Abstract
Background
Human epidermal growth factor receptor 2 (HER2) fluorescence in situ hybridization (FISH) is a quantitative assay for selecting breast cancer patients for trastuzumab therapy. However, current HER2 FISH procedures are labor intensive, manual methods that require skilled technologists and specialized fluorescence microscopy. Furthermore, FISH slides cannot be archived for long term storage and review. Our objective was to develop an automated brightfield double in situ hybridization (BDISH) application for HER2 gene and chromosome 17 centromere (CEN 17) and test the assay performance with dual color HER2 FISH evaluated breast carcinomas.
Methods
The BDISH assay was developed with the nick translated dinitrophenyl (DNP)-labeled HER2 DNA probe and DNP-labeled CEN 17 oligoprobe on the Ventana BenchMark® XT slide processing system. Detection of HER2 and CEN 17 signals was accomplished with the silver acetate, hydroquinone, and H2O2 reaction with horseradish peroxidase (HRP) and the fast red and naphthol phosphate reaction with alkaline phosphatise (AP), respectively. The BDISH specificity was optimized with formalin-fixed, paraffin-embedded xenograft tumors, MCF7 (non-amplified HER2 gene) and BT-474 (amplified HER2 gene). Then, the BDISH performance was evaluated with 94 routinely processed breast cancer tissues. Interpretation of HER2 and CEN 17 BDISH slides was conducted by 4 observers using a conventional brightfield microscope without oil immersion objectives.
Results
Sequential hybridization and signal detection for HER2 and CEN 17 ISH demonstrated both DNA targets in the same cells. HER2 signals were visualized as discrete black metallic silver dots while CEN 17 signals were detected as slightly larger red dots. Our study demonstrated a high consensus concordance between HER2 FISH and BDISH results of clinical breast carcinoma cases based on the historical scoring method (98.9%, Simple Kappa = 0.9736, 95% CI = 0.9222 – 1.0000) and the ASCO/CAP scoring method with the FISH equivocal cases (95.7%, Simple Kappa = 0.8993%, 95% CI = 0.8068 – 0.9919) and without the FISH equivocal cases (100%, Simple Kappa = 1.0000%, 95% CI = 1.0000 – 1.0000).
Conclusion
Automated BDISH applications for HER2 and CEN 17 targets were successfully developed and it might be able to replace manual two-color HER2 FISH methods. The application also has the potential to be used for other gene targets. The use of BDISH technology allows the simultaneous analyses of two DNA targets within the context of tissue morphological observation.
Background
The human epidermal growth factor receptor 2 (HER2) oncogene, located on the long arm of chromosome 17 (17q12-q21), is over-expressed or amplified in approximately 20% of breast carcinoma cases [1,2]. HER2 status in breast cancer is used as a prognostic factor, a predictive factor, and a therapy selection factor [3] for the humanized monoclonal antibody trastuzumab (Herceptin®; Genentech), which is an FDA approved drug for use as monotherapy or combined chemotherapy for treatment of breast cancer patients with amplified HER2 status. Trastuzumab adjuvant treatment for early HER2 positive breast cancer is effective for improving patient survival and cost-effectiveness analyses of such treatment have shown acceptable ratios [4-7]. However, there is a negative aspect to trastuzumab therapy, namely cardiac toxicity [3], which is possibly due to myocardial HER2 gene over-expression associated with anthracycline treatment [8] and substantial cost.
Quantitative HER2 fluorescence in situ hybridization (FISH) analyses for detecting HER2 gene amplification and semi-quantitative HER2 immunohistochemistry (IHC) analyses for detecting over-expressed HER2 protein are performed to determine the HER2 status of breast cancer patients. The optimal scoring method for determination of HER2 gene status is the use of chromosome 17 centromere (CEN 17) enumeration for calculating the HER2/CEN 17 ratio [9]. One study showed that chromosome 17 polysomy (13%) and chromosome 17 monosomy (2%) were confirmed among 147 breast cancer cases with 2+ and 3+ HER2 IHC scores [10]. Also, chromosome 17 polysomy is a key prognosis indicator for breast cancer patients. Patients with chromosome 17 polysomy and no HER2 gene amplification have better prognosis compared to patients with HER2 gene amplification [11]. Dual color FISH for HER2 and CEN 17 targets is recommended especially for borderline IHC cases [12]. However, there are additional drawbacks to conducting HER2 FISH assays beyond the requirement for a specialized fluorescence microscope and the difficulty of preserving FISH signal during a long term storage. For example, HER2 FISH testing has exhibited a higher assay failure rate in the hands of some investigators when compared to HER2 IHC testing (5% vs. 0.08%), the FISH assay procedure time is longer than the IHC assay (36 hours vs. 4 hours), and the FISH interpretation time is longer than IHC interpretation time (7 minutes vs. 45 seconds) [1]. Another disadvantage of the FISH assay is the difficulty of correlating cytomorphological aspects of the tissue sample with the gene status [13]. Furthermore, tissue slides for the dual color FISH test are still processed manually in most laboratories, which practice can introduce human errors during the lengthy assay. In fact, FISH assays may not always be performed accurately [14]. An international HER2 proficiency testing study showed that there was 20% (4 out of 20 samples) discordance with HER2 FISH testing among 5 experienced laboratories [15]. On the other hand, some reports using proficiency testing surveys conducted by the College of American Pathologists have demonstrated a much higher concordance for FISH [16].
There are alternatives to FISH for determining HER2 gene status. The chromogenic in situ hybridization (CISH) assay using a DAB chromogen and H2O2 substrate system for horseradish peroxidase (HRP)-based signal detection has been evaluated and the value of this assay for assessing HER2 status has been demonstrated [12,17-22]. CISH slides can be interpreted using an ordinary brightfield microscope without oil-immersion lenses and can provide visible tissue morphology for correlation with the HER2 gene signal. However, with the current CISH method, the assessment of the HER2/CEN 17 ratio is conducted by enumerating HER2 and CEN 17 separately using two different tissue sections.
Other brightfield microscopy in situ hybridization (ISH) methods use autometallography and enzyme metallography: 1) Nanogold® with gold enhancement in situ hybridization (GOLDFISH) [23,24] and 2) enzyme metallography or silver in situ hybridization (SISH) [25-27]. The GOLDFISH procedure utilizes the tyramide signal amplification principle and produces large clusters for amplified HER2 gene signal. On the other hand, the SISH method produces discrete metallic silver black signals. Horseradish peroxidase (HRP) of the detection system reacts with silver acetate, hydroquinone, and H2O2 and deposits metallic silver particles at the reaction site. The reaction product can be seen as discrete black dots under a brightfield microscope. Advantages of SISH include the high sensitivity for detection of single gene copies, the high resolution for quantifying DNA targets, and the high contrast with tissue counterstaining for visual separation of the signal and tissue morphology [27].
