Abstract
It has been suggested that carbonate radical anions are biologically important because they may be produced during the inflammatory response. The carbonate radicals can selectively oxidize guanine in DNA and RNA by one-electron transfer mechanisms and the guanine radicals thus formed decay by diverse competing pathways with other free radicals or nucleophiles. Using a photochemical method to generate CO3•− radicals in vitro, we compare the distributions of products initiated by the one-electron oxidation of guanine in the trinucleotides 5’-r(GpCpU) and 5’-d(GpCpU) in aqueous buffer solutions (pH 7.4). Similar distributions of stable end-products identified by LC-MS/MS methods were found in both cases. The guanine oxidation products include the diastereomeric pair of spiroiminodihydantoin (Sp) and 2,5-diamino-4H-imidazolone (Iz). In addition, intrastrand cross-linked products involving covalent bonds between the G and U bases (G*CU*) were also found, although with different relative yields in the 2’-deoxy- and the ribotrinucleotides. The positive ion MS/MS spectra of the 5’-r(G*pCpU*) and 5’-d(G*pCpU*) products clearly indicate the presence of covalently linked G*-U* products that have a mass smaller by 2 Da than the sum of the G and U bases in both types of trinucleotides. The 5’-d(G*CU*) cross-linked product was further characterized by 1D and 2D NMR methods that confirm its cyclic structure in which the guanine C8-atom is covalently linked to the uracil N3-atom.
Keywords: Free radicals, DNA, RNA, Oxidation, Carbonate radical anion, Guanine, Uracil, Cross-linked DNA
Introduction
Inflammation is believed to play an important role in the etiology of a number of human cancers [1]. The overproduction of free radicals associated with chronic inflammation is suspected to induce enhanced cell proliferation and malignant cell transformation [2]. Among the oxyl radicals that are believed to be generated at sites of inflammation [3–5], hydroxyl radicals and carbonate radical anions can directly react with and irreversibly damage DNA molecules [6, 7]. The hydroxyl radical is an extremely strong electrophile that rapidly and unselectively reacts with DNA mostly via the addition to double bonds of the nucleobases and by abstraction of hydrogen atoms from either the 2-deoxyribose moieties or the methyl group of thymine [6, 8, 9]. In contrast, CO3•− radicals with a reduction potential of 1.59 V vs NHE [10] selectively oxidize biomolecules by one-electron abstraction mechanisms [11]. It has been suggested that in vivo, CO3•− radicals are formed via homolysis of nitrosoperoxycarbonate derived from the combination of carbon dioxide and peroxynitrite, an important byproduct of the inflammatory response [4, 5, 12].
Our own laser flash photolysis experiments have shown that CO3•− radicals induce site-selective one-electron oxidation of guanine bases in 2’-deoxyribooligonucleotides in either the single- or double-stranded forms [7, 13, 14]. Neutral guanine radicals, G(−H)•, directly monitored via their UV absorption maximum at 315 nm, give rise to a series of relatively stable end products [13, 15, 16] including mostly the diastereomeric pair of spiroiminodihydantoin (Sp), the guanidinohydantoin (Gh) lesion, and the intrastrand cross-link between G* and T* bases, as well as minor quantities of 2,5-diamino-4H-imidazolone (Iz) lesions. Analysis of the intrastrand cross-linked products by NMR and mass spectrometry methods indicate that the G* and T* bases are covalently linked via the C8 atom of guanine and the N3-atom of thymine. These unusual cross-links are formed in single-stranded DNA with 5’-GCnT and 5’-TCnG sequences with n = 0, 1, 2 and 3, and in double-stranded DNA containing the 5’-GT dinucleotide sequence [16]. The cross-linking mechanism involves the nucleophilic addition of the thymine N3 to the C8 position of guanine radical followed by the removal of the unpaired electron by an O2 molecule. Different kinds of cross-links involving a covalent bond between the guanine C8 and thymine C5 methyl group of the adjacent G and T bases in DNA produced by radiation-induced •OH radicals in deoxygenated solutions have been observed [17, 18]. The formation of these cross-links requires H-atom abstraction from the methyl group of thymine and is not initiated by the one-electron oxidation of guanine as in the case of C8 – N3 cross-links between G* and T bases.
In this work, we hypothesized that cross-linking triggered by electron abstraction from guanine might occur between the guanine C8 atom and the N3 position of uracil. The latter is a demethylated form of thymine (with an H-atom instead of the methyl at C5), in which the N3(H) group remains available for the cross-link formation with the C8 site of guanine. In contrast to DNA where T pairs with A, in RNA uracil is the complementary base to adenine. In cellular RNA, bases are often unpaired in single-stranded regions and the bases are chemically more reactive than in double-stranded regions of RNA [19]. Although oxidatively generated damage to RNA has received much less attention than DNA damage, there is growing evidence that the enhanced oxidation of RNA is associated with early events in the development of neurodegenerative diseases [20, 21]. Recently, enhanced levels of 8-oxo-7,8-dihydroguanosine (8-oxoGuo) have been found in RNA isolated from brains of deceased patients with Alzheimer disease [22], as well as in patients suffering from mild cognitive impairment, a transition between normal aging and dementia [23]. The 8-oxoGuo lesion, a major product of guanine oxidation, is significantly more abundant in RNA than 8-oxodGuo in DNA exposed to Fenton reagents (H2O2/ascorbate/Fe3+ or Cu2+) in vitro, or in rat liver after doxorubicin administration in vivo [24]. However, it has not yet been demonstrated whether other guanine oxidation products such as Iz, Sp, Gh, occur in RNA as they do in DNA exposed to oxidizing agents [4, 5]. Here, we investigated whether these lesions, as well as cross-links between G-C8 and U-N3, are formed in 5’-r(GpCpU), and compared the relative proportions of these products with those formed in the analogous single-stranded 2’-deoxyribotrinucleotide 5’-d(GpCpU) exposed to carbonate radicals. These trinucleotide sequences were selected because in previous experiments we showed that the yields of cross-linked products are maximal in the 5’-d(GCT) sequence context in which the interacting G and T are separated by one cytosine residue [16]. The carbonate radicals were generated by a continuous illumination method that produces relatively low, steady-state concentrations of these radicals that are more relevant to biological conditions than the laser pulse methods previously employed for generating carbonate radicals [16]. We found that the oxidation of 5’-r(GpCpU) by CO3•− radicals generates basically the same major lesions including Sp, Iz and 5’-G*CU* products with the C8 – N3 cross-links between the G* and T* bases as in the cases of 5’-d(GpCpU). However, the relative proportions of end-products were different in the 2’-deoxyribo- and ribonucleotide sequences.
Materials and methods
Materials
All chemicals (analytical grade) were used as received. The 5’-d(GpCpU) from Sigma Genosys (Woodlands, TX), and 5-r(GpCpU) from Bio-Synthesis (Lewisville, TX) were purified and desalted using reversed-phase HPLC. The integrity and composition of the oligonucleotides were confirmed by LC-MS techniques.
Oxidation of trinucleotides by CO3•− radicals
The CO3•− radicals were generated by the photochemical method used in our previous experiments [7, 13, 14, 25]. This method involves the photodissociation of peroxodisulfate to sulfate radical anions, followed by the one-electron oxidation of HCO3− anions by SO4•− radicals:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
In order to minimize the contribution of the direct oxidation of trinucleotides by SO4•− radicals (reaction 3), we employed high concentrations of HCO3− (300 mM) and much lower concentrations of oligonucleotides (0.1 mM). Under these conditions, the contribution of the direct oxidation of trinucleotides by SO4•− radicals, determined from the ratio of the pseudo first-order rate constants, k3[GCU]/(k4[HCO3−] + k3[GCU]), does not exceed ~2% [7, 13, 14, 25].
Trinucleotides (10 nmol) were dissolved in 1 mL of air-equilibrated solutions containing 300 mM NaHCO3 and 10 mM Na2S2O8 and the pH was adjusted to 7.5 with 1 M NaH2PO4.The solutions were irradiated with light from a 100 W high pressure Xenon lamp that was reflected from a dichroic mirror with the reflectance in the 300 – 340 nm spectral range that delivered an incident energy of ~100 mW/cm2 to the sample. After irradiation for 10 – 30 s, the sample was immediately subjected to HPLC analysis.
