Abstract
Background
Dichlorodiphenyltrichloroethane (DDT) is a persistent estrogenic organochlorine pesticide that is a rodent hepatic tumor promoter, with inconclusive carcinogenicity in humans. We have previously reported that o, p'-DDT elicits primarily PXR/CAR-mediated activity, rather than ER-mediated hepatic responses, and suggested that CAR-mediated effects, as opposed to ER-mediated effects, may be more important in tumor promotion in the rat liver. To further characterize species-specific hepatic responses, gene expression analysis, with complementary histopathology and tissue level analyses were investigated in immature, ovariectomized C57BL/6 mice treated with 300 mg/kg o, p'-DDT, and compared to Sprague-Dawley rat data.
Results
Rats and mice exhibited negligible histopathology with rapid o, p'-DDT metabolism. Gene expression profiles were also similar, exhibiting PXR/CAR regulation with the characteristic induction of Cyp2b10 and Cyp3a11. However, PXR-specific target genes such as Apoa4 or Insig2 exhibited more pronounced induction compared to CAR-specific genes in the mouse. In addition, mouse Car mRNA levels decreased, possibly contributing to the preferential activation of mouse PXR. ER-regulated genes Cyp17a1 and Cyp7b1 were also induced, suggesting o, p'-DDT also elicits ER-mediated gene expression in the mouse, while ER-mediated effects were negligible in the rat, possibly due to the inhibitory effects of CAR on ER activities. In addition, o, p'-DDT induced Gadd45a, Gadd45b and Cdkn1, suggesting DNA damage may be an additional risk factor. Furthermore, elevated blood DHEA-S levels at 12 h after treatment in the mouse may also contribute to the endocrine-related effects of o, p'-DDT.
Conclusion
Although DDT is known to cause rodent hepatic tumors, the marked species differences in PXR/CAR structure, expression patterns and ligand preference as well as significant species-specific differences in steroidogenesis, especially CYP17A1 expression and activity, confound the extrapolation of these results to humans. Nevertheless, the identification of potential modes of action as well as species-specific responses may assist in the selection and further development of more appropriate models for assessing the toxicity of DDT to humans and wildlife.
Background
Dichlorodiphenyltrichloroethane (DDT) is a persistent organochlorine pesticide that is a hepatic tumor promoter in the mouse and rat [1], with inconclusive carcinogenicity in the human liver [2,3]. Technical grade DDT is a mixture of p,p'-DDT, o, p'-DDT and their metabolites such as 1,1-dichloro-2,2-bis(p-chlorophenyl) ethylene (DDE) and l,l-dichloro-2,2-bis(p,p-chlorophenylethane) (DDD) [4]. o, p'-DDT exerts estrogenic effects in responsive tissues such as the uterus [5] by binding to estrogen receptor (ER)α and β subtypes [6]. In addition, o, p'-DDT is an agonist for pregnane X receptor (PXR) and constitutive androstane receptor (CAR) [7,8]. Thus, it may elicit complex responses through multiple nuclear receptors.
Several structurally diverse estrogenic compounds elicit ER-mediated responses in the liver [9,10]. We have previously reported that o, p'-DDT elicits primarily PXR/CAR-mediated, rather than ER-mediated hepatic responses, and suggested that CAR-mediated effects, as opposed to ER-mediated effects, may be more important in DDT-induced tumor promotion in the rat liver [11].
Although the rat is a preferred toxicological model because of the accumulated background knowledge regarding chemical-induced toxicity compared to mice, several studies report significant species-specific differences in response to chemical exposures. For example, 1,4-bis-[2-(3,5,-dichloropyridyloxy)] benzene (TCPOBOP) acts as potent phenobarbital-type enzyme inducer in the mouse liver but not in the rat or human. This is due to a substitution of Thr350 in mice with Met in the rat and human CAR [12-14]. Alternatively, another phenobarbital-type enzyme inducer 2,4,6-triphenyldioxane-1,3 induces hepatic CYP2B in rats but not in mice [15]. Such differences may affect CAR-mediated hepatic drug metabolism and disposition following xenobiotic exposure. In addition, rat genome annotation lags behind the mouse and human in maturity, confounding a more comprehensive assessment of rat gene expression data. Furthermore, targeted gene-disruption can be used in mice to further elucidate the roles of PXR, CAR and ER in o, p'-DDT elicited effects. Thus, we were motivated to investigate hepatic gene expression in the mouse following o, p'-DDT treatment to identify species-specific and conserved responses. Overall, the hepatic gene expression profiles were comparable, however there were marked species-specific differences in genes regulated by ER, which may involve species-difference in CAR activation. Furthermore, blood levels of DHEA-S, a precursor of androgen and estrogen, exhibited mouse-specific elevation, which may also contribute to endocrine system disruption. Thus, the present study further elucidates the dynamics of o, p'-DDT elicited responses in the rodent, and identifies species-specific responses that may be important to o, p'-DDT exposure for humans and wildlife.
Methods
Husbandry
Female C57BL/6 mice, ovariectomized on postnatal day 20 were obtained from Charles River Laboratories (Raleigh, NC) on day 25. Mice were housed in polycarbonate cages containing cellulose fiber chip bedding (Aspen Chip Laboratory Bedding, Northeastern Products, Warrensberg, NY) and maintained at 40–60% humidity and 23°C in a room with a 12 h dark/light cycle (7 am-7 pm). Animals were allowed free access to de-ionized water and Harlan Teklad 22/5 Rodent Diet 8640 (Madison, WI), and acclimatized for 4 days prior to dosing.
Treatments and Necropsy
Mice (n = 5) were orally gavaged once or once daily for three consecutive days with 300 mg/kg b.w. o, p'-DDT (99.2% purity, Sigma-Aldrich, St Louis, MO) in 0.1 ml of sesame oil (Sigma-Aldrich) vehicle. An equal number of time-matched vehicle control animals (n = 5) were also treated in the same manner. Mice receiving one dose were sacrificed 2, 4, 8, 12, 18, and 24 h after treatment. Mice receiving three daily doses were sacrificed 24 h after the third treatment (72 h). All procedures were performed with the approval of the Michigan State University All-University Committee on Animal Use and Care. Animals were sacrificed by cervical dislocation and animal body weights were recorded. Whole liver weights were recorded and sections of the left lateral lobe (approximately 0.1 g) were snap-frozen in liquid nitrogen and stored at -80°C. The right lateral lobe was placed in 10% neutral buffered formalin (NBF, VWR International, West Chester, PA) for histopathology and stored at room temperature.
Histopathology
Following fixation of the right lateral lobe for at least 24 h in 10% NBF, the samples were embedded in paraffin according to standard techniques. Five μm sections were mounted on glass slides and stained with hematoxylin and eosin. All embedding, mounting and staining of tissues were performed at the Histology Laboratory, (Department of Physiology, Michigan State University). The histopathology of each liver section was scored according to the NTP Pathology guidelines.
Measurement of blood DHEA-S and androstenedione
Rat plasma samples were obtained from a previous study [11]. Mouse serum and rat plasma samples were used for determining dehydroepiandrosterone sulfate (DHEA-S) and androstenedione levels using a DHEA-S ELISA kit (Calbiotech Inc., Spring Valley, CA) and Androstenedione ELISA kit (Genway Biotech Inc., San Diego, CA), respectively. The detection limits, defined by the vendors, are 0.02 μg/ml of DHEA-S and 0.043 ng/ml of androstenedione, respectively.
