Abstract
Background
Geobacter species are δ-Proteobacteria and are often the predominant species in a variety of sedimentary environments where Fe(III) reduction is important. Their ability to remediate contaminated environments and produce electricity makes them attractive for further study. Cell motility, biofilm formation, and type IV pili all appear important for the growth of Geobacter in changing environments and for electricity production. Recent studies in other bacteria have demonstrated that signaling pathways homologous to the paradigm established for Escherichia coli chemotaxis can regulate type IV pili-dependent motility, the synthesis of flagella and type IV pili, the production of extracellular matrix material, and biofilm formation. The classification of these pathways by comparative genomics improves the ability to understand how Geobacter thrives in natural environments and better their use in microbial fuel cells.
Results
The genomes of G. sulfurreducens, G. metallireducens, and G. uraniireducens contain multiple (~70) homologs of chemotaxis genes arranged in several major clusters (six, seven, and seven, respectively). Unlike the single gene cluster of E. coli, the Geobacter clusters are not all located near the flagellar genes. The probable functions of some Geobacter clusters are assignable by homology to known pathways; others appear to be unique to the Geobacter sp. and contain genes of unknown function. We identified large numbers of methyl-accepting chemotaxis protein (MCP) homologs that have diverse sensing domain architectures and generate a potential for sensing a great variety of environmental signals. We discuss mechanisms for class-specific segregation of the MCPs in the cell membrane, which serve to maintain pathway specificity and diminish crosstalk. Finally, the regulation of gene expression in Geobacter differs from E. coli. The sequences of predicted promoter elements suggest that the alternative sigma factors σ28 and σ54 play a role in regulating the Geobacter chemotaxis gene expression.
Conclusion
The numerous chemoreceptors and chemotaxis-like gene clusters of Geobacter appear to be responsible for a diverse set of signaling functions in addition to chemotaxis, including gene regulation and biofilm formation, through functionally and spatially distinct signaling pathways.
Background
Chemotaxis is a trait shared by many bacteria that enables cells to move toward chemical attractants and away from repellents. The chemotaxis system of E. coli regulates flagellar-based motility; it has been studied in the great detail and has served as a paradigm for chemotactic motility [1,2]. However, it is now apparent from genomic, genetic and biochemical studies conducted with other bacteria that a diversity of pathway functions and purposes exist well beyond the E. coli paradigm [3-5].
The E. coli chemotaxis pathway includes 11 genes, most of which are organized in a cluster near the flagellar genes [6]. This cluster contains two of the five genes for the transmembrane chemoreceptors, which are also known as methyl-accepting chemotaxis proteins (MCPs), and a single gene for each of the chemotaxis signaling proteins: the autophosphorylating histidine kinase (CheA), a scaffold protein (CheW), a methyltransferase (CheR), a methylesterase (CheB), a response regulator (CheY), and a CheY phosphatase (CheZ). The other three MCP genes are distantly located in the genome. Chemotactic signals are detected by a membrane array of MCPs, to which CheW and CheA are bound, and regulate CheA-mediated phosphorylation of CheY and CheB. By binding to the flagellar motor protein, FliM, CheY phosphate (CheY~P) induces swimming E. coli to tumble, which has the effect of reorienting the direction of swimming. CheB~P reduces kinase activity by demethylating the MCPs, which reduces the rate of CheY~P (and CheB~P) formation, and consequently reduces the cell tumbling frequency. The tumble-promoting activity of CheY~P is also extinguished by the action of CheZ. Overall, this stimulus-response pathway biases swimming motion of E. coli toward attractants and away from repellents. Adaptation to stimuli, mediated by the reversible methylation of MCPs in the process catalyzed by CheR and CheB, allows cells to remain sensitive to small changes in chemoeffector concentration over a large range [7,8].
Analyses of bacterial genome sequences show that homologs of chemotaxis genes are widespread [3,7]. From these surveys, it is apparent that the MCP and che genes in E. coli are relatively few in number, which may plausibly reflect modest requirements for sensory transduction in the environment that E. coli inhabits. By comparison, the chemotaxis-like systems in other bacteria are greater in number and diversity [4,5]. The copies of the 'core' genes, e.g. cheAWY, are clustered in multiple distinct locations and additional genes are present (cheC, cheD, cheV and cheX) that generate greater mechanistic diversity [5]. For example, Armitage and colleagues have shown that two chemotaxis clusters in the genome of Rhodobacter sphaeroides play a role in chemotaxis [9], an observation that plausibly reflects the greater need for different signaling pathways in complex environments. Pertinent to the analysis that we present below, is the fact that Geobacter sp. also occupies complex ecological niches in sedimentary environments. The published genome of Geobacter sulfurreducens has 34 MCP genes and six major che gene clusters [10]; these pathways are likely to play an important role in environmental adaptation.