Recently, an automated HER2 SISH assay was evaluated for assessing the inter-observer interpretative reproducibility of the HER2 gene status of 99 clinical cases when compared against the reference standard FISH results [28]. Overall concordance between dual color HER2 FISH and single color HER2 SISH was 96.0% (kappa = 0.754, 95% CI = 0.518–0.993) and the discrepancies were mainly observed among tumors with the heterogeneity of tumor cell populations [28]. Advantages of the SISH assay compared to the CISH assay include that the SISH assay produces signal clusters and separately visualized discrete black dots that are easier to count in the majority of clinical samples. With SISH, the endogenous gene copies present in non-neoplastic stromal cells are also routinely and reproducibly visualized. However, like the CISH assay, the detection of CEN 17 signal cannot be performed on the same tissue section. Thus, it would be ideal to visualize both HER2 and CEN 17 on the same tissue section like two-color FISH assays. Dual ISH staining for HER2 gene and CEN 17 would be beneficial for analyzing chromosome 17 aneusomy and for delineation of cases displaying genotypic intratumoral heterogeneity.
One prerequisite for testing HER2 status reproducibly is the use of automation for conducting the test in the same manner among different laboratories located in different parts of the world. Thus, as a step toward standardizing HER2 testing, our objective was to develop an automated brightfield double in situ hybridization (BDISH) assay for simultaneous detection of HER2 and CEN 17 DNA targets on formalin-fixed, paraffin-embedded breast cancer tissue samples. Using this method, HER2 status testing can be conducted in a simplified manner for more accurately identifying the patients who are eligible for trastuzumab therapy and potentially leading to the improvement of breast cancer patient care in the future.
Methods
Tissue samples
MCF7 and BT-474 xenograft tumors were utilized for optimizing the BDISH assay. MCF7 is a breast adenocarcinoma cell line with non-amplified HER2 status and BT-474 is a breast ductal carcinoma with amplified HER2 status (50–60 copies of HER2) and chromosome 17 polysomy [29]. Paraffin sections (4 μm) containing tissue cores of formalin-fixed, paraffin-embedded MCF7 and BT-474 xenograft tumors were placed onto Superfrost® Plus glass slides (Erie Scientific Company, Portsmouth, New Hampshire).
Ninety-four (94) breast cancer cases were used from the Cleveland Clinic Foundation and the Cleveland Clinic Lerner College of Medicine, Cleveland, OH, USA under IRB approved protocol. Tissue samples were routinely processed for paraffin-embedding after fixing with an alcoholic formalin fixative. All breast cancer cases had been previously tested for HER2 status by FISH using the PathVysion® HER-2 DNA Probe Kit (Abbott Molecular, Des Plaines, Illinois) at the Cleveland Clinic Foundation. However, it should be noted that non-consecutive tissue sections were used for FISH and BDISH analyses.
Brightfield in situ hybridization
The BenchMark® XT automated slide processing system (Ventana Medical Systems, Inc., Tucson, Arizona) was used for the optimization and performance evaluation of the BDISH assay for HER2 and CEN 17 DNA targets. A protocol was established so that the entire assay procedure consisting of baking, deparaffinization, pretreatment, hybridization, stringency wash, signal detection, and counterstaining was completed as a one-step fully automated assay. Paraffin tissue sections on glass slides were baked at 65°C for 20 minutes prior to the deparaffinization step with EZ Prep™ (Ventana) at 75°C for 16 minutes. Deparaffinized tissue sections were pretreated with a combination of heat treatment with Reaction Buffer (Tris-based pH 7.6 solution, Ventana) and ISH Protease 2 or ISH Protease 3 (Ventana) to unmask DNA targets. Pretreatment conditions were chosen for optimal signal to noise ratio and tissue morphology preservation for the xenograft control slides as well as clinical case tissue slides.
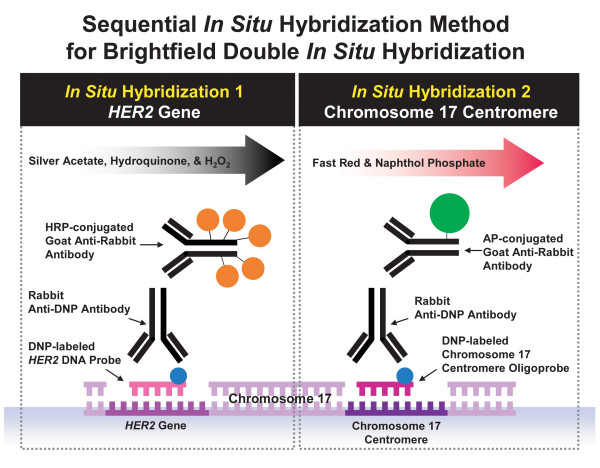
Sequential ISH procedures for HER2 and CEN 17 signal detection were conducted for a complete BDISH assay (Figure 1). Reaction Buffer was used for the washing steps during immunological detection. Liquid Coverslip™ (LCS, a hydrophobic reagent, Ventana) was used for controlling liquid evaporation throughout the assay. For HER2 gene detection, the INFORM® HER2 DNA Probe (Ventana), a dinitrophenyl (DNP)-labeled, nick-translated repeat deleted DNA probe was applied to the glass slide for co-denaturing the probe and target at 95°C. Then, the hybridization step was conducted at 52°C for 2 hours. After 3 stringency wash steps were performed at 72°C with 2× SCC (Ventana), tissue sections were incubated with monoclonal rabbit anti-DNP antibody (Ventana) for 20 minutes and then with HRP-conjugated anti-rabbit antibody for 16 minutes at 37°C. The metallic silver deposit for HER2 ISH signal was developed using silver acetate, hydroquinone, and H2O2 reaction in the presence of HRP using the ultraView™ SISH Detection Kit (Ventana). For CEN 17 detection, the INFORM Chromosome 17 Probe (Ventana), a DNP-labeled oligoprobe, was applied to the tissue sections, denatured at 95°C and hybridized at 44°C for 2 hours. Then, after 3 stringency wash steps at 59°C with 2× SSC, tissues were incubated with rabbit monoclonal anti-DNP antibody for 20 minutes and then with an alkaline phosphatase (AP)-conjugated anti-rabbit antibody for 12 minutes at 37°C. Finally, the signal for CEN 17 was visualized with a fast red and naphthol phosphate reaction using ultraView Red ISH Detection Kit. Diaminobenzidine (DAB) chromogen and H2O2 substrate reagents from the ultraView Universal DAB Detection Kit (Ventana), 5-bromo-4 chloro-3-indolyl phosphate (BCIP) substrate and nitro blue tetrazolium (NBT) oxidant reagents from the ISH iVIEW™ Blue Detection Kit (Ventana), and a ready-to-use tetramethyl benzidine (TMB) solution (Fitzgerald Industries International, Concord, Massachusetts) were also evaluated for CEN 17 signal detection of the BDISH application. Because both HER2 and CEN 17 probes were labeled with the same DNP hapten, CEN 17 signal detection was completed without DNP-labeled CEN 17 probe after HER2 signal detection to ensure that the anti-DNP antibody of CEN 17 detection didn't recognize the DNP hapten of HER2 probe.