Protein preparation and Western analysis
Total protein was extracted from liver samples from all the time points using the Total Protein Extraction kit (Chemicon International Inc., Temecula, CA). Protein concentrations were determined using Bicinchoninic Acid Protein Assay Kit (Sigma-Aldrich). Total protein (100 μg) was resolved on a 11% denaturing SDS-polyacrylamide gel, and transferred to Hybond-ECL membrane (GE Healthcare, Waukesha, WI). Blots were incubated with blocking buffer (0.1% Tris-buffered saline pH 7.4 containing 1% low-fat dry milk) for 10 min at room temperature. Goat anti-CYP17A1, goat anti-actin antibodies and donkey horseradish peroxidase-conjugated anti-goat IgG were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Immunochemical staining was performed as described previously [16], with dilution of 1:500 (anti-CYP17A1), 1:500 (anti-actin) and 1:10000 (anti-goat IgG) using blocking buffer, respectively. SuperSignal West Dura substrate (Thermo Fisher Scientific, Inc., Waltham, MA) was used for signal detection. The Western analysis was performed on three independent biological replicates.
RNA isolation
Total RNA was isolated from left lateral liver sections using TRIZOL Reagent (Invitrogen, Carlsbad, CA) and resuspended in The RNA Storage Solution (Ambion, Austin, TX). RNA concentrations were determined by spectrophotometry (A260) and purity was assessed by the A260:A280 ratio and by visual inspection of 3 μg on a denaturing gel.
Microarray analysis
Temporal changes in gene expression were assessed using an independent reference design in which o, p'-DDT-treated samples were co-hybridized with time-matched vehicle controls using 3 biological replicates and 2 independent labelings of each sample (i.e. dye swap) for each time point. Whole Mouse Genome 4 × 44 K Oligo Microarray Kit (Agilent Technologies, Inc, Santa Clara, CA) was used for global gene expression analysis. All the reagents and enzymes were provided by Agilent Technologies, and the microarray analysis was performed according to the vendor's protocol. The microarrays were scanned at 635 nm (Cy5) and 532 nm (Cy3) using a GenePix 4000B microarray scanner (Molecular Devices, Union City, CA). Images were analyzed for feature and background intensities using GenePix Pro 6.0 (Molecular Devices). All data were managed in the toxicogenomic information management system dbZach relational database [17].
Microarray data normalization and statistical analysis
Data were normalized using a semi-parametric approach [18]. Model-based t-values were calculated from normalized data, comparing treated and vehicle responses per time-point. Empirical Bayes analysis was used to calculate posterior probabilities (p1 [t]-value) of activity on a per gene and time-point basis using the model-based t-value [19]. Genes were filtered for activity based on the p1(t)-value. p1(t) values approaching one indicate changes in gene expression which are more robust. In this study, unique genes with a p1(t) > 0.999 for a minimum of two time points and absolute value fold change ≥ 1.5-fold compared to time-matched vehicle control for at least one time point were considered differentially expressed.
Hierarchical clustering
A total of 996 orthologous rat and mouse genes were in common on the rat cDNA, mouse cDNA, and Agilent mouse oligonucleotide microarrays based on HomoloGene http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene. Differentially expressed orthologs (|fold change| ≥ 1.5 for at least one time point in either species) for ethynylestradiol (EE)-treated mouse liver, o, p'-DDT treated rat liver and o, p'-DDT-treated mouse liver, were hierarchical clustered using Cluster 3.0 and TreeView software http://rana.lbl.gov/eisen/. In addition, differentially expressed genes known to be regulated by PXR, CAR and ER agonists [20-23], were identified from the literature and their expression profiles were subjected to hierarchical clustering.
Correlation analysis
Correlation analysis was performed using relaxed criteria to allow for more inclusive comparison between mouse and rat data sets. Genes with a p1(t) > 0.99 and absolute fold change ≥ 1.5-fold at one or more time points in the o, p'-DDT-treated mouse liver samples were selected and used for correlation analysis. This analysis involved a multivariate correlation-based visualization application [24] that has been used to compare two independent gene expression profile datasets [11,25]. It calculates the temporal correlations between gene expression (fold change) and significance values (p1 [t]-value) for orthologous o, p'-DDT-treated mouse (this study) and o, p'-DDT-treated rat genes [11], and summarizes the results in a scatter plot.
Quantitative Real-Time PCR (QRT-PCR)
Expression levels of mouse Car (Nr1i3), Cyp17a1, Cyp2b10, Cyp3a11, Cyp7b1, Gadd45a, Gadd45b, Gapdh, Gclm, Pxr (Nr1i2), and Srebf1, and rat Gapdh and Cyp7b1 were measured by QRT-PCR. For each sample, 2 μg of total RNA was reverse transcribed by SuperScript II using an anchored oligo-dT primer as described by the manufacturer (Invitrogen). The resultant cDNA (1.0 μl) was used as the template in a 30 μl PCR reaction containing 0.1 μM each of forward and reverse gene-specific primers designed using Primer3 [26], 3 mM MgCl2, 1.0 mM dNTPs, 0.025 IU AmpliTaq Gold and 1 × SYBR Green PCR buffer (Applied Biosystems, Foster City, CA). PCR amplification was conducted in MicroAmp Optical 96-well reaction plates (Applied Biosystems) on an Applied Biosystems PRISM 7000 Sequence Detection System using the following conditions: initial denaturation and enzyme activation for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A dissociation protocol was performed to assess the specificity of the primers and the uniformity of the PCR generated products. Each plate contained duplicate standards of purified PCR products of known template concentration covering eight orders of magnitude to interpolate relative template concentrations of the samples from the standard curves of log copy number versus threshold cycle. The copy number of each unknown sample for each gene was standardized to that of Gapdh gene to control for differences in RNA loading, quality and cDNA synthesis. Primer sequences and amplicon sizes are provided as Additional file 1.
Statistical analysis
Body weight, relative liver weight, hepatic concentrations of o, p'-DDT, o, p'-DDD, and o, p'-DDE and QRT-PCR data are presented as the mean ± SE. Statistical analysis was performed with two-way ANOVA followed by pairwise comparisons using Tukey's Honestly Significant Difference post hoc test to control Type I error (α = 0.05). For QRT-PCR data, the relative expression levels of target genes were scaled such that the standardized expression level of the time-matched vehicle control group was equal to 1 for graphing purposes. All statistics were performed using SAS 9.1.3 software (SAS Institute Inc., Cary, NC).
Results
Body Weight, Relative Liver Weight, and Histopathology
Relative liver weight was significantly (p < 0.05) increased at 72 h compared to vehicle-treated mice (Fig. 1A). No effect on body weight was observed. These results are similar to those previously observed in the rat o, p'-DDT study [11]. There were no significant histological changes by o, p'-DDT treatment, except for two animals exhibiting cell death of small numbers of hepatocytes at 4 h or 72 h, where the nuclei were shrunken and hyperchromatic.
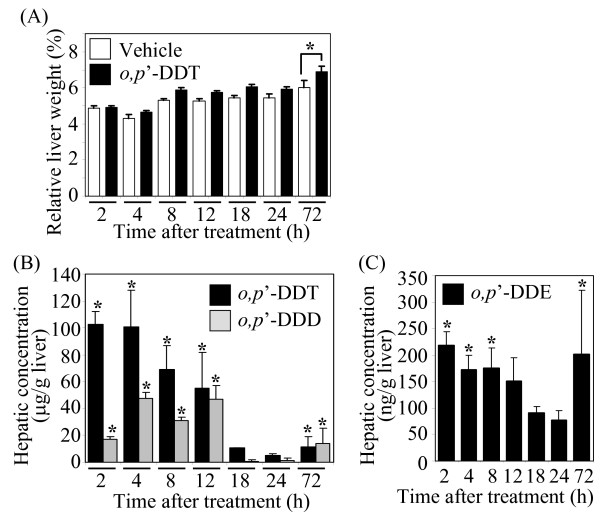
Figure 1.