Reversed-phase HPLC separation of the oxidation products
The end products produced by photochemical oxidation were separated using an Agilent 1200 Series LC system (quaternary LC pump with degasser, thermostated column compartments, and diode array detector) equipped with an analytical (250 × 4 mm i.d.) Varian, Microsorb C18 column. Typically, HPLC separations of the oxidation products were performed employing a 1 – 40% linear gradient of methanol in 20 mM phosphate (for NMR experiments) or acetate (for LC/MS/MS analysis) buffer solutions, pH 7 for 45 min at a flow rate of 1 mL/min.
LC-MS/MS experiments
The oxidation products were identified using an Agilent 1100 Series capillary LC/MSD Ion Trap XCT mass spectrometer equipped with an electrospray ion source as described previously [16]. Typically, 1 – 8 µL of the sample solution were injected in a narrow bore (50 × 1 mm i. d.) Zorbax SB-C8 column and eluted with an isocratic mixture (55% methanol in water with 0.1% formic acid) at a flow rate of 0.2 mL/min. The mass spectra were recorded in the positive mode.
NMR spectra of the oligonucleotide irradiation products
The 1D 1H spectra, and 2D, HSQC (heteronuclear single quantum correlation), HMBC (heteronuclear multiple bond correlation), and DQF (double quantum filtered) - COSY (correlation spectroscopy) spectra were recorded in D2O using a 500 MHz Bruker NMR instrument and a sample concentration of 4.6 mM. The chemical shifts cited are relative to sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS).
Results
Identification of the end products generated by CO3•− radicals
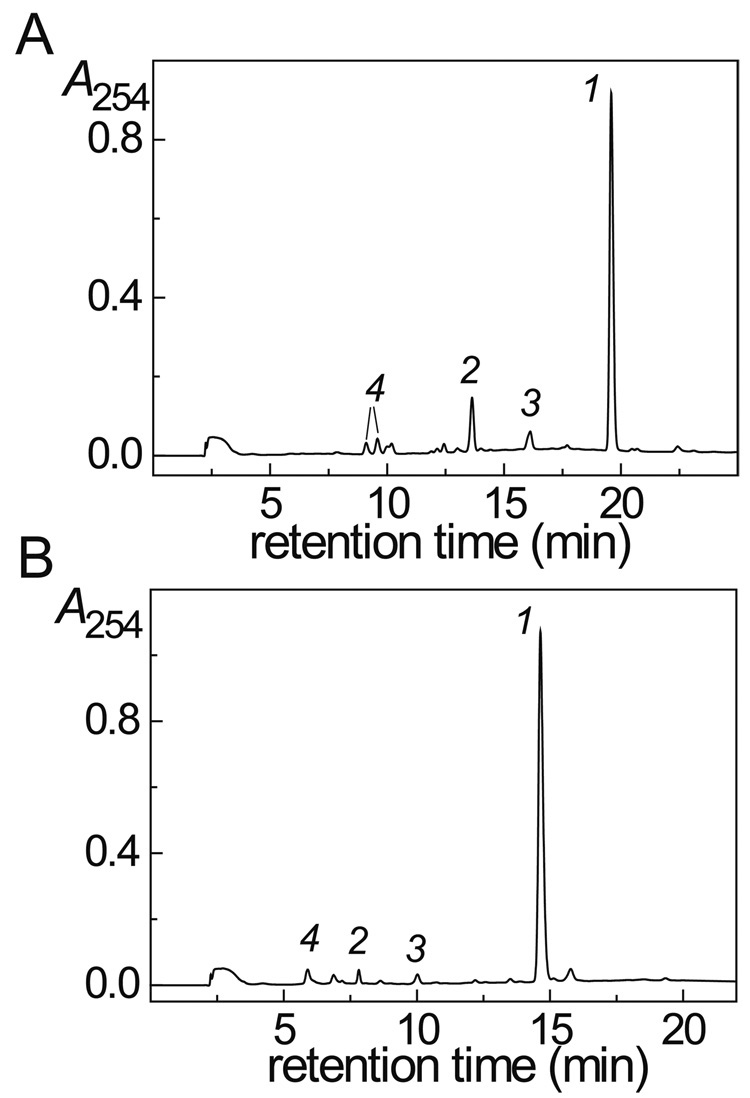
The 5’-d(GpCpU) and 5’-r(GpCpU) trinucleotides were oxidized by CO3•− radicals generated by the one-electron oxidation of HCO3− anions with SO4•− radicals derived from the photodissociation of peroxodisulfate [7, 13, 15]. The oxidation products were isolated by reversed-phase HPLC (Fig. 1), and identified by LC-MS/MS analysis (Fig. 2 and 3). We found that oxidation of the 5’-r(GpCpU) and 5’-d(GpCpU) trinucleotides by CO3•− radicals yields the same major products including trinucleotides in which the single guanine base is oxidized to Iz and Sp lesions (Fig. 2) and intrastrand cross-linked products (Fig. 3). Due to the presence of an additional hydroxyl group in the sugar residues of the 5’-r([Sp]pCpU) sequence, the two diastereomeric Sp could not be separated (Fig. 1B), although they were separable in the 5’-d([Sp]pCpU) case (Fig. 1A). Although, the major products are the same (Fig. 1), their relative yields in DNA and RNA sequences are different. In the case of 5’-d(GpCpU), the major product is the cross-linked 5’-d(G*pCpU*) (15.9%), and the yields of both the 5’-d([Sp]pCpU) (7.3%) and the 5’-d([Iz]pCpU) (7.5%) are lower by a factor of 2 (Fig. 1A). In the case of 5’-r(GpCpU), the major product is 5’-r([Sp]pCpU) (4.6%) and the yields of 5’-r(G*pCpU*) (2.5%) and 5’-r([Iz]pCpU) (1.8%) trinucleotides are approximately 2 times lower (Fig. 1B).
Fig. 1.
Reversed-phase HPLC elution profiles of the end-products derived from the oxidation of the 5’-d(GpCpU) (panel A) and 5’-r(GpCpU) (panel B) trinucleotides by CO3•− radicals. HPLC elution conditions (detection at 254 nm): 1 – 40% linear gradient of methanol in 20 mM phosphate buffer solution, pH 7 for 45 min at a flow rate of 1 mL/min. Panel A: the intact 5’-d(GpCpU) (1) elutes at 19.5 min, the 5’-d(G*pCpU*) (2) cross-linked product at 13.6 min, 5’-d([Iz]pCpU) (3) elutes at 16.1 min, and the two 5’-d([Sp]pCpU) diastereomers (4) elute at 9.1 min, and 9.6 min. Panel B: the intact 5’-r(GpCpU) (1) elutes at 14.6 min, the 5’-r(G*pCpU*) (2) cross-linked product at 6.9 min, 5’-r([Iz]pCpU) (3) elutes at 10.0 min, and the mixture of the two 5’-r([Sp]pCpU) isomers (4) elutes at 5.9 min as an unseparated mixture of the diastereomers.
