Abstract
Liquid chromatography (LC) coupled with mass spectrometry (MS) and database assignment methods have been used to conduct a large-scale proteome survey of the R6/2 mouse model of Huntington’s disease (HD). Although the neuropathological mechanisms of HD are not known, the mutant huntingtin gene that causes the disease is thought to alter gene transcription, leading to a cascade of neurotoxic events. In this report, we have focused on characterizing changes in the brain proteome associated with HD pathophysiology. Differences in the relative abundances of proteins (R6/2 compared to wild type) in brain tissue from the striatum and cortex, two primary loci of dysfunction in HD, were assessed by using a label-free approach based on calibrations to internal standards. In total, assignments were made for ~400 proteins. A set of criteria was used to establish 160 high confidence assignments, ~30% of which appear to show differences in expression relative to WT (wild type) animals. Many of the proteins that were differentially expressed are known to be associated with neurotransmission, and likely play key roles in HD etiology. This study is the first to report that the majority of differentially expressed proteins in the striatum are up-regulated, while the majority of the expressed proteins in the cortex are down-regulated. The differentially expressed proteins identified in this proteomic screen may be potential biomarkers and drug targets for HD, and may further our understanding of the disease pathology.
Keywords: Huntington’s disease, liquid chromatography, mass spectrometry, brain proteome, label-free quantification, striatum, cortex
Introduction
Huntington’s disease (HD) is caused by the expansion of a polyglutamine (CAG) repeat (>35 resulting in HD) in the coding region of the huntingtin gene resulting in dysfunction and degeneration of the striatum and corticostriatal pathway.1 Symptoms, including adventitious movements, cognitive deficits, and emotional impairment typically appear in middle age and progress until death. Despite the discovery of the mutated huntingtin gene, its pathogenic contribution to HD pathophysiology is still poorly understood. The HD involvement of several pathogenetic mechanisms, however, has been proposed including huntingtin protein misfolding and aggregation,2 oxidative stress, altered mitochondrial metabolism,3, 4 excitotoxicity, and impairment of the ubiquitin proteasome system (UPS).5 These pathological mechanisms likely arise from alterations in the proteome caused by dysfunctional gene transcription.
Research on HD has been greatly facilitated by the development of transgenic mouse models. These models make it possible to explore HD pathology by employing direct examination of tissue and more invasive sampling methods than are possible in human studies. The R6/2 mouse line, the most characterized HD model, expresses a portion of the human HD gene that may express as many as 150 CAG repeats.6 R6/2 mice develop a progressive behavioral and neurological phenotype by 6–8 weeks of age, which closely mimics juvenile-onset HD in humans.7
To better understand the neuropathological pathways of HD, early attempts have focused on studying differences in gene expression associated with HD by using DNA microarray techniques.8, 9 More recently, high resolution NMR spectroscopy has been used to examine metabolite profiles of R6/2 lines.10 Although important advances have been made, interrogation of HD pathophysiology at the level of the proteome is likely to provide more direct insight about pathological mechanisms. To date, only a few studies have examined the proteomes of HD mouse models;11, 12 this work used two-dimensional (2D) gel electrophoresis, and although this approach provides insight into the proteome profile, limitations associated with the methodology13–16 leaves much unknown.
In the last decade, efforts have been made to develop analytical proteome platforms,17–25 which hold promise for improving proteome coverage. Here we use liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) to determine relative abundance changes in the brain proteome of 8-week-old symptomatic R6/2 mice compared to their wild-type (WT) controls. Of particular focus in this work, is the striatum and cortex as they are the primary loci of dysfunction in HD. These two regions were isolated and the extracted proteins were subjected to LC-MS/MS analysis with peptide and protein identification by database search. We have identified several hundred proteins from a single LC-MS/MS experiment. A significant fraction of these proteins were found to be differentially regulated in disease samples relative to WT. This comprehensive analysis of the R6/2 striatal and cortical proteome provides a preliminary survey of protein expression changes in the HD mouse, and gives insight to further interrogations of the altered HD proteome.
Materials and methods
General strategy for comparative studies of mouse brain proteome
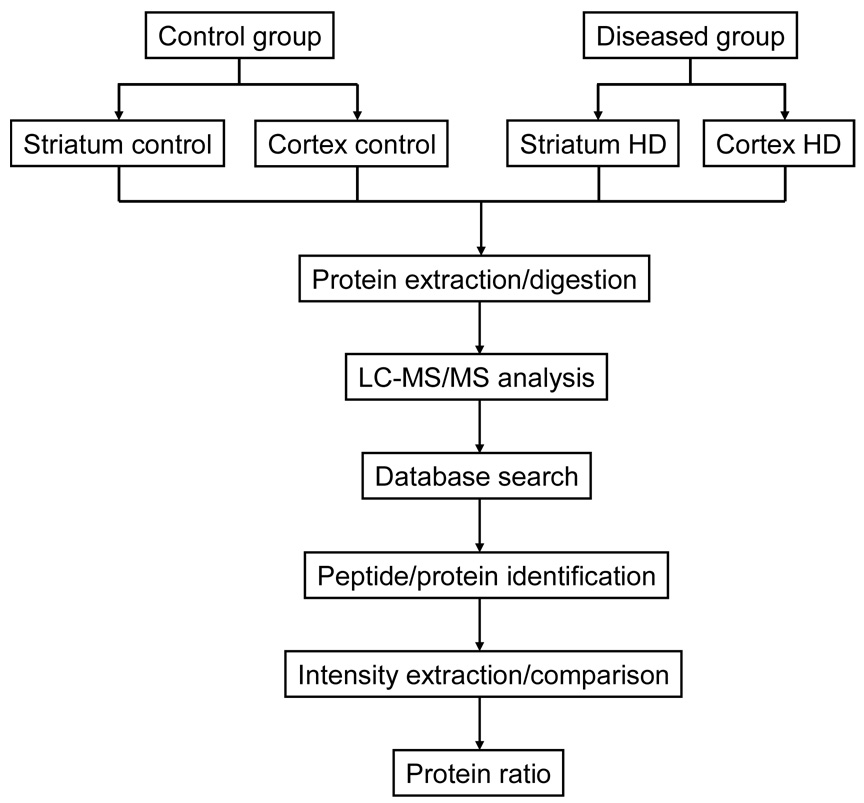
The general experimental approach used in this study for comparative mouse brain proteome profiling is described in Figure 1. The R6/2 mouse model of HD and WT controls were used in order to examine differentially expressed proteins associated with HD. The striatum and cortex were isolated from the rest of the mouse brain, followed by protein extraction using the protocols described below. Tissue dissection was performed with extreme caution to avoid cross contamination with other brain tissue. The extracted proteins were then reduced, alkylated and digested with trypsin. Subsequently, the resulting tryptic peptides were subjected to LC-MS/MS analysis for peptide and protein identification as well as relative quantification. Here we note that additional peptide fractionation (e.g., strong cation exchange chromatography) has not been used for the sake of simplicity and experimental throughput. However, utilization of multi-dimensional separations is desirable in the future for a more comprehensive proteome profiling.
