Abstract
Uveal melanoma is the most common primary cancer of the eye, and often results not only in vision loss, but also in metastatic death in up to half of patients. For many years, the details of the molecular pathogenesis of uveal melanoma remained elusive. In the past decade, however, many of these details have emerged to reveal a fascinating and complex story of how the primary tumor evolves and progresses. Early events that disrupt cell cycle and apoptotic control lead to malignant transformation and proliferation of uveal melanocytes. Later, the growing tumor encounters a critical bifurcation point, where it progresses along one of two genetic pathways with very distinct genetic signatures (monosomy 3 vs 6p gain) and metastatic propensity. Late genetic events are characterized by increasing aneuploidy, most of which is nonspecific. However, specific chromosomal alterations, such as loss of chromosome 8p, can hasten the onset of metastasis in susceptible tumors. Taken together, this pathogenetic scheme can be used to construct a molecularly based and prognostically relevant classification of uveal melanomas that can be used clinically for personalized patient management.
Keywords: aneuploidy, cell-cycle deregulation, cell survival, inhibition of apoptosis, metastasis, phenotypic bifurcation, prognosis, uveal melanoma
Clinical, histopathologic & genetic features in uveal melanoma
Uveal melanoma is a malignant tumor that arises from neural crest-derived melanocytes of the uveal tract, which includes the iris, ciliary body and choroid of the eye (Figure 1). Uveal melanoma occurs in approximately 4–7 individuals per million in the USA, and the incidence is approximately the same in other countries with large Caucasian populations [1,2]. The most common age at diagnosis is 50–60 years, but uveal melanoma can occur at any age. There have been a few families with bona fide Mendelian inheritance of uveal melanoma [3], but this is rare. Options for treating the primary tumor include enucleation (eye removal), various forms of radiation therapy, laser hyperthermia and surgical resection [4]. The 5-year local-tumor control rates in most specialized treatment centers exceed 90%. However, despite successful treatment of the primary tumor, metastasis occurs by hematogenous spread in up to half of all patients. The most common site of metastasis is the liver, which is involved in over 90% of patients. The reason for this peculiar hepatic tropism is not known. Importantly, regional lymphatic dissemination does not occur, owing to an absence of lymphatic drainage of the ocular interior. Metastasis is uniformly fatal, usually within a few months of diagnosis despite systemic therapy [5,6].
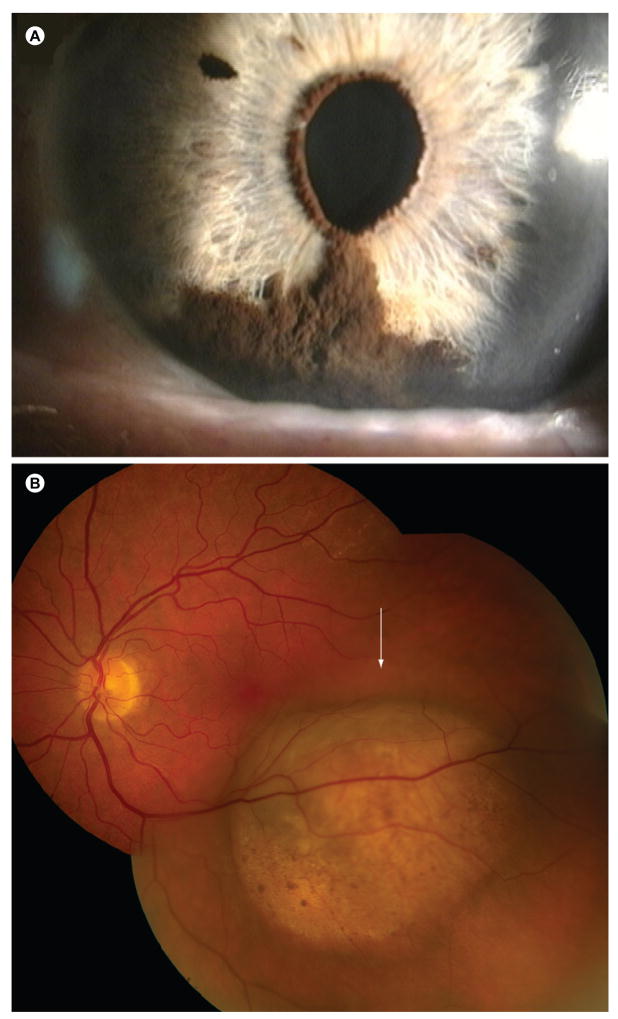
Figure 1. Clinical appearance of uveal melanomas.
(A) Iris melanoma.
(B) Choroidal melanoma. Photograph taken with a fundus camera through the dilated pupil, showing the posterior interior of the eye.