Relative liver weight and tissue level analysis. (A) Relative liver weight. Immature ovariectomized C57BL/6 mice were orally administered 300 mg/kg o, p'-DDT or sesame oil vehicle at time 0, 24 and 48 h. Mice were sacrificed 2, 4, 8, 12, 18, 24 or 72 h after the initial dose. The relative liver weight was significantly increased at 72 h. (B) Hepatic concentration of o, p'-DDT and o, p'-DDD. (C) Hepatic concentration of o, p'-DDE. Hepatic tissue levels of o, p'-DDT, o, p'-DDD and o, p'-DDE were determined using high-resolution gas chromatograph/HRMS from three randomly selected mice orally gavaged with 300 mg/kg o, p'-DDT. The data are presented as mean ± SE. The asterisk (*) indicates a significant (p < 0.05) difference from the vehicle controls.
Hepatic o, p'-DDT, o, p'-DDD and o, p'-DDE concentration
Animals treated with o, p'-DDT showed significantly greater levels of o, p'-DDT, from 2 to 18 h compared to vehicle controls (Fig. 1B). The levels of the metabolites o, p'-DDD and o, p'-DDE were significantly greater than controls from 2 to 12 h, and at 72 h (Figs. 1B and 1C), indicating that o, p'-DDT is readily metabolized in the liver, and that much of the o, p'-DDT and its metabolites remain primarily distributed to the liver from 2 to 12 h, similar to the rat [11]. Note that there was a statistical outlier in the 72 h treated group, which lead to significant o, p'-DDE and o, p'-DDD levels, but was still included in the analysis as there was no technical or biological justification for its removal.
Microarray Data
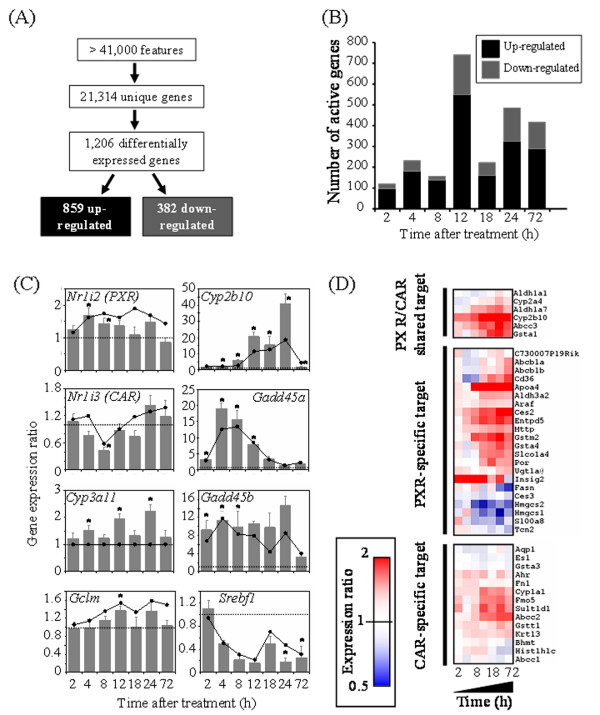
Gene expression was assessed using the 4 × 44 K Mouse Agilent array containing approximately 44,000 oligonucleotide probes, representing 34,204 annotated genes including approximately 21,000 unique genes. Model-based t-values that compared treated and vehicle responses on a per time-point basis followed by Empirical Bayes analysis identified 1,206 differentially expressed genes across the time course based on a p1(t) > 0.999 for a minimum of two time points and absolute fold change ≥ 1.5-fold compared to time-matched vehicle control for at least one time point (Fig. 2A). Overall, 859 genes were induced while 382 genes were repressed. Changes in gene expression ranged from 36.4-fold induction for Cyp2c55 gene to -13.0-fold repression for gene Car3. The 12 h time point exhibited the greatest number of differentially expressed genes (Fig. 2B). All of the gene expression data as well as the identified differentially regulated genes are provided in Additional files 2 and 3, respectively.
Figure 2.
Microarray and QRT-PCR results. (A) Identification of differentially regulated genes. (B) Number of differentially expressed genes following o, p'-DDT treatment in the mouse liver. Differentially expressed genes were selected based on a p1(t) ≥ 0.999 at two or more time points and an absolute fold change ≥ 1.5 at one or more time points relative to time-matched vehicle controls. All differentially expressed genes are listed in Additional file 3. (C) Verification of microarray results by QRT-PCR. QRT-PCR results relative to time-matched vehicle controls are shown as bar and presented as mean ± SE. Microarray results are represented as lines. The dashed line indicates the expression level of the time-matched vehicle control. The asterisk (*) indicates a significant (p < 0.05) difference from the time-matched vehicle controls for QRT-PCR, n = 5. (D) Hepatic expression of PXR/CAR-target genes in o, p'-DDT-treated mouse. A heat map of o, p'-DDT elicited microarray expression profiles for selected PXR-, CAR-specific and PXR/CAR-shared target genes identified in the literature [20-23]. While some CAR-regulated genes such as Cyp1a1, Fmo5, Sult1d1 or Abcc2 were moderately induced, several PXR-target genes, including ApoA4, Ces2, Gstm2 or Insig2, exhibited strong induction. However, other PXR-target genes such as Hmgcs1 and Hmgcs2 were down-regulated.
A number of phase I drug metabolizing enzyme genes were induced such as Cyp1a1 (~2.2-fold), Cyp2b9 (~5.0-fold), Cyp2b10 (~18.4-fold), Cyp2b13 (~4.1-fold), Cyp2c39 (~2.1-fold), and Cyp2c55 (~36.4-fold) (Table 1). The induction of Cyp2b10 and Cyp3a11 were verified by QRT-PCR (Fig. 2C). The steroid metabolism genes, Cyp17a1 (~6.2-fold) and Cyp7b1 (~5.8-fold), were also induced, while Cyp51 and Cyp7a1 were down-regulated. Steroidogenic Cyp11a1 mRNA level was not detected by microarray or QRT-PCR (data not shown). Note that the hepatic induction of Cyp17a1 and Cyp7b1 by o, p'-DDT was observed in the mouse but not in the rat [11]. Several phase II genes (Gsta1 (~3.4-fold), Gstm2 (~3.2-fold), Ugt2b35 (~3.4-fold)) were also induced.
Table 1.
