of fragmentation on both ends. This strongly suggests that an interaction between two Cbs located in the two subtelomeric region is a necessary prerequisite for the fragmentation process to occur. On the basis of these and our earlier results, we would like to suggest a working hypothesis for the mechanism of DNA processing in Stylonychia (Figure 7), which is a modification and extension to our previously suggested model [24]. The six-base inverted repeat occurring in the subtelomeric regions required for DNA fragmentation could determine a loop structure stabilized by DNA-binding proteins and probably by the appropriate chromatin structure. An endonuclease could bind to this complex or to the six-base inverted repeat cutting the stucture at the base of the loop. As the 3'-inverted repeat is always found within the first 60 bp of the subtelomeric region, whereas the position of the 5'-inverted repeat seems quite variable, this would imply that initially a precise cut is made only at the 3'-end and parts of the 5'-subtelomeric regions would become lost. We continously observed a proofreading mechanism at the 5' end, which we cannot explain at present but excluded that these observations are due to an experimental artefact. This machinery could be responsible for filling the gap at the 5' end. Similar proofreading mechanisms have been postulated recently by Herbert and Rich [30] and comparable phenomena were described in Paramecium [31]. It still has to be determined, however, whether these processes also occur with the endogenous precursor sequences. Finally, de novo addition of telomeric sequences to the resulting gene-sized DNA molecules occurs [27]. Although this model is still very hypothetical, using our experimental approach it should be possible to verify it experimentally and also eventually to characterize the postulated proofreading machinery.
Figure 7.
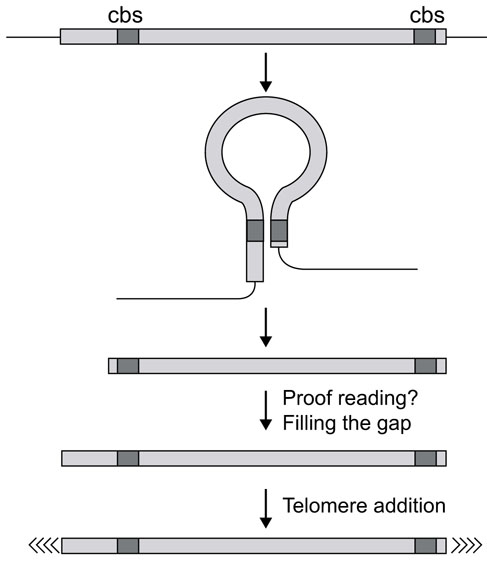
Model for DNA processing during macronuclear development in hypotrichous ciliates. Inverted repeats acting as cis-acting DNA sequences required for DNA processing are localized in both subtelomeric regions but not necessarily at similar positions. By interaction of these sequences, a loop-like structure is formed, which is then resolved on its base by a specific nuclease. This implies that a precise cut is made only at the 3' end, the 5' end has to be filled in by a so far unknown proofreading mechanism. Telomeric sequences are then added de novo.
Conclusions
Our data indicate that no micronuclear specific DNA sequences are required for specific DNA fragmentation during macronuclear development of the hypotrichous ciliate Stylonychia. Instead they are found in both subtelomeric regions of macronuclear precursor sequences, although they do not have to be localized at identical positions with respect to the breakage site. We show that these sequences are not only required but also sufficient for DNA fragmentation. Moreover, a functional analysis of an inverted repeat found in this region revealed that it functions as a St-Cbs.
Materials and methods
Stylonychia lemnae were grown in neutral Pringsheim solution and fed daily with the alga Chlorogonium elongatum [11]. To achieve conjugation, cells of two different mating types were mixed and the stages of macronuclear development were determined by phase microscopy. DNA at concentrations between 5-20 μg/ml of the various constructs was injected into the developing macronucleus of the polytene chromosome stage using the procedure described earlier [25,27]. Cells were allowed to finish macronuclear development and, after 15-20 cell divisions, DNA was isolated from vegetative macronuclei. Sometimes DNA was isolated from exconjugant cells only 6-10 h after injection. To characterize the DNA of injected cells, PCR analyses were performed. For these analyses, total cellular DNA isolated from 30-40 cells [25] was dissolved in 40 μl and aliquots of 10 μl were used for each PCR reaction carried out as described by Saiki et al. [32]. The PCR program used was described in Wen et al. [25]. Following PCR, the samples were separated on 1% agarose gels. The gels were blotted onto nylon membranes (Amersham Pharmacia Biotech) and hybridized with random primed probes [33] labeled with DIG-oxigenin-dUTP (Boehringer Mannheim).