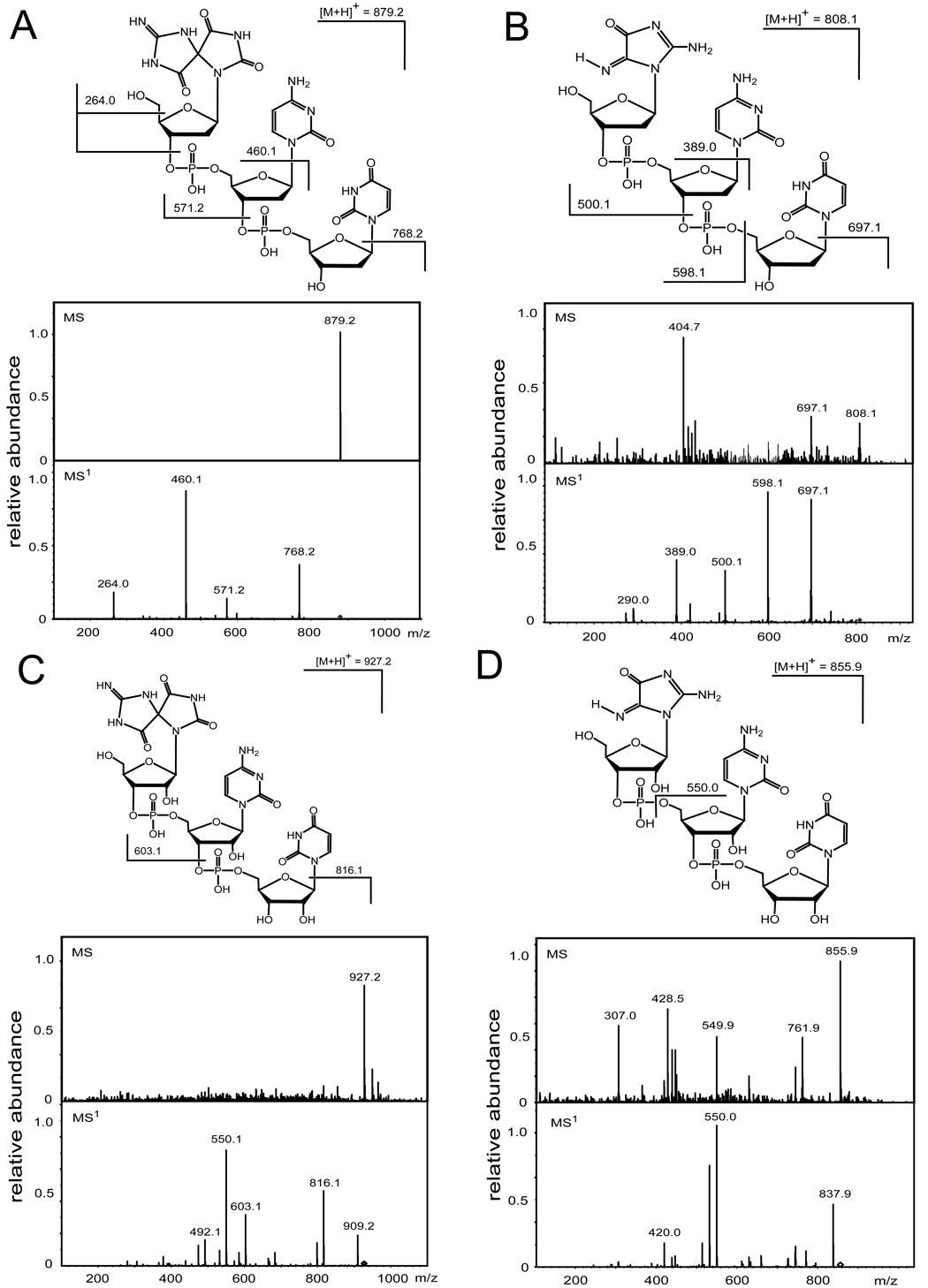
Fig. 2.
Positive ion spectra (MS/MS) of 5’-d([Sp]pCpU) (A), 5’-d([Iz]pCpU) (B), 5’-r([Sp]pCpU) (C), and 5’-r([Iz]pCpU) (D). MS – spectra of the molecular ions, [M + H]+; MS1 – product ion spectra obtained by fragmentation of [M + H]+.
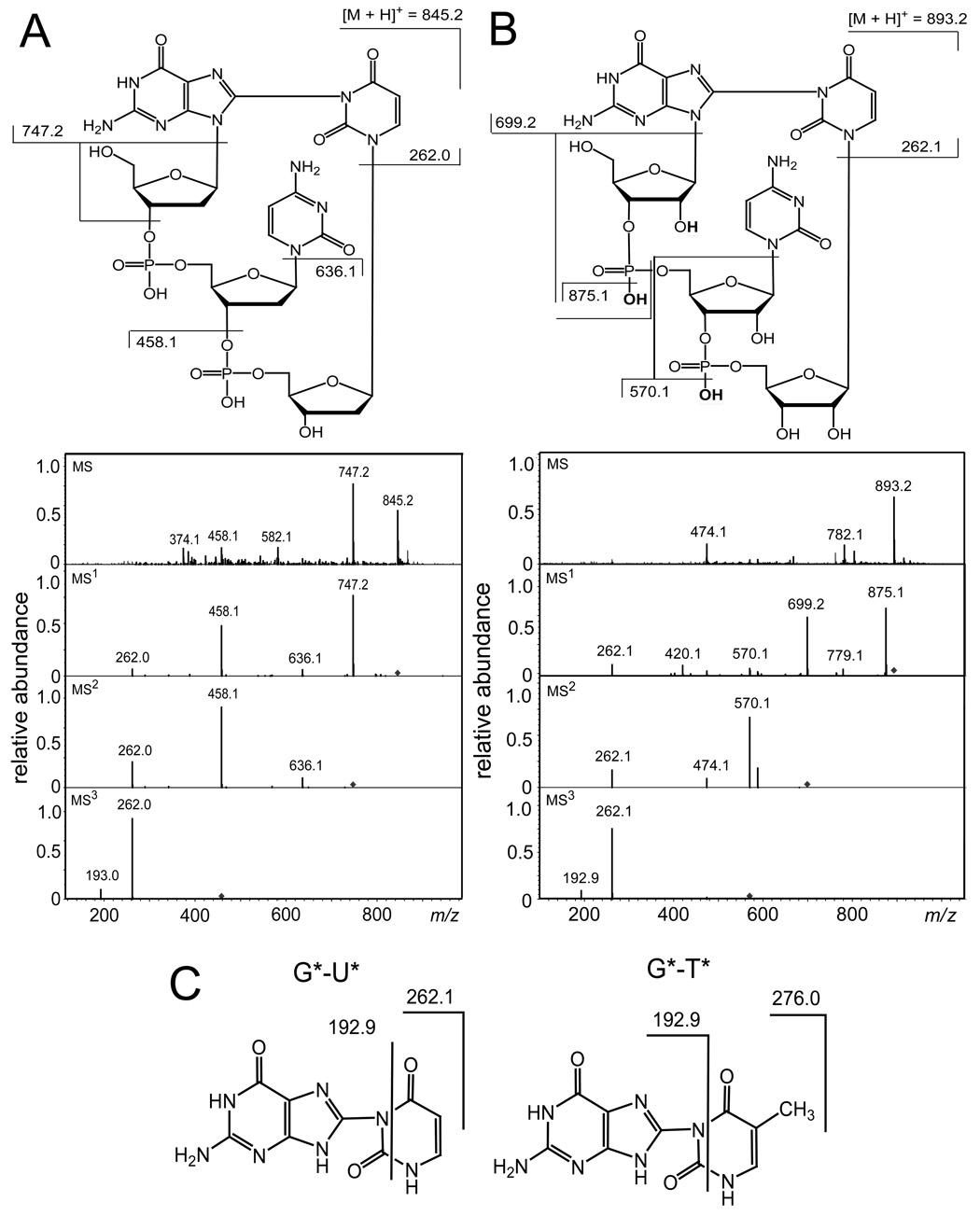
Fig. 3.
Positive ion spectra (MS/MS) of the 5’-G*pCpU* cross-linked products. Panel A: 5’-d(G*pCpU*). MS: spectrum of the molecular ion, [M + H]+ at m/z 845.2. MS1: product ion spectrum obtained by fragmentation of the molecular ion, [M + H]+ at m/z 845.2. MS2: product ion spectrum obtained by fragmentation of the ion, [M + H − 98]+ at m/z 747.2. MS3 product ion spectrum obtained by fragmentation of the ion, [M + H − 387]+ at m/z 458.1. Panel B: 5’-r(G*pCpU*). MS: spectrum of the molecular ion, [M + H]+ at m/z 893.2. MS1: product ion spectrum obtained by fragmentation of the ion, [M + H ]+ at m/z 893.2. MS2: product ion spectrum obtained by fragmentation of the molecular ion, [M + H − 194]+ at m/z 699.2. MS3: product ion spectrum obtained by fragmentation of the ion, [M + H − 323]+ at m/z 570.1. Panel C: Fragmentation patterns of the G*-U* and G*-T* dimer fragments.