Biochemical, genetic and physiological investigations of chemotaxis-like signaling pathways in bacteria other than E. coli have led to the realization that some of these pathways carry out functions distinct from the well-established role in regulating flagellar motor rotation. These functions include regulation of type IV pili-dependent motility, the expression of the motility apparatus (both flagella and type IV pili) and biofilm formation. As examples, Pseudomonas aeruginosa, Rhodospirillum centenum, Myxoccocus xanthus, and Synechocystis sp. all have multiple chemotaxis-like operons that have provided new insight into their diverse functions. P. aeruginosa has four major che clusters; two are involved in chemotaxis with different suggested roles, a third that regulates type IV pili motility and biosynthesis, and the fourth is involved in biofilm formation [11-16]. R. centenum has three che clusters; one mediates chemotaxis, a second regulates cyst development, and a third regulates flagellar synthesis [17-19]. M. xanthus has eight clusters; the functions for four clusters have been identified to date [20]. Each cluster regulates a different function, including cell motility, biosynthesis of the motility apparatus, or regulation of developmental genes [21-25]. The functions for two of the three clusters found in the genome of Synechocystis PCC6803 have been identified: one regulates type IV-dependent motility, the other pilus biosynthesis [26]. In a final example, only one of three che clusters in the Vibrio cholera genome regulates chemotaxis. Mutations in the two remaining clusters do not affect chemotaxis; their functions are yet to be identified [27].
Geobacter species are Gram-negative δ-Proteobacteria and are predominant in the Fe(III)-reduction zone of sedimentary environments. The ability to remediate subsurface environments contaminated with organics or metals and to produce electricity from waste organic matter makes Geobacter attractive species for further study [28,29]. Geobacter species are facultative anaerobes that can oxidize organic compounds completely to CO2 by using metal ions, e.g. Fe(III), Mn(IV) and U(VI), or electrodes as electron acceptors [30]. Most of the electron acceptors for Geobacter species are insoluble under environmental conditions. To overcome this constraint, they have developed mechanisms of electron transfer from the cell interior to the electron acceptors outside the cell. Shewanella and Geothrix use either chelators that solubilize metal oxides, or electron-shuttling compounds that transfer electrons from the cell surface to the insoluble acceptors [31,32]. Geobacter species use a different mechanism of electron transfer, in which the cells make direct contact with insoluble electron acceptors [33]. Cell motility and other processes that involve the synthesis of extracellular structures to mediate electron transfer (type IV pili and extracellular matrix materials) are critical for Geobacter species survival in the environment, and could potentially be regulated by chemotaxis-like pathways.
Clearly, flagella and pili play important roles in Geobacter physiology. When grown with insoluble metal oxides, G. metallireducens produce flagella and pili [34]. Moreover, it is postulated that flagellar-based motility and chemotaxis bring G. metallireducens cells to the metal oxide surfaces more efficiently, and that pili promote attachment and/or transfer electron [34]. In G. sulfurreducens, it has been demonstrated that the cells make direct contact with insoluble oxides via nanowires, pili that are electrically conductive and are essential for oxide reduction [33], and recent evidence suggests that this mechanism is more widespread than first thought [35]. Other than a direct role in electron conduction, pili, and the other extracellular matrix molecules that are involved in biofilm formation, are being studied for the role they play in efficient electricity production in microbial fuel cells [36,37].
The diverse functions of chemosensory systems in other bacteria suggest intriguing roles for the Geobacter chemotaxis and chemotaxis-like pathways. Therefore, we conducted an analysis of the chemotaxis gene homologs in three Geobacter species with completed genome sequences: G. sulfurreducens, G. metallireducens and G. uraniireducens as an initial step to understand their cellular functions better. All three genomes were found to contain an abundance of che gene homologs, which were organized into six to seven gene clusters and subdivided into predicted operons. The chemoreceptor (MCP) genes have a different organization. While several are located in che clusters, most are dispersed throughout the genome – this organization is typical of genomes that contain numerous MCP genes, e.g. M. xanthus [20]. We predict the functions of a number of the Geobacter gene clusters by comparisons to clusters of known function in other species. We have also found clusters, which are, to our knowledge, unique to the Geobacter species; these may be particularly important to Geobacter physiology.
