Summary
Pontin and reptin belong to the AAA+ family and they are essential for the structural integrity and catalytic activity of several chromatin remodeling complexes. They are also indispensable for the assembly of several ribonucleoprotein complexes, including telomerase. Here, we propose a structural model of the yeast pontin/reptin complex based on a cryo-electron microscopy reconstruction at 13 Å. Pontin/reptin hetero-dodecamers were purified from in vivo assembled complexes forming a double ring. Two rings interact through flexible domains projecting from each hexamer, constituting an atypical asymmetric form of oligomerization. These flexible domains and the AAA+ cores reveal significant conformational changes when compared to the crystal structure of human pontin that generate enlarged channels. This structure of endogenously assembled pontin/reptin complexes is different to previously described structures, suggesting that pontin and reptin could acquire distinct structural states to regulate their broad functions as molecular motors and scaffolds for nucleic acids and proteins.
Keywords: cryo-EM, Rvb1, Rvb2, Tip48, Tip49, pontin, reptin, Ino80, AAA+
Introduction
Chromatin remodeling enzymes modify chromatin structure to control the access of the DNA repair and transcriptional machinery to the site where they need to operate. Several large multi-subunit ATP-dependent chromatin remodeling complexes have been discovered and among them the Ino80 complex (Ino80.com), found in yeast and humans, comprises more than 12 distinct subunits (Carrozza et al., 2003; Gallant, 2007). Ino80.com and related remodeling complexes contain two proteins of the AAA+ superfamily of ATP-binding proteins with an approximate molecular weight of 54 kDa each and named Rvb1p (or pontin) and Rvb2p (or reptin) (Gallant, 2007; Wood et al., 2000). Rvb1p and Rvb2p are essential for the incorporation of Arp5 into and the catalytic activity of Ino80.com (Jonsson et al., 2001; Jonsson et al., 2004). Recent results link the remodeling functions of human Rvb1 with the cellular response after DNA damage (Jha et al., 2008). Pontin and reptin also participate in the maturation of small nucleolar RNAs as part of multiprotein complexes involved in small nucleolar ribonucleoprotein assembly (Boulon et al., 2008; McKeegan et al., 2007; Watkins et al., 2004; Zhao et al., 2008). Interestingly, a recent report reveals that pontin and reptin are essential for the assembly of telomerase, another ribonucleoprotein complex (Venteicher et al., 2008). Therefore, these proteins could have a general role in the assembly and regulation of several large protein complexes (Gallant, 2007; Kim et al., 2006).
Both human and yeast Rvb1/pontin and Rvb2/reptin proteins have been found to form a ~650 kDa complex containing equimolar amounts of Rvb1 and Rvb2 (Puri et al., 2007). An 1:6:6 stoichiometry of the Ino80 protein to Rvb1 and Rvb2 was observed in purified yeast Ino80 complexes, but it has not been defined if there is a direct interaction between Ino80 and Rvb1p or Rvb2p (Jonsson et al., 2001; Jonsson et al., 2004). Also, the two bacterially expressed human recombinant proteins can assemble into a double hexameric ring complex in vitro whose low resolution structure was solved using electron microscopy of negatively stained samples (Puri et al., 2007). All these results strongly indicate that the dodecamer is likely one of the possible functional forms of the proteins. Still, each protein seems to have specific roles and some data suggests subunit specific functions performed by the individual proteins without being part of a pontin/reptin complex (Gallant, 2007). Although pontin and reptin assemble into double-ring dodecamers, it is still unknown if the rings are homo or hetero-oligomeric. Several lines of evidence suggest that homo-oligomeric rings can exist.
Recombinant pontin forms hexameric rings, indicating that at least pontin does not need reptin to oligomerize (Matias et al., 2006). Under certain conditions human reptin can also form a ~400 kDa homo-oligomer (Puri et al., 2007). On the other hand, a recent work suggests that yeast pontin and reptin exist as a hetero-oligomeric single ring (Gribun et al., 2008). Therefore, a clear view is lacking about whether the pontin/reptin complex forms single/double rings and homo-/ hetero-oligomeric rings.
The ATPase activity of both yeast pontin and reptin is required for the viability of the cell whereas this seems dispensable for the catalytic activity of the Ino80.com (Jonsson et al., 2004). Actually, there are conflicting reports about the levels of ATPase activity of these proteins depending on the experimental context. Recombinant yeast pontin/reptin complex displays enhanced ATPase activity when compared to the individual proteins in some experiments (Gribun et al., 2008) whereas no detectable activity is observed by others (Jonsson et al., 2004). The recombinant human proteins purified from E\coli can assemble a dodecamer in vitro with weak ATPase activity, which is not affected by DNA (Puri et al., 2007). On the other hand a recombinant human pontin preparation that crystallized in its ADP-bound form showed very low activity and it seemed that the actual conformation of the protein blocked the removal of ADP from its binding pocket (Matias et al., 2006). Thus, ATP binding and hydrolysis is indispensable for at least some of the biological roles of the Rvb proteins in vivo, but their precise contribution to their functions is unclear (Jonsson et al., 2004). A likely possibility to reconcile all these observations is that the in vivo ATPase activity of pontin and reptin could be regulated in the context of larger multi-protein complexes.
Pontin (Rvb1) and reptin (Rvb2) are homologous to the prokaryotic RuvB protein (Yamada et al., 2002), an AAA+ motor required for Holliday junction migration. Bacterial RuvB forms homo-oligomeric hexameric rings on DNA (Chen et al., 2002) and each subunit contains the characteristic Walker A and Walker B motifs that constitute the active ATP-binding site. Surprisingly, the linear structure of the eukaryotic Rvb proteins revealed that the Walker A and B motifs that form the hallmark of these ATP-binding proteins were separated by a long sequence (Supplementary Figure S1). This paradox was solved recently after the resolution of the atomic structure of the human pontin (Rvb1) hexamer using X-ray crystallography (Matias et al., 2006). Each monomer comprises three well-defined domains, two of them highly reminiscent of those of the bacterial RuvB protein and containing the nucleotide binding site, whereas a third domain is inserted in the middle of the structure. This new domain (ATPase-insert domain from now on) is unique among AAA+ proteins, protrudes out of the hexamer and it has been shown to bind double and single stranded DNA (Matias et al., 2006). Interestingly, the central channel in the pontin ring has a diameter of around 18 Å, which is not wide enough to allow dsDNA to go through, but its negative electrostatic potential is similar to other helicases that bind ssDNA.