Several clinical, histopathological and genetic factors related to the prognosis of uveal melanoma have been identified. Clinical features associated with metastatic death include larger tumor size, involvement of the ciliary body and increased patient age [7]. Histopathologic features associated with poor prognosis include epithelioid cytology, tumor infiltration by macrophages and/or lymphocytes, mitotic figures, heterogeneous nucleolar size and extracellular matrix patterns [8–10]. Genetic features associated with metastasis include monosomy 3 and gain of chromosome 8q [11–14]. Gene-expression signatures have also been identified that accurately distinguish tumors at low metastatic risk (class 1 signature) and high metastatic risk (class 2 signature) [15,16]. More recently, oncogenic mutations in GNAQ, a Gαq stimulatory subunit involved in activating the RAF/MEK/ERK pathway in melanocytes, have been identified in both class 1 and 2 tumors [17,18], suggesting that this is an early or initiating event that occurs before the divergence of these two signatures. This review summarizes recent research regarding molecular events associated with tumor progression in uveal melanoma.
Cell-cycle deregulation in uveal melanoma
It has been known for several years that disruption of the retinoblastoma tumor suppressor pathway is common in uveal melanoma. The retinoblastoma protein inhibits cell-cycle progression through the G1–S phase transition point, so inactivation of retinoblastoma leads to unregulated proliferation. In uveal melanoma, retinoblastoma is inactivated by hyperphosphorylation, allowing cells to re-enter the cell cycle [19,20]. One mechanism for retinoblastoma hyperphosphorylation is via methylation and inactivation of the INK4A gene, which encodes the p16Ink4a tumor suppressor that activates retinoblastoma by blocking its phosphorylation by cyclin D/CDK4. We have shown that INK4A is a direct transcriptional target of the master melanocyte differentiation factor MITF, and that loss of p16Ink4a allows melanocytes to escape MITF-induced growth inhibition and to re-enter the cell cycle [19]. This occurs in approximately a third of uveal melanomas [21]. In the other two-thirds, retinoblastoma hyperphosphorylation appears to be the result of cyclin D overexpression [22–25].
The mechanism of cyclin D overexpression in uveal melanoma is not clear. In some types of cancer, this can be due to gene amplification, but this does not appear to be the case for uveal melanoma. Cyclin D overexpression can also occur through oncogenic mutations that constitutively activate the RAF/MEK/ERK pathway and its downstream target cyclin D. In skin melanoma, for example, such mutations occur commonly in BRAF, RAS and KIT [26,27]. There is increasing evidence that this pathway is also activated in uveal melanoma [28], but mutations in BRAF, RAS and KIT have rarely been found [28–32]. Recently, however, oncogenic mutations were discovered in GNAQ, which could account for the cyclin D overexpression [17]. Since this mutation is found in tumors of all sizes and molecular classes [18], it is most likely an early or initiating event in uveal melanoma. This is a potentially important discovery for providing the first opportunity for targeted molecular therapy in uveal melanoma.
Cell survival & inhibition of apoptosis in uveal melanoma
Normally, tumor suppressor mechanisms eliminate damaged cells that have sustained oncogenic mutations through senescence or apoptosis [33]. In order for an evolving neoplasm to progress, it must establish mechanisms for circumventing these tumor suppressor mechanisms. Uveal melanoma cells appear to exploit several pathways to avoid apoptosis and to promote survival.
The p53 pathway recognizes many forms of oncogenic insults, including DNA damage and redox abnormalities, and responds by triggering cell-cycle arrest or apoptosis [34]. The p53 gene is mutated in more than half of all human cancers, but it is rarely mutated in uveal melanoma [35]. Instead, p53 appears to be functionally inhibited by overexpression of its inhibitor HDM2 [24,36]. Maintenance of HDM2 overexpression is important for survival of uveal melanoma cells, as evidenced by the massive apoptosis induced by the introduction of a small molecular inhibitor of HDM2 in uveal melanoma cell lines [37].
The Bcl2 family of apoptosis regulators represents another level of control over cell survival that is often deregulated in cancer [38]. The BCL2 gene is a direct transcriptional target of MITF, thus it is expressed at high levels in melanocytes [39]. Another antiapoptotic family member, Bcl-xL, can be inactivated by deamidation [40]. Uveal melanoma cells often fail to deamidate Bcl-xL in response to DNA damage [36]. Thus, uveal melanomas often harbor defects at multiple points in the Bcl2 pathway, thereby contributing to the apoptosis-resistant phenotype of these tumors.
The PI3K–AKT pathway is a major mediator of cell survival and is activated in many uveal melanomas [41,42]. The tumor suppressor PTEN is a negative regulator of the PI3K–AKT pathway and is downregulated or completely inactivated in many uveal melanomas [43]. Recent work from our laboratory indicates that PTEN down-regulation occurs in tumors with greater aneuploidy, suggesting that it is a late event in tumor progression [44]. Taken together, these findings suggest that activation of the PI3K–AKT prosurvival pathway is a common strategy used by uveal melanoma cells to avoid apoptosis.
The insulin-like growth factors (IGFs) regulate cell proliferation, differentiation and apoptosis through their interaction with the IGF-1 receptor (IGF1R), leading to activation of the RAF/MEK/ERK and PI3K–AKT pathways [45]. The IGF1R is strongly expressed in many uveal melanomas, and inhibition of this receptor induces growth arrest and cell death in uveal melanoma cell lines [46,47]. Compounds that inhibit IGF1R are being evaluated as potential agents in treating metastatic uveal melanoma [48].