Methods
For protein sequence similarity searches, NCBI protein BLAST and position-specific-iterated-BLAST (blastp and psi-blast, respectively, http://blast.ncbi.nlm.nih.gov/) [38] were used with default parameter values against the genomes of G. sulfurreducens PCA, G. metallireducens GS-15, and G. uraniireducens Rf4 (GenBank accession numbers AE017180.1, CP000148.1 and CP000698.1, respectively, http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). To identify the Geobacter homologs of chemotaxis genes, E. coli, B. subtilis and Thermotoga maritima chemotaxis proteins were used as the test sequences, because these proteins are well-studied representatives, and are listed in the curated databases [39-41]. The following sequences were used: the E. coli aspartate receptor methyl-accepting (MA) domain (residues 267–514) (gi|16129838), the complete sequences of E. coli CheA (gi|1788197), CheB (gi|16129835), CheR (gi|16129836), CheW (gi|16129839), CheY (gi|16129834) and CheZ (gi|16129833); the complete sequences of B. subtilis CheC (gi|2634017), CheD (gi|2634018) and CheV (gi|2633772), and the complete sequence of T. maritima CheX (gi|81553634). ClustalW http://www.ebi.ac.uk/clustalw/ was used with default values for the parameters to conduct multiple sequence alignments to determine percent identities and to establish the class membership of the methyl-accepting domains [42]. TMHMM2 [43], TmPred [44], and TopPred [45] were used (with parameters set to default values) to predict the number of transmembrane helices in the putative methyl-accepting chemotaxis proteins. A polypeptide segment was designated a transmembrane α-helix when at least two of the three programs identified the same polypeptide segment as a transmembrane helix. Phylip (version 3.6) was used to construct CheA and CheY phylogenetic trees by the neighbor-joining method [46,47], as implemented in NEIGHBOR. SEQBOOT was used to generate 1000 bootstrap replicates and pairwise distances were estimated with PRODIST. The JTT model was used with no among-site variation. The trees were left unrooted.
The organization of che gene operons in the Geobacter sp. was predicted with FGENESB (Softberry Inc., http://www.softberry.com). FGENESB identifies protein-coding genes with Markov chain models of coding regions and translation start and termination sites, and annotates them with information from public databases. The sequence parameters (coding content, oligonucleotide composition, and gene length distribution) were estimated in FGENESB for each genome separately through an iterative procedure with the minimum ORF length set to 100 nt. Additional features, e.g. tRNA and rRNA, σ70 family promoters, and rho-independent terminators, were predicted from sequence similarity, linear discriminant analysis, or modeling approaches. FGENESB-based operon predictions were generated from the directions of adjacent genes, the distribution of intergenic distances, the presence or absence of predicted promoter and terminator regions, and the conservation of pairs of adjacent genes across microbial genomes (V. Solovyev, personal communication). The operon annotation of the G. sulfurreducens genome used in this study has been described previously [48], and is available online http://www.geobacter.org/research/gsel/[49].
σ54-regulated promoters were predicted from a search of the G. sulfurreducens genome with PromScan [50]. This software assigns a score representing the Kullback-Leibler distance, based on 186 known sites from 47 bacterial species [51]. The G. sulfurreducens genome was found to contain 110 predicted σ54-regulated promoters with a score equal to or greater than 80 (the default value) in noncoding regions upstream of genes and operons. The current accuracy of prediction is 78%, an estimate obtained from experiments that positively identified 14 RpoN-dependent regulation out of 18 predicted sites (J. Krushkal, C. Leang, M. Puljic, T. Ueki, R. Adkins, and D. Lovley, unpublished results). In addition, PromScan was used to look for σ54-regulated promoters upstream of the major che clusters in the G. metallireducens and G. uraniireducens genomes. Finally, putative σ28-regulated promoters upstream of the flagellar filament gene (fliC) and the major che clusters in the genomes of G. sulfurreducens, G. metallireducens and G. uraniireducens were identified with Virtual Footprint [52] and the Neural Network Promoter Prediction software for bacterial species [53]. Five hundred base pairs upstream of the putative initiation codons of genes of interest were analyzed using default parameters.