Selected microarray results
| Gene symbol | Entrez GeneID | Gene expression ratio (h)a | ||||||
| 2 | 4 | 8 | 12 | 18 | 24 | 72 | ||
| Drug metabolizing enzyme | ||||||||
| Cyp1a1 | 13076 | 1.22 | 1.22 | 1.22 | 2.16 | 1.78 | 1.92 | 1.42 |
| Cyp1a2 | 13077 | 1.16 | 1.26 | 1.30 | 1.61 | 1.50 | 2.29 | 1.30 |
| Cyp2b9 | 13094 | 1.78 | 1.90 | 2.41 | 4.11 | 3.75 | 4.97 | 2.23 |
| Cyp2b10 | 13088 | 2.19 | 2.12 | 2.69 | 11.37 | 12.47 | 18.42 | 4.29 |
| Cyp2b13 | 13089 | 1.49 | 1.36 | 1.46 | 3.79 | 3.54 | 4.09 | 2.04 |
| Cyp2c39 | 13098 | 1.25 | 1.41 | 1.23 | 1.51 | 1.60 | 2.13 | 1.72 |
| Cyp2c55 | 72082 | 1.71 | 3.19 | 7.43 | 15.18 | 30.38 | 36.40 | 7.70 |
| Reductive reaction | ||||||||
| Gsta1 | 14857 | 1.30 | 0.91 | 1.39 | 1.76 | 3.00 | 3.42 | 2.37 |
| Gsta2 | 14858 | 1.53 | 0.83 | 2.23 | 3.52 | 7.01 | 6.60 | 2.91 |
| Gstm2 | 14863 | 1.03 | 1.14 | 2.20 | 2.06 | 2.44 | 3.19 | 1.90 |
| Gstt3 | 103140 | 1.22 | 1.17 | 1.81 | 2.31 | 2.18 | 2.65 | 1.62 |
| Ugt2b35 | 243085 | 1.13 | 1.17 | 1.94 | 2.73 | 2.30 | 3.38 | 2.53 |
| DNA damage/apoptosis | ||||||||
| Casp9 | 12371 | 1.01 | 0.67 | 0.62 | 0.61 | 0.64 | 0.75 | 0.83 |
| Cdkn1a | 12575 | 2.53 | 15.36 | 3.37 | 2.27 | 0.75 | 1.54 | 1.00 |
| Fas | 14102 | 0.94 | 0.89 | 0.78 | 0.80 | 0.85 | 0.66 | 0.72 |
| Gadd45a | 13197 | 2.96 | 12.58 | 13.54 | 8.52 | 3.16 | 1.41 | 2.29 |
| Gadd45b | 17873 | 4.55 | 8.35 | 6.66 | 7.83 | 5.12 | 6.80 | 3.32 |
| Tnfrsf19 | 29820 | 1.22 | 1.75 | 5.86 | 4.29 | 1.67 | 1.66 | 2.21 |
| Metabolism | ||||||||
| ApoA4 | 11808 | 1.15 | 1.05 | 4.31 | 5.03 | 5.74 | 3.54 | 3.34 |
| Ces2 | 234671 | 1.16 | 1.46 | 1.68 | 2.22 | 2.11 | 3.36 | 3.05 |
| Cyp17a1 | 13074 | 1.43 | 1.27 | 3.97 | 6.24 | 5.52 | 3.68 | 1.88 |
| Cyp51 | 13121 | 0.98 | 1.02 | 0.85 | 0.39 | 0.72 | 0.35 | 0.49 |
| Cyp7a1 | 13122 | 1.07 | 0.58 | 0.25 | 0.44 | 0.66 | 0.51 | 1.09 |
| Cyp7b1 | 13123 | 0.98 | 1.51 | 3.05 | 5.77 | 4.15 | 5.44 | 3.41 |
| Insig2 | 72999 | 2.80 | 5.00 | 4.40 | 4.63 | 1.45 | 2.36 | 0.81 |
| Srebf1 | 20787 | 0.94 | 0.51 | 0.31 | 0.21 | 0.70 | 0.47 | 0.31 |
| Cell proliferation | ||||||||
| Ccnb2 | 12442 | 0.94 | 0.84 | 0.82 | 0.49 | 0.68 | 0.86 | 0.83 |
| Ccnd1 | 12443 | 0.71 | 1.17 | 1.08 | 0.85 | 2.32 | 0.78 | 0.66 |
| Ccng2 | 12452 | 1.26 | 1.66 | 1.39 | 0.71 | 0.34 | 1.03 | 0.95 |
| Mdm2 | 17246 | 1.13 | 1.19 | 1.16 | 1.11 | 0.97 | 1.09 | 1.28 |
| Stmn1 | 16765 | 0.74 | 1.00 | 0.69 | 0.35 | 0.59 | 0.84 | 0.60 |
| Oxidative stress | ||||||||
| Gclm | 14630 | 1.06 | 1.15 | 1.36 | 1.54 | 1.33 | 1.59 | 1.50 |
| Gsr | 14782 | 1.02 | 1.40 | 2.03 | 2.47 | 2.39 | 2.54 | 2.33 |
| Hmox2 | 15369 | 1.18 | 1.55 | 1.44 | 1.39 | 1.40 | 1.52 | 1.43 |
| Nqo1 | 18104 | 0.99 | 1.10 | 1.71 | 1.97 | 2.57 | 2.35 | 2.43 |
| Transcription factor/signal transuction | ||||||||
| Ahr | 11622 | 1.20 | 1.43 | 1.29 | 1.50 | 0.96 | 1.47 | 1.15 |
| Nr1i2 (PXR) | 18171 | 1.16 | 1.63 | 1.74 | 1.62 | 1.90 | 1.68 | 1.44 |
| Nr1i3 (CAR) | 12355 | 1.12 | 1.20 | 0.58 | 0.91 | 1.18 | 1.29 | 1.38 |
a Highlighted values indicate expression ratio where p1(t) > 0.999.
A number of DNA damage-related genes such as Cdkn1a (~15.4-fold), Gadd45a (~13.5-fold), Gadd45b (~8.4-fold) and Tnfrsf19 (~5.9-fold) were induced following o, p'-DDT treatment, while Fas and Casp9 were down-regulated (~2.0-fold). The oxidative stress-responsive genes Gclm (~1.6-fold), Gsr (~2.5-fold), Hmox2 (~1.6-fold) and Nqo1 (~2.6-fold) exhibited induction. There was no clear induction for the cell proliferation-related genes such as Ccnd1, Ccnb2, Mdm2 or Stmn1, while their orthologs were induced by o, p'-DDT in the rat liver [11]. mRNA levels for Ahr and Pxr (Nr1i2) were slightly elevated, whereas Car (Nr1i3) mRNA levels decreased following o, p'-DDT treatment. Srebf1 which was previously shown to be down-regulated by o, p'-DDT in the rat liver [11], was also down-regulated in the mouse liver. Overall, the QRT-PCR results closely paralleled the expression pattern seen in the microarray data (Fig. 2C).
We also examined the expression of genes regulated by CAR, PXR or ER that were selected based on the null animal studies [20-23]. While CAR-specific targets such as Cyp1a1 (~2.2-fold), Fmo5 (~2.3-fold), Sult1d1 (~2.8-fold) and Abcc2 (~2.7-fold) showed relatively weak induction, several PXR-regulated genes were induced more strongly including ApoA4 (~5.8-fold), Ces2 (~3.4-fold), Gstm2 (~3.2-fold) or Insig2 (~5.0-fold) genes (Fig. 2D). However, other PXR genes, such as Hmgcs1 and Hmgcs2, were unexpectedly down-regulated.
Comparative Analysis of Gene Expression
Hepatic gene expression profiles of EE-treated mice, o, p'-DDT-treated rats and o, p'-DDT-treated mice were compared by hierarchical clustering focusing on the 538 orthologs represented on both microarray platforms that satisfied the |fold change| ≥ 1.5 for at least one time point in any data set. The filtering criteria used here were relaxed compared to those used in Figs. 2A and 2B to include more orthologs in the correlation analysis, and thus be more informative of the overall similarity between the two data sets. Despite the differences in microarray platform (i.e., cDNA vs. oligonucleotide), o, p'-DDT elicited gene expression profiles in the rat and mouse that were more similar to each other than to the EE-treated mouse profile (Fig. 3A).
Figure 3.