Here, we have purified yeast pontin/reptin complexes and analyzed their structure using electron microscopy under liquid nitrogen temperatures (cryo-EM) at ~13 Å resolution. The significantly improved resolution and the absence of staining agents have allowed us to provide a structural model for the architecture of the pontin/reptin complex. We show that each protein forms a hexameric ring with a similar arrangement in the core domains, but each ring displays a unique conformation due to their interaction using the ATPase-insert domains. Significant changes in the conformation of the AAA+ core and the ATPase-insert domains are found compared to the crystal structure of human pontin (Matias et al., 2006). This 3D structure is different to that shown in previous works for pontin/reptin complexes assembled in vitro from purified proteins (Gribun et al., 2008; Puri et al., 2007), suggesting that pontin and reptin could interact to form structurally distinct complexes.
Results
Yeast pontin and reptin purify as an equimolar complex that hydrolyses ATP
Yeast pontin/reptin complex was expressed and purified using baculovirus and an N-terminal His-tagged reptin for affinity purification as described before (Jonsson et al., 2004). Dynamic light scattering of the purified material revealed a major species of ~600 kDa (data not shown). Prior to any further structural or functional analysis the purified protein was subjected to a gel filtration chromatography in order to ensure the isolation of a homogeneous assembly for electron microscopy (EM) analysis. The sample resolved in four distinct peaks with the first three containing pontin and reptin as revealed in SDS electrophoresis of all the fractions (Figure 1A, only the fractions from the peaks are selected for display). Peak 2 contained equimolar amounts of both proteins and it was compatible with a roughly 650 kDa complex according to the calibration of the column. Although gel filtration chromatography cannot be used to estimate the exact molecular weight of the complex, the analysis of this peak by EM (see later) was only compatible with a dodecameric complex. Peak 1 corresponded to a larger aggregate or assembly, whereas peak 3 could be compatible with a predominance of monomeric forms (~50 kDa) which contained mostly reptin. Since the chromatographic column was loaded with an affinity purified pontin/His-reptin complex, monomeric reptin could derive from a pool of free reptin protein or from the disassembly of the pontin/reptin complex. In addition, fractions from the chromatography were tested for the presence of endogenous DNA or RNA using radio labelled [γ-32P]ATP, and no residual polynucleotides were detected bound to the pontin/reptin complexes after size exclusion chromatography (data not shown).
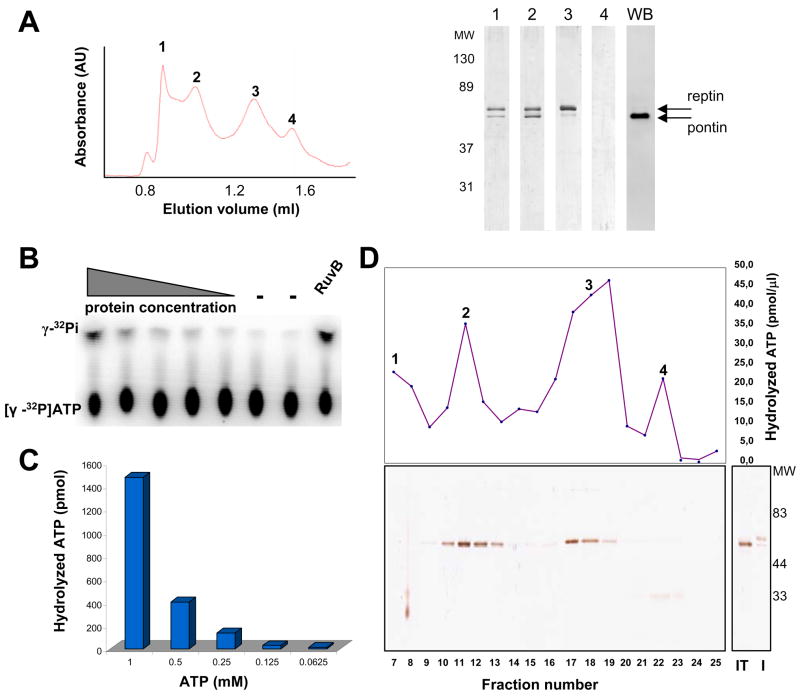
Figure 1. Purification and ATPase activity of pontin/reptin.
(A) Left panel: Elution profile of the size-exclusion chromatography fractions from the purification of pontin/reptin. Right panel: a fraction from each of the four peaks (labeled accordingly) was analyzed by SDS-page and silver staining. The lane at the far right shows the western blot of fraction 2 using polyclonal serum against pontin. “MW” stands for molecular weight standards.
(B) Effect of protein concentration on the ATPase activity. ATP hydrolysis was assayed at 1 mM ATP. Protein concentrations range from 4 μM to 4 nM monomer concentration. Control reactions were perfomed without protein (−) and with B. subtilis RuvB. Free phosphate produced by ATP hydrolysis was separated from ATP by TLC.
(C) ATPase activity as a function of ATP concentration.
(D) Top panel: The pontin/His-reptin complex was processed with TEV protease, loaded in a gel filtration column as in (A) and the fractions tested for its ATPase activity. Bottom panel: SDS gel and silver staining analysis of each of the fractions tested. “I” and “IT” stand for the input material before and after cleavage with TEV, respectively. Note that reptin runs similar to pontin after removing the His-tag.
Our preparation of pontin/His-reptin was capable of hydrolyzing ATP as revealed by the ATPase activity of the purified complex at varying protein (Figure 1B) and ATP concentrations (Figure 1C). ATPase assays at an optimum ATP concentration were then also performed in the presence of several types of single and double stranded DNA substrates, but in none of the cases a clear stimulation of the ATPase activity was observed (data not shown). Pontin/His-reptin was also processed with the TEV protease to remove the His tag, run in a gel filtration chromatography and the ATPase activity of each fraction measured (Figure 1D). After removal of the His-tag, the sample behaved in the chromatography identically to the tagged protein (Figure 1D, note that this time, reptin without the His-tag ran closer to pontin in the SDS electrophoresis, but both proteins were detected by western blotting (data not shown)). ATPase activity was assayed for aliquots of fractions 7 to 25, which, despite the low protein concentration of the fractions, revealed a significant although weak ATPase activity in those fractions corresponding with both the pontin/reptin complex (peak 2) and the free subunits (peak 3).