Figure 1.
Schematic flow diagram of the experimental approach used for the comparative proteomics analysis of mouse brain samples.
Animals
Male transgenic R6/2 mice (B6CBA-TgN [HDexon1] 62Gpb) and WT littermate controls were purchased from the Jackson Laboratory (Bar Harbor, ME). The R6/2 and WT mice were grouped by polymerase chain reaction (PCR) tests (rather than phenotype) performed by Jackson Laboratory. We tested animals at 8 weeks of age, at which time the HD phenotype is observable. Three pairs of R6/2 and WT mice were used in this work. Each pair was obtained from a different litter born on different dates. All mice were housed individually in the departmental animal colony under standard conditions (12 hr light/dark cycle with lights on at 07:30 h) with access to food and water ad libitum. Both the housing and experimental use of animals followed the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee.
Brain tissue collection
At 8 weeks of age, the mice were sacrificed without anesthesia and the whole brain was dissected. Subsequently, the striatum and cortex were isolated and placed in separate Eppendorf tubes. We found no appreciable difference in brain weights of R6/2 and those of WT mice and the striatum and cortex typically weigh ~20 mg and ~40 mg, respectively. The samples were immediately frozen in liquid nitrogen and transferred to a −80 °C freezer until further analysis.
Protein extraction and digestion
The protocol used for protein extraction from mouse brain tissue is similar to methods reported previously.26–30 Briefly, for each tissue sample, 100 µL extraction buffer containing 5 mM phosphate (pH = 7.0) was added to the Eppendorf tube. The sample was then crushed and homogenized with a pestle driven by an electric motor. Another 200 µL phosphate buffer was used to wash the pestle afterwards and was combined with previous suspensions. The samples were sonicated in an ice bath for 5 min and subsequently incubated at 25 °C for 1 h. Trifluoroethanol (300 µL) was added and the samples were sonicated again in an ice-water bath and then allowed to incubate at 37 °C for 2 h. In order to remove non-soluble tissue debris from the suspensions, the samples were centrifuged at 13,000 rpm for 30 min. The supernatants were transferred into separate tubes and protein concentration was determined by Bradford assay experiments. Typically, extracted proteins from the striatum and cortex were estimated to be ~1 mg and ~2 mg respectively. Protein disulfide bonds were reduced with the addition of dithiothreitol (DTT) at a molar ratio of 40:1 (DTT:protein). After 2 h incubation at 37 °C, the sample was cooled and allowed to react with iodoacetamide (IAM) at a molar ratio of 80:1 (IAM:protein). The reaction was carried out on ice in complete darkness for another 2 h. Upon alkylation, a 40 fold excess of cysteine was added (at 25 °C for 30 min) in order to react with any residual DTT and IAM. Subsequently, the sample was diluted 5 fold with 2.4 mL 50 mM NH4HCO3 (PH = 8.0) prior to addition of 2% (w/w) TPCK-treated trypsin. The digestion was allowed to occur at 37°C for 24 h. Tryptic peptides in the mixture were purified further by passing the sample through Oasis HLB cartridges (Waters Inc., Milford, MA) and vacuum dried before further analysis.
Nanoflow LC-MS/MS analysis
The mixture of tryptic peptides was reconstituted in water (HPLC-grade) and a standard protein digest [cytochrome c (bovine and horse) prepared and characterized in separate experiments] was spiked into the sample. Because the concentration of the spiked protein is known, this can be used as an internal standard for peptide intensity normalization. The reverse phase capillary column. (75 µm × 130 mm) was packed in house with a MeOH slurry of 5 µm, 100 Å Magic C18AQ (Microm BioResources Inc., Auburn, CA). Nanoflow LC separation (carried out with UltiMate 3000 LC Systems, Dionex, Sunnyvale, CA) coupled online to a hybrid LTQ-FTICR (linear ion trap Fourier transform ion cyclotron resonance) mass spectrometer (ThermoElectron, San Jose, CA) was used for MS and MS/MS analysis. Depending on the length of gradient used in this study and the average peak width (~20–30 sec) under the conditions employed, the LC peak capacity is estimated to be ~ 300 to 400. The mobile phase was comprised of: solvent A, 97% H2O, 3% ACN, and 0.1 % formic acid (FA); and, solvent B, 100% acetonitrile (ACN) and 0.1 % FA. The following gradient was used for LC separation: B was increased up from 6% to 20% in 100 min and then increased to 28% in 20 min; at this point, B was rapidly increased to 90% and maintained for 15 min before 100% A was used to equilibrate the column. A 6.4 µL of the sample (containing about 300 ng tryptic peptides) was loaded onto a µ-precolumn cartridge (300 µm i.d.×5 mm, C18 PepMap100, 5 µm, 100 Å) for each LC separation. The precolumn is commercially available from LC Packings (Dionex, Sunnyvale, CA). The instrument was operated in a data-dependent mode for peptide identification and set up to perform one full MS scan with the FT analyzer followed by five MS/MS scans with the ion trap mass spectrometer. The commercial dynamic exclusion algorithm was utilized in order to preclude multiple analyses of the same abundant precursor ion.
Each sample was run in triplicate; however, the third experiment was carried out under MS-only mode. Acquisition of MS-only data is beneficial in that it allows more accurate precursor ion intensities to be extracted. Information from LC-MS/MS experiments was used only for the purpose of peptide and protein identification.
Protein assignment and peptide intensity extraction
A suite of commercial algorithms were used for much of the data processing. This includes the creation of MS/MS files (SEQUEST DTA files by using the Bioworks Browser, ThermoElectron, San Jose, CA) which are submitted to a search engine (MASCOT, Matrix Science Ltd. London, UK) for a database search against the SWISS-PROT protein database (http://ca.expasy.org). Peptide assignments from the MASCOT search were parsed to keep only those identifications with scores above extensive homology. An algorithm written in house was utilized to obtain extracted ion chromatograms for precursor ions. This algorithm integrates intensities associated with peaks for the monoisotopic ions of specific peptides; these integrated values can be use used to infer information about the relative abundances of ions. This code also provides information about peptide retention times, which can be coupled with MASCOT search results to improve confidences associated with relative abundance assessment. Thus, our dataset for each peptide assignment contains a report of precursor ion intensity and retention time, as well as all standard information associated with the MASCOT search.
As noted above, all intensity values are extracted from MS-only experiments. Peptides identified in the MS/MS scans were searched against MS-only scans for a possible match within certain m/z and retention time windows. An m/z tolerance of 0.02 is utilized given the high mass accuracy of the FTICR mass spectrometer. Searches are allowed for a relatively large retention time window (typically ~2 min) and a local maximum is established as the peak center. Then peak integration was performed using a much narrower retention time window (10% base peak height which is typically ~30 s).