The positive ion spectra (MS/MS) of 5’-d(G*pCpU*) and 5’-r(G*pCpU*) provide a further characterization of these unusual lesions (Fig. 3). The mass of 5’-d(G*pCpU*) with molecular ion, [M + H]+ detected at m/z = 845.2 (Fig. 3A) is smaller by 2 Da than the mass of the unmodified 5’-d(GpCpU) recorded at m/z = 847.2. The fragmentation of [M + H]+ occurs via typical pathways for protonated 2’-deoxyribooligonucleotides including cleavage of the guanine N-glycosidic bond associated with H-atom transfer from the sugar residue to the guanine base and the formation of a double-bond in the 2-deoxyribose moiety, followed by cleavage of the phosphodiester C3’–O3’ bond with the concomitant transfer of an H-atom from the sugar residue to the phosphate and release of a furan-type residue [26–31]. The [M + H − 98]+ formed is detected at m/z = 747.2. Further cleavage of [M + H − 98]+ results in the ion [M + H − 209]+ at m/z = 636.2 arising from a loss of the cytosine base. The ions, [M + H − 387]+ at m/z = 458.1 and [M + H − 583]+ at m/z = 262.1, are produced by cleavage of the C3’-O3’ bond in [M + H − 209]+ and the N-glycosidic bond in [M + H − 387]+, respectively. The [M + H − 583]+ ion at m/z = 262.1 is assigned to the G*-U* fragment and its mass is equal to the sum of the masses of G and U minus 2 Da. These results suggest that the 5’-d(G*pCpU*) product derived from oxidation of 5’-d(GpCpU) by CO3•− radicals contains an intrastrand cross-link between the G and U bases which is similar to 5’-d(G*pCpT*) found in our previous work [16].
The mass of 5’-r(G*pCpU*) with molecular ion, [M + H]+ detected at m/z = 893.2 (Fig. 3B) is also smaller by 2 Da than the mass of the unmodified 5’-r(GpCpU) recorded at m/z = 895.2. However, the fragmentation patterns of the ribooligonucleotides (Fig. 3B) are quite different than those observed in the case of their 2’-deoxy analogues (Fig. 3A). Indeed, the presence of the 2’-hydroxyl group increases the stability of the N-glycosidic bond and neither its cleavage nor cleavage of phosphodiester C3’–O3’ bond associated with the release of a furan-type residue are the primary fragments of [M + H]+. The main fragmentation modes of the protonated ribooligonucleotides include loss of neutral water from the phosphate group and cleavage of the 5’-P–O bond followed by cleavage of the N-glycosidic bond [26, 32, 33]. Indeed, the fragmentation of [M+H]+ at m/z = 893.2 leads to the [M + H − 18]+ ion at m/z = 875.1 corresponding to the loss of H2O and the [M + H − 194]+ ion at m/z = 699.2 arising from cleavage of the cytidine 5’-P–O bond and the guanine N-glycosidic bond. Further fragmentation of [M + H − 194]+ at m/z = 699.2 generates the [M + H − 323]+ ion at m/z = 570.1 derived from the loss of H2O and cytosine. Cleavage of the uracil N-glycosidic bond in [M + H − 323]+ results in the formation of the ion at m/z 262.1 that represents the G*-U* fragment which has the same mass as the fragment derived from cleavage of the 5’-d(G*pCpU*) cross-linked product (Fig. 3A).
Further fragmentation of G*-U* dimers derived from either 5’-d(G*pCpU*) (Fig. 3A) or 5’-r(G*pCpU*) (Fig. 3B) generates the ion detected at m/z = 192.9 (Fig. 3C). The ion at m/z = 192.9 has been observed in the fragmentation of the cross-linked 5’-d(G*pCpT*) product [16], that can be considered a support for fragmentation mode associated with cleavage of the pyrimidine ring of U and T bases in the dimers (Fig. 3C).
Thus, the LC-MS/MS analysis provides evidence that the oxidation of 5’-d(GpCpU) and 5’-r(GpCpU) trinucleotides by CO3•− radicals yields the cyclic intrastrand cross-linked products in which G and U are linked to one another and are separated by a single C residue (Fig. 3).
Role of molecular oxygen in cross-link formation
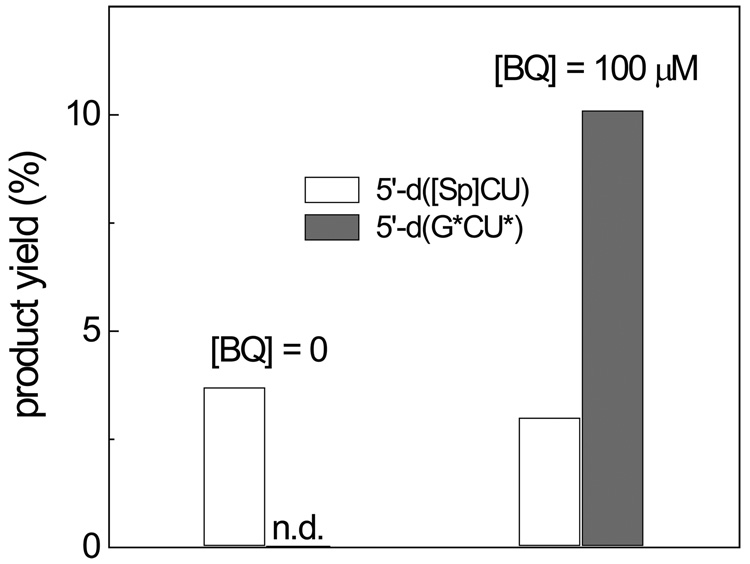
Our previous experiments have shown that the yields of the 5’-d(G*CT*) cross-linked products become negligible in deoxygenated solutions [16]. Although, the cross-linked product does not contain additional oxygen atoms in comparison with the parent 5’-d(GCT), oxygen is required for the oxidation of the intermediates. To obtain insights into the mechanism of these reactions, we explored what oxidant can replace oxygen in the cross-link formation. Here, we found that addition of 1,4-benzoquinone at a 100 µM level to the solutions extensively purged with argon restores formation of 5’-d(G*CT*) (data not shown) and 5’-d(G*CU*) (Fig. 4) cross-linked products. The 1,4-benzoquinone is a moderate one-electron oxidant with the redox potential, E˚(BQ/BQ•−) = 0.078 V vs NHE [34] and it is not surprising that it can successfully substitute oxygen, which is a weaker one-electron oxidant (redox potential, E˚(O2/ O2•−) = − 0.16 V vs NHE [35]) in the cross-linking reaction.
Fig. 4.
Yields of the 5’-d(G*pCpU) and 5’-d([Sp]pCpU) products generated by CO3•− radicals in deoxygenated solutions containing 0 and 100 µM 1,4-benzoquinone (BQ). The yields were calculated from the integrated peak areas in the HPLC elution profiles and the molecular absorptivities at 260 nm (n.d. – not detected).