Primers for construction of the vectors and for analysis of the DNA from injected cells are summarized in Table 1. Their positions are indicated in Figure 2 and Figure 6a. All deletion constructs based on pCE5 were PCR fragments cloned into pGEM-7Zf+ (Promega) (pCE12; pCE18), in pCR2.1-Topo (Invitrogen) (pCE19) and in pUC57(T) (Fermentas) (pCE20). stGFP was constructed by ligation of the leader and trailer sequences (including telomeres) of the α1-tubulin gene of Stylonychia [28] with the open reading frame of the GFP (S65T) as described previously [29]. From this construct, a PCR fragment was amplified using P27 and P28 (Table 1 and Figure 6a) as primers. This PCR fragment was cloned into pUC57(T) (Fermentas), resulting in the construct α1-GFPtel-.
Table 1.
Primers used for construction of the deletion vectors and for PCR analysis
| Different deletion constructs | ||||||
| Primer | Sequence | pCE12 | pCE18 | pCE19 | pCE20 | α1-GFPtel- |
| P4 | 5'-GGATCCATCAGATAACTCGCAAC | 5' | ||||
| P548 | 5'-CTGCAGGTCGACTCTAGAGCTC | |||||
| P7 | 5'-CAGATACAACGTCCCTCAAC | |||||
| P8 | 5'-GCGGGATCCATCTTCATTTAAACTAGATG | 3' | 3' | 3' | ||
| P9 | 5'-GGCTCGAGTTGCTACTCTCAGATATTC | |||||
| P15 | 5'-CGGAGGTACCCCGATATTTAAAATCATTAATC | 3' | ||||
| P20 | 5'-CCGCAGGATCCTTGAGAGTCTGCCATTTAAC | 5' | ||||
| P23 | 5'-GTTAAATGGCAGACTCTCAAGAAGAAATGC | |||||
| P25 | 5'-CCGGATCCAGACTTTCAGAGCGATATCTAG | 5' | ||||
| P26 | 5'-GCGGATCCCTCTCAAAGTGTGACATATTCC | 5' | ||||
| P27 | 5'-CGCAGGATCCAGAACAGTGGATTCGGAGGGAAATC | 5' | ||||
| P28 | 5'-CCGAGGTACCGAGATAGAATCGATTAAATAATGGG | 3' | ||||
| pGFP2 | 5'-CGCGAATTCTCACTTGTACAGCTCCTCCATG | |||||
| pUCP1 | 5'-GTCGACCTGCAGGCATGCAAGCTT | |||||
| pUCP2 | 5'-CGGGTACCGAGCTCGAATTC | |||||
| pGEMP1 | 5'-CGCATGCTCCTCTAGACTCGAGGAATTCGG | |||||
| pGEMP2 | 5'-GCTATGCATCCAACGCGTTGGGAGCTCTCC | |||||
| PMUT2 | 5'-GGTCAGAAATACTAGTTGATTCAGATAAGAAAG | |||||
Sequences of the primers used for construction of the different deletion vectors and for PCR analysis after injection of these constructs into the developing macronucleus. The 5' and 3' primers used for construction of each deletion vector are indicated.
For mutagenesis of the sequence 5'-TTGAAA present in the 3'-subtelomeric region of the 1.3 kb macronuclear precursor sequence, a megaprimer was generated in a PCR reaction using the primer combination P20/PMUT2 (Table 1 and Figure 2b). This PCR product was then used as megaprimer for amplifying the whole mutagenized pCE5mut using pCE5 as a template. By simultaneously inserting a second mutation in pCE5mut, it was possible to identify the mutagenized product by restriction analysis with BcuI. To ensure correct amplification the PCR reactions were performed using Pfu-Polymerase (Stratagene).
Acknowledgments
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft and the Alfried Krupp von Bohlen und Halbach Foundation. We thank N.K. Jakob for his work on the stGFP construct and Sabine Feiler for technical assistance.