Relative protein quantification
We focused only on those peptide assignments associated with high MASCOT scores because of a limited number of samples from each brain region (two sets of samples with MS/MS and one set with MS-only for detailed intensity analysis). For example, although the homology score for peptide identification is 30, only peptides with scores >60 were considered for intensity comparisons. This conservative approach should minimize spurious peptide assignments.
Label-free relative quantification was carried out by comparing peptide ion intensities between WT and R6/2 samples. For proteins identified with multiple peptide assignments, ion intensities were summed as total ion current prior to comparison. The relative protein abundance was then normalized using information associated with the internal standard, a known protein digest. In this approach, we average the peptide intensity ratios obtained for the internal standard and then the mean ratio is used as normalization factor when the protein abundance ratio of interest is determined by comparing R6/2 and WT samples. Because the spiked protein digest is added in equal amounts to both samples, the normalization factor is expected to be ~1.
Results
Observed proteins in LC-MS/MS experiments
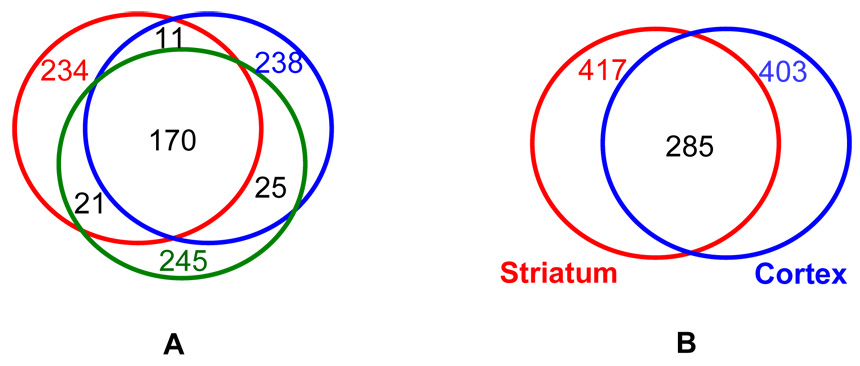
To assess the reproducibility of LC-MS/MS experiments, triplicate analyses of the same sample (R6/2 striatum) were performed and the number of proteins identified in individual runs were compared. As shown in Figure 2A, a similar number of proteins (234, 238 and 245 respectively) were obtained from replicate analyses of the diseased striatum sample. Additionally, a relatively high percentage (~70%) of the proteins was observed across all three runs indicating good reproducibility in LC-MS/MS identification.
Figure 2.
Venn diagrams showing the protein overlap in different experiments. Part A shows a Venn diagram representing the number of proteins observed for each of the triplicate runs of the diseased striatum sample, as well as the protein overlap. Part B shows a Venn diagram summary of the proteins identified from striatum and cortex by LC-MS/MS analysis. In total, 957 and 967 peptides have been observed in striatum and cortex respectively, corresponding to 417 and 403 protein identifications, respectively.
An abbreviated example of the format of summarized peptides and proteins is provided in Table 1; a complete dataset with peptide sequence and other information (such as score, m/z, and intensity information) is provided in supplemental material. In total, 957 and 967 peptides were observed in striatum and cortex corresponding to 417 and 403 protein identifications, respectively. Such information is represented graphically in Figure 2B, which also shows that a considerable number of proteins (285) are common to both striatum and cortex. Due to the nature of the extraction buffer we used in sample preparation, cytosolic proteins (soluble components) are favored in this study for all samples. No attempts to solubilize insoluble components were made here. We expect such an analysis (e.g., membrane fraction examination) to provide complementary information and will report such studies in a future paper. Peptides associated with structural proteins such as actin, tubulin and spectrin are observed most frequently. In addition, peptides associated with enzymes involved in energy metabolism, such as sodium/potassium-transporting ATPase, ATP synthase, malate dehydrogenase, creatine kinase and fructose-bisphosphate aldolase, were observed. If one ignores differences in the sizes of different proteins and assumes similar ionization efficiencies for all peptides, then the large numbers of peptides associated with individual proteins reflects relatively high abundance of these proteins in the proteome.
Table 1.
A list of identified proteins as well as number of peptides in striatum.
| accession # | protein name | # of peptidesa | scoreb | Sequence coverage(%) | |
|---|---|---|---|---|---|
| 1 | Q6PIC6 | Sodium/potassium-transporting ATPase alpha-3 chain | 15 | 116 | 22 |
| 2 | P56480 | ATP synthase beta chain, mitochondrial precursor | 14 | 111 | 35 |
| 3 | Q03265 | ATP synthase alpha chain, mitochondrial precursor | 12 | 86 | 27 |
| 4 | P63017 | Heat shock cognate 71 kDa protein | 11 | 132 | 23 |
| 5 | Q62261 | Spectrin beta chain, brain 1 | 11 | 121 | 7 |
| 6 | P02089 | Hemoglobin beta-2 chain | 10 | 105 | 88 |
| 7 | P68372 | Tubulin beta-2C chain | 10 | 88 | 33 |
| 8 | P16546 | Spectrin alpha chain, brain | 9 | 104 | 5 |
| 9 | P60710 | Actin, cytoplasmic 1 | 9 | 107 | 34 |
| 10 | Q8VDN2 | Sodium/potassium-transporting ATPase alpha-1 chain precursor | 8 | 112 | 10 |
The number of unique peptides observed with scores above MASCOT homology.
Highest MASCOT score obtained for the observed peptides associated with a given protein.
A substantial proportion of proteins (~150) were also detected; however, these are represented by fewer peptides suggesting lower abundances. The large number of proteins in this category is unlike what is observed in the plasma proteome, in which very high-abundance proteins mask signals of lower-abundance species necessitating the use of abundant protein depletion techniques.31,32–35 This intermediate concentration range of tissue proteins includes a number of protein families that are known to have important biological functions. These include heat shock proteins, as well as synaptic and vesicle related proteins.
Label-free quantification of proteins expressed in mouse striatum and cortex
As we have shown above, ~400 proteins have been assigned from the LC-MS/MS analyses. Using only the most confident assignments (MASCOT scores above 60, as described in the methods) ~160 proteins were considered for further quantification studies (using the label-free approach based on comparison of peptide ion intensities, also described in the methods). For proteins assigned with multiple peptides, summed precursor ion intensities were used for the abundance-ratio calculation. Alternatively, the average intensity ratios of individual peptides of a given protein can be used. Most observed proteins with multiple peptides show good agreement (usually on the order of ± 30% relative standard deviation, in cases where it is possible to quantify) among individual peptide ratios. Some proteins have been observed with moderate deviations in peptide-abundance ratios. Contributions to such deviations have been discussed in detail by Smith and coworkers.36 In these cases, the latter approach (averaging individual peptide ratios) would be problematic as the mean value might not reflect accurately the actual protein abundance ratio. With summed peptide ion intensities, the resulting ratio would be biased to that of more abundant peptide ions. Such a bias could be beneficial as our data indicates that integration of peak intensity is more reliable with respect to relatively abundant peptide ions. Thus, total peptide ion intensities are used to calculate the protein abundance ratio in this work.