The pontin/reptin complex assembles as a double hexameric ring
Freshly collected fractions from the column were directly applied to carbon-coated grids for observation in the electron microscope after negative staining. Visual inspection of the EM fields corresponding to peak 2 revealed that the majority of the views were suggestive of a four petal flower (Figure 2A, within circles) and similar images were found regardless of the presence or absence of the His-tag (data not shown). Interestingly, this shape was very closely reminiscent of the electron microscopy images taken for double hexameric ring complexes of the related prokaryotic RuvB helicase (Chen et al., 2002), whereas they strikingly contrasted those reported for the recombinant human Rvb1/Rvb2 complex observed by EM (Puri et al., 2007). The comparison between our images and the atomic (Yamada et al., 2002) and EM structures (Chen et al., 2002) of bacterial RuvB indicated that the four petal images corresponded to side views of a double-ring pontin/reptin complex, with every two petals comprising each of the rings. This assumption was supported by the identification of hexameric circular top views under different experimental conditions (see below). The abundance of these double-layer side views confirmed that the pontin/reptin complex in peak 2 formed a double-ring. Intriguingly, a more careful inspection revealed two broad types of side views, one showing four petals of fairly similar size and other with a remarkable asymmetry between the two rings (Figure 2B). In the asymmetric view one of the rings seemed wider and rounded while the other appeared narrower. This configuration contrasted with the images from the prokaryotic homo-oligomeric double ring (Chen et al., 2002) and indicated that the conformation of each ring in the pontin/reptin oligomer could be different. On the other hand the large molecular weight peak 1 (Figure 1A) was found to comprise larger aggregates of several shapes and sizes suggesting a non-specific aggregation of the proteins without functional significance (Figure 2C).
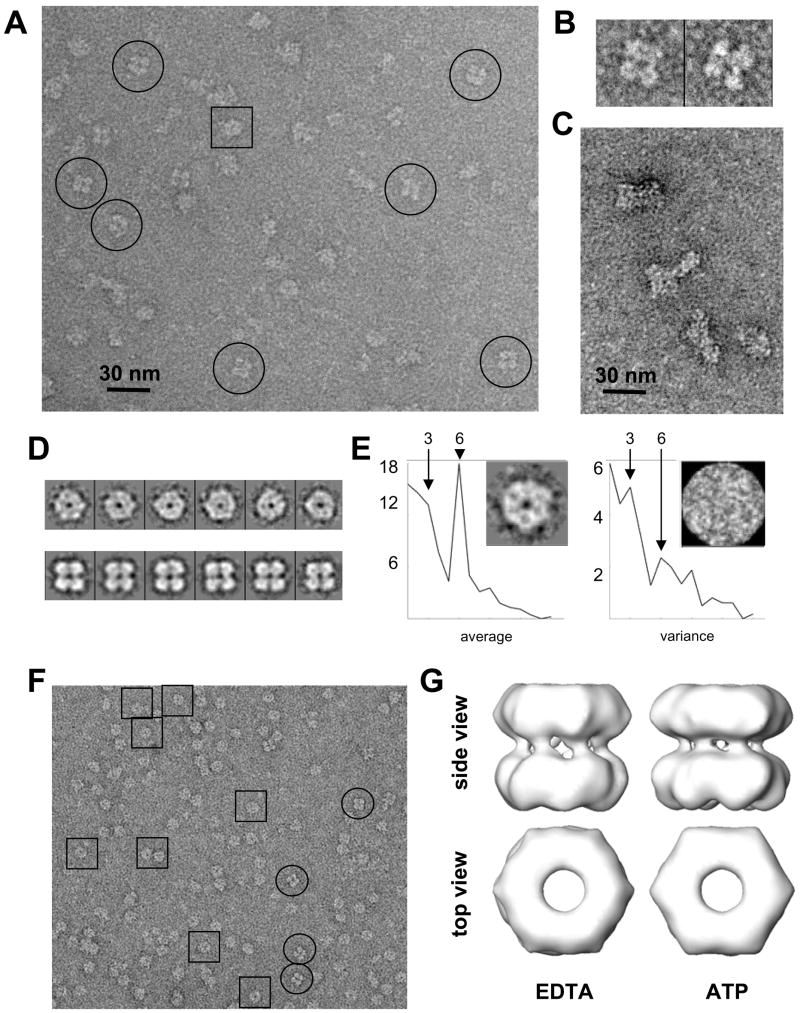
Figure 2. Negatively stained structure of pontin/reptin.
(A) Electron micrograph of chromatographic peak 2 using negative staining agents. Typical side views are highlighted within circles. Scale bar represents 30 nm.
(B) Two typical side views to highlight the asymmetry between the two rings.
(C) Electron micrograph of chromatographic peak 1 using negative staining agents. Scale bar represents 30 nm.
(D) Gallery of reference-free 2D averages obtained for the images in (A) and corresponding to top (top lane) and side views (bottom lane) of the complex.
(E) Analysis of the rotational symmetry of the top particles. The left panel displays the average power-spectrum and the 2D average whereas the right panel shows the 2D variance. Note that the scale of the variance is much lower.
(F) Electron micrograph of a chromatographic fraction similar to that shown in (A), but when the sample was incubated and the column was calibrated with ATP-Mg2+. Selected side and top views are highlighted within circles and squares respectively.
(G) 3D structure of negatively stained pontin/reptin complexes purified by size exclusion chromatography in the presence of EDTA or ATP.
Several thousand images from the micrographs obtained for the double-ring oligomer after negative staining were computationally selected to analyze more systematically these views by a meticulous classification using a combination of reference-free averaging, rotational symmetry analysis and neural networks methods (Supplementary Figure S2; see methods for details). We obtained a collection of reference-free class averages that divided the data-set into circular images and four-petal square-shaped averages (Figure 2D). Circular views showed a high component of 6-fold rotational symmetry (with low variance at this symmetry) (Figure 2E) confirming that the pontin/reptin oligomers were arranged as hexamers. Top views were nevertheless a patent minority in this dataset which showed a marked preponderance of side views.
We studied the low resolution 3D structure of the pontin/reptin complex using negative staining. For this purpose, the sample was extensively dialyzed against a buffer containing EDTA and then loaded in a gel filtration chromatography equilibrated with the same buffer. In another experiment, the EDTA dialyzed sample was dialyzed again in a buffer containing Mg2+, incubated with ATP and run in a size exclusion column equilibrated with ATP- Mg2+. The peaks for the oligomeric fraction were observed in the microscope. Interestingly, whereas the EDTA-dialyzed sample produced images similar to the original preparation (not shown), the ATP-incubated complexes revealed a vast increase in the number of top views, being now side views a minority (Figure 2F). This is a strong indication that conformational changes have been induced upon nucleotide binding that modify the view preference during adsorption. Thousands of side view images in both experimental conditions were extracted and subjected to angular refinement, where an initial template volume was built using average top and side views and assuming a 6-fold rotational symmetry. Only the side views were used for the refinement, the standard methodology for image processing of double-ring oligomers. The refined reconstructions revealed that each ring was made of a compact density without clear separation between subunits and a flat top surface (Figure 2G), characteristics shared by the atomic structure of the human pontin hexamer (Matias et al., 2006). No significant conformational differences were found after incubation with ATP at low resolution, at least as reflected in the side views of double rings.