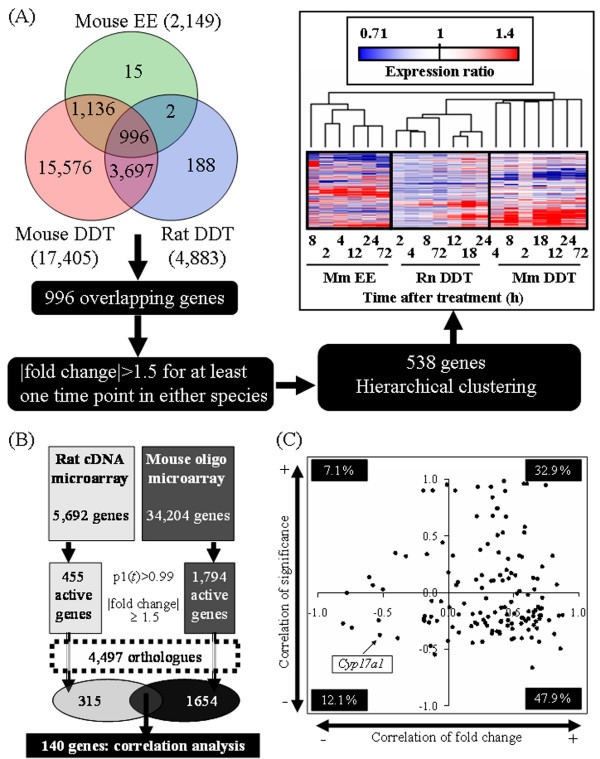
Comparative analysis of global gene expression profiles elicited by o, p'-DDT. (A). Comparative gene expression analysis between EE-treated mouse, o, p'-DDT-treated rat and o, p'-DDT-treated mouse. A total of 996 orthologs were represented on the rat cDNA microarray, mouse cDNA microarray and mouse Agilent oligonucleotide microarrays determined by HomoloGene http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene. 538 of these orthologs showed a |fold change| ≥ 1.5 for at least one time point in either species. These 538 differentially expressed orthologs were subjected to hierarchical clustering. The dendrogram illustrates that mouse o, p'-DDT gene expression profiles are more similar to rat o, p'-DDT gene expression profiles than the mouse EE gene expression profiles. (B) Correlation analysis using differentially expressed orthologous genes. The temporal profiles of o, p'-DDT-treated mouse liver (current study) and those of the o, p'-DDT-treated rat liver [11] were compared by determining the Pearson's correlation of the temporal gene expression (fold change) and significance (p1 [t] value) between orthologs. Both studies used comparable study designs and data analysis methods, although different platforms were used (i.e., rat cDNA microarray and mouse Agilent oligonucleotide microarray). 140 genes were identified as differentially expressed orthologs. (C) Scatter plot of the 140 differentially expressed orthologous genes. Correlations for gene expression and significance approaching 1.0 indicate that the behavior or the orthologous genes are similar and would fall in the upper right quadrant. Orthologs tended to localize in upper- or lower-right quadrant (32.9% and 47.9% of total number of spots, respectively), indicating that temporal gene expression changes for o, p'-DDT-treated mouse and rat liver are comparable. However, poor correlations between the temporal p1(t) values and gene expression fold changes would fall within the lower left quadrant. For example, Cyp17a1 fell into this quadrant suggesting that significant differences exist between the rat and mouse ortholog expression profiles.
Correlation Analysis
One hundred and forty orthologs were identified as differentially expressed on both rat cDNA and mouse oligo microarrays (Fig. 3B), which were further examined by correlation analysis to determine if their gene expression profiles were comparable. Again, the filtering criteria used here were relaxed compared to those used in Figs. 2A and 2B to include more orthologs in the correlation analysis. The scatter plot shows that a majority of the spots localized to either the upper-right (32.9%) or lower-right (47.9%) quadrants (Fig. 3C), indicating that the gene expression profiles between o, p'-DDT-treated rat and mouse were highly similar in terms of their gene expression pattern. Temporal correlation of the p1(t)-value (statistical significance) showed relatively low correlation between the two data sets, with 60% of spots located in the lower-right or lower-left quadrants. All correlation analyses results are provided in Additional file 4.
For example, Cyp17a1, a key enzyme gene for steroidogenesis (Fig. 4A) fell into the lower left quadrant (Fig. 3C) suggesting divergent gene expression profiles in the mouse and rat. Fig. 4B clearly demonstrates that Cyp17a1 is significantly induced in the mouse and is non-responsive in the rat in both microarray and QRT-PCR data. Cyp7b1, another steroid metabolism gene, exhibited a similar species-specific gene expression profile (Fig 4B). Moreover, hepatic CYP17A1 protein induction was evident at 18 and 24 h in the mouse liver (Fig. 4C).
Figure 4.
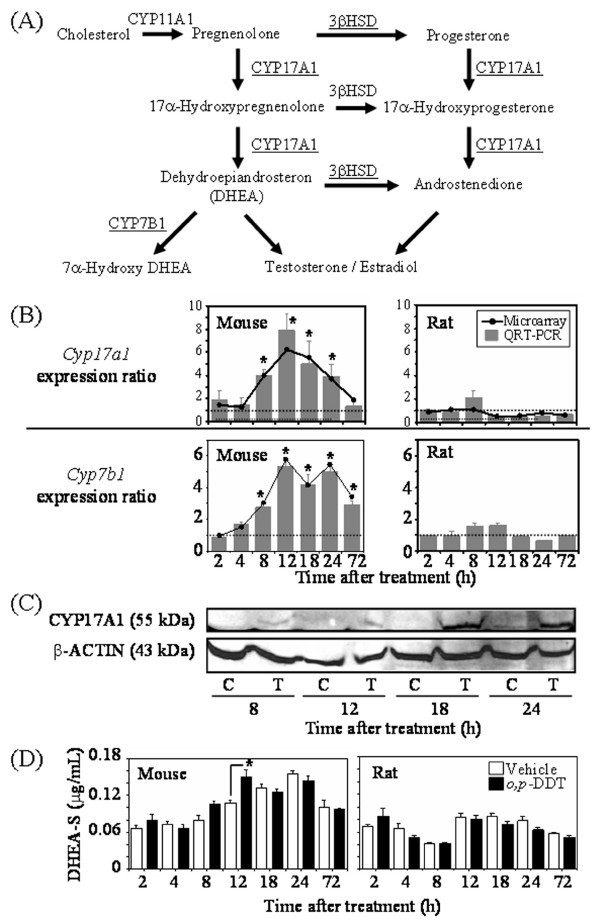
Species-specific regulation of steroid hormone metabolism elicited by o, p'-DDT. (A) Overview of the role of CYP17A1 and CYP7B1 in steroid metabolism. CYP17A1 metabolizes pregnenolone and progesterone to produce DHEA and androstenedione, respectively. Hepatic CYP7B1 is involved in bile acid biosynthesis, and also responsible for 7α-hydroxylation of DHEA. (B) Hepatic Cyp17a1 and (C) Cyp7b1 mRNA levels in the o, p'-DDT-treated mouse and rat. QRT-PCR results relative to time-matched vehicle controls are shown as bars and presented as mean ± SE. Microarray results are represented as lines. o, p'-DDT induced Cyp17a1 and Cyp7b1 mRNAs in the mouse liver, while it did not affect in the rat liver [11]. The dashed line indicates the expression level of the time-matched vehicle control. The asterisk (*) indicates a significant (p < 0.05) difference from the time-matched vehicle controls for QRT-PCR, n = 5. (C) Representative Western analysis result for hepatic CYP17A1 protein in o, p'-DDT-treated mouse liver. CYP17A1 protein levels were induced at 18 and 24 h. Western analyses were performed on 3 independent biological replicates to verify the consistency of the results. C, control; T, 300 mg/kg o, p'-DDT. (D) Blood DHEA-S levels. DHEA-S level was significantly higher at 12 h following o, p'-DDT treatment compared to time-matched controls in the mouse, while it did not change in rats.