Utilization of peak intensity in LC-MS mode for better quantification
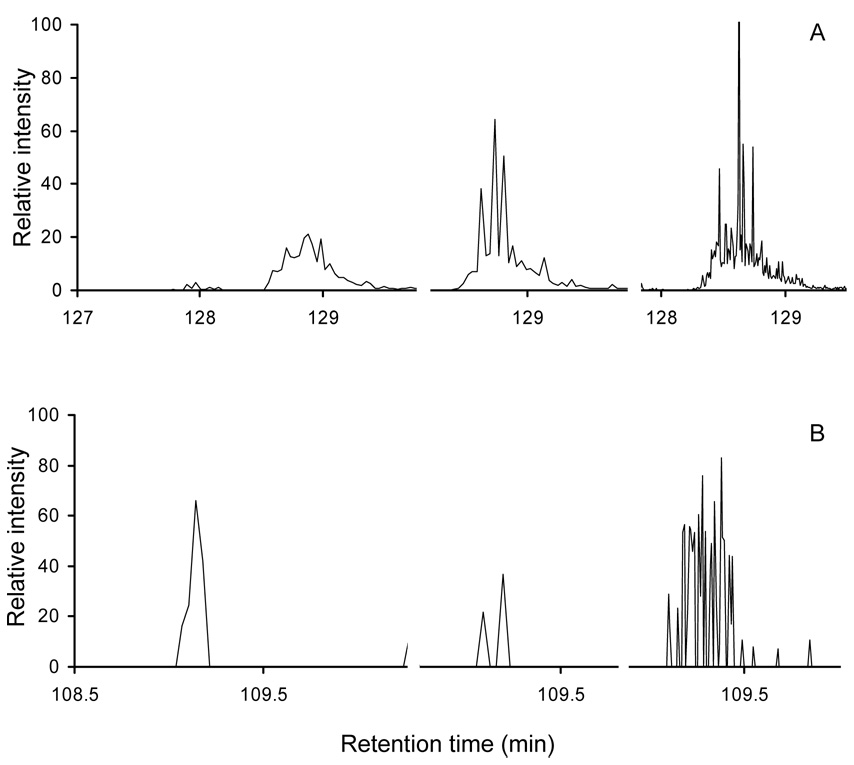
Once peptides are identified by LC-MS/MS it is useful to record MS-only scans (under LC-MS mode) in order to more reliably determine the intensities of peaks. This is shown in Figure 3A, which shows extracted ion chromatograms of the cytochrome c peptide ion [GITWGEETLMEYLENPKK+2H]2+ obtained from triplicate runs acquired using the LC-MS/MS and the LC-MS modes. For abundant proteins, measured integrated peak intensities of 4.2×107, 3.2×107, and 3.7×107 for the two LC-MS/MS and the LC-MS analysis, respectively, are in relatively close agreement (a relative range of 13.5%). For less abundant proteins with significantly weaker signals, however, intensity values in LC-MS/MS experiments are often misleading. For example, Figure 3B shows extracted ion chromatograms of a low-signal peptide ion [TSVNVVGDSFGAGIVYHLSK+2H]2+. The first two peaks from the LC-MS/MS experiments have integrated values of 2.5×105 and 6.5×105 (differing by more than a factor of two and clearly providing a limited sampling of the peak). As seen in the third trace (LC-MS mode, MS-only), more scan points (afforded by the LC-MS analysis) allow a better peak shape to be obtained (and thus more accurate intensity values, in this case, a value of 1.1×106).
Figure 3.
Part A shows plots of extracted ion chromatograms for the peptide ion [GITWGEETLMEYLENPKK+2H]2+ obtained from triplicate runs with the left and center plots acquired in LC-MS/MS mode and the right plot acquired in LC-MS mode. Part B shows extracted ion chromatograms for [TSVNVVGDSFGAGIVYHLSK+2H]2+ obtained from triplicate runs with the left and center plots acquired in LC-MS/MS mode and the right plot acquired in LC-MS mode. We note that for both plots only a partial view of the ion chromatogram (with the peak of interest) is shown such that three peaks of the same ion can be displayed side by side within a single plot for better visual comparison.
Using peptide peak intensities extracted from LC-MS experiments, we calculate initial protein-abundance ratios for those signals that are identified by LC-MS/MS. An additional step was the normalization of peak intensities based on internal standards (i.e., the signal associated with peaks for the spiked peptides). Normalization corrects for small run-to-run instrumentation variations. In this work, cytochrome c (bovine and horse) digests were added in equal amount to both the WT and R6/2 samples prior to LC-MS/MS analysis. Table 2 lists several observed peptides from cytochrome c as well as their precursor ion intensities. Peptide ratios were calculated by dividing intensities in R6/2 by those in WT samples. The mean ratio is 0.98 ± 0.17 with good agreement over 3 orders of magnitude variation in peptide intensities (106 – 109).
Table 2.
Cytochrome c peptides and precursor ion intensities.a
| peptide sequence | charge state | m/z (exp.)b | intensity (control) | intensity (HD) | ratio c |
|---|---|---|---|---|---|
| GITWGEETLMEYLENPKK | +2 | 1069.53 | 3.50·107 | 3.10·107 | 0.89 |
| GITWGEETLMEYLENPKK | +3 | 713.36 | 6.08·107 | 5.27·107 | 0.87 |
| GITWGEETLMEYLENPK | +2 | 1005.49 | 1.54·107 | 1.10·107 | 0.71 |
| GITWGEETLMEYLENPK | +3 | 670.66 | 2.89·106 | 2.34·106 | 0.81 |
| EETLMEYLENPKK | +2 | 812.40 | 6.22·108 | 6.98·108 | 1.12 |
| EETLMEYLENPKK | +3 | 541.94 | 6.75·108 | 8.26·108 | 1.22 |
| EETLMEYLENPK | +2 | 748.35 | 1.05·109 | 1.06·109 | 1.01 |
| EETLMEYLENPK | +1 | 1495.70 | 6.45·107 | 7.57·107 | 1.17 |
| GITWKEETLMEYLENPK | +2 | 1041.02 | 1.06·106 | 1.12·106 | 1.06 |
Observed peptides from cytochrome c (bovine and horse) digests as well as their precursor ion intensities from one pair of WT and R6/2 samples. Peptide abundance ratios are listed as well.
These are experimentally determined values from LC-MS/MS experiments.
Ratios are calculated as intensity of R6/2 over that of WT samples.