The 3D cryo-EM reconstruction of pontin/reptin reveals an asymmetric double ring enclosing a large cavity
In order to define a higher resolution description of the pontin/reptin oligomer, fresh fractions collected from the chromatographic peak 2 when performed in the absence of any nucleotide (Figure 1A) were rapidly frozen in liquid ethane and observed at liquid nitrogen temperatures (cryo-EM) (Figure 3A). The sample revealed in cryo-EM similar views to those found using staining agents with a clear preponderance of four-petal side views (Figure 3A, within circles). More than 9000 particles were selected and prepared for angular refinement methods (following the scheme summarized in Supplementary Figure S3). Initially, the 6-fold rotational symmetry (c6) found in top views and in the previously resolved EM and X-ray structures was assumed during image processing to build an initial template volume, which was the input for further refinement. Unexpectedly, the resulting reconstruction revealed a double-ring protein where each ring displayed a significant dissimilar degree of resolution and each ring behaved differently in response to changing the threshold for rendering. We suspected this could be a reflection of heterogeneity in our data set, either from genuine structural conformational variability or data quality, leading us to explore the source of this heterogeneity. First, we extensively evaluated the possibility of other symmetries in the complex as the roots of such heterogeneity, such as d6 (this is, the two rings showing identical structure), c3 and c2 or even no symmetry at all. Also, we tested whether each ring could display a different rotational symmetry by performing several supervised classifications (results not shown). After all these analyses, we found that only 6-fold rotational symmetry (c6) was compatible with the data set, prompting us to look for other possible causes of heterogeneity. To this end the whole data set was processed allowing the sorting of the images of the particles into three possible output reconstructions. Once the muti-reference refinement converged, particles associated with each model were split and processed independently from scratch. This procedure resulted in the sorting of particles into three classes containing 33.5 %, 34.5 % and 32 % of the total images. Class 1 mainly comprised particles corresponding with un-conventional views in between a “typical” top and a “typical” side view (Figure 3B). Class 3 included particles with a very low signal to noise ratio. Classes 1 and 3 seemed to correspond with images with a high possibility of being incorrectly classified and aligned, since they gave rise to different solutions depending on the bias introduced by the initial references (results not shown). Hence, these particles seemed responsible for the defects in the initial reconstruction performed with the whole data set. Conversely, class 2 was enriched in the well characterized four-petal views corresponding to rotations along the longitudinal axis of the complex. These particles were then separated from the rest of the data and processed completely independently from scratch (Figure 3C) to derive a 3D reconstruction which did not show any of the defects previously observed. Therefore, this classification procedure allowed us to isolate a self-consistent sub-set of the data by removing either noisy images or those of molecules with conformations different to the major species (further controls detailed in Methods section).
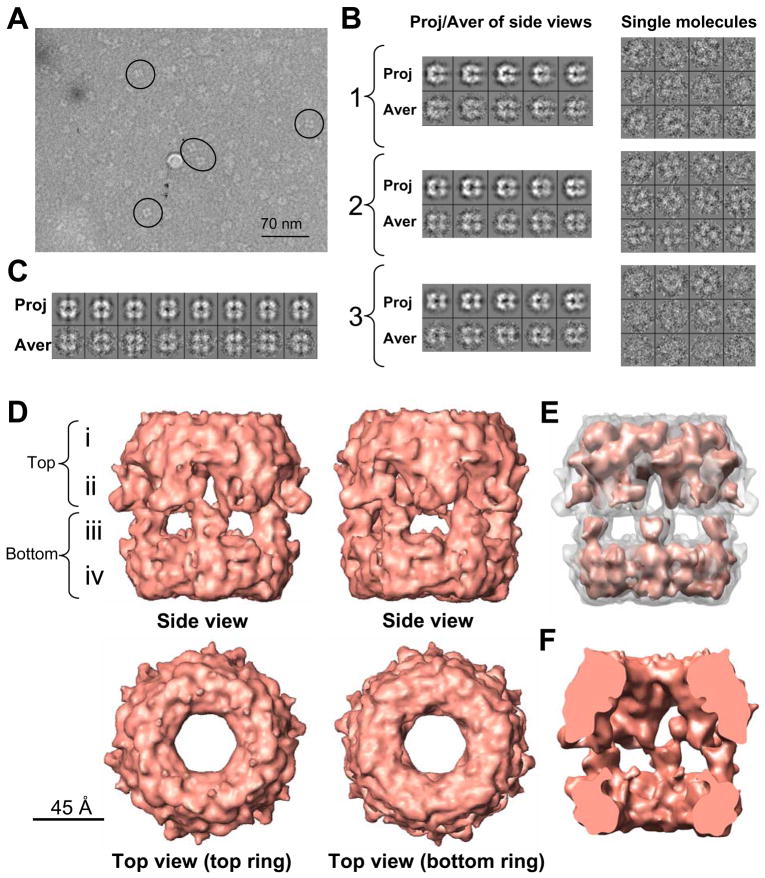
Figure 3. Cryo-EM structure of pontin/reptin.
(A) Cryo-electron micrograph with typical side views within circles.
(B) Typical projections and 2D averages obtained after refinement of each of the three subsets obtained after splitting the total data set (see text). A gallery of selected particles corresponding to the class averages displayed for each sub-group is shown at the right panels.
(C) Gallery of projections and 2D averages obtained after the independent refinement of the data from the selected sub-group (number 2).
(D) Two side views of the cryo-EM structure of pontin/reptin rotated around their longitudinal axis (top row) and top views of the reconstruction (bottom row). The reconstruction has been rendered to a threshold representing roughly 75 % of its total mass in order to highlight the structural features. Scale bar represents 4.5 nm.
(E) Volume rendered at the threshold used in (D) as a white transparency but containing the same volume rendered at a higher threshold representing around 25% of the protein mass.
(F) Cut open images of the side view displayed above.
The 3D reconstruction of the pontin/reptin complex at ~13 Å resolution (Supplementary Figure S4) revealed a dodecamer containing two rings, each showing a distinct conformation, the top ring being slightly wider than the bottom ring (Figure 3D). The complex showed a height of ~130 Å and a diameter ranging from ~90 Å at the narrower end and ~135 Å at the widest region. Any rendering threshold maintaining the contact between the protein densities produced a volume compatible with a ~650 kDa complex, assuming an average density for proteins of 1.35 g/ml. Both rings defined a large internal closed cavity (Figure 3F) communicating with the outside by central holes in each ring and by lateral entrances distributed around the equator of the double-ring. When this reconstruction was inspected from its side, the double hexamer revealed four differentiated layers of density, i–iv (Figure 3D). The outer top and bottom layers (i and iv) each consisted of a compact ring likely comprising the core domains of two Rvb hexamers whereas the centre of the oligomer resolved in two distinct regions (ii and iii). Layer iii was mainly made of lateral holes separated by six thin protein densities projecting towards the top while layer ii contained six large masses separated by small holes. Therefore, the two Rvb rings were interacting back to back in an apparent asymmetric fashion. This asymmetry and the distinct morphology of each ring was genuine since it appeared during the first steps of refinement even if complete symmetry between the two rings was used as bias in the initial template reference (d6 symmetry). Rotations along the longitudinal axis of the double-ring would be responsible of the asymmetric and symmetric side views detected by negative staining (compare the two side views in Figure 3D).