References
- Klar AJ, Hicks JB, Strathern JN. Directionality of yeast mating-type interconversion. Cell. 1982;28:551–561. doi: 10.1016/0092-8674(82)90210-0. [DOI] [PubMed] [Google Scholar]
- Borst P. Molecular genetics of antigenic variation. Immunol Today. 1991;12:A29–A33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- Nash TE, Mowatt MR. Characterization of a Giardia lamblia variant-specific surface protein (VSP) gene from isolate GS/M and estimation of the VSP gene repertoire size. Mol Biochem Parasitol. 1992;51:219–227. doi: 10.1016/0166-6851(92)90072-r. [DOI] [PubMed] [Google Scholar]
- Beermann S. The diminution of heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda). Chromosoma. 1977;60:297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- Tobler H. The differentiation of germ and somatic cell lines in nematodes. Results Probl Cell Differ. 1986;13:1–69. doi: 10.1007/978-3-540-39838-7_1. [DOI] [PubMed] [Google Scholar]
- Gerbi SA. Unusual chromosome movements in sciarid flies. Results Probl Cell Differ. 1986;13:71–104. doi: 10.1007/978-3-540-39838-7_2. [DOI] [PubMed] [Google Scholar]
- Lieber M. Immunoglobulin diversity: rearranging by cutting and repairing. Curr Biol. 1996;6:134–136. doi: 10.1016/s0960-9822(02)00443-8. [DOI] [PubMed] [Google Scholar]
- Harriman W, Volk H, Defranoux N, Wabl M. Immunoglobulin class switch recombination. Annu Rev Immunol. 1993;11:361–384. doi: 10.1146/annurev.iy.11.040193.002045. [DOI] [PubMed] [Google Scholar]
- Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammermann D, Steinbruck G, Berger LV, Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974;45:401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- Kraut H, Lipps HJ, Prescott DM. The genome of hypotrichous ciliates. Int Rev Cytol. 1986;99:1–28. doi: 10.1016/s0074-7696(08)61422-9. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Jahn CL. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991;1:397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Wen J, Maercker C, Lipps HJ. Sequential excision of internal eliminated DNA sequences in the differentiating macronucleus of the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 1996;24:4415–4419. doi: 10.1093/nar/24.22.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- Prescott DM. Origin, evolution, and excision of internal elimination segments in germline genes of ciliates. Curr Opin Genet Dev. 1997;7:807–813. doi: 10.1016/s0959-437x(97)80044-5. [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Eder C. Macronucleus structure and macronucleus development in hypotrichous ciliates. Int J Dev Biol. 1996;40:141–147. [PubMed] [Google Scholar]
- Yao MC, Zheng K, Yao CH. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell. 1987;48:779–788. doi: 10.1016/0092-8674(87)90075-4. [DOI] [PubMed] [Google Scholar]
- Yao MC, Yao CH, Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- Yao MC. Programmed DNA deletions in Tetrahymena : mechanisms and implications. Trends Genet. 1996;12:26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- Baird SE, Fino GM, Tausta SL, Klobutcher LA. Micronuclear genome organization in Euplotes crassus : a transposonlike element is removed during macronuclear development. Mol Cell Biol. 1989;9:3793–3807. doi: 10.1128/mcb.9.9.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol Cell. 1999;4:695–704. doi: 10.1016/s1097-2765(00)80380-9. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Gygax SE, Podoloff JD, Vermeesch JR, Price CM, Tebeau CM, Jahn CL. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus [published erratum appears in Nucleic Acids Res 1999 Feb 15;27(4):following 1222]. Nucleic Acids Res. 1998;26:4230–4240. doi: 10.1093/nar/26.18.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maercker C, Lipps HJ. Analysis of the subtelomeric regions of macronuclear gene-sized DNA molecules of the hypotrichous ciliate Stylonychia lemnae: implications for the DNA fragmentation process during macronuclear development? Dev Genet. 1993;14:378–384. doi: 10.1002/dvg.1020140507. [DOI] [PubMed] [Google Scholar]
- Wen JP, Eder C, Lipps HJ. The processing of macronuclear-destined DNA sequences microinjected into the macronuclear anlagen of the hypotrichous ciliate Stylonchia lemnae. Nucleic Acids Res. 1995;23:1704–1709. doi: 10.1093/nar/23.10.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder C, Maercker C, Meyer J, Lipps HJ. The processing of macronuclear DNA sequences during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Int J Dev Biol. 1993;37:473–477. [PubMed] [Google Scholar]
- Jönsson F, Wen JP, Fetzer CP, Lipps HJ. A subtelomeric DNA sequence is required for correct processing of the macronuclear DNA sequences during macronuclear development in the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 1999;27:2832–2841. doi: 10.1093/nar/27.14.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985;13:415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrück G, Jakob NK, Gallert K-C, Hüls C, Müllner S. Vektoren zur heterologen Proteinexpression in eukaryontischen Protisten, vorzugsweise Ciliata und deren Transformation. [A vector for protein expression in eukaryotic protists, preferentially ciliates and the transformation of ciliates] Deutsche Patentanmeldung. 1998;198:48 486.0–41. [Google Scholar]
- Herbert A, Rich A. RNA processing and the evolution of eukaryotes. Nat Genet. 1999;21:265–269. doi: 10.1038/6780. [DOI] [PubMed] [Google Scholar]
- Meyer E, Duharcourt S. Epigenetic programming of developmental genome rearrangements in ciliates. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]