Differential expressed proteins in striatum and cortex
It is worthwhile to compare protein expression among three pairs because it is necessary to consider the biological variability among them. Table 3 lists several proteins observed across all three pairs (both striatal and cortical tissues) with corresponding abundance ratios. In general, most of the proteins show the same direction of regulation. An exception is elongation factor 1-alpha 1 that has abundance ratios that indicate elevated expression in two R6/2 striatal tissues, whereas the opposite is seen in the third disease sample. With respect to those proteins showing the same trend in expression, a difference in the extent of altered regulation was also observed. A few proteins such as alpha-synuclein and excitatory amino acid transporter 2 have abundance ratios with fairly small deviations (from each other) in all three pairs, suggesting a similar extent of altered expression. Several mutant versions of alpha-synuclein have been thought to be directly involved in Parkinson’s disease.37 Some evidence also suggests that alpha-synuclein is recruited into huntingtin aggregates in HD.38 Variation of protein ratios, however, could be large in some cases, as seen by hemoglobin beta-2 chain, spectrin alpha chain and ATP synthase beta chain. Large standard deviations in protein ratios could certainly be attributed, at least in part, to the biological variations among different mouse pairs. Another factor that may contribute to observed changes is associated with the experimental design. The database search method used in this study for the purpose of peptide and protein assignments does not differentiate different variants of proteins (e.g., different chains of a protein). For example, common peptides to spectrin alpha chain and spectrin beta chain could be assigned to both proteins. Coincidently, all three proteins in Table 3 bearing large variations among different pairs are those species with multiple variants such as alpha and beta chains. Despite the variations, however, general trends of protein regulation are still shown across all three pairs of mice.
Table 3.
Example abundance ratios for R6/2 and WT pairs.
| ratioa Ib | ratio II | ratio III | average ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|
| accession # | protein name | striatum | cortex | striatum | cortex | striatum | cortex | striatum | cortex |
| O55042 | Alpha-synuclein | 2.18 | 0.33 | 4.58 | 0.38 | 3.31 | 0.44 | 3.36± 1.20 | 0.38± 0.06 |
| P02089 | Hemoglobin beta-2 chain | 3.30 | 1.30 | 9.56 | 2.74 | 13.90 | 1.47 | 8.92± 5.33 | 1.84± 0.79 |
| P08249 | Malate dehydrogenase, mitochondrial | 1.70 | 0.50 | 1.63 | 0.17 | 1.63 | 0.37 | 1.65± 0.04 | 0.35± 0.17 |
| P10126 | Elongation factor 1-alpha 1 | 0.57 | 1.35 | 2.25 | 2.90 | 2.76 | 1.64 | 1.86± 1.15 | 1.96± 0.82 |
| P16546 | Spectrin alpha chain | 1.35 | 0.16 | 2.14 | 0.43 | 7.20 | 0.55 | 3.56± 3.17 | 0.38± 0.20 |
| P17183 | Gamma enolase | 2.42 | 0.33 | 4.86 | 0.66 | 3.18 | 0.25 | 3.49± 1.25 | 0.41± 0.22 |
| P43006 | Excitatory amino acid transporter 2 | - | 0.15 | - | 0.20 | - | 0.19 | - | 0.18± 0.03 |
| P56480 | ATP synthase beta chain | 1.62 | 0.15 | 2.77 | 0.28 | 5.74 | 0.23 | 3.38± 2.13 | 0.22± 0.07 |
Abundance ratios are calculated as intensity of R6/2 over WT samples.
Ratio I, II and III refer to individual values determined from three pairs of mice, respectively.
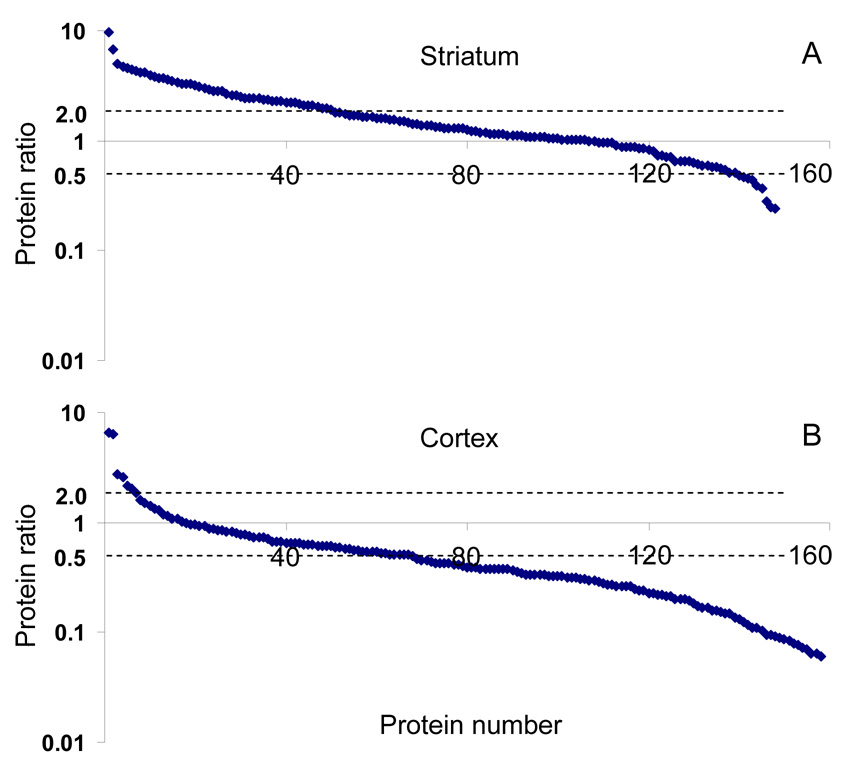
Only highly confident protein assignments were considered for quantification. In total, 162 and 181 proteins were quantified from the striatum and cortex samples, respectively. Figure 4 shows protein abundance ratios plotted against protein identifications (represented by a series of numbers) from one of the pairs, which is representative of the overall regulation pattern of all groups. The number of proteins is slightly lower than that of the quantified proteins because some have been observed in only one of the sample pairs (either in the WT or R6/2 sample). For those proteins observed in striatum (shown in Figure 4A), the majority show only slight changes in expression between normal and R6/2 tissues as two-thirds of the proteins have ratios less than 2 (located between two dashed lines). The remaining third differ significantly (a factor >2) in expression. It is noteworthy that most of these differentially expressed proteins are up-regulated in striatum as seen in Figure 4A.
Figure 4.
Observed protein abundance ratios in striatum (A) and cortex (B). Positive ratios have been calculated by dividing intensity of R6/2 samples by that of WT samples and plotted on a logarithmic scale. Proteins (represented by a series of numbers) are sorted by descending order of abundance ratios. See text for discussion.
Interestingly, the opposite trend was observed for proteins in the cortex as demonstrated by Figure 4B, where most of the differentially expressed proteins are down-regulated in R6/2 samples. Additionally, a larger fraction (~2/3) of cortex proteins showed more than a 2 fold change in abundance. We also observed that of those quantified species, 119 proteins were common to both the striatum and the cortex. This suggests that some proteins that overlap may have very different regulation patterns in R6/2 samples between the striatum and the cortex, and in some cases even opposite directions. For example, several hemoglobins (alpha, beta-1, and beta-2) were more abundant in the R6/2 striatum by a factor 5 to 9, while in the cortex they were almost equal in abundance between WT and R6/2 samples. The same is true for proteins from the tubulin family, which are highly abundant in both the striatum and the cortex. It is intriguing that although most tubulins in striatum show minimal differences between WT and R6/2 samples, cortex tubulins are significantly down-regulated in R6/2 samples by a factor of 3 to 6. Different spatial patterns of protein expression are not unexpected because proteomic changes in disease are likely localized, and striatum and cortex are two distinct brain regions implicated in HD. Whole brain analysis, therefore, would lack this important level of sensitivity.