Pontin and reptin can form homo-oligomers
The homo or hetero-oligomeric nature of the pontin/reptin rings remains a major unsolved issue. While performing gel filtration analysis of a pontin/His-reptin preparation kept for long time on storage, we found out that the chromatographic peak corresponding to assembled oligomers was enriched in pontin (Figure 4A). Very importantly, gel filtration could discriminate between assembled oligomers and free subunits and the oligomers appeared in the microscope as typical “four-petal” side views and also top views (Figure 4B). Since only the His-tag in reptin was used for purification, pontin and reptin were initially equimolar in the preparation (Figure 1A, peak 2). Therefore, the most likely explanation for the preponderance of pontin in the oligomeric fraction is the disassembling of some double-rings into single-rings and free subunits. The gel filtration column had not sufficient resolving power to discriminate double and single rings as perfectly separated peaks, but double rings could be unambiguously defined by the aspect of their “four-petal” side view in the microscope (Figure 4B, circles). On the other hand, top views could belong to either double or single ring oligomers (Figure 4B, squares). Furthermore, views were found in the micrographs with dimensions compatible with a putative side view of a single-ring oligomer (Figure 4B–4C). The much greatest abundance of pontin in the oligomeric fraction could only be consistent with the existence of homo-oligomers. Hetero-oligomeric rings containing equimolar pontin and reptin would always produce equimolar bands in the SDS gel of size-exclusion purified oligomers, regardless of being single or double rings. On the other hand, homo-oligomeric rings can produce a non-equimolar fraction after the disruption of the initial double-ring complex if some protein stays as free subunits, as we observed (Figure 4A).
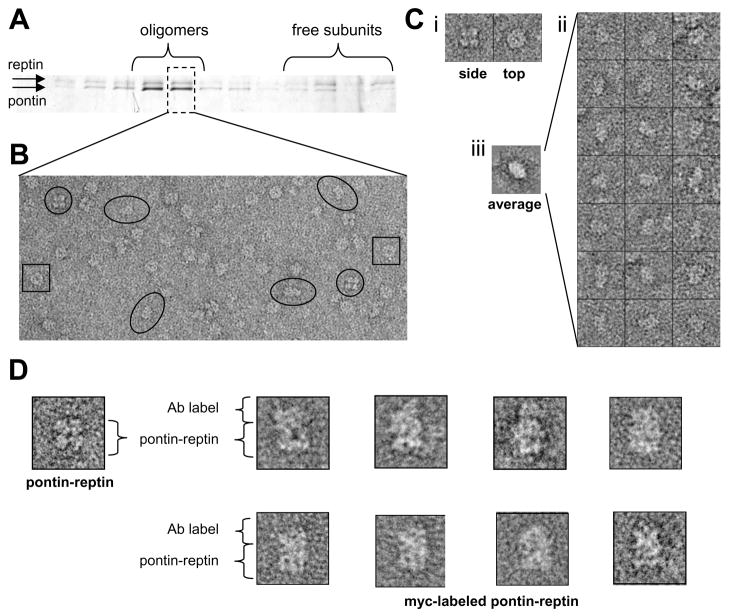
Figure 4. Pontin and reptin can form homo-oligomers.
(A) Elution profile of the size-exclusion chromatography of a pontin/reptin preparation after long-term storage. Similar results were found for columns calibrated with EDTA (not shown) and ATP (shown).
(B) EM field of a selected fraction of the chromatography. The sample incubated with ATP produced a better distribution of distinct views in the microscope, where “four-petal” side views (circles), top views (squares) and putative side views of single rings (ellipsoids) were detected.
(C) Analysis of the views present in a fraction of the chromatography corresponding to oligomers revealed both top and side views (i). Some particles showed dimensions compatible with side views for a single-ring oligomer (ii), for which a reference-free 2D average was obtained (iii).
(D) Selected particles of pontin/reptin-myc complexes labeled with an anti-myc antibody
To further investigate this issue, we performed a labeling experiment to locate reptin in either one or the two rings of the double-ring complex, to indicate the presence of homo or hetero-oligomeric rings respectively. The N-terminal His-tag was not adequate for such approach because it locates at the center of the complex according to our model (see following section below), not allowing a discrimination of the label between the two rings. We therefore purified a pontin/reptin complex containing a C-terminal myc-tag in reptin which, according to the model proposed below, should be pointing outwards. We incubated these complexes with saturating amounts of purified anti-myc antibody and the immune-complexes were repurified and observed in the microscope. Whereas only a few characteristic undistorted double rings images were observed, these were always decorated with putative antibody density in just one end of the double-ring side view, suggesting that all reptin monomers locate at the same ring (Figure 4D).
The core domains of pontin/reptin show conformational differences with the atomic structure of human pontin
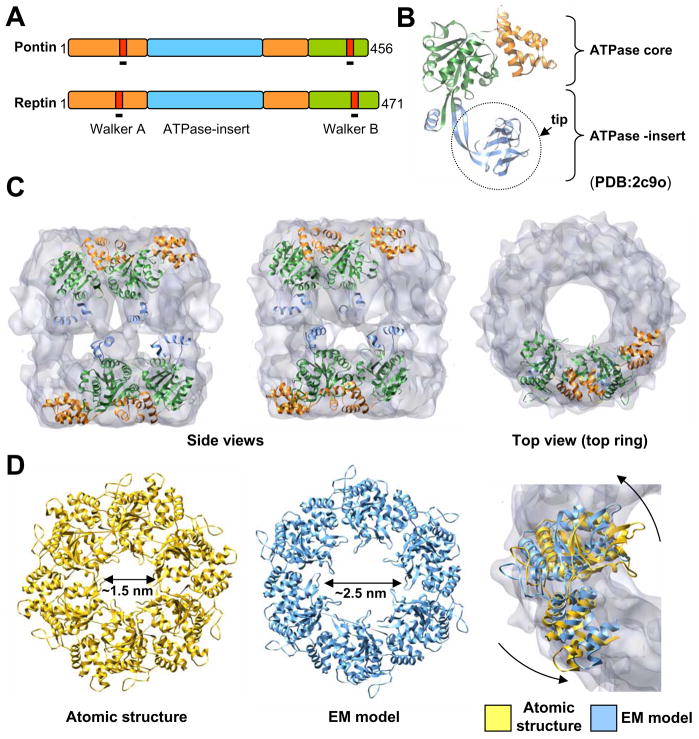
Due to the high homology between the human and yeast proteins (Supplementary Figure S1), we selected the atomic structure of human pontin (Figure 5A–5B) (Matias et al., 2006) to fit into the 3D reconstruction. The “tips” of the ATPase-insert domains (residues 123–237) (Figure 5B) were removed of the analysis due to its expected flexibility. Initially we performed a rigid-body fit using the hexameric atomic structure of pontin for an exhaustive 6-dimensional search to explore all its possible orientations within the whole EM map (Garzon et al., 2007). The best solutions were then further refined allowing the monomers to rotate and translate inside the EM density but maintaining the 6-fold symmetry (see Methods). These experiments unambiguously located the core domains of the Rvb proteins within the ends of the molecule (cross-correlation coefficients 0.86 and 0.79 for each ring) (Figure 5C, only two monomers per ring shown for clarity). The complete human pontin structure could not fit the EM density since the conformation of the ATPase-insert domains in the crystal structure did not match that of the pontin/reptin complex. The comparison between the EM density and the fitted atomic structure did not permit the definition of the absolute handedness of the map based on differences in correlation coefficients and one of the hands was selected for representation (similar results were obtained with the other hand which is shown in Supplementary Figure 5).