Figure 1.
Brightfield double in situ hybridization (BDISH) signal detection scheme with a sequential in situ hybridization method. HER2 gene signal was detected with a DNP-labeled nick translated DNA probe hybridization followed by silver signal detection system (silver acetate, hydroquinone, and H2O2 reaction). Then, chromosome 17 centromere (CEN 17) signal was detected with a DNP-labeled CEN 17 oligoprobe hybridization followed by fast red and naphthol phosphate reaction signal detection system.
Single or double stained tissue sections for HER2 and/or CEN 17 targets were counterstained with Hematoxylin II (Ventana) for 4 or 8 minutes and Bluing Reagent (Ventana) for 4 minutes. Counterstained slides were first rinsed with distilled water containing DAWN® (Proctor & Gamble Company, Cincinnati, Ohio) for removing LCS from slides and then rinsed with distilled water until soap was removed completely from the slide. Slides were blotted very gently with paper towels and completely dried at 45°C or 65°C in the oven for at least 15 minutes. One drop of Cytoseal™ 60 (Richard-Allen Scientific) was applied onto a dried slide and a glass coverslip was carefully placed onto the slide. Excess mounting media was removed from the slides by gently pressing the slides against paper towels. Different coverslipping methods were also evaluated for preserving the fast red staining during the assay development. BDISH results were observed with a Nikon ECLIPSE 90i microscope (Nikon Instruments Inc., Melville, New York) equipped with Nikon digital camera DXM1200F (Nikon) without oil immersion objective lenses, up to 60×. However, for presentation purposes, mainly comparing to FISH images taken with a 100× objective lens, brightfield photographs contained in this report were obtained using a 100× oil immersion objective lens.
FISH
PathVysion HER-2 DNA Probe Kit was used for the FISH test for HER2 and CEN 17 targets of xenograft tumor controls as previously described [30]. Photographs of FISH images were taken with Zeiss Axioplan 2 microscope (Carl Zeiss MicroImaging, Inc., Thornwood, New York) with Metasystems JAIM4+ CCD1 Charge Coupling Imaging Camera (MetaSystems Group Inc., Watertown, Massachusetts) at 100× using an oil-immersion lens.
BDISH performance test
Performance of the BDISH assay was compared to FISH results as the reference standard using the historical criteria for HER2 amplification with the PathVysion assay (Negative: HER2/CEN 17 < 2.0 and Positive: HER2/CEN 17 ≥ 2.0) and using the ASCO/CAP guideline criteria (Negative: HER2/CEN 17 > 1.8, Equivocal: 1.8 ≤ HER2/CEN 17 ≤ 2.2, and Positive: HER2/CEN 17 > 2.2) with or without the equivocal cases. Scoring BDISH slides was conducted by 4 observers (MK, MD, FPL, and RRT), who were experienced with scoring HER2 FISH slides. Scoring occurred at different sites and at different occasions using different microscopes. Each individual observer evaluated the set of slides at their own pace and judgement. No scores were provided by the observers when the staining quality was deemed not adequate. There was no communication among observers regarding their scoring experience of the BDISH slides. Concordance data of FISH scores vs. consensus BDISH scores among 4 observers and FISH scores vs. individual BDISH scores by 4 observers were determined using SAS 9.1 (SAS Institute Inc, Cary, North Carolina) in calculating frequency tables and Kappa statistics. The consensus among observers was defined as the agreement of three or more observers on a given observation. Scoring of BDISH assays was also analyzed for the sensitivity and specificity against FISH scores with the historical scoring method and the ASCO/CAP scoring method without the equivocal cases. Discordant cases were investigated by a non-observer (HN) for possible causes using BDISH slides.
Results
BDISH assay optimization
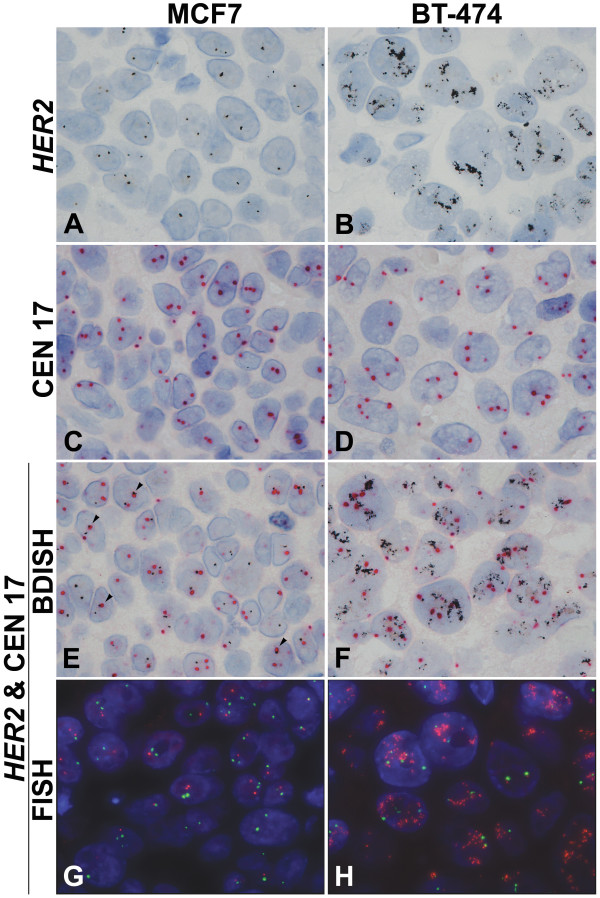
Images of HER2 single ISH, CEN 17 single ISH, and HER2 and CEN 17 BDISH results with formalin-fixed, paraffin-embedded xenograft tumor sections are presented in Figure 2. Single copies of HER2 signal were recognized as black discrete dots in the nuclei with MCF7 xenograft tumor (Figure 2A) while amplified HER2 gene signals were visualized as either an increased number of HER2 signals, clusters of black dots with BT-474 tumor, and/or both (Figure 2B). Single CEN 17 copies were observed as red dots that were slightly larger than the black dots for HER2 genes in the nuclei with MCF7 tumor (Figure 2C) and BT-474 tumor (Figure 2D). After single staining for HER2 gene or CEN 17 was optimized, the BDISH application with sequential detection for HER2 targets followed by CEN 17 targets was tested on xenograft tumors. Single copies of HER2 genes and CEN 17 were stained in the nuclei of MCF7 tumor cells (Figure 2E) and amplified HER2 genes and single copies of CEN 17 were visualized in the nuclei of BT-474 tumor cells (Figure 2F). Because of the size difference and color contrast of black dots for HER2 gene and red dots for CEN 17, they could be visually separated even when red and black signals were co-localized in the nuclei of MCF7 tumor cells (arrowheads, Figure 2E). When CEN 17 probe was omitted from the complete BDISH assay, there was no fast red staining on xenograft tumor sections (data not shown). Thus, the anti-DNP antibody used for CEN 17 signal detection (the second ISH detection) didn't recognize the DNP-hapten of HER2 probe signal (the first ISH detection) even though the same hapten was used for the sequential hybridization method. For image comparison of BDISH and FISH for HER2 gene and CEN 17, FISH images with MCF7 tumor and BT-474 tumor are presented in Figure 2G and Figure 2H, respectively. HER2 genes are seen as red-orange dots and CEN 17 targets are seen as green dots.