Results and discussion
Geobacter Chemotaxis Genes: Numbers and Organization
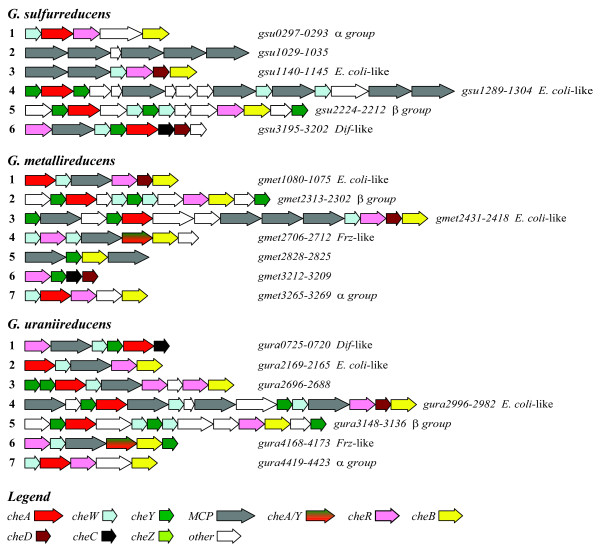
BLAST analysis of the G. sulfurreducens, G. metallireducens, and G. uraniireducens genomes identified multiple copies of the chemotaxis genes; over 60 genes in each species were homologous to the known che and mcp genes in E. coli, B. subtilis and T. maritima (Table 1). Homologs of all the che genes from E. coli were present in the Geobacter species, except cheZ, which is found much more frequently in genomes of β- and γ-proteobacteria in comparison to the genomes of α-, ε-, and especially δ-proteobacteria [54]. The Geobacter genomes also contained cheC, cheD, cheV, and cheX homologs. With the exception of the genes for the chemoreceptors – the methyl-accepting chemotaxis proteins (MCPs), which were dispersed throughout the genomes, most of the che genes were clustered, as shown in Figure 1. In some cases, additional genes encoding hypothetical proteins of unknown function or annotated proteins with functions not known to be involved in chemotaxis-related signaling pathways were located in these clusters. There are six major chemotaxis-related gene clusters in G. sulfurreducens, and seven major clusters each in G. metallireducens and G. uraniireducens; their physical arrangements are depicted in Figure 1. None of these clusters is located close to the flagellar gene clusters.
Table 1.
Numbers of che gene homologs in E. coli, B. Subtilis and Geobacter sp.a
| Species | |||||
| Gene | E. coli | B. subtilis | G. met. | G. sul. | G. ura. |
| cheAb | 1 | 1 | 5 | 4 | 7 |
| cheB | 1 | 1 | 8 | 4 | 5 |
| cheR | 1 | 1 | 9 | 5 | 10 |
| cheW | 1 | 1 | 8 | 10 | 10 |
| cheYc | 1 (1) | 1 (3) | 10 (21) | 7 (25) | 10 (25) |
| cheZ | 1 | 0 | 0 | 0 | 0 |
| cheC | 0 | 1 | 2 | 1 | 1 |
| cheD | 0 | 1 | 3 | 3 | 2 |
| cheX | 0 | 0 | 1 | 1 | 1 |
| cheV | 0 | 1 | 1 | 1 | 1 |
| mcp | 5 | 10 | 18 | 34 | 24 |
| Total | 11 | 17 | 65 | 70 | 71 |
| No. of che clustersd | 1 | 1 | 7 | 6 | 7 |
aThe numbers of homologs were determined by blastp searches (with default values for the parameters).
bThe number of cheA homologs in the genomes of G. met. and G. ura. includes a contribution from one cheAY fusion.
cThe numbers are cheY genes in the major clusters. Numbers in parentheses also includes genes that encode for singleton CheY-like receiver domain proteins.
dChemotaxis gene clusters are defined to contain three or more che genes.
Figure 1.
Physical arrangement of the major Geobacter chemotaxis-like gene clusters. Affiliations with che clusters of known function are indicated after the clusters, as E. coli-like, Dif-like and Frz-like (both from M. xanthus), and the α and β groups. These assignments were made by the relative agreements between che gene content, the physical arrangement in the cluster and the percent identities. The α and β group designations refer to che clusters that are unique to the Geobacteraceae and the δ-Proteobacteria, respectively.