Figure 5. Location and conformational changes in the core domains of pontin/reptin.
(A) Primary structure and domains present in pontin and reptin. Color codes for domains are maintained throughout the figure.
(B) Atomic structure of human pontin (PDB file 2c9o) (Matias et al., 2006). Color codes as in (A).
(C) Fitting of the core domains of the atomic structure of Rvb1p into the EM density. For clarity, only two consecutive monomers are shown.
(D) Conformational differences between the atomic structure of human pontin (yellow color) and the atomic model after fitting into the EM density (blue color). At the right, the two structures are represented superimposed on to the EM density of one of the rings.
These fitting experiments revealed that the monomers of each ring had significantly rotated outwards and counter clockwise around the longitudinal axis by ~5 to ~10 degrees compared to the crystal structure (Figure 5D). Such movements were found to impact the shape and dimensions of the internal cavity, but more importantly the central openings of the top and bottom rings were now slightly larger in the EM structure (> 2.5 nm in diameter) than in the crystal structure (around 1.5 nm in diameter) (Matias et al., 2006).
Pontin/reptin double ring is maintained by interactions between the ATPase-insert domains of opposite rings
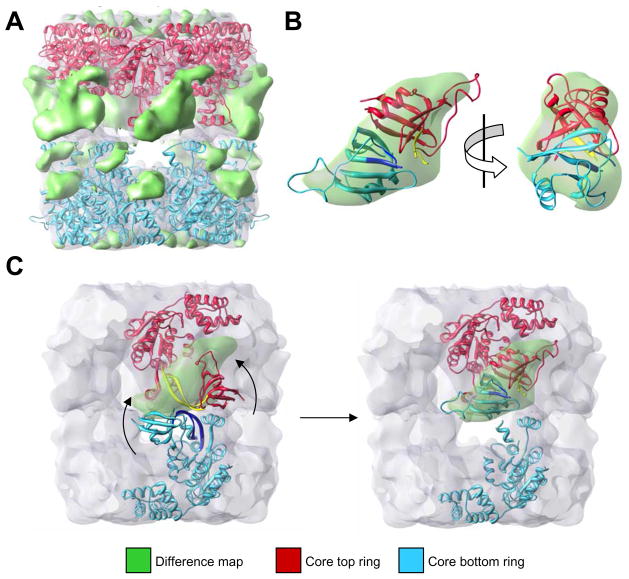
To identify a more precise location of the ATPase-insert domains, the density of the fitted atomic core domains (Figure 5) was subtracted from the experimental EM map after adequate normalization and scaling. The difference map obtained mostly represented those densities of the whole pontin/reptin complex not accounted by the core domains of both rings (Figure 6A and Supplementary Figure S5, green color). Differences present at the ends of the rings were minor and their location suggested that they corresponded with areas of the protein not solved in the atomic structure of pontin. Importantly, the bulk of the differences were located in the equatorial regions of the top ring, and these densities could easily accommodate the ends of ATPase-insert domains from both rings by manual fitting (Figure 6B). Relatively simple motions of the ATPase-insert domains would be sufficient to account for this rearrangement (Figure 6C).
Figure 6. Location of the ATPase-insert domains in the pontin/reptin double-ring.
(A) Difference map between the EM reconstruction and the core domains of the atomic structure of pontin after fitting within the EM structure, shown as a green solid density over the EM reconstruction in transparency. The fitting results of the atomic model for the core domains are also displayed.
(B) Two orthogonal views of the difference map density of one pair of top and bottom subunits where the tips of the ATPase-insert domains from both rings have been fitted.
(C) Modeling of the speculative conformational changes required to adapt the conformation of the tips of the ATPase-insert domains as observed in the crystal structure (Matias et al., 2006) to the experimental EM density. The proposed region acting as hinge is colored in dark blue and yellow.
Discussion
We now propose a high resolution structural model of the pontin/reptin complex and its implications in the functional properties of this chromatin remodeling factor. The complex is formed by the back to back interaction between two hexameric rings. The core domains of each ring are facing outwards and they probably have a very similar structure in both rings. The integrity of the complex is maintained through interactions between the ATPase-insert domains of each ring. The monomers have rotated outwards compared to the crystal structure of human pontin (Matias et al., 2006), leaving a larger central hole which could now be sufficient to accommodate both a single or double stranded DNA. These conformational changes could be caused by the structural strain introduced as a result of the close interaction between the two rings mediated by the ATPase-insert domains (Figure 6C).
The cryoEM structure of the pontin/reptin complex reveals that each ring displays a distinct conformation. Importantly, this asymmetry between rings is not caused by large differences in the conformation of the core domains, but it is mostly due to the way the two rings interact. The tips of the ATPase-insert domains of both rings appear in contact in the proximity of one of the rings that consequently adopts an apparently wider profile (layer ii) whereas the proximity of the other ring (layer iii) contains six lateral holes. Relatively minor torsions of the ATPase-insert domains around a flexible beta strand pair functioning as a hinge would be certainly plausible since these domains seem inherently flexible (Matias et al., 2006).
Interestingly, the ATPase-insert domain of eukaryotic pontin/reptin was proposed to act as a novel DNA binding module based on its close resemblance with Replication Protein A (RPA), a ssDNA binding protein, and it was shown to bind both single and double-stranded DNA in electrophoretic mobility shift assays (Matias et al., 2006). The configuration we find in the pontin/reptin complex implies that some regulation of DNA binding could be potentially bound to the formation of the double-ring. Nevertheless, it will not be surprising if the DNA binding activity of pontin/reptin is also regulated by the oligomerization state of these proteins in the context of the complete remodeling complex.