Structural characterization of the 5’-d(G*pCpU*) lesion by 1D and 2D NMR methods
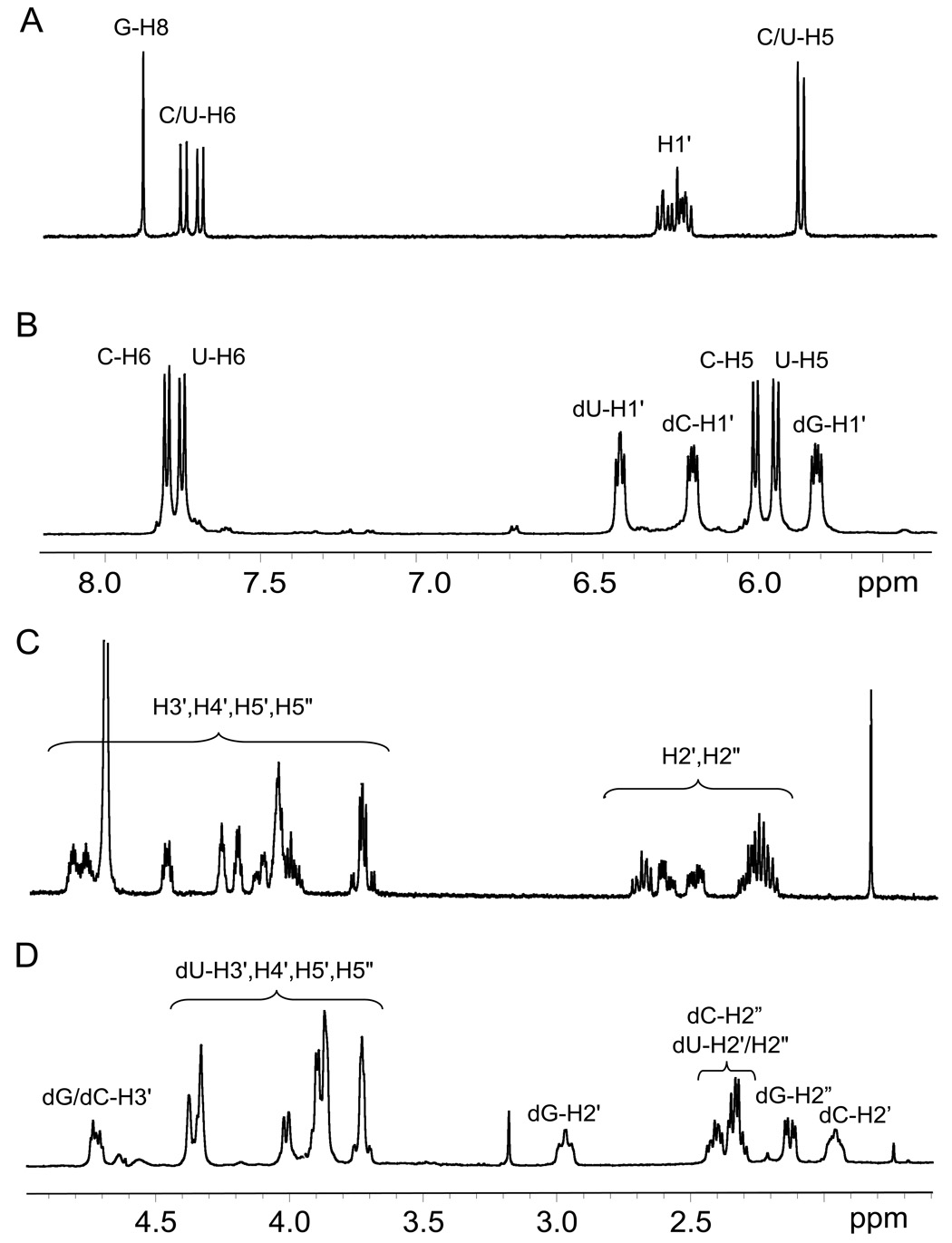
In order to identify the sites of guanine and uracil bases involved in the formation of the covalent bond in the 5’-(G*pCpU*) cross-linked product, we synthesized 5’-d(G*pCpU*) in amounts sufficient for analysis by 1D and 2D NMR methods. This approach has provided rich structural information on the cross-linked products generated by the oxidation of the 5’-d(GpCpT) trinucleotide by CO3•− radicals [16]. The 1D proton NMR spectra of 5’-d(G*pCpU*) and the intact 5’-d(GpCpU) were recorded in D2O solutions where the resonances of the exchangeable imino protons are not detectable (Fig. 5). The 5 – 8 ppm portions of the intact 5’-d(GpCpU) and crosslinked 5’-d(G*pCpU*) trinucleotides are shown in Figs 5A and 5B, respectively. In the intact 5’-d(GpCpU), the guanine H8 proton is identified as the singlet at 7.9 ppm, whereas three H1’ protons are assigned to a group of multiplets in the 6 – 6.2 ppm region. Both cytosine and uracil have H5 and H6 protons and the more downfield pair of indistinguishable H6 protons appears as two distinct doublets in the 7.6 – 7.8 ppm region. In contrast, the C/U-H5 protons are overlapped and show two peaks at 5.7 and 5.75 ppm. The spectrum of 5’-d(G*pCpU*) cross-linked product (Fig. 5A) differs from the spectrum of the intact 5’-d(GpCpU) trinucleotide (Fig. 5B). There is no longer a signal for guanine H8 and this in itself suggests that this position of guanine is involved in the bond with uracil. It is also evident that the C/U-H5 protons are now resolved into 2 distinct doublets in the 5.8 – 6.0 ppm region while the C/U-H6 protons appear again as two doublets in the 7.7 – 7.9 ppm region. The presence of signals for these 4 protons shows that neither C/U carbons 5 or 6 are involved in the bonding to guanine. It is also of interest that in 5’-d(G*pCpU*) the 2-deoxyribose H1’ protons are now resolved into three distinct multiplets and it is this fact which allows for a more complete assignment of the proton NMR signals.
Fig. 5.
The 1D proton NMR spectra of the intact 5’-d(GpCpU) (A and C) and 5’-d(G*pCpU*) (B and D).
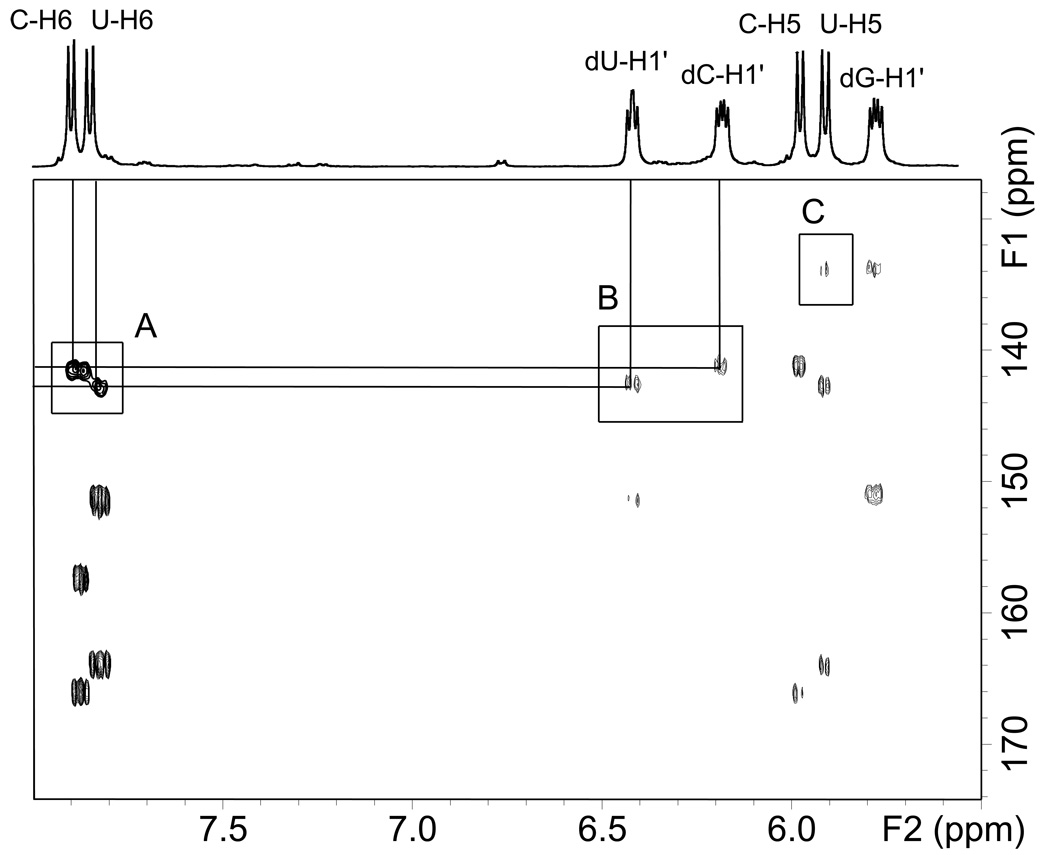
The individual H1’ resonances were assigned using a combination of 2D heteronuclear HSQC and HMBC experiments (Fig. 6), and homonuclear phase sensitive DQF-COSY experiments (Fig. 7). In the HSQC spectrum of 5’-d(G*pCpU*), the C/U-H6 protons show strong correlations to their respective carbon signals allowing the assignment of these carbons as a C/U pair (box A in Fig. 6). Thus, by carrying out a HMBC experiment, it was possible to assign the cross-peaks associated with the correlation of these carbons to their associated H1’ protons. In this manner, the multiplet at 6.3 – 6.4 ppm represents one of the dC/dU-H1’ protons, and the multiplet at 6.05 – 6.15 ppm represents the other dC/dU-H1’ proton (box B in Fig. 6). Therefore by elimination, the dG-H1’ resonance can be assigned to the multiplet at 5.65 – 5.75 ppm.