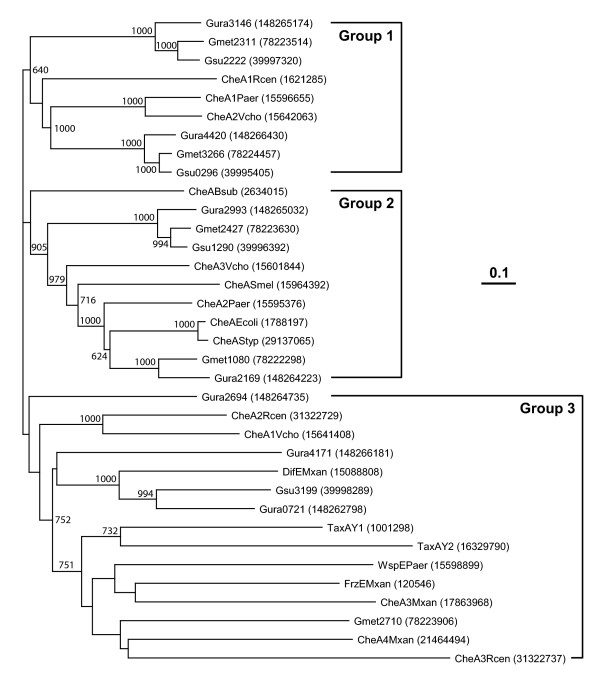
The genomes of G. sulfurreducens, G. metallireducens and G. uraniireducens code four, five and seven predicted cheA genes, respectively. The homologs encoded by the cheA genes are clustered in three groups of the phylogenetic tree (Figure 2), demonstrating that the multiple cheA genes did not result from recent gene duplication events, but are paralogs that have been evolving separately for some time, which suggests that they play distinct cellular roles. Each CheA homolog, together with the other cognate che gene products, is likely to regulate a separate chemotaxis-like pathway. The presence of multiple che homologs and clusters are a strong indication of different pathways that raise intriguing questions about function, and whether or not the pathways are redundant or exhibit crosstalk. By comparing the gene order and the percent identities of the gene products from other bacteria, in which chemotaxis and chemotaxis-like pathways are studied extensively, we were able to predict the functions for many of the Geobacter che clusters. From this analysis, it seems unlikely that different clusters constitute redundant pathways; instead, each pathway has a distinct function. In addition, plausible mechanisms to reduce unwanted crosstalk between pathways emerged.
Figure 2.
Neighbor-joining tree of putative CheA homologs of Geobacters and CheAs from other well studied species. These include Escherichia coli (Ecoli), Bacillus subtilis (Bsub), Pseudomonas aeruginosa (Paer), Sinorhizobium meliloti (Smel), Rhodospirillum centenum (Rcen), Vibrio cholerae (Vcho), Myxococcus xanthus (Mxan), Salmonella typhimurium (Styp) and Synechocystis sp. strain PCC6803 (Syne). The GenInfo Identifier protein sequence numbers are displayed in parentheses at right. All positions with gaps in the aligned sequences were excluded. Bootstrap values from 1000 replicates of >600 are shown in respective nodes. The tree figure was generated with TreeView, version 1.6.6 [96].
The Geobacter genomes are predicted to have large numbers of standalone response regulators proteins that are comprised only of the receiver domain [55]; we refer to these as CheY-like proteins. The G. metallireducens, G. sulfurreducens, and G. uraniireducens genomes have 21, 25, and 25 homologs, respectively, but the majority is probably not involved in chemotaxis-like signaling [54]. Only 38% of the homologs are located in the major che or flagellar gene clusters (Table 1), the remainder (11, 18 and 15, respectively) are located elsewhere on the chromosome. Of those we suspect to play a role in chemotaxis-like signaling, i.e. the cheY genes that are located in the major che or flagellar gene clusters, about 50% reside in a branch of the phylogenetic tree with E. coli and Salmonella CheY (four apiece from G. metallireducens and G. sulfurreducens; five from G. uraniireducens, Figure S1) [see Additional file 1]. These CheY homologs are most likely to have response regulator functions as the substrates of CheA-mediated phosphorylation in chemotaxis pathways. The Geobacter CheY-homologs located elsewhere in the tree (relatively distant to E. coli and Salmonella CheY), but are situated in che or flagellar gene clusters on the chromosome, probably also function in chemotaxis-like pathways, perhaps in some other manner. By contrast, the genes encoding the most distantly-related CheY-like proteins, i.e. located outside che clusters, away from the flagellar genes, and are (relatively) distant to E. coli and Salmonella CheY in the phylogenetic tree (Figure S1) [see Additional file 1], probably function in other two-component pathways. For instance, B. subtilis and Nostoc CheY-like homologs, which are not in the che clusters, are involved in two-component pathways unrelated to chemotaxis [56,57]. Therefore, we postulate that the standalone receiver proteins encoded within the Geobacter che and flagellar gene clusters plausibly represent predicted CheYs with chemotaxis-like pathway function.