Discussion
Down-regulation of glutamate transporters in cortex
Recently, a considerable amount of attention has been paid to differential brain proteomics.27, 39, 40 Brain tissue is considered the target of choice for studying neurodegenerative as well as other human brain diseases. It is believed that proteomic alterations in such diseases occur more selectively in the brain (physically near the disease pathology) rather than other tissues or body fluids (e.g., plasma). Thus, changes in protein abundance would be more noticeable in the brain relative to other types of tissue. Although obtaining brain tissue is invasive, the establishment of mouse models allows easy access to such tissue samples. This investigation used the R6/2 mouse model of HD (the most extensively studied model system of HD) to examine proteomic alterations associated with the disease.
Among the proteins that have been identified and quantified, the two glutamate transporter proteins [the excitatory amino acid transporter 1 (GLAST) and the excitatory amino acid transporter 2 (GLT1)] are responsible for the clearance of synaptic glutamate. Examination of these proteins along with their expression, located on both neurons and glia, may be instrumental to understanding the mechanisms underlying HD and other neurodegenerative diseases.
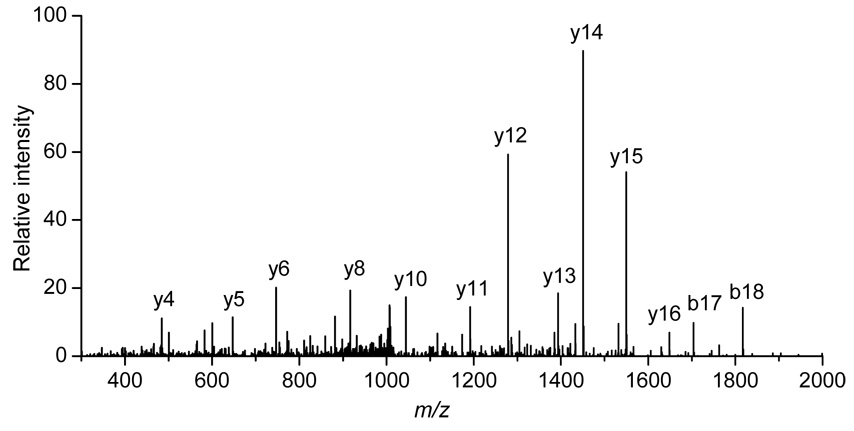
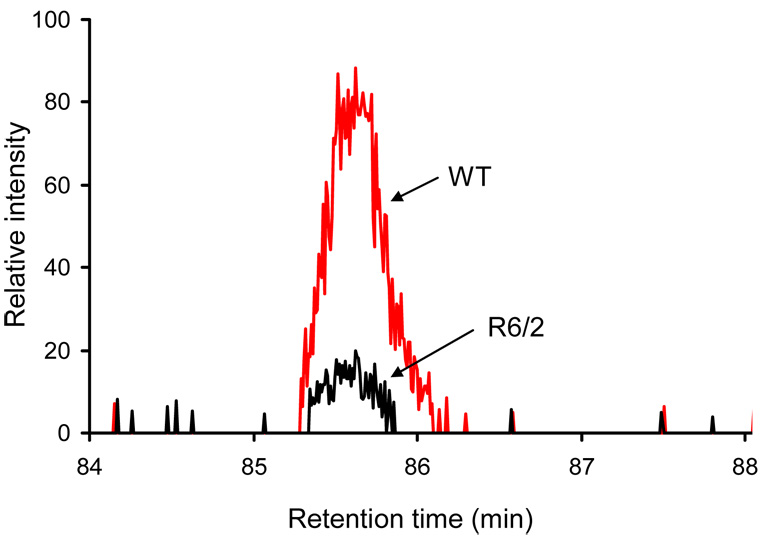
In this work, GLT1 was assigned by the identification of the peptide ion [TSVNVVGDSFGAGIVYHLSK+2H]2+ from its MS/MS spectrum shown in Figure 5. A strong continuous y-ion series was observed during collision-induced dissociation (CID). A subsequent protein database search (MASCOT) of this spectrum resulted in a confident assignment of this peptide (with a homology score of 135). Integration of the precursor ion intensities yielded significantly lower values in R6/2 relative to WT samples with a ~6-fold variation. Such a marked difference is evident in the extracted ion chromatograms of this peptide ion for both the WT and R6/2 samples (Figure 6).
Figure 5.
Collision-induced dissociation spectrum leading to the assignment of the [TSVNVVGDSFGAGIVYHLSK+2H]2+ peptide ion from GLT1. A MASCOT score of 135 is obtained from these data. See text for details.
Figure 6.
Extracted ion chromatograms of the GLT1 peptide ion [TSVNVVGDSFGAGIVYHLSK+2H]2+ from the WT (denoted by red line) and the R6/2 (denoted by black line) samples. See text for details.
GLAST was also confidently assigned by the detection of the peptide ion [TTTNVLGDSLGAGIVEHLSR+2H]2+. Like GLT1, GLAST is significantly down-regulated in HD cortex (~6-fold variation, data not shown here). Underexpression of both glutamate transporters can lead to excess synaptic glutamate resulting in excitotoxicity. In fact, an overactive glutamate input to striatum may underlie excessive firing of striatal neurons in R6/2 mice.41 Although down-regulation of GLT1 has been previously observed through Western blotting as well as immunohistochemistry in R6/2 mouse models,42, 43 our study extends these findings by showing down-regulation of GLAST, as well as GLT1.
Other biologically important proteins and their abundance changes
Although glutamate transporter proteins are found to be associated with neurotransmission and excitotoxicity, a number of other observed proteins are also of considerable interest in understanding HD pathology. Table 4 lists several groups of such proteins and their abundance changes in striatum and cortex. Many of these proteins are observed only in WT or R6/2 samples and a ratio is not available. We therefore use “up” or “down” to indicate the direction of regulation.
Table 4.