Phenotypic bifurcation & metastatic potential in uveal melanoma
Uveal melanomas metastasize almost exclusively by hematogenous dissemination, primarily to the liver, but also to other sites [49]. Approximately half of all primary uveal melanomas are capable of metastasis, and this inexorably leads to death within a few months [50]. A major focus of recent research has been to identify primary tumors that have acquired metastatic competency. By and large, the molecular alterations described previously are observed in metastasizing and non-metastasizing uveal melanomas, suggesting that they occur relatively early in tumor progression, prior to the acquisition of metastatic competence. Other molecular changes tend to occur selectively, either in metastasizing or non-metastasizing tumors, suggesting that they occur relatively late in progression of the primary tumor. Gain of chromosome 6p occurs mainly in non-metastasizing tumors, whereas loss of one copy of chromosome 3 (monosomy 3) occurs mostly in metastasizing tumors. These two events are almost completely mutually exclusive, which has been suggested to represent a bifurcation in tumor progression [44,51,52].
In general, tumors with monosomy 3 contain greater numbers of chromosomal abnormalities (aneuploidy) than tumors with disomy 3 [44], suggesting that loss of chromosome 3 leads to increased genomic instability. Downregulation of the tumor suppressor PTEN has been linked to increasing aneuploidy and worse clinical outcome [43,44], suggesting that this may be a late event in the evolution of monosomy 3 tumors. Gain of copies of chromosome 8q occurs commonly and has been associated with poor outcome [53]. Several potential oncogenes on chromosome 8q have been specifically implicated as having possible pathogenetic significance, including MYC, NBS1 and DDEF1 [54–56]. It seems likely that chromosome 8q gain is not an independent risk factor for metastasis, but that it is more common in tumors with monosomy 3, which is a strong metastatic risk factor [44]. A late chromosomal alteration with independent prognostic significance is loss of chromosome 8p; on average, monosomy 3 tumors with 8p loss will metastasize faster than those without 8p loss [57]. A minimally deleted region has been identified that contains six potential metastasis suppressor genes. The most interesting of these genes is LZTS1, which is a known tumor suppressor and undergoes promoter methylation on the one remaining allele in metastasizing uveal melanomas. We have shown that loss of LZTS1 is associated with increased invasion and migration of uveal melanoma cells [57].
Gene-expression profiling of primary uveal melanomas has revealed two major transcriptomic subgroups with prognostic significance: class 1 (low metastatic risk) and class 2 (high metastatic risk) tumors [15,58]. When compared with clinical, pathologic and chromosomal factors (including monosomy 3), the accuracy of the gene-expression signatures for predicting metastasis is far superior [59]. Consequently, the gene-expression profile has been modified and improved so that it requires as few as three genes to retain full predictive accuracy and can be used on fine-needle biopsy samples and archival specimens [16].
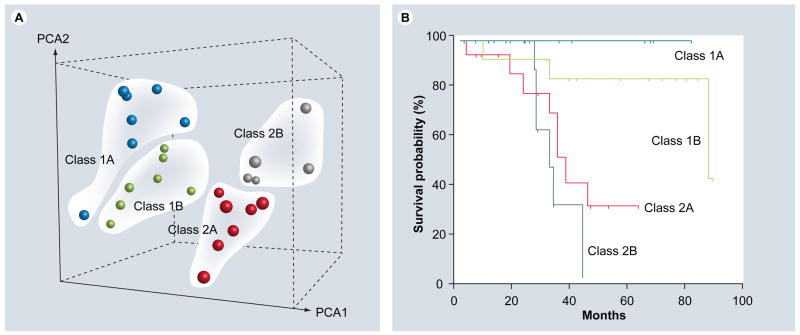
More recent work has allowed us to subdivide these molecular classes into four prognostically significant subclasses (1A, 1B, 2A and 2B), based on gene-expression profiling (Figure 2). The actuarial metastasis-free survival is longest for class1A and shortest for class 2B. The subclass 1B signature corresponds closely to gain of chromosome 6p, and the subclass2B signature corresponds closely to loss of chromosome 8p. Currently, we employ a molecular predictive test that incorporates both RNA-based gene-expression profiling and DNA-based assessment of these chromosomal arms, and this test is being validated for predictive accuracy in a prospective, multicenter, international study.
Figure 2. Molecular classification of uveal melanomas based on transcriptomic and chromosomal features.
(A) Unsupervised principal component analysis, showing natural clustering of uveal melanomas into four groups according to gene-expression profile and status of chromosomes 3, 6p and 8p. Class 1A – minimal aneuploidy (blue spheres); class 1B – 6p gain (green spheres); class 2A – monosomy 3 (red spheres) and class 2B – monosomy 3 and 8p loss (gray spheres). (B) Kaplan–Meier survival analysis showing that molecular classification accurately predicts metastatic death. PCA: Principle component analysis.