Number and Diversity of Geobacter MCPs
The three Geobacter genomes investigated in this study were found to have significant numbers of genes for MCPs: 34 in G. sulfurreducens, 18 in G. metallireducens, and 24 in G. uraniireducens (Table 1). These putative MCPs were identified through the presence of the highly conserved methyl-accepting (MA) domain, which was first assigned a biochemical function in the E. coli chemoreceptors as the domain methylated in a CheR-dependent process [58,59]. The large number of MCP-coding genes in the Geobacter genomes, by comparison to either E. coli or B. subtilis, plausibly reflects a greater need to detect sensory stimuli in the subsurface environment. With the exception of the aerotaxis receptor, all E. coli MCPs have periplasmic ligand-binding domains that detect the external chemoeffector concentrations, two transmembrane (TM) helices, and the (juxtamembrane) HAMP and the methyl-accepting (MA) domains located in the cytoplasm. The sequences of the predicted Geobacter MCPs reveal significantly greater diversity in the domain organization and architecture of the sensing domains (Figure S2) [see Additional file 1].
The N-terminal regions of MCPs sense various environmental stimuli through diverse means, because the length and heterogeneity of these regions are greater, compared to the cytoplasmic domains, which are mostly organized like the E. coli MCPs (a single HAMP domain followed by the MA domain). Domain architectures of representative Geobacter MCPs are shown in Figure S2 and Table S1 [see Additional file 1]. With respect to transmembrane (TM) segments, the Geobacter MCPs fall into three groups according to the number of predicted TM helices (zero, one or two). Of the 76 predicted MCPs found in the genomes of G. metallireducens, G. sulfurreducens and G. uraniireducens, ninety percent have two TM helices. Most of these, 80%, have periplasmic domains that are ~150–200 amino acid residues (aa) in length, which are most similar in size to the periplasmic domains of the major E. coli MCPs. Three percent of the Geobacter MCPs have larger periplasmic domains (~250–430 aa), while the others have a significantly smaller domain (<100 aa). MCPs with the Tar-like and larger periplasmic domains probably detect signals through these domains by ligand binding. While the structures of most of these MCPs are not known, the sensory domains of two MCPs (Gsu0935 and Gsu0582) are PAS domains with covalently-bound hemes [60]. Interestingly, this places redox-active sensing domains in the periplasm. By contrast, the MCPs with small periplasmic domains are more likely to detect signals via associations with other proteins, as in the case of DifA of M. xanthus [24,61], or detect intracellular signals when the MCPs have no TM segments [4].
MCP MA domains were recognized to belong to a superfamily based on a multiple sequence alignment first conducted by Le Moual and Koshland [59]. A more recent analysis of approximately 2000 MCPs identified seven classes (named 24 H, 28 H, 34 H, 36 H, 38 H, 40 H and 44 H), which are defined by the number of heptad repeats (H) in the cytoplasmic domain [62]. The most well characterized MCPs of E. coli, Tar and Tsr, belong to class 36 H, and the MCPs from T. maritima (TM1143) and B. subtilis (McpA and McpC) belong to class 44 H. Multiple-sequence-alignments of the Geobacter MCPs revealed that G. metallireducens has MCPs in classes 24 H, 34 H, 36 H and 40 H; G. sulfurreducens has MCPs in classes 24 H, 34 H, 40 H and 44 H, and G. uraniireducens has MCPs in classes 24 H, 34 H, 36 H, 40 H and 44 H. The majority of the MCPs are members of class 34 H (17, 24, and 21% in G. metallireducens, G. sulfurreducens and G. uraniireducens, respectively) and class 40 H (61, 71 and 46%, respectively). [Additional file 2 is the multiple sequence alignment of the Geobacter MCPs.] G. metallireducens and G. uraniireducens each have one MCP in class 36 H (Gmet1078, Gura2167), and G. sulfurreducens and G. uraniireducens each have one MCP in class 44 H (Gsu3196, Gura0724).