Abundance changes for several groups of proteins.a
| Swiss-Prot accession # | protein name | ratio in striatumb | ratio in cortex |
|---|---|---|---|
| Glutamate related proteins | |||
| P56564 | Excitatory amino acid transporter 1 | NAc | 0.17± 0.06 |
| P43006 | Excitatory amino acid transporter 2 | NA | 0.18± 0.02 |
| Q9D6M3 | Mitochondria glutamate carrier 1 | 1.64± 0.91 | 0.15± 0.04 |
| P05201 | Aspartate aminotransferase | 1.17± 0.07 | 0.33± 0.04 |
| Heat shock proteins | |||
| P07901 | Heat shock protein HSP 90-alpha | 2.40± 1.55 | NA |
| P11499 | Heat shock protein HSP 90-beta | 3.35± 1.66 | 1.18± 0.29 |
| P63017 | Heat shock cognate 71 kDa protein | 1.35± 0.49 | 0.62± 0.31 |
| P63038 | 60 kDa heat shock protein | 4.40± 2.75 | 0.24± 0.07 |
| Q64433 | 10 kDa heat shock protein | 2.13± 1.58 | 1.40± 0.49 |
| Q9CQN1 | Heat shock protein 75 kDa | 2.57± 1.36 | 0.86± 0.07 |
| Ubiquitin proteasome system (UPS) related proteins | |||
| P62991 | Ubiquitin | 4.92± 3.39 | 0.39± 0.11 |
| P61089 | Ubiquitin-conjugating enzyme E2 | Up | 0.41± 0.01 |
| Q7TMY8 | E3 ubiquitin protein ligase | down | NA |
| Q7TQI3 | Ubiquitin thiolesterase protein | NA | down |
| Q9R0P9 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 3.01± 1.89 | 0.38± 0.07 |
| Q9Z2U1 | Proteasome subunit alpha type | 2.71d | 0.38 |
| Synaptic and vesicle related proteins | |||
| O08547 | Vesicle trafficking protein | NA | Up |
| O08599 | Syntaxin binding protein | 3.88± 1.95 | 0.26± 0.11 |
| O35526 | Syntaxin-1A | NA | 0.26± 0.06 |
| O88935 | Synapsin-1 | NA | 0.22± 0.12 |
| P46460 | Vesicle-fusing ATPase | 1.34 | 0.08± 0.02 |
| P60879 | Synaptosomal-associated protein 25 | 3.37± 1.46 | 3.55± 2.64 |
| P61264 | Syntaxin-1B2 | 2.33 | 0.26± 0.10 |
| Q62277 | Synaptophysin | 3.12 | 1.04± 0.07 |
| Q64332 | Synapsin-2 | 1.39± 0.38 | 0.26± 0.06 |
| Q9ER00 | Syntaxin-12 | NA | 0.80 |
| Q9WV55 | Vesicle-associated membrane protein-associated protein A | NA | down |
| Q9QY76 | Vesicle-associated membrane protein-associated protein B | NA | 0.12 |
These groups of proteins are believed to bear important biological functions.
Abundance ratios are calculated as intensity of R6/2 over WT samples.
Data not available (proteins not observed).
Ratios without standard deviations are obtained from a single pair of mice.
In the first group of proteins, mitochondrial glutamate carrier 1, which transports glutamate across the inner mitochondrial membrane, is observed with decreased expression in R6/2 cortex, whereas it is slightly up-regulated in R6/2 striatum. Another glutamate-related protein, aspartate aminotransferase, catalyzes the conversion of aspartate to glutamate and is also less abundant in HD cortex while no appreciable difference is observed in HD striatum. A slight overexpression (a factor of 1.8) of this protein in R6/2 striatum was reported previously.11 Additionally, we identified many heat shock proteins (HSP), which are molecular chaperones that help other proteins achieve proper folding.44 Improper folding can lead to protein aggregation and it is well known that several neurodegenerative diseases including HD are characterized by aggregates formed by misfolded proteins.45 Although most of these HSPs exhibited insignificant differences between WT and R6/2 samples, 60 kDa heat shock protein is ~4-fold less abundant in HD cortex while ~4 times more abundant in HD striatum.
Another class of proteins that shows differential expression is associated with the ubiquitin proteasome system (UPS), including ubiquitin, ubiquitin carboxyl-terminal hydrolase isozyme, ubiquitin-conjugating enzyme, E3 ubiquitin protein ligase, ubiquitin thiolesterase protein, and proteasome subunit alpha type. Several UPS proteins were observed to be up-regulated in striatum but down-regulated in cortex of R6/2 compared to WT mice. UPS plays an important role in degrading misfolded, damaged, or unwanted proteins that could otherwise form potentially toxic aggregates. Impairment of UPS function is thought to be involved in HD pathology as well as other neurodegenerative diseases.5 Precise roles of those dysregulated proteins in UPS pathways as well as implication in HD progression are not yet clear at this point. Moreover, a number of identified proteins are involved in neurotransmission across synaptic regions such as synapsin, syntaxin, synaptophysin, synaptotagmin, and vesicle-fusing ATPase. While many of these proteins in striatum show elevated expression in R6/2 samples, several proteins present in the cortex, such as synapsin-1, vesicle-fusing ATPase and vesicle-associated membrane protein-associated protein B, are observed to be significantly down-regulated in R6/2 relative to WT mice. It might be argued that the primary drivers of the differentially expressed proteins in R6/2 mice are due to the influence of neuron atrophy and gliosis.6 Further interrogation of biological pathways of these proteins is needed to provide additional insight regarding altered expression in HD.
Protein identification by the isotope-coded affinity tag approach
The very recent work by Chern and co-workers46 used isotopic labeling techniques to examine protein expression in R6/2 striatum. Although we found only ~30% proteins that overlapped with the 203 protein list of Chern and co-workers, such a low percentage is not surprising given that they focused on a nucleus-enriched fraction of the striatum, whereas we analyzed the total cell lysate without subcellular enrichment. Additionally, ICAT tends to examine cysteine-containing peptides, which could contribute to the observation of different proteins in LC-MS/MS experiments.27
Another point of contrast is that unlike our data, which show up-regulation of a majority of differentially expressed proteins in striatum, Chern and co-workers46 reported the opposite trend consistent with previous work on gene expression.47 Comparisons of the overall regulation patterns, however, should be made cautiously because they depend on what proteins (or genes) are observed. Among the 68 altered proteins reported by Chern and co-workers, only 13 appeared in our dataset. Overall regulation pattern is also affected by the cutoff of the abundance ratio used to assess significant variation; slightly higher or lower values may influence the bulk number that is up or down regulated. It is also noteworthy that we sampled mice at an earlier age (8 weeks versus 10.5). Thus, comparison of our data with data obtained from isotopic labeling techniques will require a systematic investigation of comparable variables.
Summary
We presented a comprehensive comparative proteome profiling of HD-mouse models using a LC-MS/MS-based approach. Several hundred proteins were observed and their relative abundances were assessed by a label-free protein quantification method. A number of proteins, moreover, were found to be differentially expressed in R6/2 striatum and cortex. In particular, we note two glutamate transporter proteins were significantly down-regulated in cortex, which could result in excitotoxicity, a well-known pathological process thought to be involved in neurodegenerative diseases. Our findings may play an important role in understanding the underlying mechanisms of HD pathology, thus contributing to potential treatments. Future work will focus on several different areas such as studies of age-dependant disease progression, exploration of other methods for tissue protein extraction (e.g., enrichment of membrane proteins39) and plasma biomarker discovery (a tissue to plasma strategy).