Fig. 6.
Superimposed portions of the HSQC and HMBC NMR spectra of 5’-d(G*pCpU*). The regions of the spectra shown represent correlations of the aromatic base / 2-deoxyribose H1’ protons and (1) their directly bonded carbons (HSQC) and (2) carbons separated by multiple bonds. The cross-peaks in the box marked A are HSQC correlations between C/U-H6 and their corresponding C/U-C6 atoms. The cross-peaks in the box marked B are HMBC correlations between dC/dU-H1’ and the C/U-C6 atoms. The box marked C shows a long-range 4-bond correlation from U-H5 to G-C8 (see the text).
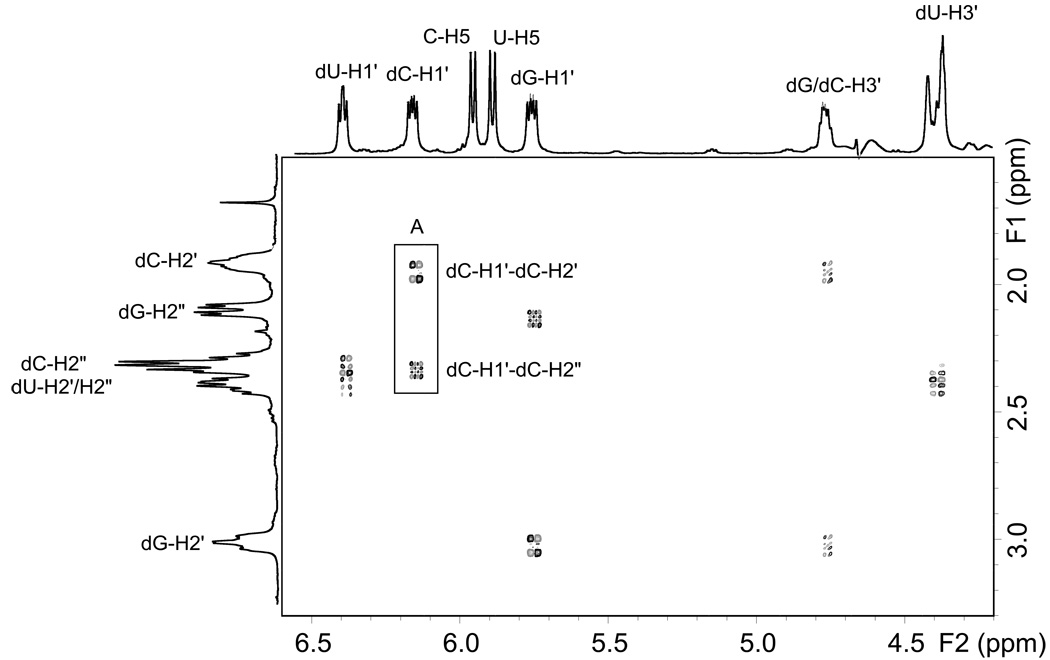
Fig. 7.
Portion of the phase sensitive DQF-COSY spectrum of 5’-d(G*pCpU*). The regions shown focus on the 2-deoxyribose scalar correlations between H1’ and H2’/H2” (5.5–6.5ppm) and H3’ and H2’/H2”(4.3–4.9ppm). The highlighted box shows the difference in correlation patterns between dC-H1’-H2’ and dC-H1’-H2”.
Differentiation between the cytidine and uridine signals was made by examination of the phase sensitive DQF-COSY spectrum (Fig 7). The proton NMR signals of the H-3’ protons of oligonucleotides are affected by the internucleotide phosphate group. In the case of 5’-d(G*pCpU*), the terminal dU-H3’ has no attached phosphate and resonates further upfield than the corresponding dG/dC-H3’ protons. Three crosspeaks in Fig. 6 between 4–5 ppm represent COSY correlations between the H3’ protons and the H2’/H2” protons. The dU-H3’ proton can thus be assigned within the furthest upfield multiplet at 4.36–4.41 ppm. Also the overlapping dG/dC-H3’ signals can be assigned to the multiplet at 4.74–4.78 ppm. It is now possible to distinguish between the dU/dC 2-deoxyribose protons, and thus we can use the scalar correlations between H3’ and H2’/H2” to assign the dC-H2’/H2” and dU-H2/H2” protons. In a similar manner, examination of the H2’/H2” to H1’ scalar correlations allows the assignment of dC-H1’ as the 6.12–6.15 ppm multiplet and dU-H1’ as the 6.36–6.38 ppm multiplet.
Examining the pattern of the correlations between each H1’ and the corresponding H2’/H2” pair, allows us to differentiate between the H2’/H2” signals. Chazin et al. [36] have shown that in a DQF-COSY experiment there is a difference in the pattern of cross-peaks for H1’-H2’ compared to those for H1’-H2”. The former show cross-peaks with 4 lobes while the H2’-H2” cross-peaks show 8–16 lobes depending on spectral resolution. This is clearly seen in the highlighted box of Fig. 7 and occurs due to the differences in coupling of the H2’/H2” protons with H1’ and H3’ protons. In this manner, dU-H2’, dU-H2” and dC-H2” can be assigned to the multiplet at 2.33–2.46 ppm, dC-H2’ is the multiplet at 1.99–2.02 ppm, dG-H2” is the multiplet at 2.15–2.19 ppm and finally dG-H2’ is the downfield multiplet at 2.99–3.04 ppm. The fact that dG-H2’ is shifted downfield relative to the other H2’/H2” signals suggests that the G base is in a syn conformation due to its bonding to uracil. Cadet et al. [37] have shown that when thymine is substituted by 6-methyluracil in a β-2’-deoxyribonucleoside, there is a shift in conformation from anti to syn and this results in the positioning of the 2-keto group of 6-methyluracil over the 2-deoxyribose ring causing a downfield shift of the H2’ signal. In the case of 5’-d(G*pCpU*), it is likely that the rigid nature of the G*-U* crosslink forces the guanine to adopt a syn conformation placing the base over the sugar ring and causing a similar downfield shift of the dG-H2’ signal.
Now that the individual H1’ protons have been assigned, it is possible to unambiguously assign the C/U H5/H6 protons using a combination of the HSQC/HMBC spectra as shown in Fig. 6 and it is also possible to see HMBC correlations of G-H1’ to G-C4 and G-C8. It is interesting that there appears to be a cross-peak between G-C8 and U-H5 as shown in Fig. 6, box C. Such long-range 4 bond proton carbon couplings can be seen and their appearance is more likely within a coupling pathway involving unsaturated bonds or a planar zig-zag (w-coupling) configuration [38–40]. The rigid nature of the G*-U* crosslink and the unsaturation between U-H5 and G-C8 thus allows this long-range correlation to be seen and this is further evidence that the bonding that occurs between guanine and uracil involves the position 8 in guanine and the uracil N3 position.
DISCUSSION
Two mechanisms of M-2 tandem lesion formation
The existence of intrastrand cross-links between adjacent G-T bases in oligonucleotides (the so-called tandem lesions) have been discovered by Box and co-workers [41–44]. These tandem lesions are generated by the oxidation of oligonucleotides with GT sequences by radiation-induced •OH radicals in deoxygenated solutions. In the intrastrand cross-links formed the bases are linked by a covalent bond between the guanine C8 and thymine C5 methyl group. The mass of these products is smaller by 2 D than the mass of the precursor G-T sequences and the mechanism involves the formation of the C-centered 5-(2’-deoxyuridinyl)methyl radical (U-•CH2) intermediate [17, 18]. Indeed, it has been shown that the independent generation of U-•CH2 radicals by photolysis of 5-(phenylthiomethyl)-2’-deoxyuridine results in the formation of these G-T tandem lesions. The U-•CH2 radicals rapidly react with O2, thus suppressing the formation of the covalent bond between U-•CH2 and G-C8 [45]. On the other hand, oxygen does not prevent the formation of the M-2 tandem lesions between adjacent A and T bases [46, 47]. Greenberg and co-workers proposed that the reaction of the U-•CH2 radicals with O2 is reversible [k(O2) = 2 × 109 M−1s−1, and k−(O2) = 3.4 s−1] and the small transient concentrations of these radicals present in oxygenated solutions can account for the formation of the tandem lesions [18, 46, 47]. The formation of the 5-(2’-deoxyuridinyl)methyl radical intermediates suggests that the initial cross-linking step can occur via the oxidation of thymine [17, 18] mediated by •OH radicals induced by radiation [41–44], or derived from Fenton reactions [48]).