Effects on DHEA-S Levels
Species-specific induction of CYP17A1 mRNA and protein levels suggests that steroid metabolism may also be differentially affected. More specifically, since CYP17A1 metabolizes pregnenolone and 17α-hydroxypregnenolone to produce DHEA (Fig. 4A), blood levels of DHEA-S were measured using an enzyme-linked immunosorbent assay. Serum DHEA-S levels exhibited a significant (p < 0.05) increase relative to vehicle at 12 h in the mouse, (Fig. 4D). In contrast, plasma DHEA-S was not significantly different between treated and control rats. Mouse serum androstenedione levels were below the detection level (data not shown).
Discussion
o, p'-DDT elicits phenobarbital-type activities in the mouse liver, characterized by the increase in relative liver weight or induction of Cyp2b10, Cyp3a11 and GST mRNA levels [27,28]. In the rat, we previously reported that o, p'-DDT elicits PXR/CAR-mediated responses, with negligible ER-mediated activity [11], despite the responsive of the liver to estrogens [9,29], and the association of estrogenic activity with hepatic tumorigenesis [30]. Consequently, PXR/CAR-mediated responses to DDT may have a more significant role in hepatic tumor promotion [11]. In order to further investigate o, p'-DDT elicited gene expression mediated by PXR/CAR, we investigated its hepatic expression profile using a comparable mouse model, study design and data analysis [11]. Comparative analysis indicated that the hepatic gene expression profiles were similar between rat and mouse. However, correlation analysis of orthologous genes revealed that hepatic Cyp17a1 mRNA and protein levels as well as serum DHEA-S were only elevated in the mouse following o, p'-DDT treatment.
Cyp17a1 exhibits dose-dependent induction by EE in the mouse liver [10], suggesting ER-mediated induction. In addition, induction of Cyp7b1 and down-regulation of Cyp7a1 by EE are abolished in ER alpha-null mice but not in ER beta-, Fxr-, Pxr- or Car-null mice [23]. Collectively, these results indicate that Cyp7a1 mRNA levels are regulated by ERα in the mouse liver. Considering the strong inductions of Cyp17a1 and Cyp7b1 observed only in the mouse liver as well as down-regulation of Cyp7a1 in the mouse liver, o, p'-DDT may elicit ERα-mediated activity in the mouse liver but not in the rat liver.
In addition, the mouse profile suggests that o, p'-DDT elicited gene expression was predominately PXR regulated based on genes known to be regulated by PXR-, CAR- or PXR/CAR [20-23]. In part, this could be due to the repression of mouse Car mRNA levels by o, p'-DDT compared to its induction in the rat liver [11]. Furthermore, several genes associated with cell proliferation (Ccnb1, Ccnb2, Mdm2 and Stmn1) were induced in the rat liver [11], but not in the mouse. Ccnd1, a known CAR-regulated cell proliferation-related gene [31], was not significantly induced in the mouse, further supporting the hypothesis that o, p'-DDT preferentially activated PXR.
CAR also has an inhibitory effect on ER-mediated gene expression [32]. Therefore, the availability of CAR may inhibit ER-mediated effects elicited by o, p'-DDT in the rat liver [11], while the down regulation of Car mRNA in the mouse and the preferential activation of PXR may facilitate more ER-mediated gene expression. Consequently, ER-mediated responses may be important when assessing DDT-elicited hepatic responses in the mouse in addition to PXR/CAR-mediated response when compared to the rat.
CYP17A1 is an important enzyme in steroid biosynthesis that metabolizes pregnenolone and progesterone to produce DHEA and androstenedione, respectively [33,34]. Therefore, induction of CYP17A1 mRNA and protein levels may affect DHEA metabolism. o, p'-DDT treatment increased DHEA-S levels in the mouse whereas no change was detected in the rat. Although the liver is usually not a major steroidogenesis organ, microsomes are capable of participating in steroidogenesis [35]. In addition, hepatic microsomal CYP17A1 exhibits age-dependent expression, with higher expression and activity in immature rats [35,36]. o, p'-DDT elicited changes in other steroidogenic enzymes (e.g., Cyp11a1) and metabolites (e.g., androstenedione) were below the level of detection. Collectively, alterations in steroid levels following o, p'-DDT treatment could affect steroid levels, since peripheral tissues can use circulating DHEA and DHEA-S to produce androgens and estrogens [37]. Furthermore, the 7α-hydroxylation of DHEA and pregnenolone by o, p'-DDT induced CYP7B1 could also facilitate elimination of DHEA or the synthesis of neuro-active hormones [38,39].
In addition to PXR-, CAR- or ER-mediated gene expression changes, o, p'-DDT induced Gadd45a, Gadd45b and Cdkn1, all of which are DNA damage-responsive genes [40,41]. Consistent with this are the reports of the DNA damaging potential of DDT [42-44]. Consequently, DNA damage may be an additional risk factor for tumor initiation/promotion following o, p'-DDT exposure in addition to PXR/CAR- and ER-mediated activities. Considering that the induction of DNA damage-responsive genes precedes Cyp2b10 or Cyp3a11 induction, the DNA damage may not be caused by oxidative stress derived from enzyme induction. Moreover, Gclm and Hmox, both known oxidative stress-responsive genes [45,46], exhibited relatively weak induction compared to rats [11], suggesting that oxidative stress was not strongly induced.
Conclusion
In conclusion, o, p'-DDT elicits a broad spectrum of species-conserved and specific effects. This includes PXR/CAR- and ER-mediated responses, altered steroidogenesis, oxidative stress, and DNA damage (Fig. 5). Although DDT is known to cause liver tumors in both mice and rats, the marked species differences in PXR/CAR structure, expression patterns and ligand preference as well as significant species-specific differences in steroidogenesis, especially CYP17A1 expression and activity, confound the extrapolation of these results to humans. Nevertheless, the identification of potential modes of action as well as species-specific responses may assist in the development or selection of more appropriate models for assessing the toxicity of DDT to humans and wildlife.
Figure 5.
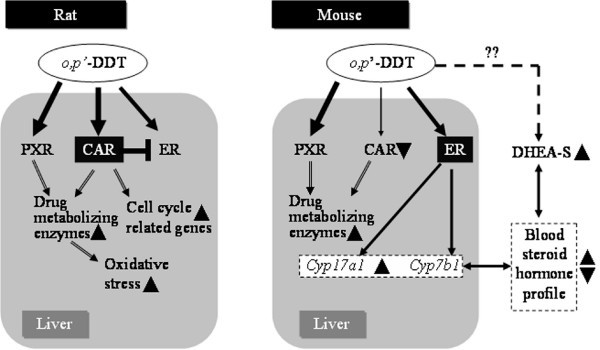
Potential species-specific responses to o, p'-DDT. In the rat liver, o, p'-DDT elicits PXR/CAR-mediated response, with negligible ER-mediated effects despite the established estrogenicity of o, p'-DDT in other target tissues (e.g., uterus). The absence of ER-mediated gene expression may be due, in part, to the inhibitory effects of CAR on ER. In contrast, o, p'-DDT elicits not only PXR/CAR-mediated responses but also ER-mediated effects such as the induction of Cyp17a1 and Cyp7b1 in the mouse liver. Gene expression profiling suggests that o, p'-DDT preferentially activates PXR in the mouse liver. Therefore, the inhibitory effects of CAR on ER-mediated signaling would be reduced in the mouse liver, allowing the activation of ER-mediated gene expression by o, p'-DDT. In addition, only mouse DHEA-S levels were increased, which could affect both endocrine and liver physiology. The induction of Cyp17a1 and Cyp7b1 may be involved in either the metabolism or disposition of DHEA, or the conversion of DHEA into other metabolites including neurosteroids.