Figure 2.
Brightfield in situ hybridization and dual color fluorescence in situ hybridization (FISH) for HER2 and CEN 17. HER2 and CEN 17 detection with formalin-fixed, paraffin-embedded xenograft tumors, MCF7 (non-amplified HER2 gene and chromosome 17 polysomy) (A, C, E, G) and BT-474 (amplified HER2 gene and chromosome 17 polysomy) (B, D, F, H). Normal HER2 gene signal is seen as black dots in the nuclei of MCF7 xenograft tumor (A) while amplified HER2 gene signal is seen as clusters of black dots in the nuclei of BT-474 tumor (B). CEN 17 signal is detected as red dots that are slightly larger than silver black dots (C, D). Double staining of HER2 gene and CEN 17 is obtained with silver grains and red dots (E, F). Individual HER2 gene and CEN 17 signals can be still recognized when both targets are co-localized (arrow heads, E). HER2 FISH signal is red-orange and CEN 17 FISH signal is green in the blue nuclei counterstained with DAPI (G, H). 100×.
One successful way to preserve the fast red staining was the use of a toluene-based Cytoseal 60 mounting medium placed onto completely dried tissue sections prior to coverslipping with glass coverslips. We also confirmed that the red signal was successfully preserved with the Tissue-Tek® film coverslipper method (Sakura Finetek Japan, Tokyo, Japan) after air-drying slides (data not shown). The most common method of coverslipping tissue sections stained with fast red is the use of an aqueous mounting medium. However, this method did not produce crisp fast red staining for quantitative analyses of BDISH signals (data not shown). For the second color for the BDISH application, DAB, BCIP/NBT, and TMB detection systems that produce brown, blue, and green to blue final product, respectively, were evaluated. However, they did not provide sufficient contrast against the HER2 ISH black signal (data not shown).
BDISH assay performance
After optimizing the BDISH assay for HER2 gene and CEN 17 with formalin-fixed, paraffin-embedded xenograft tumor sections, we applied the assay to 94 breast carcinoma cases and scoring BDISH slides was conducted by 4 observers (MK, MD, FPL, and RRT). The consensus among observers was defined as the agreement of three or more observers on a given observation. With the historical scoring method (Negative: HER2/CEN 17 < 2 and Positive: HER2/CEN 17 ≥ 2.0) (Table 1), the consensus concordance rate was 98.9% (Simple Kappa = 0.9736, 95% CI = 0.9222 – 1.0000), the sensitivity was 96.3%, and the specificity was 100%. Individual concordance ranges were between 97.8% (Simple Kappa = 0.9466, 95% CI = 0.8736 – 1.0000) and 100% (Simple Kappa = 01.0000, 95% CI = 1.0000 – 1.0000). With the ASCO/CAP scoring method (Negative: HER2/CEN 17 > 1.8, Equivocal: 1.8 ≤ HER2/CEN 17 ≤ 2.2, and Positive: HER2/CEN 17 > 2.2) (Table 2), the consensus concordance rate was 95.7% (Simple Kappa = 0.8993%, 95% CI = 0.8068 – 0.9919). Individual concordance ranges were between 92.5% (Simple Kappa = 0.8275, 95% CI = 0.7102 – 0.9448) and 95.7% (Simple Kappa = 0.9069, 95% CI = 0.8206 – 0.9933). With the ASCO/CAP scoring method without the FISH equivocal cases (Table 3), the consensus concordance rate was 100% (Simple Kappa = 1.0000%, 95% CI = 1.0000 – 1.0000). The sensitivity was 100% and the specificity was also 100%. Individual concordance ranges were between 97.7% (Simple Kappa = 0.9442, 95% CI = 0.8678 – 1.0000) and 100% (Simple Kappa = 1.0000, 95% CI = 1.0000 – 1.0000).
Table 1.
Performance of brightfield double in situ hybridization (BDISH) with clinical samples based on the historic scoring method
| FISH | Total | |||
| Positive | Negative | |||
| BDISH | Positive | 26 | 0 | 26 |
| Negative | 1 | 66 | 67 | |
| Total | 27 | 66 | 93 | |
Frequency missing = 1
Sensitivity 96.3%
Specificity 100%
Concordance 98.9%
Kappa 0.9736
Table 2.
Performance of brightfield double in situ hybridization (BDISH) with clinical samples based on the ASCO/CAP method with FISH equivocal cases
| FISH | Total | ||||
| Positive | Equivocal | Negative | |||
| BDISH | Positive | 25 | 1 | 0 | 26 |
| Equivocal | 0 | 0 | 0 | 0 | |
| Negative | 0 | 3 | 63 | 66 | |
| Total | 25 | 4 | 63 | 92 | |
Frequency missing = 2
Concordance 95.7%
Kappa 0.8993
Table 3.
Performance of brightfield double in situ hybridization (BDISH) with clinical samples based on the ASCO/CAP method without 4 FISH equivocal cases
| FISH | Total | |||
| Positive | Negative | |||
| BDISH | Positive | 25 | 0 | 25 |
| Negative | 0 | 63 | 63 | |
| Total | 25 | 63 | 88 | |
Frequency missing = 2
Sensitivity 100%
Specificity 100%
Concordance 100%
Kappa 1.0000
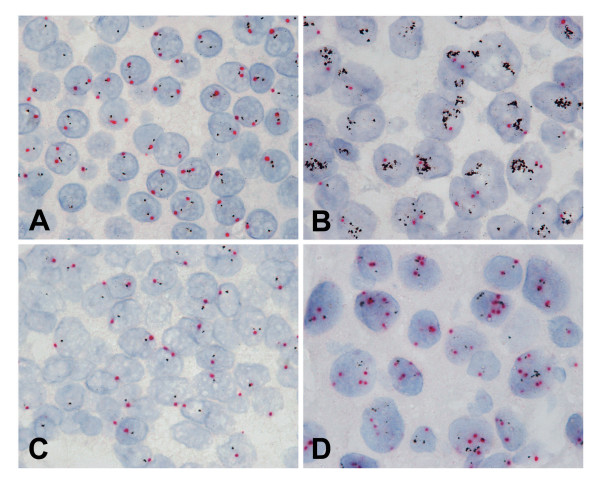
Representative images of BDISH staining on clinical samples are presented in Figure 3. Cancer cells were easily identified based on the tissue morphology and assessments of HER2/CEN 17 ratios could be readily conducted. Non-amplified HER2 gene cases showed 0–4 copies of HER2 genes and 0–4 copies of CEN 17 depending on cell cycle stage and how each cell was cut within a tissue section (Figure 3A), while amplified HER2 gene cases showed multiple copies or clusters of HER2 genes and a few copies of CEN 17 (Figure 3B). Besides non-amplified and amplified HER2 cases, cases with a single copy of HER2 gene due to centromere 17 monosomy or a monoallelic deletion of HER2 gene (Figure 3C) and multiple copies of CEN 17 due to chromosome 17 polysomy (Figure 3D) were also observed.