When independently analyzed for micro-RNA expression profiles [60] and global DNA methylation profile [Onken MD. Harbour JW, Washington University School of Medicine, MO, USA. Unpublished Data], primary uveal melanomas cluster into two groups that are identical to the class 1 and class 2 groups identified by gene-expression profiling. This suggests that the two groups are not only prognostically relevant, but reflect a fundamental underlying biological structure that is different between class1 and class 2 tumors. Consequently, we have used the gene-expression classes as a point of reference for exploring the underlying pathobiology of uveal melanomas and the molecular basis of the phenotypic bifurcation. A functional analysis of the differentially expressed genes in class 1 versus class 2 tumors revealed that class 1 tumors strongly resemble normal uveal melanocytes, with strong expression of neural crest and melanocyte lineage genes [61]. In contrast, these genes were downregulated in class 2 tumors, which instead expressed high levels of genes found in neural and ectodermal stem cells [61,62]. One reasonable explanation for these findings is that class 1 tumors, which are not competent for metastasis, are capable of melanocytic differentiation, whereas class 2 tumors, which are competent for metastasis, have a relative block to melanocyte differentiation. This hypothesis is supported by recent work from our laboratory showing that class 2 tumor cells exhibit more stem cell-like properties and are multipotent for lineage differentiation [Landerville S, Agapova O and Harbour JW, Washington University School of Medicine, MO, USA. Unpublished Data].
How can this hypothesis be reconciled to the class 1 versus class 2 bifurcation? One explanation is that growing uveal melanocytic neoplasms reach a critical restriction point where limited resources, such as oxygen, prevent further tumor progression without the acquisition of growth advantages conveyed by chromosome 6p gain or chromosome3 loss. This could explain why virtually all uveal melanomas acquire one or the other of these chromosomal alterations. Consistent with hypoxia being an important selective pressure in this process, expression of the hypoxia inducible factor 1-α correlates significantly with the class 2 gene-expression signature [62]. If this hypothesis is correct, then 6p gain may lead to a phenotype that remains competent for melanocytic differentiation (class 1), whereas monosomy 3 may result in at least a relative block to melanocyte differentiation (class 2). The differentiated, more genomically stable class 1 tumor cells may be more resistant to further accumulation of oncogenic lesions compared with class 2 tumor cells [44].
Much less is known about uveal melanoma progression following successful metastasis to distant sites, owing largely to the difficulty obtaining such tissue for analysis. The most extensive genetic analysis to date is by Meir et al., who used gene-expression profiling to implicate the NF-κB pathway in this process [63].
Conclusion & future perspective
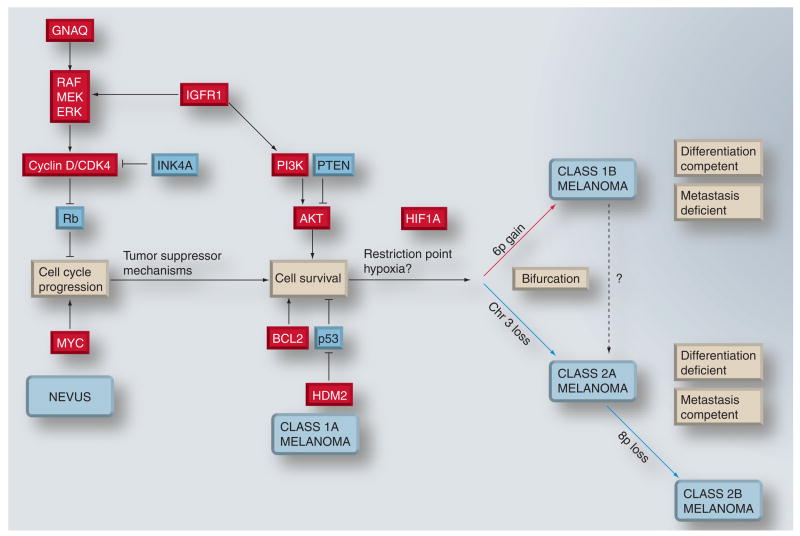
Even though many details are still not known, a coherent picture of uveal melanoma progression is starting to emerge (Figure 3). Early changes subvert the normal melanocyte regulatory machinery to deregulate the cell cycle and lead to an accumulation of low-grade transformed cells. Further alterations are necessary to prevent apoptosis and promote survival. If tumor suppressor mechanisms win this ‘arms race’, the result is growth arrest and senescence, manifest clinically as a nevus. If the tumor succeeds in developing appropriate escape mechanisms, the tumor continues to grow to become a class 1 uveal melanoma. Eventually, the tumor grows to a point where further obstacles, or selective pressures, lead to further growth and survival. The result is a bifurcation in further tumor progression, with gain of chromosome 6p or loss of chromosome 3 somehow relieving this selective pressure. Gain of chromosome 6p leads to long-term stabilization, differentiation competence and metastasis incompetence. Loss of chromosome 3 leads to further genomic instability, accumulation of further aneuploidy, a loss of differentiation competence and a gain of metastatic competence. The genetic progression from 1A–1B–2A to 2B probably represents an adaptation to multiple selective pressures, including hypoxia [62,64], immune response [65,66] and other factors.