In contrast, our experiments have shown that the one-electron abstraction from guanine bases can trigger the formation of a new type of intrastrand cross-link, which also has a mass (M-2), i.e., 2 Da lower than the mass of the intact oligonucleotide (M), but that has another structure [16]. Indeed, extensive LC-MS/MS, 1D and 2D NMR studies have shown that in these M-2 lesions, the C8-atom of guanine in G*-T* is linked to the N3 atom of thymine. These G*-T* lesions have been detected in single-stranded G*CnT* oligonucleotides in which G and T bases can be separated by n = 0, 1, and 2 cytosine bases, and in double-stranded DNA with adjacent G and T bases [16]. The formation of these G*-T* lesions can be triggered by the one-electron oxidation of guanine by oxidants with appropriate reduction potentials, such as CO3•− and SO4•− radicals, and photoexcited riboflavin [16]. The C8-centered G(−H)• radicals formed react further with thymine. The deprotonation of thymine N3(H) with pKa = 9.67 [49] greatly enhances its nucleophilicity that explains the remarkable large yield of the G*-T* lesion at pH 10 [25]. The radical adduct arising from the nucleophilic addition of the N3-atom of T to the C8 position of the G(−H)• radical can be oxidized by O2 which abstracts the second electron that must be removed in order to form the stable C8/G*-N3/T* cross-linked product. Indeed, the yields of these cross-linked products are negligible in the absence of O2 [16]. However, oxygen is not a unique oxidant for the second electron abstraction reaction, and efficient formation of the G*CT* and G*CU* lesions was detected in deoxygenated solutions containing 1,4-benzoquinone (Fig 4). Similar mechanisms have been proposed by Perrier et al. for the formation of Nε-(guanin-8-yl)-lysine cross-links [50]. In this work, we found that uracil, which also has a relatively labile proton at N3 (pKa = 9.18 [49]) can replace thymine in these cross-linking reactions. High resolution NMR studies show that in the case of cross-linked tandem lesions, the product 5’-d(Gp*CpU*) is generated by the oxidation of the parent 5’-d(GpCpU) by CO3•− radicals. The formation of the intrastrand covalent bonds between G and T (or U) does not involve the H-atom abstraction from the methyl group (or C5) or C6 positions of thymine or uracil. These observations, coupled with the observed loss in mass of 2 Da upon formation of the cross-linked products, is consistent with covalent bond formation between the C8-atom of guanine and the N3 atom of uracil.
Biological implications
Oxidatively generated damage to DNA associated with overproduction of free radicals in inflammatory tissues has been implicated in the etiology of many human cancers [1, 2, 4, 5]. In contrast, oxidatively generated damage to RNA has received much less attention because it is generally assumed that degradation of damaged RNA is too rapid to play a role in the synthesis of abnormal proteins [51]. Nevertheless, significant fractions of RNA bases adopt secondary, non-Watson-Crick type secondary structures in which they are more susceptible to oxidatively generated damage than in double-stranded regions. Hence, the levels of oxidatively generated RNA damage under persistent oxidative stress associated with the inflammatory response may be significantly higher than in the case of DNA [20, 21, 52] Although, 8-oxo-7,8-dihydroguanine is the only oxidized base that has been reported in RNA, the RNA counterparts of many other guanine lesions can also be produced under conditions of oxidative stress [53]. In this work, we found for the first time that the oxidation of guanine in a RNA fragment by the biologically important CO3•− radicals generates the same major products, including Sp, Iz, and the intrastrand cross-linked C8/G*-N3/U* products. Although these cross-linked lesions have not yet been observed in cellular DNA, our hypothesis is that these may arise in significant levels under conditions of oxidative and nitrosative stress that lead to the formation of peroxynitrite and nitrosoperoxycarbonate, and ultimately the carbonate radical [4, 5, 12]. The formation of the C8/G*-N3/U* products is initiated by the one electron-abstraction step involving the CO3•− radical (“single hit”), while the subsequent steps occur via the nucleophilic N3/U or N3/T addition mechanism and the oxidation of the resulting intermediate by molecular oxygen, which is abundant in cells.
Overall, our results indicate that the final oxidation products arising from an initial one-electron abstraction from guanine in DNA by carbonate radicals is very different from the product distributions generated by •OH radicals induced by radiation or derived from Fenton reactions [54, 55]. Our identification of the ultimate stable guanine oxidation products in DNA and RNA could serve as biomarkers for determining the participation of the carbonate radical or other one-electron oxidants of guanine in in vivo environments during oxidative stress. Thus, our in vitro results could have an important impact on confirming the putative role of carbonate radicals in tissues subjected to inflammatory conditions.
Acknowledgements
We thank Dr. C. H. Lin for insightful discussion of the NMR experiments. This work was supported by the National Institute of Environmental Health and Sciences (5 R01 ES 011589-07). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health and Sciences or the National Institutes of Health. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from Research Facilities Improvement (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the 500MHz spectrometer and ion trap were supported by the National Science Foundation (CHE-0116222 and CHE-0234863, respectively). We would like to acknowledge the highly useful and constructive suggestions of the anonymous reviewers.