Figure 3.
Brightfield double in situ hybridization (BDISH) for HER2 and chromosome 17 centromere (CEN 17) on formalin-fixed, paraffin-embedded clinical breast cancer cases. Examples of normal HER2 gene (A), amplified HER2 gene (B), single HER2 gene (C), and chromosome 17 polysomy (D) cases were shown. 100×.
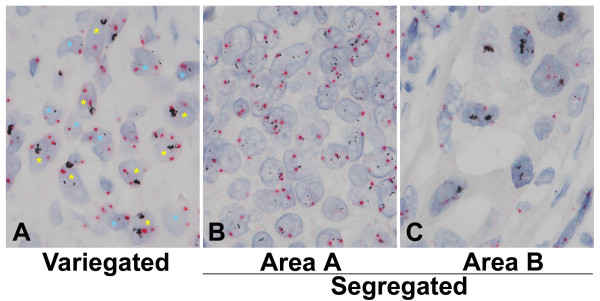
All discordant cases, that we defined it even by one observer disagreement between the BDISH and FISH scores, were re-examined for possible causes of conflicting results and there were 9 discordant cases. All nine (9) discordant cases of the BDISH slides presented at least some degree of the genotypic heterogeneity of tumor cell populations. In general, there were two types of the tumor cell heterogeneity with HER2 gene status within the same tissue section: 1) variegated different genotype tumor cell populations in the same area of tissue section (Figure 4A) and 2) segregated tumor populations in different areas of tissue section (Figures 4B&C). Breast cancers with obvious tumor cell heterogeneity are shown as examples in Figure 4. However, the subtle genotype heterogeneity of tumor cell populations is often seen among the equivocal cases, and it also can be seen in Figures 3D &4B which show less obvious variegated tumor cell heterogeneity. Three (3) of 9 discordant cases demonstrated the segregated tumor cell heterogeneity while the other 6 cases showed various degrees of the variegated tumor cell heterogeneity.
Figure 4.
The heterogeneity of breast tumor cell populations. The heterogeneity of breast cancer cells was demonstrated with brightfield double in situ hybridization (BDISH) stained tissue sections. In general, there were 2 types of the tumor cell heterogeneity: 1) variegated tumor populations of, as an example, non-amplified (blue asterisk) and amplified (yellow asterisk) HER2 gene cells in the same area (A) and 2) segregated tumor populations of, as an example, discrete (B) and clustered (C) HER2 gene cells in different areas. The tumor cell heterogeneity with different appearance of HER2 and CEN 17 is also seen among the non-large clustered HER2 cells (B). 100×.
Discussion
Accurate HER2 status testing is important for identifying breast cancer patients who may benefit from receiving trastuzumab therapy. Currently, in the United States, HER2 IHC methods are most commonly used for primary screening for HER2 status, and borderline cases are subjected to dual FISH for HER2 and CEN 17 to determine the HER2/CEN 17 ratio. Because the discordance rate between local and central/reference HER2 status testing with IHC and FISH is significantly high [14,31-33], the standardization of diagnosing breast cancer cases is recognized as a very important task for improving personalized cancer patient care [3,34]. The American Society of Clinical Oncology and the College of American Pathologists has published a guideline recommendation for testing HER2 status in breast cancer [3] and the Canadian National Consensus has updated the Canadian HER2/neu testing guideline [35]. Two potential solutions for improving the standardization of HER2 status testing include: 1) automating the entire process for slide staining [36] and slide reading [36-39] and 2) consolidating the HER2 testing process within experienced laboratories and pathologists that perform large numbers of HER2 tests [15].
One way to improve the accuracy of HER2 status testing is to automate the assay procedure for HER2 IHC and HER2 FISH assays so that human errors can be diminished. HER2 IHC assays can be performed using an automated slide staining system, but HER2 FISH assays remain technically challenging and time consuming manual molecular diagnostic assays in most laboratories. An evaluator of FISH slides must have access to specialized fluorescence microscopy in a dark room. Because of unstable FISH staining characteristics, the signals of FISH slides can be bleached easily, even while reviewing and enumerating signals. Furthermore, digital images of the FISH slide need to be captured with a sensitive camera system for each patient case for the HER2 gene status record. Therefore, it is desirable to automate a tissue-based HER2 gene status test that can be observed with a regular brightfield microscope and that produces stained slides that can be archived.
While the concept of multi-color brightfield ISH applications was published in 1990's [40,41], it was a recent achievement to visualize HER2 and CEN 17 targets within the same nuclei of tissue sections with a manual dual brightfield ISH application [42]. This dual ISH application utilized TMB chromogen for HER2 gene staining. However, based on published images [28,42], TMB staining does not provide discrete signals when compared to the SISH application. The advantages of the BDISH application for HER2 gene and CEN 17 presented in the current study are: 1) the automation of the ISH application; 2) the visualization of both HER2 gene and CEN 17 targets in the nuclei of the same cell; 3) the generation of discrete HER2 gene signals; 4) the ability to reproducibly detect endogenous HER2 and CEN 17 signals in the stromal tissues and lymphocytes as a reliable internal assay control; 5) the ability to visualise signal with brightfield microscopy with non-oil immersion lenses; and 6) the capability to permanently archive the slides.
HER2 and CEN 17 probes are co-hybridized for dual color HER2 FISH. However, for the BDISH assay, because the stringency conditions for the nick-translated HER2 probe and the CEN 17 oligoprobe were different, it was necessary to conduct sequential ISH staining steps for HER2 gene and CEN 17 targets. For CEN 17 ISH, we have optimized a new detection system with an alkaline phosphatase-conjugated antibody and fast red chromogen and naphthol phosphate substrate reaction. The fast red-based detection was selected to obtain a good contrast of CEN 17 ISH signal against the discrete black dots of HER2 SISH signal. DAB, BCIP/NBT, and TMB detection systems did not provide sufficient contrast against HER2 ISH black signal (data not shown). Because fast red precipitate is soluble in organic solvents, in general, aqueous mounting medium is used for coverslipping. However, the standard coverslipping method with aqueous mounting medium on wet tissue sections did not produce tissue sections with high resolution and therefore detailed tissue structure could not be observed (data not shown). A successful method to preserve fast red staining for CEN 17 and high resolution tissue morphology was, after completely dry the slides, to apply a toluene-based tissue mounting medium (Cytoseal 60) for coverslipping with cover glass or to use a film coverslipper (Tissue-Tek® film coverslipper). Incomplete drying resulted in faint red background staining particularly around the fast red precipitate sites with this method. Interestingly, the use of aqueous mounting medium onto the dried tissue slides produced yellowish background staining on tissue sections and this method did not produce satisfactory results (data not shown).