Figure 3. Provisional model for malignant progression in uveal melanoma.
Early events probably include those that lead to cell-cycle deregulation. Such mutations trigger tumor suppressor mechanisms that lead to apoptosis and/or senescence. In most cases, these mechanisms succeed in either eliminating the tumor or arresting it at the stage of a benign nevus. The tumor can continue to progress if these mechanisms can be overcome by further mutations that block apoptosis and promote survival. Eventually, this progression leads to a lesion that would correspond clinically to uveal melanoma. Early uveal melanomas are relatively uniform in their gene-expression signature, with minimal aneuploidy (class 1A). Tumor progression continues as additional genetic lesions accumulate, and tumors either undergo gain of chromosome 6p and retain a class 1 gene-expression signature (class 1B), or they acquire a class 2 gene-expression signature, usually associated with loss of chromosome 3. As class 2A tumors acquire further genetic lesions and genomic instability, they can progress to a class 2B stage, associated with 8p loss and more rapid onset of metastasis. Class 1 tumors exhibit melanocytic differentiation and a low metastatic risk, whereas class 2 tumors show defective differentiation and high metastatic risk. Chromosomal gain and gene upregulation are indicated in red. Chromosomal loss and gene downregulation are indicated in blue.
An understanding of this process will be critical for designing strategies and developing targeted therapeutic compounds for delaying or preventing metastasis. For example, a strategy aimed at inhibiting early events in the RAF/MEK/ERK pathway may have little or no effect on metastasizing tumor cells that have accumulated other mutations that render the early mutations irrelevant for further tumor progression. Along these lines, we remain sceptical of the concept of oncogene addiction (that describes the need for sustained activity of oncogenes for initiation and maintenance of cancers [67]) as a basis for developing compounds to treat metastatic disease. Rather, the key events in metastatic capacity appear to involve differentiation competence. Therefore, research focusing on the mechanisms of differentiation in normal melanocytes and in melanoma cells may lead to new targeted therapeutic strategies.
Executive summary
Clinical, histopathologic & genetic features in uveal melanoma
Uveal melanoma is the most common primary cancer of the eye and results in metastatic death in up to half of all patients.
The main clinical and histopathologic features associated with metastatic death include larger tumor size, increased patient age, epithelioid cytology and extracellular matrix pattern.
Genetic features associated with metastasis include monosomy 3 and class 2 gene-expression signature.
Cell-cycle deregulation in uveal melanoma
The retinoblastoma tumor suppressor pathway is disrupted in virtually all tumors, usually by elevated expression of cyclin D.
The RAF/MEK/ERK pathway is constitutively activated.
Activating mutations in GNAQ occur in approximately half of all primary uveal melanomas and can activate the RAF/MEK/ERK pathway.
Cell survival & inhibition of apoptosis in uveal melanoma
The p53 pathway is functionally blocked by its inhibitor HDM2.
Defects in the Bcl2 pathway contribute to apoptosis resistance.
The PI3K–AKT prosurvival pathway is constitutively activated.
IGF1R is often upregulated and can activate the PI3K–AKT pathway.
Phenotypic bifurcation & metastatic potential in uveal melanoma
Molecular changes that differentiate metastasizing and non-metastasizing primary uveal melanomas occur relatively late in tumor progression.
Gain of chromosome 6p occurs mainly in non-metastasizing tumors, whereas loss of chromosome 3 occurs mostly in metastasizing tumors.
Monosomy 3 leads to further genomic instability, accumulation of aneuploidy, a loss of differentiation competence and a gain of metastatic competence.
Silencing of a metastasis modifier locus on chromosome 8p is associated with shorter metastasis-free survival.
Gene-expression profiling reveals two basic molecular groups of primary uveal melanomas: class 1 (low metastatic risk) and class 2 (high metastatic risk).
Tumors can be further divided into genetically and prognostically relevant subgroups: class 1A (minimal aneuploidy), class 1B (6p gain), class 2A (diploid for 8p) and class 2B (8p loss).
The phenotype of class 1 tumors is very similar to differentiated uveal melanocytes, whereas that of class 2 tumors is more similar to neural and ectodermal progenitor cells, suggesting a defect in differentiation.
Very little genetic information is available to date on metastatic tumors.
Conclusion & future perspective
A provisional model for tumor progression in primary uveal melanomas can be constructed based on current genetic information.
Current research is focusing on the use of genetic and genomic information to identify patients at high risk of metastasis and to treat them pre-emptively with targeted adjuvant agents prior to the development of overt metastatic disease.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Solange Landreville, Solange Landreville, PhD, Department of Ophthalmology & Visual Sciences, Washington University School of Medicine, 660 South Euclid Avenue, Camous Box 8096, St Louis, MO 63110, USA, Tel.: +1 314 747 0088, Fax: +1 314 747 5073, landrevilles@vision.wustl.edu.