Many AAA+ proteins are typically organized in the form of double rings containing the AAA+ rings facing outwards to provide a mechanical motor whereas functional domains face inwards (Yamada et al., 2002). The structural model we propose for the pontin/reptin complex agrees with this general structural framework. Still, there is not a clear function for these ATPase hexameric rings within their remodeling complexes. Mounting evidence is accumulating to suggest that maybe a major role of pontin/reptin could be the function as a platform for several protein-protein and nucleic acid-protein interactions (Jonsson et al., 2004; McKeegan et al., 2007; Watkins et al., 2004). Rvb1p has been shown to recruit Arp5 to yeast Ino80.com (Jonsson et al., 2004) and recent reports have added further evidence on this direction revealing a role of these proteins in the assembly of small nucleolar RNAs (Boulon et al., 2008; McKeegan et al., 2007; Zhao et al., 2008), in the recruitment of Tip49 to complexes with histone acetylase transferase activity (Jha et al., 2008) and in the assembly of the telomerase holoenzyme (Venteicher et al., 2008).
The double ring pontin/reptin complex contains equimolar amounts of each of the two proteins and the possibility that each protein forms an independent hexamer or that each ring of the complex contains three monomers of each protein should be considered. The high homology between pontin and reptin implies that they cannot be distinguished at the resolution of these EM studies. Homo-oligomeric hexamers of human pontin have been assembled in vitro and their structure solved at atomic resolution proving that this protein can by itself form a hexameric oligomer (Matias et al., 2006). Several of the experiments we have performed here suggest that each ring is composed of just one of the proteins forming homo-oligomeric hexamers, although a fully definitive and conclusive proof is still lacking. A recent report has proposed that recombinant yeast pontin and reptin assemble into a hetero-oligomeric single-ring (Gribun et al., 2008). If analyzed carefully, the original experiments by these authors described that only single-rings were assembled in vitro when incubating both proteins together, but a formal proof of the hetero-oligomeric nature of such rings was missing. Given the close sequence similarity between pontin and reptin, these proteins could potentially interact into homo- or hetero-oligomeric single or double rings depending on the actual experimental conditions. Such behaviors have been for instance described for the thermosome, a chaperon built by two highly related subunits (α, β) (Nitsch et al., 1997). Nevertheless, it is maybe possible that the double and single hexameric rings, and even homo and hetero-oligomers, could represent different functional states of these proteins (see later).
A low resolution structure of the human pontin/reptin complex derived from negatively stained images has been reported recently (accession code EMD-1317 at http://www.ebi.ac.uk/msd-srv/emsearch/form) (Matias et al., 2006), which is strikingly distinct to our reconstruction. The four-petal EM images of the yeast pontin/reptin complex (this work) are reminiscent of the images from bacterial RuvB (Chen et al., 2002) whilst they strongly differ from the EM images of the published human double-ring complex (Puri et al., 2007). The atomic structure of human pontin reveals a continuous ring with tightly packed monomers and whose top surface is remarkably flat (Matias et al., 2006). These features are shared by both rings of the yeast pontin/reptin we have solved here (Figure 3D) whereas they are incompatible with the published human pontin/reptin complex, and such difference between both structures cannot be only explained by the disparity in resolution.
A possibility to reconcile all the different structures published could be the existence of several conformations of the pontin/reptin complexes where double and single rings and homo and hetero-oligomers can form. Several functions have been ascribed by several authors to free subunits, single and double-rings of the Rvb proteins. Given the complex set of events that are controlled by pontin and reptin, it is likely these proteins transit through several functional states comprising either different conformations of the proteins or different forms of oligomerization. The different 3D structures solved for the pontin/reptin complex, including ours, could represent distinct structural transitions helping these proteins integrate the functions of the AAA+ motor domains, nucleic acid binding activities and a surface for protein-protein interactions into one complex.
Experimental Procedures
Purification of the pontin/reptin complex
Yeast pontin/His-reptin complexes were cloned and expressed using recombinant baculovirus and a His-tag at the N-terminus of reptin (Jonsson et al., 2004). Size exclusion chromatography was performed using a Superdex 200 PC 3.2/30 column equilibrated with 25mM TrisHCl pH8.0, 125mM NaCl. All purification steps were performed in the absence of nucleotides or magnesium. In a different set of experiments, the pontin/His-reptin complex was dialyzed against a buffer containing 2mM EDTA and loaded in the same gel filtration column. Also, the EDTA dialyzed sample was again dialyzed against a buffer containing magnesium, incubated with ATP and purified in a size exclusion column equilibrated with 15mM MgCl2 and 1mM ATP.
ATPase activity of pontin/reptin
ATPase assays with the purified sample contained variable amounts of protein (from 4 μto 4 nM monomer concentration) and were performed in a buffer containing 50 mM Tris-HCl (pH 7.5), 75 mM NaCl, 10 mM Mg(AcOH)2, 0.05 mg/ml BSA, 1 mM DTT, 1 mM ATP and 100 nM [γ-32P]ATP (specific activity 3000 Ci/mmol), for 1 hr at 25°C. Reactions were stopped by adding 2 μl of 0.5 M EDTA (pH 8.0). ATP hydrolysis was determined by thin-layered chromatography (TLC). Aliquots (1 μl) from each reaction were spotted onto PEI cellulose TLC plates and the substrate ([γ-32P]ATP) was separated from the product (γ-32Pi) with ATPase running buffer (200 mM potassium-phosphate buffer, pH 7.5). Plates were dried and quantified on a Phosphorimager. In a separate experiment, pontin/reptin complexes were processed with TEV protease to remove the His tag and separated by gel filtration. The ATPase activity of aliquots of the fractions (4μl of 50μl of total volume) was assayed similarly but reactions were performed in a 10 μl volume for 16 hrs at 25°C.
Electron microscopy and 3D reconstruction of negatively stained pontin/reptin
Fractions corresponding with oligomers were applied to carbon-coated grids after glow-discharge and negatively stained with 2% uranyl acetate or 1% uranyl formate. Observations were performed in a JEOL 1230 electron microscope operating at 100kV and micrographs were recorded under low dose conditions at a nominal magnification of 50,000. Micrographs were digitized using a MINOLTA Dimage Scan Multi Pro scanner at 2400 dpi and averaged to 4.2 Å/pixel. The contrast transfer function of the microscope for each micrograph was estimated using CTFFIND3 (Mindell and Grigorieff, 2003) and corrected. Several thousands of individual particles for each experiment were extracted using the “boxer” program implemented in EMAN (Ludtke et al., 1999). The raw data was analyzed using reference-free alignment and classification methods implemented in EMAN (Ludtke et al., 1999) and maximum likelihood (Scheres et al., 2005). Neuronal networks classification and analysis of the rotational symmetry in the data set were performed using procedures in XMIPP (Sorzano et al., 2004). An initial 3D template for refinement was built using an average of the top and side views of the complex and the 6-fold symmetry of the molecule.