The specificity of single ISH for HER2 gene or CEN 17 and BDISH for both targets was evaluated with xenograft tumors. HER2 and CEN 17 copy numbers have been documented previously using the FISH assay [29]. MCF-7 cells are characterized as non-amplified HER2 and chromosome 17 polysomy (3 copies of chromosome 17 per nucleus) while one of chromosome 17 with HER2 deletion (2 HER2 copies per nucleus) [29]. BT-474 cell line presents HER2 amplification with 50–60 copies of HER2 genes and 4–6 copies of CEN 17 per nucleus [29]. Amplified HER2 genes are located not only on chromosome 17, but also are translocated on other chromosomes [29]. HER2 and CEN 17 copy numbers produced with the single target ISH and BDISH methods matched with previously reported results. As both probes are labeled with the same DNP hapten, our first concern was to determine if detecting specific signal for each probe was feasible. We confirmed that the fast red chromogen detection reagents did not produce red signal when the fast red ISH was performed without the CEN 17 probe after detection of HER2 by SISH (data not shown). Thus, the SISH detection and the fast red detection can be combined to perform a sequential double ISH assay with 2 probes labeled with the same hapten. Because the sequential BDISH application uses 2 specific stringency conditions based on the length and sequences of 2 probes, it is not necessary to design 2 probes that require the same stringency for co-hybridization, like double color FISH assays.
Concordance rates between a set of gold standard dual color HER2 FISH scores and HER2 and CEN 17 BDISH scores by 4 observers were calculated for assessing the performance of the BDISH assay. There were 9 discordant cases (9.6% of the total cases) based on BDISH score disagreement with FISH scores, even by one observer. We have found that the number of equivocal cases influences the concordance rate with the ASCO/CAP scoring method. There were 4 equivocal cases based on FISH scores and all cases showed the BDISH score disagreement by at least 2 observers. A similar observation was reported with an international HER2 testing proficiency study [15]. In their study, the discordant cases (20%) were caused by the specimen having FISH HER2/CEN 17 ratios between 1.7 and 2.3 that are close to the 'equivocal' defined by ASCO/CAP HER2 scoring method (1.8 – 2.2). They also stated "equivocal cases are difficult to interpret, even highly experienced and validated laboratories" [15]. In one study, when the FISH assay was used as the primary test for HER2 status assessment of breast carcinoma cases, heterogeneity of HER2 gene status was observed in 40 of 742 cases (5%) [43]. It has been speculated that genomic and phenotypic heterogeneity of tumor cells is the main reason for the inconsistency of HER2 testing results [44]. With current study, all of our discordant cases (9/9 or 100%) displayed the tumor cell population heterogeneity: three samples showed significant segregated tumor cell population heterogeneity (Figures 4B&C) and other cases showed subtle heterogeneity of tumor cell populations that are seen among equivocal cases (4/4 or 100%). Nonetheless, since consecutive tissue sections were not used for the FISH and BDISH analyses, one can speculate that the tissue sections for the FISH and BDISH tissue sections contained tumor cell populations with different HER2 status. Further clinical evaluations of HER2 and CEN 17 BDISH application with patient treatment outcome data are required for more accurate HER2 status assessment of breast cancer patients to be obtained.
Conclusion
We have successfully developed an automated BDISH application for HER2 gene and CEN 17 targets in formalin-fixed, paraffin-embedded tissue sections that is highly concordant to the FISH and is reproducibly interpreted among observers. Assessment of HER2 gene status can be conducted without the use of a specialized fluorescence microscope and the time required for completing HER2 gene status assessment can be shortened significantly. Furthermore, this application has the potential to be used for other gene targets, any combination of a gene and its chromosome centromere, and tissue section-based gene assessment tests including gene translocation studies. The use of BDISH technology allows the simultaneous analyses of two DNA targets within the context of tissue morphology observation.
Competing interests
HN, BHW, ML, AEM, FG, MF, EW, TM are employed by Ventana Medical Systems, Inc. RRT and MD received grant support and honorarium for speaking from Ventana Medical Systems, Inc.
Authors' contributions
LW, ML, JP, and RRT were responsible for identifying and prequalification of the clinical cases used in this study. HN and TMG were responsible for the BDISH assay development and feasibility studies, staining the clinical samples, and preparing the manuscript draft and image data. BHW and ML were responsible for the final assay development. AEM conducted all statistical analyses for the performance of BDISH assay. FG was the study coordinator and contributed intellectual content of the study. MF designed the probes. EW, MK, MD, RRT, and TMG critiqued the assay performance with their molecular histology expertise during the assay development. FPL, MK, MD, and RRT were the observers for scoring the clinical samples. All authors contributed intellectual inputs to the study. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Vu Nguyen for instrument maintenance and Drs. Raymond B. Nagle and Guadalupe Manriquez for slide reviewing during the assay development.
Contributor Information
Hiroaki Nitta, Email: hiro.nitta@ventana.roche.com.
Beatrice Hauss-Wegrzyniak, Email: beatrice.wegrzyniak@ventana.roche.com.
Megan Lehrkamp, Email: megan.lehrkamp@ventana.roche.com.
Adrian E Murillo, Email: adrian.murillo@ventana.roche.com.
Fabien Gaire, Email: fabien.gaire@ventana.roche.com.
Michael Farrell, Email: mike.farrell@ventana.roche.com.
Eric Walk, Email: eric.walk@ventana.roche.com.
Frederique Penault-Llorca, Email: fpenault@cjp.fr.
Masafumi Kurosumi, Email: mkurosumi@cancer-c.pref.saitama.jp.
Manfred Dietel, Email: manfred.dietel@charite.de.
Lin Wang, Email: wangle2@ccf.org.
Margaret Loftus, Email: loftusm@ccf.org.
James Pettay, Email: pettayj@ccf.org.
Raymond R Tubbs, Email: tubbsr@ccf.org.
Thomas M Grogan, Email: tom.grogan@ventana.roche.com.