Olga A Agapova, Olga A Agapova, PhD, Department of Ophthalmology & Visual Sciences, Washington University School of Medicine, 660 South Euclid Avenue, Camous Box 8096, St Louis, MO 63110, USA, Tel.: +1 314 747 0088, Fax: +1 314 747 5073, agapova@vision.wustl.edu.
J William Harbour, J William Harbour, MD, Department of Ophthalmology & Visual Sciences and Siteman Cancer Center, Washington University School of Medicine, 660 South Euclid Avenue, Campus Box 8096, St. Louis, MO 63110, USA, Tel.: +1 314 362 3315, Fax: +1 314 747 5073, harbour@wustl.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Inskip PD. Frequent radiation exposures and frequency-dependent effects: the eyes have it. Epidemiology. 2001;12(1):1–4. doi: 10.1097/00001648-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 2▪.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18(1):75–84. doi: 10.1016/j.ohc.2004.07.002. Overview of several clinical, histopathologic, cytologic, cytogenetic and molecular genetic factors that are related to the prognosis of uveal melanoma. [DOI] [PubMed] [Google Scholar]
- 3.Easton DF, Steele L, Fields P, et al. Cancer risks in two large breast cancer families linked to BRCA2 on chromosome 13q12–13. Am J Hum Genet. 1997;61(1):120–128. doi: 10.1086/513891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbour JW. Clinical overview of uveal melanoma: introduction to tumors of the eye. In: Albert DM, Polans A, editors. Ocular Oncology. Marcel Dekker; NY, USA: 2003. pp. 1–18. [Google Scholar]
- 5.Bedikian AY, Legha SS, Mavligit G, et al. Treatment of uveal melanoma metastatic to the liver: a review of the M. D. Anderson Cancer Center experience and prognostic factors. Cancer. 1995;76(9):1665–1670. doi: 10.1002/1097-0142(19951101)76:9<1665::aid-cncr2820760925>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Kujala E, Makitie T, Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44(11):4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 7.Augsburger JJ, Gamel JW. Clinical prognostic factors in patients with posterior uveal malignant melanoma. Cancer. 1990;66(7):1596–1600. doi: 10.1002/1097-0142(19901001)66:7<1596::aid-cncr2820660726>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Seddon JM, Polivogianis L, Hsieh CC, Albert DM, Gamel JW, Gragoudas ES. Death from uveal melanoma. Number of epithelioid cells and inverse SD of nucleolar area as prognostic factors. Arch Ophthalmol. 1987;105(6):801–806. doi: 10.1001/archopht.1987.01060060087039. [DOI] [PubMed] [Google Scholar]
- 9.Augsburger JJ, Schroeder RP, Territo C, Gamel JW, Shields JA. Clinical parameters predictive of enlargement of melanocytic choroidal lesions. Br J Ophthalmol. 1989;73(11):911–917. doi: 10.1136/bjo.73.11.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooy CM, De Jong PT. Prognostic parameters in uveal melanoma: a review. Surv Ophthalmol. 1996;41(3):215–228. doi: 10.1016/s0039-6257(96)80024-5. [DOI] [PubMed] [Google Scholar]
- 11.Horsman DE, Sroka H, Rootman J, White VA. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet Cytogenet. 1990;45(2):249–253. doi: 10.1016/0165-4608(90)90090-w. [DOI] [PubMed] [Google Scholar]
- 12.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J Natl Cancer Inst. 1990;82(22):1765–1769. doi: 10.1093/jnci/82.22.1765. [DOI] [PubMed] [Google Scholar]
- 13.Sisley K, Rennie IG, Cottam DW, Potter AM, Potter CW, Rees RC. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes Cancer. 1990;2(3):205–209. doi: 10.1002/gcc.2870020307. [DOI] [PubMed] [Google Scholar]
- 14.Scholes AG, Damato BE, Nunn J, Hiscott P, Grierson I, Field JK. Monosomy 3 in uveal melanoma: correlation with clinical and histologic predictors of survival. Invest Ophthalmol Vis Sci. 2003;44(3):1008–1011. doi: 10.1167/iovs.02-0159. [DOI] [PubMed] [Google Scholar]
- 15.Tschentscher F, Husing J, Holter T, et al. Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res. 2003;63(10):2578–2584. [PubMed] [Google Scholar]
- 16▪▪.Onken MD, Worley LA, Davila RM, Char DH, Harbour JW. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. J Mol Diagn. 2006;8(5):567–573. doi: 10.2353/jmoldx.2006.060077. Feasibility of transcription-based classification of uveal melanomas using fine-needle aspirates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher DE, Medrano EE, McMahon M, et al. Meeting report: Fourth International Congress of the Society for Melanoma Research. Pigment Cell Melanoma Res. 2008;21(1):15–26. doi: 10.1111/j.1755-148X.2007.00437.x. [DOI] [PubMed] [Google Scholar]
- 18.Onken M, Worley LA, Long MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008 doi: 10.1167/iovs.08-2145. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol. 2005;168(1):35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delston RB, Harbour JW. Rb at the interface between cell cycle and apoptotic decisions. Curr Mol Med. 2006;6(7):713–718. doi: 10.2174/1566524010606070713. [DOI] [PubMed] [Google Scholar]