Supplementary Material
Acknowledgements
We gratefully acknowledge support from the National Institutes of Health (R01 AG 024547 and R01 NS 35663) and the Analytical and Neuroscience Nodes of the Indiana University METACyt Initiative (funded by a grant from the Lilly Endowment). The authors thank Stephen Valentine and Brent Williams for helpful discussion and contributions to data analysis.
References
- 1.Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded an unstable on Huntington's disease chromosomes. Cell. 1993;72:971–993. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Bates GP. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 3.Browne SE, Ferrante RJ, Beal MF. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne SE, Beal MF. Neurochem. Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 5.Bence NF, Sampat RM, Kopito RR. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 6.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 7.Carter RJ, Lione LA, Humby T, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. J. Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desplats PA, Kass KE, Gilmartin T, Stanwood GD, Woodward EL, Head SR, Sutcliffe JG, Thomas EA. J. Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- 9.Crocker SF, Costain WJ, Robertson HA. Brain Res. 2006;1088:176–186. doi: 10.1016/j.brainres.2006.02.102. [DOI] [PubMed] [Google Scholar]
- 10.Tsang TM, Woodman B, Mcloughlin GA, Griffin JL, Tabrizi SJ, Bates GP, Holmes E. J. Proteome Res. 2006;5:483–492. doi: 10.1021/pr050244o. [DOI] [PubMed] [Google Scholar]
- 11.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, Cini C, Marco CD, Butterfield DA. Mol. Cell. Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Zabel C, Chamrad DC, Priller J, Woodman B, Meyer HE, Bates GP, Klose J. Mol. Cell. Proteomics. 2002;1:366–375. doi: 10.1074/mcp.m200016-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Romijin EP, Krijgsveld J, Heck AJR. J. Chromat. A. 2003;1000:589–608. doi: 10.1016/s0021-9673(03)00178-x. [DOI] [PubMed] [Google Scholar]
- 14.Hancock WS, Apffel AJ, Chakel JA, Hahnenberger KC, Choudhary G, Traina J, Pungor E. Anal. Chem. 1999;71:742A–748A. doi: 10.1021/ac9907641. [DOI] [PubMed] [Google Scholar]
- 15.Figeys D. Anal. Chem. 2003;75:2891–2905. doi: 10.1021/ac030142m. [DOI] [PubMed] [Google Scholar]
- 16.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 17.Venable JD, Dong M, Wohlschlegel J, Dillin A, Yates JR. Nature Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 18.Washburn MP, Wolters DA, Yates JR. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, Jacobs JM, Camp DG, II, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Anal. Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 21.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci., U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 23.Garcia BA, Siuti N, Thomas CE, Mizzen CA, Kelleher NL. Int. J. Mass Spectrom. 2007;259:184–196. [Google Scholar]
- 24.Myung S, Lee YL, Moon MH, Taraszka JA, Sowell R, Koeniger SL, E HA, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MV, Ewing M, E CD. Anal. Chem. 2003;75:5137–5145. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 25.Valentine SJ, Kulchania M, Srebalus Barnes CA, Clemmer DE. Int. J. Mass Spectrom. 2001;212:97–109. [Google Scholar]
- 26.Wang H, Qian W, Mottaz HM, Clauss TRW, Anderson DJ, Moore RJ, Camp DG, II, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. J. Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Qian W, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp DG, II, Khan AH, Smith DJ, Smith RD. J. Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taraszka JA, Gao X, Valentine SJ, Sowell RA, Koeniger SL, Miller DF, Kaufman TC, Clemmer DE. J. Proteome Res. 2005;4:1238–1247. doi: 10.1021/pr050037o. [DOI] [PubMed] [Google Scholar]
- 29.Taraszka JA, Kurulugama R, Sowell R, Valentine SJ, Koeniger SL, Arnold RJ, Miller DF, Kaufman TC, Clemmer DE. J. Proteome Res. 2005;4:1223–1237. doi: 10.1021/pr050038g. [DOI] [PubMed] [Google Scholar]
- 30.Xun Z, Sowell RA, Kaufman TC, Clemmer DE. J. Proteome Res. 2006;6:348–357. doi: 10.1021/pr060488o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson NL, Anderson NG. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Plasencia M, Ragg S, Valentine SJ, Clemmer DE. Brief. Funct. Genomic. Proteomic. 2004;3:177–186. doi: 10.1093/bfgp/3.2.177. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Valentine SJ, Plasencia M, Trimpin S, Naylor S, Clemmer DE. J. Am. Soc. Mass Spectrom. 2007 doi: 10.1016/j.jasms.2007.04.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentine SJ, Liu X, Plasencia M, Hilderbrand AE, Kurulugama RT, Koeniger SL, Clemmer DE. Expert Rev. Proteomics. 2005;2:553–565. doi: 10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 35.Valentine SJ, Plasencia M, Liu X, Krishnan M, Naylor S, Udseth HR, Smith RD, Clemmer DE. J. Proteome Res. 2006;5:2977–2984. doi: 10.1021/pr060232i. [DOI] [PubMed] [Google Scholar]
- 36.Qian W, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, II, Smith RD. Mol. Cell. Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vila M, Wu DC, Przedborski S. Trends Neurosci. 2001;24:S49–S55. doi: 10.1016/s0166-2236(00)01984-6. [DOI] [PubMed] [Google Scholar]
- 38.Charles V, Mezey E, Reddy PH, Dehejia A, Young TA, Polymeropoulos MH, Brownstein MJ, Tagle DA. Neuroscience Letters. 2000;289:29–32. doi: 10.1016/s0304-3940(00)01247-7. [DOI] [PubMed] [Google Scholar]
- 39.Bihan TL, Goh T, Stewart II, Salter AM, Bukhman YV, Dharsee M, Ewing R, Wiśniewski JR. J. Proteome Res. 2006;5:2701–2710. doi: 10.1021/pr060190y. [DOI] [PubMed] [Google Scholar]
- 40.Pierson J, Norris JL, Aerni H, Svenningsson P, Caprioli RM, Andrén PE. J. Proteome Res. 2004;3:289–295. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 41.Rebec GV, Conroy SK, Barton SJ. Neuroscience. 2006;137:327–336. doi: 10.1016/j.neuroscience.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 42.Liévens JC, Woodman B, Mahal A, Spasic-Boscovic O, Samuel D, Goff LK, Bates GP. Neurobiol. Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- 43.Behrens PF, Franz P, Woodman B, Lindenberg KS, Landwehrmeyer GB. Brain. 2002;125:1908–1922. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- 44.Sharp S, Workman P. Adv. Cancer Res. 2006;95:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 45.Taylor JP, Hardy J, Fischbeck KH. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 46.Chiang M, Juo C, Chang H, Chen H, Yi EC, Chern Y. Mol. Cell. Proteomics. 2007;6:781–797. doi: 10.1074/mcp.M600356-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, Ross CA, Borchelt DR, Tapscott SJ, Young AB, Cha J-HJ, Olson JM. Hum. Mol. Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.