References
- Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, Gralow JR, Livingston RB, Gown AM. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291:1972–1977. doi: 10.1001/jama.291.16.1972. [DOI] [PubMed] [Google Scholar]
- Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5:63–69. doi: 10.3816/CBC.2004.n.011. [DOI] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, Vijver M van de, Wheeler TM, Hayes DF. American society of clinical oncology/college of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- Garrison LP, Jr, Lubeck D, Lalla D, Paton V, Dueck A, Perez EA. Cost-effectiveness analysis of trastuzumab in the adjuvant setting for treatment of HER2-postive breast cancer. Cancer. 2007;110:489–498. doi: 10.1002/cncr.22806. [DOI] [PubMed] [Google Scholar]
- Norum J, Olsen JA, Wist EA, Lønning PE. Trastuzumab in adjuvant breast cancer therapy. A model based cost-effectivemess analysis. Acta Oncol. 2007;46:153–164. doi: 10.1080/02841860601096841. [DOI] [PubMed] [Google Scholar]
- Fagnani F, Colin X, Arveux P, Coudert B, Misset JL. Cost/effectiveness analysis of adjuvant therapy with trastuzumab in patients with HER2 positive early breast cancer. Bull Cancer. 2007;94:711–720. [PubMed] [Google Scholar]
- Millar JA, Millward MJ. Cost effectiveness of trastuzumab in the early breast cancer: a lifetime model. Pharmocoeconomics. 2007;25:429–442. doi: 10.2165/00019053-200725050-00006. [DOI] [PubMed] [Google Scholar]
- de Korte MA, de Vries EG, Lub-de Hooge MN, Jager PL, Gietema JA, Graaf WT van der, Sluiter WJ, van Veldhuisen DJ, Suter TM, Sleijfer DT, Perik PJ. 111Indium-trastuzumab visualizes myocardial human epidermal growth factor receptor 2 expression shortly after anthracycline treatment but not during heart failure: a clue to uncover the mechanisms of trastuzumab-related cardiotoxicity. Eur J Cancer. 2007;43:2046–2051. doi: 10.1016/j.ejca.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Dal Lago L, Durbecq V, Desmedt C, Salgado R, Verjat T, Lespagnard L, Ma Y, Veys I, Di Leo A, Sotiriou C, Piccart M, Larsimont D. Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol Cancer Ther. 2006;5:2572–2579. doi: 10.1158/1535-7163.MCT-06-0129. [DOI] [PubMed] [Google Scholar]
- Salido M, Tusquets I, Corominas JM, Suarez M, Espinet B, Corzo C, Bellet M, Fabregat X, Serrano S, Solé F. Polysomy of chromosome 17 in breast cancer tumors showing an overexpression of ERBB2: a study of 175 cases using fluorescence in situ hybridization and immunohistochemistry. Breast Cancer Res. 2005;7:R267–R273. doi: 10.1186/bcr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrisi R, Rotmensz N, Bagnardi V, Viale G, Curto BD, Dell'orto P, Veronesi P, Luini A, D'Alessandro C, Cardillo A, Goldhirsch A, Colleoni M. HER2 status in early breast cancer: relevance of cell staining patterns, gene amplification and polysomy 17. Eur J Cancer. 2007;43:2339–2344. doi: 10.1016/j.ejca.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Cayre A, Mishellany F, Lagarde N, Penault-Llorca F. Comparison of different commercial kits for HER2 testing in breast cancer: looking for the accurate cut-off for amplification. Breast Cancer Res. 2007 doi: 10.1186/bcr1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros MB, Simpson PT, Jones C, Natrajan R, Westbury C, Steele D, Savage K, Mackay A, Schmitt FC, Ashworth A, Reis-Filho JS. Unlocking pathology archives for molecular genetic studies: a realizable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest. 2006;86:398–408. doi: 10.1038/labinvest.3700390. [DOI] [PubMed] [Google Scholar]
- Perez EA, Suman VJ, Davidson NE, Martino S, Kaufman PA, Lingle WL, Flynn PJ, Ingle JN, Visscher D, Jenkins RB. HER2 testing by local, central, and reference laboratories in specimens from the north central cancer treatment group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24:3032–3038. doi: 10.1200/JCO.2005.03.4744. [DOI] [PubMed] [Google Scholar]
- Dowsett M, Hanna WM, Kockx M, Penault-Llorca F, Rüschoff J, Gutjahr T, Habben K, Vijver MJ van de. Standardization of HER2 testing: results of an international proficiency-testing ring study. Mod Pathol. 2007;20:584–591. doi: 10.1038/modpathol.3800774. [DOI] [PubMed] [Google Scholar]
- Persons DL, Tubbs RR, Cooley LD, Dewald GW, Dowling PK, Du E, Mascarello JT, Rao KW, Wilson KS, Wolff DJ, Habegger-Vance G. HER-2 fluorescence in situ hybridization: results from the survey program of the College of American Pathologists. Arch Pathol Lab Med. 2006;130:325–331. doi: 10.5858/2006-130-325-HFISHR. [DOI] [PubMed] [Google Scholar]
- Arnould L, Denoux Y, MacGrogan G, Penault-Llorca F, Fiche M, Treilleux I, Mathieu MC, Vincent-Salomon A, Vilain MO, Couturier J. Agreement between chromogenic in situ hybridization (CISH) and FISH in the determination of HER2 status in breast cancer. Br J Cancer. 2003;88:1587–1591. doi: 10.1038/sj.bjc.6600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wu R, Au A, Marquez A, Yu Y, Shi Z. Determination of HER2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma. Mod Pathol. 2002;15:657–665. doi: 10.1038/modpathol.3880582. [DOI] [PubMed] [Google Scholar]
- Peiró G, Mayr D, Hillemanns P, Löhrs U, Diebold J. Analysis of HER-2/neu amplification in endometrial carcinoma by chromogenic in situ hybridization. Correlation with fluorescence in situ hybridization, HER-2/neu, p53 and Ki-67 protein expression, and outcome. Mod Pathol. 2004;17:277–287. doi: 10.1038/modpathol.3800006. [DOI] [PubMed] [Google Scholar]
- Park K, Kim J, Lim S, Han S, Lee JY. Comparing fluorescence in situ hybridization and chromogenic in situ hybridization methods to determine the HER2/neu status in primary breast carcinoma using tissue microarray. Mod Pathol. 2003;16:937–943. doi: 10.1097/01.MP.0000086487.78558.7D. [DOI] [PubMed] [Google Scholar]
- Hanna WM, Kwok K. Chromogenic in-situ hybridization: a viable alternative to fluorescence in-situ hybridization in the HER2 testing algorithm. Mod Pathol. 2006;19:481–487. doi: 10.1038/modpathol.3800555. [DOI] [PubMed] [Google Scholar]