Abbreviations
CAR: constitutive androstane receptor (Nr1i3); Casp9: caspase 9; Ccn: cyclin; Cdkn1a: cyclin-dependent kinase inhibitor 1A; Cyp: cytochrome P450; DDT: dichlorodiphenyltrichloroethane; DHEA-S: dehydroepiandrosterone sulfate; EE: ethynylestradiol; ER: estrogen receptor; FXR: farnesoid X receptor; Gclm: glutamate-cysteine ligase modifier subunit; Gadd45: growth arrest and DNA-damage-inducible 45; Gsr: glutathione reductase; GST: glutathione S-transferase; Hmox2: heme oxygenase (decycling) 2; HMGCS: 3-hydroxy-3-methylglutaryl-Coenzyme A synthase; Hsd3b: hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase; Insig2: insulin induced gene 2; Mdm2: transformed mouse 3T3 cell double minute 2; Nqo1: NAD(P)H dehydrogenase, quinone 1; Stmn1: stathmin 1; PXR: pregnane X receptor (Nr1i2); QRT-PCR: Quantitative Real-Time PCR; Srebf1: sterol regulatory element binding factor 1; Stat: signal transducer and activator of transcription; TCPOBOP: 1,4-bis-[2-(3,5,-dichloropyridyloxy)] benzene; Tnfrsf19: tumor necrosis factor receptor superfamily member 19; UGT: UDP glycosyltransferase.
Authors' contributions
NK performed all microarray, ELISA and Western assays, conducted and statistically analyzed QRT-PCR assays, as well as compiled and interpreted the data collected and generated the primary draft manuscript. JCK designed/conducted animal studies and participated in interpretation of microarray data. LDB provided database support for the microarray data including quality control assessments and statistical analysis. ED participated in microarray data analysis. KJW carried out histology examination. CT and BC carried out tissue level analysis. TRZ conceived the study and its design and supervised its completion. All authors read and approved the final draft of the manuscript.
Supplementary Material
Primer sequence.
o, p'-DDT-treated mouse liver time-course study, full data set. Blank gene expression ratios or p1(t) values are due to lack of or inapproprate statistical data for computation.
Differentially regulated genes in the o, p'-DDT-treated mouse liver.a Highligted values indicate |fold change| > 1.5. b Highlighted values indicate p1(t) > 0.999.
Correlation analysis between microarray results of o, p'-DDT-mouse and rat.
Acknowledgments
Acknowledgements
The authors thank Anna Kopec for helpful discussion and critical review of this manuscript. This study was supported by National Institute of General Medical Sciences (GM075838), U.S. Environmental Protection Agency (RD83184701). T.R.Z. is partially supported by the Michigan Agricultural Experiment Station
Contributor Information
Naoki Kiyosawa, Email: kiyosawa.naoki.wr@daiichisankyo.co.jp.
Joshua C Kwekel, Email: kwekeljo@msu.edu.
Lyle D Burgoon, Email: burgoonl@msu.edu.
Edward Dere, Email: dereedwa@msu.edu.
Kurt J Williams, Email: will1273@msu.edu.
Colleen Tashiro, Email: colleen@well-labs.com.
Brock Chittim, Email: brockc@well-labs.com.
Timothy R Zacharewski, Email: tzachare@msu.edu.
References
- Ito N, Tsuda H, Hasegawa R, Imaida K. Comparison of the promoting effects of various agents in induction of preneoplastic lesions in rat liver. Environ Health Perspect. 1983;50:131–138. doi: 10.2307/3429543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn KA, Abnet CC, Zhang M, Sun XD, Fan JH, O'Brien TR, Wei WQ, Ortiz-Conde BA, Dawsey SM, Weber JP, et al. Serum concentrations of 1,1,1-trichloro-2,2-bis(p-chlorophenyl)ethane (DDT) and 1,1-dichloro-2,2-bis(p-chlorophenyl)ethylene (DDE) and risk of primary liver cancer. J Natl Cancer Inst. 2006;98:1005–1010. doi: 10.1093/jnci/djj266. [DOI] [PubMed] [Google Scholar]
- Cocco P, Fadda D, Billai B, D'Atri M, Melis M, Blair A. Cancer mortality among men occupationally exposed to dichlorodiphenyltrichloroethane. Cancer Res. 2005;65:9588–9594. doi: 10.1158/0008-5472.CAN-05-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DDT and its derivatives http://www.inchem.org/documents/ehc/ehc/ehc009.htm
- Shelby MD, Newbold RR, Tully DB, Chae K, Davis VL. Assessing environmental chemicals for estrogenicity using a combination of in vitro and in vivo assays. Environ Health Perspect. 1996;104:1296–1300. doi: 10.2307/3432965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Katsura E, Takeuchi S, Niiyama K, Kobayashi K. Screening for estrogen and androgen receptor activities in 200 pesticides by in vitro reporter gene assays using Chinese hamster ovary cells. Environ Health Perspect. 2004;112:524–531. doi: 10.1289/ehp.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: xenosensors of endocrine disrupters? Chem Biol Interact. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Medina-Diaz IM, Arteaga-Illan G, de Leon MB, Cisneros B, Sierra-Santoyo A, Vega L, Gonzalez FJ, Elizondo G. Pregnane X receptor-dependent induction of the CYP3A4 gene by o, p'-1,1,1,-trichloro-2,2-bis (p-chlorophenyl)ethane. Drug Metab Dispos. 2007;35:95–102. doi: 10.1124/dmd.106.011759. [DOI] [PubMed] [Google Scholar]
- Ciana P, Raviscioni M, Mussi P, Vegeto E, Que I, Parker MG, Lowik C, Maggi A. In vivo imaging of transcriptionally active estrogen receptors. Nat Med. 2003;9:82–86. doi: 10.1038/nm809. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Fertuck KC, Burgoon LD, Eckel JE, Gennings C, Zacharewski TR. Temporal- and dose-dependent hepatic gene expression changes in immature ovariectomized mice following exposure to ethynyl estradiol. Carcinogenesis. 2004;25:1277–1291. doi: 10.1093/carcin/bgh114. [DOI] [PubMed] [Google Scholar]
- Kiyosawa N, Kwekel JC, Burgoon LD, Williams KJ, Tashiro C, Chittim B, Zacharewski TR. o, p'-DDT Elicits PXR/CAR-, Not ER-, Mediated Responses in the Immature Ovariectomized Rat Liver. Toxicol Sci. 2008;101:350–363. doi: 10.1093/toxsci/kfm275. [DOI] [PubMed] [Google Scholar]
- Poland A, Mak I, Glover E. Species differences in responsiveness to 1,4-bis[2-(3,5-dichloropyridyloxy)]-benzene, a potent phenobarbital-like inducer of microsomal monooxygenase activity. Mol Pharmacol. 1981;20:442–450. [PubMed] [Google Scholar]