- 21.van der Velden PA, Metzelaar-Blok JA, Bergman W, et al. Promoter hypermethylation: a common cause of reduced p16(INK4a) expression in uveal melanoma. Cancer Res. 2001;61(13):5303–5306. [PubMed] [Google Scholar]
- 22.Mouriaux F, Casagrande F, Pillaire MJ, Manenti S, Malecaze F, Darbon JM. Differential expression of G1 cyclins and cyclin-dependent kinase inhibitors in normal and transformed melanocytes. Invest Ophthalmol Vis Sci. 1998;39(6):876–884. [PubMed] [Google Scholar]
- 23.Brantley MA, Jr, Harbour JW. Inactivation of retinoblastoma protein in uveal melanoma by phosphorylation of sites in the COOH-terminal region. Cancer Res. 2000;60(16):4320–4323. [PMC free article] [PubMed] [Google Scholar]
- 24.Brantley MA, Jr, Harbour JW. Deregulation of the Rb and p53 pathways in uveal melanoma. Am J Pathol. 2000;157(6):1795–1801. doi: 10.1016/s0002-9440(10)64817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coupland SE, Anastassiou G, Stang A, et al. The prognostic value of cyclin D1, p53, and MDM2 protein expression in uveal melanoma. J Pathol. 2000;191(2):120–126. doi: 10.1002/(SICI)1096-9896(200006)191:2<120::AID-PATH591>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 26.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 27.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 28.Weber A, Hengge UR, Urbanik D, et al. Absence of mutations of the BRAF gene and constitutive activation of extracellular-regulated kinase in malignant melanomas of the uvea. Lab Invest. 2003;83(12):1771–1776. doi: 10.1097/01.lab.0000101732.89463.29. [DOI] [PubMed] [Google Scholar]
- 29.Cruz F, 3rd, Rubin BP, Wilson D, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63(18):5761–5766. [PubMed] [Google Scholar]
- 30.Zuidervaart W, van Nieuwpoort F, Stark M, et al. Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer. 2005;92(11):2032–2038. doi: 10.1038/sj.bjc.6602598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malaponte G, Libra M, Gangemi P, et al. Detection of BRAF gene mutation in primary choroidal melanoma tissue. Cancer Biol Ther. 2006;5(2):225–227. doi: 10.4161/cbt.5.2.2429. [DOI] [PubMed] [Google Scholar]
- 32.Maat W, Kilic E, Luyten GP, et al. Pyrophosphorolysis detects B-RAF mutations in primary uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(1):23–27. doi: 10.1167/iovs.07-0722. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 34.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Ehlers JP, Harbour JW. Molecular pathobiology of uveal melanoma. Int Ophthalmol Clin. 2006;46(1):167–180. doi: 10.1097/01.iio.0000195855.31324.db. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Tran BN, Worley LA, Delston RB, Harbour JW. Functional analysis of the p53 pathway in response to ionizing radiation in uveal melanoma. Invest Ophthalmol Vis Sci. 2005;46(5):1561–1564. doi: 10.1167/iovs.04-1362. [DOI] [PubMed] [Google Scholar]
- 37.Harbour JW, Worley L, Ma D, Cohen M. Transducible peptide therapy for uveal melanoma and retinoblastoma. Arch Ophthalmol. 2002;120(10):1341–1346. doi: 10.1001/archopht.120.10.1341. [DOI] [PubMed] [Google Scholar]
- 38.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 39.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 40.Deverman BE, Cook BL, Manson SR, et al. Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell. 2002;111(1):51–62. doi: 10.1016/s0092-8674(02)00972-8. [DOI] [PubMed] [Google Scholar]
- 41.Saraiva VS, Caissie AL, Segal L, Edelstein C, Burnier MN., Jr Immunohistochemical expression of phospho-Akt in uveal melanoma. Melanoma Res. 2005;15(4):245–250. doi: 10.1097/00008390-200508000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Ye M, Hu D, Tu L, et al. Involvement of PI3K/Akt signaling pathway in hepatocyte growth factor-induced migration of uveal melanoma cells. Invest Ophthalmol Vis Sci. 2008;49(2):497–504. doi: 10.1167/iovs.07-0975. [DOI] [PubMed] [Google Scholar]
- 43.Abdel-Rahman MH, Yang Y, Zhou XP, Craig EL, Davidorf FH, Eng C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol. 2006;24(2):288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 44.Ehlers JP, Worley L, Onken MD, Harbour JW. Integrative genomic analysis of aneuploidy in uveal melanoma. Clin Cancer Res. 2008;14(1):115–122. doi: 10.1158/1078-0432.CCR-07-1825. [DOI] [PubMed] [Google Scholar]
- 45.Grimberg A. Mechanisms by which IGF-I may promote cancer. Cancer Biol Ther. 2003;2(6):630–635. [PMC free article] [PubMed] [Google Scholar]