Cryo-electron microscopy and 3D reconstruction of pontin/reptin
A few microliters of the chromatographic peak 2 (in the absence of nucleotides) freshly after collecting the fractions were applied to Quantifoil R 2/2 holy grids which had been coated with a thin layer of carbon. The sample was then vitrified and observed in a FEI Tecnai G2 operated at 200 kV and equipped with a Gatan liquid nitrogen specimen holder. Images were recorded at a nominal magnification of 50,000 and digitized to a final 2.12 Å/pixel. 9316 particles were extracted and the contrast transfer function of the microscope corrected by flipping phases. Particles were then subjected to 3D refinement using EMAN (Ludtke et al., 1999). The starting 3D reference was built using “startcsym” command and 6-fold rotational symmetry. The two rings of the initial reference were then symmetrized to avoid a bias towards an asymmetric structure. This refined reconstruction revealed significantly different levels of resolution in each ring. We suspected that the source of this difference could be particles in the data set in either a different conformation or with a low signal/noise ratio. We split the data set in three subgroups using the multi-refine command in EMAN being only one self-consistent. The final reconstruction was performed using one homogeneous class of particles, 6-fold symmetry and using only side views. We validated the classification performed by refining the class 2 sub-data set using as initial 3D template the reconstruction of the other sub-groups that provided a strong initial bias towards the incorrect conformation. In all cases, the final model matched the data set and not the reference volume used, supporting the classification. The resolution of the 3D map was estimated by Fourier Shell Correlation (FSC) to be 13.6 Å, 11.10 Å, and 9.8 Å according to a correlation coefficient of 0.5, 0.33 and 0.145, respectively. The high-frequency components of the final reconstruction above 13.6 Å resolution were eliminated by low-pass filtering.
The absolute handedness of the reconstruction could not be defined at the resolution of the map since the cross-correlation values for the fitting of atomic structure of the human pontin into the EM map were similar at both hands (correlation differences obtained for opposite hands < 0.05). The application of the methodology proposed by Rosenthal and Henderson (Rosenthal and Henderson, 2003) was evaluated and found not possible since similar cross-correlation values were obtained when comparing projections of the 3D reconstruction (filtered at the resolution of a negative stain experiment) at both possible hands.
Fitting of the atomic structure into the EM map
We performed a rigid-body fit of the human pontin hexameric ring (PDB file 2c9o) in each ring of the EM reconstruction using ADP_EM (Garzon et al., 2007) To avoid interferences of flexible segments of the molecule we removed the “tips” of the ATPase-insert domains (residues 123–237) and the terminal C-terminal helix (438–449). From the best scores found, we carried out an additional refinement step by freely moving a single monomer using a multidimensional Powel optimization routine. Subsequently, 6-fold rotational symmetry was applied to the refined monomer to complete a hexameric ring but discarding those solutions producing steric clashes with contiguous monomers. This procedure allowed the monomers to rotate and translate inside the density improving the scores from 0.72 to 0.79 and from 0.77 to 0.86 for top and bottom rings respectively.
The discrepancy between the best fitting model and the EM map were computed using standard procedures after filtering and scaling the atomic model according to the experimental map. Six densities were localized which could be attributed to the flexible tips removed for the rigid body fitting. To substantiate this hypothesis we found that the ends of the ATPase-insert domains from both rings could be manually fitted into this difference map.
Labeling of Myc-tagged reptin
A monoclonal anti-myc antibody (Sigma) was re-purified by size-exclusion chromatography to eliminate aggregates. The peak of the purified antibody was analyzed in the microscope revealing a clean preparation of antibodies showing no aggregation. Freshly purified antibody was then immediately incubated with a pontin/reptin complex containing a myc-tagged at the C-terminus of reptin (Jonsson et al., 2004), under saturating amounts of antibody for 30 min at 4°C. The incubation was then loaded in a size exclusion chromatography, and the fraction corresponding to the immuno-complex was observed in the microscope.
Supplementary Material
Acknowledgments
This work has been supported by projects SAF2005-00775 and SAF2008-00451 (OL), BFU2006-02907 (SA) and BFU2007-65977/BMC (PC) from the Spanish Ministry of Science, CAM S-BIO-0214-2006 (OL and PC) from the Autonomous Region of Madrid and CA89406 from the US National Institutes of Health (AD). Llorca’s group is additionally supported by project RD06/0020/1001 of the “Red Temática Investigación Cooperativa en Cáncer (RTICC)” and by the Human Frontiers Science Program Organization (RGP39/2008). EAP is supported by a contract from the Autonomous Region of Madrid. We are very thankful for help provided by Jaime Martin-Benito, Gabriel Piedrafita, Javier Varela, Emilia Aporta and Dolores Alonso. We are grateful to the support of the computing resources from the Galicia Supercomputer Centre (CESGA), and the Barcelona Supercomputing Centre, Spain.
Footnotes
The cryo-EM map of yeast pontin/reptin complex has been deposited in the 3D EM database (http://www.ebi.ac.uk/msd/) under accession code ****** (to be provided upon acceptance).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Yu X, Egelman EH. The hexameric ring structure of the Escherichia coli RuvB branch migration protein. J Mol Biol. 2002;319:587–591. doi: 10.1016/S0022-2836(02)00353-4. [DOI] [PubMed] [Google Scholar]
- Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Garzon JI, Kovacs J, Abagyan R, Chacon P. ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics. 2007;23:427–433. doi: 10.1093/bioinformatics/btl625. [DOI] [PubMed] [Google Scholar]
- Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376:1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, Lee MH, Choi SJ, Kim KI, Kim SI, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A Dynamic Scaffold of Pre-snoRNP Factors Facilitates Human Box C/D snoRNP Assembly. Mol Cell Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Nitsch M, Klumpp M, Lupas A, Baumeister W. The thermosome: alternating alpha and beta-subunits within the chaperonin of the archaeon Thermoplasma acidophilum. J Mol Biol. 1997;267:142–149. doi: 10.1006/jmbi.1996.0849. [DOI] [PubMed] [Google Scholar]
- Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J Mol Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Carazo JM. Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics. 2005;21(Suppl 2):ii243–244. doi: 10.1093/bioinformatics/bti1140. [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/Helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Yamada K, Miyata T, Tsuchiya D, Oyama T, Fujiwara Y, Ohnishi T, Iwasaki H, Shinagawa H, Ariyoshi M, Mayanagi K, Morikawa K. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell. 2002;10:671–681. doi: 10.1016/s1097-2765(02)00641-x. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.