- Pustylnyak VO, Gulyaeva LF, Lyakhovich VV. Induction of cytochrome P4502B: role of regulatory elements and nuclear receptors. Biochemistry (Mosc) 2007;72:608–617. doi: 10.1134/S000629790706003X. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Pustylnyak VO, Lebedev AN, Gulyaeva LF, Lyakhovich VV, Slynko NM. Comparative study of CYP2B induction in the liver of rats and mice by different compounds. Life Sci. 2007;80:324–328. doi: 10.1016/j.lfs.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Williams CE, Crawford RB, Holsapple MP, Kaminski NE. Identification of functional aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator in murine splenocytes. Biochem Pharmacol. 1996;52:771–780. doi: 10.1016/0006-2952(96)00360-7. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Zacharewski TR. dbZach toxicogenomic information management system. Pharmacogenomics. 2007;8:287–291. doi: 10.2217/14622416.8.3.287. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical bayes gene screening tool for time-course or dose-response microarray data. J Biopharm Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Vargas R, Jr, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Hess HA, Guo GL, Gonzalez FJ, Korach KS, Maronpot RR, Negishi M. Estrogen receptor alpha mediates 17alpha-ethynylestradiol causing hepatotoxicity. J Biol Chem. 2006;281:16625–16631. doi: 10.1074/jbc.M602723200. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Boutros PC, Dere E, Zacharewski TR. dbZach: A MIAME-compliant toxicogenomic supportive relational database. Toxicol Sci. 2006;90:558–568. doi: 10.1093/toxsci/kfj097. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Kwekel JC, Humes DG, Burgoon LD, Zacharewski TR. Dioxin induces an estrogen-like, estrogen receptor-dependent gene expression response in the murine uterus. Mol Pharmacol. 2006;69:1599–1606. doi: 10.1124/mol.105.019638. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Wagner M, Halilbasic E, Marschall HU, Zollner G, Fickert P, Langner C, Zatloukal K, Denk H, Trauner M. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology. 2005;42:420–430. doi: 10.1002/hep.20784. [DOI] [PubMed] [Google Scholar]
- Nagel SC, Hagelbarger JL, McDonnell DP. Development of an ER action indicator mouse for the study of estrogens, selective ER modulators (SERMs), and Xenobiotics. Endocrinology. 2001;142:4721–4728. doi: 10.1210/en.142.11.4721. [DOI] [PubMed] [Google Scholar]
- Giannitrapani L, Soresi M, La Spada E, Cervello M, D'Alessandro N, Montalto G. Sex hormones and risk of liver tumor. Ann N Y Acad Sci. 2006;1089:228–236. doi: 10.1196/annals.1386.044. [DOI] [PubMed] [Google Scholar]
- Columbano A, Ledda-Columbano GM, Pibiri M, Cossu C, Menegazzi M, Moore DD, Huang W, Tian J, Locker J. Gadd45beta is induced through a CAR-dependent, TNF-independent pathway in murine liver hyperplasia. Hepatology. 2005;42:1118–1126. doi: 10.1002/hep.20883. [DOI] [PubMed] [Google Scholar]
- Min G, Kim H, Bae Y, Petz L, Kemper JK. Inhibitory cross-talk between estrogen receptor (ER) and constitutively activated androstane receptor (CAR). CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J Biol Chem. 2002;277:34626–34633. doi: 10.1074/jbc.M205239200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yao ZX, Papadopoulos V. Cytochrome P450 17alpha hydroxylase/17,20 lyase (CYP17) function in cholesterol biosynthesis: identification of squalene monooxygenase (epoxidase) activity associated with CYP17 in Leydig cells. Mol Endocrinol. 2005;19:1918–1931. doi: 10.1210/me.2004-0271. [DOI] [PubMed] [Google Scholar]
- Brock BJ, Waterman MR. Biochemical differences between rat and human cytochrome P450c17 support the different steroidogenic needs of these two species. Biochemistry. 1999;38:1598–1606. doi: 10.1021/bi9821059. [DOI] [PubMed] [Google Scholar]
- Katagiri M, Tatsuta K, Imaoka S, Funae Y, Honma K, Matsuo N, Yokoi H, Ishimura K, Ishibashi F, Kagawa N. Evidence that immature rat liver is capable of participating in steroidogenesis by expressing 17alpha-hydroxylase/17,20-lyase P450c17. J Steroid Biochem Mol Biol. 1998;64:1–2. doi: 10.1016/S0960-0760(97)00164-7. [DOI] [PubMed] [Google Scholar]
- Imaoka S, Kamataki T, Funae Y. Purification and characterization of six cytochromes P-450 from hepatic microsomes of immature female rats. J Biochem. 1987;102:843–851. doi: 10.1093/oxfordjournals.jbchem.a122124. [DOI] [PubMed] [Google Scholar]
- Labrie F, Luu-The V, Labrie C, Simard J. DHEA and its transformation into androgens and estrogens in peripheral target tissues: intracrinology. Front Neuroendocrinol. 2001;22:185–212. doi: 10.1006/frne.2001.0216. [DOI] [PubMed] [Google Scholar]
- Muller C, Hennebert O, Morfin R. The native anti-glucocorticoid paradigm. J Steroid Biochem Mol Biol. 2006;100:95–105. doi: 10.1016/j.jsbmb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Tang W, Eggertsen G, Chiang JY, Norlin M. Estrogen-mediated regulation of CYP7B1: a possible role for controlling DHEA levels in human tissues. J Steroid Biochem Mol Biol. 2006;100:42–51. doi: 10.1016/j.jsbmb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Hollander MC, Fornance AJ., Jr Role of Gadd45 in apoptosis. Biochem Pharmacol. 2000;59:43–45. doi: 10.1016/S0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- Hassoun E, Bagchi M, Bagchi D, Stohs SJ. Comparative studies on lipid peroxidation and DNA-single strand breaks induced by lindane, DDT, chlordane and endrin in rats. Comp Biochem Physiol C. 1993;104:427–431. doi: 10.1016/0742-8413(93)90013-B. [DOI] [PubMed] [Google Scholar]
- Yanez L, Borja-Aburto VH, Rojas E, de la Fuente H, Gonzalez-Amaro R, Gomez H, Jongitud AA, Diaz-Barriga F. DDT induces DNA damage in blood cells. Studies in vitro and in women chronically exposed to this insecticide. Environ Res. 2004;94:18–24. doi: 10.1016/S0013-9351(03)00047-1. [DOI] [PubMed] [Google Scholar]
- Harada T, Yamaguchi S, Ohtsuka R, Takeda M, Fujisawa H, Yoshida T, Enomoto A, Chiba Y, Fukumori J, Kojima S, et al. Mechanisms of promotion and progression of preneoplastic lesions in hepatocarcinogenesis by DDT in F344 rats. Toxicol Pathol. 2003;31:87–98. doi: 10.1080/01926230390173941. [DOI] [PubMed] [Google Scholar]
- Bauer M, Bauer I. Heme oxygenase-1: redox regulation and role in the hepatic response to oxidative stress. Antioxid Redox Signal. 2002;4:749–758. doi: 10.1089/152308602760598891. [DOI] [PubMed] [Google Scholar]
- Kiyosawa N, Ito K, Sakuma K, Niino N, Kanbori M, Yamoto T, Manabe S, Matsunuma N. Evaluation of glutathione deficiency in rat livers by microarray analysis. Biochem Pharmacol. 2004;68:1465–1475. doi: 10.1016/j.bcp.2004.05.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequence.
o, p'-DDT-treated mouse liver time-course study, full data set. Blank gene expression ratios or p1(t) values are due to lack of or inapproprate statistical data for computation.
Differentially regulated genes in the o, p'-DDT-treated mouse liver.a Highligted values indicate |fold change| > 1.5. b Highlighted values indicate p1(t) > 0.999.
Correlation analysis between microarray results of o, p'-DDT-mouse and rat.