Abbreviations
- Sp
spiroiminodihydantoin
- Gh
guanidinohydantoin
- Iz
2,5-diamino-4H-imidazolone
- 5’-G*CU*
trinucleotide with the cross-link between the guanine C8 and thymine N3 positions
- U-•CH2
5-(2’-deoxyuridinyl)methyl radical
- HSQC
heteronuclear single quantum correlation
- HMBC
heteronuclear multiple bond correlation
- DQF
double quantum filtered
- COSY
correlation spectroscopy
- DSS
sodium 2,2-dimethyl-2-silapentane-5-sulfonate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 2.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grisham MB, Jourd'heuil D, Wink DA. Review article: chronic inflammation and reactive oxygen and nitrogen metabolism--implications in DNA damage and mutagenesis. Aliment. Pharmacol. Ther. 2000;14 Suppl 1:3–9. doi: 10.1046/j.1365-2036.2000.014s1003.x. [DOI] [PubMed] [Google Scholar]
- 4.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Cadet J, Douki T, Ravanat JL. One-electron oxidation of DNA and inflammation processes. Nat. Chem. Biol. 2006;2:348–349. doi: 10.1038/nchembio0706-348. [DOI] [PubMed] [Google Scholar]
- 6.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 7.Shafirovich V, Dourandin A, Huang W, Geacintov NE. Carbonate radical is a site-selective oxidizing agent of guanines in double-stranded oligonucleotides. J. Biol. Chem. 2001;276:24621–24626. doi: 10.1074/jbc.M101131200. [DOI] [PubMed] [Google Scholar]
- 8.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 9.Pogozelski WK, Tullius TD. Oxidative Strand Scission of Nucleic Acids: Routes Initiated by Hydrogen Abstraction from the Sugar Moiety. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 10.Huie RE, Cliftonx CL, Neta P. Electron transfer reaction rates and equilibria of the carbonate and sulfate radical anions. Radiat. Phys. Chem. 1991;38:477–481. [Google Scholar]
- 11.Neta P, Huie RE, Ross AB. Rate constants for reactions of inorganic radicals in aqueous solution. J. Phys. Chem. Ref. Data. 1988;17:1027–1284. [Google Scholar]
- 12.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joffe A, Geacintov NE, Shafirovich V. DNA lesions derived from the site-selective oxidation of guanine by carbonate radical anions. Chem. Res. Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 14.Lee YA, Yun BH, Kim SK, Margolin Y, Dedon PC, Geacintovx NE, Shafirovich V. Mechanisms of guanine oxidation in DNA by carbonate radical anion, a decomposition product of nitrosoperoxycarbonate. Chem. Eur. J. 2007;13:4571–4581. doi: 10.1002/chem.200601434. [DOI] [PubMed] [Google Scholar]
- 15.Crean C, Geacintov NE, Shafirovich V. Oxidation of guanine and 8-oxo-7,8-dihydroguanine by carbonate radical anions: Insight from oxygen-18 labeling experiments. Angew. Chem., Int. Ed. Engl. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 16.Crean C, Uvaydov Y, Geacintov NE, Shafirovich V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic. Biol. Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg MM. Elucidating DNA damage and repair processes by independently generating reactive and metastable intermediates. Org. Biomol. Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 19.Burrows CJ, Rokita SE. Recognition of guanine structure in nucleic acids by nickel complexes. Acc. Chem. Res. 1994;27:295–301. [Google Scholar]
- 20.Nunomura A, Moreira PI, Takeda A, Smith MA, Perry G. Oxidative RNA damage and neurodegeneration. Curr. Med. Chem. 2007;14:2968–2975. doi: 10.2174/092986707782794078. [DOI] [PubMed] [Google Scholar]
- 21.Sayre LM, Perry G, Smith MA. Oxidative stress and neurotoxicity. Chem. Res. Toxicol. 2008;21:172–188. doi: 10.1021/tx700210j. [DOI] [PubMed] [Google Scholar]
- 22.Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, Jones PK, Ghanbari H, Wataya T, Shimohama S, Chiba S, Atwood CS, Petersen RB, Smith MA. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001;60:759–767. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 23.Lovell MA, Markesbery WR. Oxidatively modified RNA in mild cognitive impairment. Neurobiol. Dis. 2008;29:169–175. doi: 10.1016/j.nbd.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofer T, Seo AY, Prudencio M, Leeuwenburgh C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol. Chem. 2006;387:103–111. doi: 10.1515/BC.2006.014. [DOI] [PubMed] [Google Scholar]
- 25.Crean C, Lee YA, Yun BH, Geacintov NE, Shafirovich V. Oxidation of guanine by carbonate radicals derived from photolysis of carbonatotetramminecobalt(III) complexes and the pH dependence of Intrastrand DNA cross-links mediated by guanine radical reactions. ChemBioChem. 2008 doi: 10.1002/cbic.200800105. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Specrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 27.Sannes-Lowery KA, Mack DP, Hu P, Mei H-Y, Loo JA. Positive ion electrospray ionization mass spectrometry of oligonucleotides. J. Am. Soc. Mass Spectrom. 1997;8:90–95. [Google Scholar]
- 28.Ni J, Mathews MAA, McCloskey JA. Collision-induced dissociation of polyprotonated oligonucleotides produced by electrospray ionization. Rapid Commun. Mass Spectrom. 1997;11:535–540. doi: 10.1002/(SICI)1097-0231(199704)11:6<535::AID-RCM898>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 29.Wang PP, Bartlett MG, Martin LB. Electrospray collision-induced dissociation mass spectra of positively charged oligonucleotides. Rapid Commun. Mass Spectrom. 1997;11:846–856. [Google Scholar]
- 30.Vrkic AK, Hair AJR, Foote S, Reid GE. Fragmentation reactions of all 64 protonated trimer oligodeoxynucleotides and 16 mixed base tetramer oligodeoxynucleotides via tandem mass spectrometry in an ion trap. Int. J. Mass. Spectrom. 2000;194:145–164. [Google Scholar]
- 31.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 32.Cerny RL, Tomer KB, Gross ML, Grotjahn L. Fast atom bombardment combined with tandem mass spectrometry for determining structures of small oligonucleotides. Analyt. Biochem. 1987;156:175–182. doi: 10.1016/0003-2697(87)90217-x. [DOI] [PubMed] [Google Scholar]
- 33.Tromp JM, Schuerch S. Gas-phase dissociation of oligoribonucleotides and their analogs studied by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Specrom. 2005;16:1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 34.Wardman P. Reduction potentials of one-electron couples involving free radicals in aqueous solution. J. Phys. Chem. Ref. Data. 1989;18:1637–16755. [Google Scholar]
- 35.Stanbury DM. Reduction potentials involving inorganic free radicals in aqueous solution. Adv. Inorg. Chem. 1989;33:69–138. [Google Scholar]
- 36.Chazin WJ, Wuthrich K, Hyberts S, Rance M, Denny WA, Leupin W. 1H nuclear magnetic resonance assignments for d-(GCATTAATGC)2 using experimental refinements of established procedures. J. Mol. Biol. 1986;190:439–453. doi: 10.1016/0022-2836(86)90014-8. [DOI] [PubMed] [Google Scholar]
- 37.Cadet J, Taieb C, Remin M, Niemczura WP, Hruska FE. Conformational studies of alpha- and beta-pyrimidine 2'-deoxyribonucleosides in the syn and anti conformation. Biochim. Biophys. Acta. 1980;608:435–445. doi: 10.1016/0005-2787(80)90189-6. [DOI] [PubMed] [Google Scholar]
- 38.Williamson DS, Smith RA, Nagel DL, Cohen SM. Phase-sensitive heteronuclear multiple-bond correlation in the presence of modest homonuclear coupling: application to distamycin A. J. Magn. Reson. 1989;82:605–612. [Google Scholar]
- 39.Araya-Maturana R, Gavín-Sazatornilb JA, Heredia-Moyaa J, Pessoa-Mahanaa H, Weiss-López B. Long-range correlations (njC,Hn > 3) in the HMBC Spectra of 3-(4-Oxo-4H-chromen-3-YL)-acrylic Acid Ethyl Esters. J. Braz. Chem. Soc. 2005;16:657–661. [Google Scholar]
- 40.Claridge TDW. High-resolution NMR techniques in organic chemistry. Pergamon: Oxford; 1999. [Google Scholar]
- 41.Box HC, Budzinski EE, Dawidzik JD, Wallace JC, Evans MS, Gobey JS. Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA) Radiat. Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 42.Box HC, Budzinski EE, Dawidzik JB, Gobey JS, Freund HG. Free radical-induced tandem base damage in DNA oligomers. Free Radic. Biol. Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 43.Budzinski EE, Dawidzik JB, Rajecki MJ, Wallace JC, Schroder EA, Box HC. Isolation and characterization of the products of anoxic irradiation of d(CpGpTpA) Int. J. Radiat. Biol. 1997;71:327–336. doi: 10.1080/095530097144210. [DOI] [PubMed] [Google Scholar]
- 44.Box HC, Budzinski EE, Dawidzik JB, Wallace JC, Iijima H. Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res. 1998;149:433–439. [PubMed] [Google Scholar]
- 45.Romieu A, Bellon S, Gasparutto D, Cadet J. Synthesis and UV photolysis of oligodeoxynucleotides that contain 5-(phenylthiomethyl)-2'-deoxyuridine: a specific photolabile precursor of 5-(2'-deoxyuridilyl)methyl radical. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 46.Hong IS, Greenberg MM. Efficient DNA interstrand cross-link formation from a nucleotide radical. J. Am. Chem. Soc. 2005;127:3692–3693. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
- 47.Hong IS, Ding H, Greenberg MM. Oxygen independent DNA interstrand cross-link formation by a nucleotide radical. J. Am. Chem. Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong H, Cao H, Wang Y, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion unduced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knobloch B, Linert W, Sigel H. Metal ion-binding properties of (N3)-deprotonated uridine, thymidine, and related pyrimidine nucleosides in aqueous solution. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7459–7464. doi: 10.1073/pnas.0501446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. Characterization of lysine-guanine cross-links upon one-electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J. Am. Chem. Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 51.Bellacosa A, Moss EG. RNA repair: damage control. Curr. Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 52.Bregeon D, Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005;577:293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Li Z, Wu J, Deleo CJ. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- 54.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl radicals and DNA base damage. Mutat. Res. 1999;424:9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 55.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]