- Todorović-Raković N, Jovanović D, Nesković-Konstantinović Z, Nikolić-Vukosavljević D. Prognostic value of HER2 gene amplification detected by chromogenic in situ hybridization (CISH) in metastatic breast cancer. Exp Mol Pathol. 2007;82:262–268. doi: 10.1016/j.yexmp.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Tubbs R, Pettay J, Skacel M, Powell R, Stoler M, Roche P, Hainfeld J. Gold facilitated in situ hybridization (GOLDFISH): A bright field autometallographic alternative to fluorescence in situ hybridization for detection of HER-2/neu gene amplification. Amer J Pathol. 2002;160:1589–1595. doi: 10.1016/S0002-9440(10)61106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs R, Skacel M, Pettay J, Powell R, Myles J, Hicks D, Sreenan J, Roche P, Stoler MH, Hainfeld J. Interobserver interpretative reproducibility of GOLDFISH, a first generation gold-facilitated autometallographic bright field in situ hybridization assay for HER-2/neu amplification in invasive mammary carcinoma. Am J Surg Pathol. 2002;26:908–913. doi: 10.1097/00000478-200207000-00009. [DOI] [PubMed] [Google Scholar]
- Tubbs R, Pettay J, Hicks D, Skacel M, Powell R, Grogan T, Hainfeld J. Novel bright field molecular morphology methods for detection of HER2 gene amplification. J Mol Histol. 2004;35:589–594. doi: 10.1007/s10735-004-2191-9. [DOI] [PubMed] [Google Scholar]
- Downs-Kelly E, Pettay J, Hicks D, Skacel M, Yoder B, Rybicki L, Myles J, Sreenan J, Roche P, Powell R, Hainfeld J, Grogan T, Tubbs R. Analytical validation and interobserver reproducibility of EnzMet GenePro: a second-generation bright-field metallography assay for concomitant detection of HER2 gene status and protein expression in invasive carcinoma of the breast. Am J Surg Pathol. 2005;29:1505–1511. doi: 10.1097/01.pas.0000172294.67409.4f. [DOI] [PubMed] [Google Scholar]
- Powell RD, Pettay JD, Powell WC, Roche PC, Grogan TM, Hainfeld JF, Tubbs RR. Metallographic in situ hybridization. Hum Pathol. 2007;38:1145–1159. doi: 10.1016/j.humpath.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Dietel M, Ellis IO, Höfler H, Kreipe H, Moch H, Dankof A, Kölble K, Kristiansen G. Comparison of automated silver enhanced in situ hybridization (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch. 2007;451:19–25. doi: 10.1007/s00428-007-0424-5. [DOI] [PubMed] [Google Scholar]
- Grushko TA, Blackwood MA, Schumm PL, Hagos FG, Adeyanju MO, Feldman MD, Sanders MO, Weber BL, Olopade OI. Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res. 2002;62:1481–1488. [PubMed] [Google Scholar]
- Hicks DG, Tubbs RR. Assessment of the HER2 Status in Breast Cancer by Fluorescence In-Situ Hybridization (FISH): a Technical Review with Interpretative Guidelines. Hum Pathol. 2005;36:250–261. doi: 10.1016/j.humpath.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Paik S, Bryant J, Tan-Chiu E, Romond E, Hiller W, Park K, Brown A, Yothers G, Anderson S, Smith R, Wickerham DL, Wolmark N. Real-world performance of HER2 testing-National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst. 2002;94:852–854. doi: 10.1093/jnci/94.11.852. [DOI] [PubMed] [Google Scholar]
- Roche PC, Suman VJ, Jenkins RB, Davidson NE, Martino S, Kaufman PA, Addo FK, Murphy B, Ingle JN, Perez EA. Concordance between local and central laboratory HER2 testing in the breast intergroup trial N9831. J Natl Cancer Inst. 2002;94:855–857. doi: 10.1093/jnci/94.11.855. [DOI] [PubMed] [Google Scholar]
- Dowsett M, Hanby AM, Laing R, Walker R, National HER2 Consultation Steering Group HER2 testing in the UK: consensus from a national consultation. J Clin Pathol. 2007;60:685–689. doi: 10.1136/jcp.2006.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JS, Symmans WF, Pusztai L, Hortobagyi GN. Standardizing slide-based assays in breast cancer: hormone receptors, HER2, and sentinel lymph nodes. Clin Cancer Res. 2007;13:2831–2835. doi: 10.1158/1078-0432.CCR-06-2522. [DOI] [PubMed] [Google Scholar]
- Hanna W, O'malley FP, Barnes P, Berendt R, Gaboury L, Magliocco A, Pettigrew N, Robertson S, Sengupta S, Têtu B, Thomson T. Undated recommendation from the Canadian National Consensus Meeting on HER2/neu testing in breast cancer. Curr Oncol. 2007;14:149–153. doi: 10.3747/co.2007.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs RR, Pettay JD, Swain E, Roche PC, Powell W, Hicks DG, Grogan T. Automation of manual components and image quantification of direct dual label fluorescence in situ hybridization (FISH) for HER2 gene amplification: a feasibility study. Appl Immunohistochem Mol Morphol. 2006;14:436–440. doi: 10.1097/01.pai.0000213101.26193.f1. [DOI] [PubMed] [Google Scholar]
- Ciampa A, Xu B, Ayata G, Baiyee D, Wallace J, Wertheimer M, Edmiston K, Khan A. HER-2 status in breast cancer: correlation of gene amplification by FISH with immunohistochemistry expression using advanced cellular imaging system. Appl Immunohistochem Mol Morphol. 2006;14:132–137. doi: 10.1097/01.pai.0000150516.75567.13. [DOI] [PubMed] [Google Scholar]
- Theodosiou Z, Kasampalidis IN, Karayannopoulou G, Kostopoulos I, Bobos M, Bevilacqua G, Aretini P, Starita A, Lyroudia K, Pitas I. Evaluation of FISH image analysis system on assessing HER2 amplification in breast carcinoma cases. Breast. 2007;17:80–84. doi: 10.1016/j.breast.2007.07.041. [DOI] [PubMed] [Google Scholar]
- Stevens R, Almanaseer I, Gonzalez M, Caglar D, Knudson RA, Ketterling RP, Schrock DS, Seemayer TA, Bridge JA. Analysis of HER2 gene amplification using an automated fluorescence in situ hybridization signal enumeration system. J Mol Diagn. 2007;9:144–150. doi: 10.2353/jmoldx.2007.060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speel EJ, Jansen MP, Ramaekers FC, Hopman AH. A novel triple-color detection procedure for brightfield microscopy, combining in situ hybridization with immunocytochemistry. J Histochem Cytochem. 1994;42:1299–1307. doi: 10.1177/42.10.7930513. [DOI] [PubMed] [Google Scholar]
- Hopman AHN, Claessen S, Speel EJM. Multi-colour brightfield in situ hybridization on tissue sections. Histochem Cell Biol. 1997;108:291–298. doi: 10.1007/s004180050168. [DOI] [PubMed] [Google Scholar]
- Laakso M, Tanner M, Isola J. Dual-colour chromogenic in situ hybridization for testing HER-2 oncogene amplification in archival breast tumors. J Pathol. 2006;210:3–9. doi: 10.1002/path.2022. [DOI] [PubMed] [Google Scholar]
- Tubbs RR, Hicks DG, Cook J, Downs-Kelly E, Pettay J, Hartke MB, Hood L, Neelon R, Myles J, Budd GT, Moore HC, Andresen S, Crowe JP. Fluorescence in situ hybridization (FISH) as primary methodology for the assessment of HER2 status in adenocarcinoma of the breast. Diagn Mol Pathol. 2007;16:207–210. doi: 10.1097/PDM.0b013e318064c72a. [DOI] [PubMed] [Google Scholar]
- Tuma RS. Inconsistency of HER2 test raises questions. J Natl Cancer Inst. 2007;99:1064–1065. doi: 10.1093/jnci/djm075. [DOI] [PubMed] [Google Scholar]