- 46.All-Ericsson C, Girnita L, Seregard S, Bartolazzi A, Jager MJ, Larsson O. Insulin-like growth factor-1 receptor in uveal melanoma: a predictor for metastatic disease and a potential therapeutic target. Invest Ophthalmol Vis Sci. 2002;43(1):1–8. [PubMed] [Google Scholar]
- 47.Economou MA, All-Ericsson C, Bykov V, et al. Receptors for the liver synthesized growth factors IGF-1 and HGF/SF in uveal melanoma: intercorrelation and prognostic implications. Invest Ophthalmol Vis Sci. 2005;46(12):4372–4375. doi: 10.1167/iovs.05-0322. [DOI] [PubMed] [Google Scholar]
- 48.Economou MA, Andersson S, Vasilcanu D, et al. Oral picropodophyllin (PPP) is well tolerated in vivo and inhibits IGF-1R expression and growth of UVEAL melanoma. Invest Ophthalmol Vis Sci. 2008;49(6):2337–2342. doi: 10.1167/iovs.07-0819. [DOI] [PubMed] [Google Scholar]
- 49.Ramaiya KJ, Harbour JW. Current management of uveal melanoma. Exp Rev Ophthal. 2007;2(6):939–946. [Google Scholar]
- 50.Kath R, Hayungs J, Bornfeld N, Sauerwein W, Hoffken K, Seeber S. Prognosis and treatment of disseminated uveal melanoma. Cancer. 1993;72(7):2219–2223. doi: 10.1002/1097-0142(19931001)72:7<2219::aid-cncr2820720725>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 51.Parrella P, Sidransky D, Merbs SL. Allelotype of posterior uveal melanoma: implications for a bifurcated tumor progression pathway. Cancer Res. 1999;59(13):3032–3037. [PubMed] [Google Scholar]
- 52.Kilic E, van Gils W, Lodder E, et al. Clinical and cytogenetic analyses in uveal melanoma. Invest Ophthalmol Vis Sci. 2006;47(9):3703–3707. doi: 10.1167/iovs.06-0101. [DOI] [PubMed] [Google Scholar]
- 53.Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19(1):22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 54.Parrella P, Caballero OL, Sidransky D, Merbs SL. Detection of c-myc amplification in uveal melanoma by fluorescent in situ hybridization. Invest Ophthalmol Vis Sci. 2001;42(8):1679–1684. [PubMed] [Google Scholar]
- 55.Ehlers JP, Harbour JW. NBS1 expression as a prognostic marker in uveal melanoma. Clin Cancer Res. 2005;11(5):1849–1853. doi: 10.1158/1078-0432.CCR-04-2054. [DOI] [PubMed] [Google Scholar]
- 56.Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin Cancer Res. 2005;11(10):3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- 57▪.Onken MD, Worley L, Harbour JW. A metastasis modifier locus on human chromosome 8p in uveal melanoma identified by integrative genomic analysis. Clin Cancer Res. 2008;14(12):3737–3745. doi: 10.1158/1078-0432.CCR-07-5144. Identification of a region on chromosome 8p that modifies metastatic efficiency in uveal melanoma. [DOI] [PubMed] [Google Scholar]
- 58▪▪.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. First demonstration that gene-expression signatures accurately distinguish uveal melanoma tumors at low metastatic risk (class 1) and high metastatic risk (class 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59▪▪.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13(5):1466–1471. doi: 10.1158/1078-0432.CCR-06-2401. Comparaison of gene-expression profile versus monosomy 3 for predicting metastasis in uveal melanoma. [DOI] [PubMed] [Google Scholar]
- 60.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18(3):184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 61▪.Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006;66(9):4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. Id2 loss in primary uveal melanoma triggers upregulation of E-cadherin, promoting anchorage-independent cell growth, a likely antecedent to metastasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪▪.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18(3):191–200. doi: 10.1097/CMR.0b013e3283005270. Overview of the more accurate biomarkers to predict metastasis and association of class 2 signature with a primitive neural/ectodermal stem cell-like phenotype. [DOI] [PubMed] [Google Scholar]
- 63▪▪.Meir T, Dror R, Yu X, et al. Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci. 2007;48(11):4890–4896. doi: 10.1167/iovs.07-0215. The most extensive genetic analysis of metastatic uveal melanomas by gene-expression profiling. [DOI] [PubMed] [Google Scholar]
- 64.Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26(2):333–339. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- 65.Niederkorn JY, Wang S. Immunology of intraocular tumors. Ocul Immunol Inflamm. 2005;13(1):105–110. doi: 10.1080/09273940490518586. [DOI] [PubMed] [Google Scholar]
- 66.Chen PW, Ksander BR. Influence of immune surveillance and immune privilege on formation of intraocular tumors. Chem Immunol Allergy. 2007;92:276–289. doi: 10.1159/000099278. [DOI] [PubMed] [Google Scholar]
- 67.Weinstein IB. Cancer. Addiction to oncogenes – the Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]