MCPs and Che proteins form specific clusters. In E. coli, all the MCPs and most of the Che proteins are found in clusters that are often located at the cell poles [63-65]. When bacteria have two or more chemotaxis (or chemotaxis-like) gene clusters, the clusters are observed to have distinct locations and compositions [66-68]. We speculate that MCP class membership is a contributing factor of cluster specificity. According to this reasoning, MCPs in the same class are more likely to belong to the same cluster, and conversely, MCPs in different classes are likely to segregate. Cluster formation, in part, is generated by contacts between the MCP MA domains. The MA domain is a coiled-coil hairpin that dimerizes to form a long four helix bundle [69,70], where the bundle length is determined by the number of heptad repeats, ~210 Å for class 36 H MCP (E. coli Tsr) and ~260 Å for class 44 H MCP (T. maritima TM1143). We postulate that class-specific MCP clusters are more likely to form for the following reason: two different MCPs, which contain MA domains belonging to the same class, are more likely to engage in the interactions that lead to the formation of clusters than two MCPs that contain MA domains from different classes (and therefore different MA-domain lengths).
The localization of P. aeruginosa and R. sphaeroides protein clusters provide some support for class-specific cluster formation. P. aeruginosa McpB and WspA, which are found in distinct signaling clusters in distinct locations (polar and lateral, respectively), belong to different classes (36 H and 40 H, respectively) [66,67]. R. sphaeroides McpG and TlpT (a soluble MCP) belong to different MA classes (34 H and 36 H, respectively) and locate in different clusters (polar membrane and cytoplasmic locations, respectively) [68]. We anticipate that the multiple classes of MCPs present in the Geobacter species contribute to the formation of segregated MCP signaling clusters. On the other hand, MA class membership is certainly not the only factor to consider. For example, this mechanism cannot easily explain the localization of MCPs that do not belong to any class [62]. In addition, the compositions of signaling clusters are influenced undoubtedly by the specificity of interactions between the different MA domain and Che protein homologs. These effects (and others), considered together, can contribute to the assembly of specific signaling units, which function in the same cell without unwanted crosstalk.
The Prevalence and Specificity of CheR Tethering Segments
The role of a semi-conserved pentapeptide at the C-terminus of some MCPs, and first observed in the E. coli high abundance receptors Tar and Tsr (NWETF) [71], has a well established role in sensory adaptation by mediating efficient receptor methylation and demethylation [7,71-74]. In the process of receptor methylation, the pentapeptide NWETF binds to the β-subdomain of CheR at a location that is distinct from the active site – methylation site interaction [75]; this interaction tethers CheR near the methylation sites of clustered receptors. It is plausible to expect that all MCPs containing the C-terminal NWETF or a pentapeptide similar to NWETF provide adaptational assistance via the mechanism established in E. coli [73,76]. MCPs that contain the CheR-binding pentapeptide are restricted primarily to the Proteobacteria; the genomes of bacteria in other phyla reveal few, if any, MCPs that contain a recognizable CheR-tethering segment, as defined previously [7]. In such species – for example B. subtilis and T. maritima, methylation operates through a different, pentapeptide-independent mechanism [77]. Less than 10% of the ~2500 MCPs listed in the SMART database of completed bacterial genomes contain a recognizable CheR tethering segment; this segment always follows the MA domain (SM00283) in the primary sequence of the MCP, which then ends in a pentapeptide that binds CheR [7]. Therefore, many MCPs are probably methylated and demethylated via a pentapeptide-independent mechanism.
Closer analysis of all the MCPs that contain the NWETF pentapeptide or a similar pentapeptide, reveal a restricted class membership, either to class 34 H or to class 36 H [62]. 85% of these MCPs belong to class 36 H and contained the class-specific xWxxF pentapeptide motif; 15% belong to class 34 H and contained the class-specific xF/YxxF/Y motif for the pentapeptide [7]. In contrast to the kingdom-wide percentages, most pentapeptide-containing Geobacter MCPs belonged to class 34 H (100%, 75%, and 80% for G. sulfurreducens, G. metallireducens and G. uraniireducens, respectively). G. metallireducens and G. uraniireducens have one mcp gene apiece in the class 36 H with a C-terminal DWKEF pentapeptide, a sequence more similar to the E. coli consensus (NWETF). Using the pentapeptide-containing MCPs as one criterion, we defined the che clusters to which these mcp genes belong as 'E. coli-like' clusters (Figure 1).
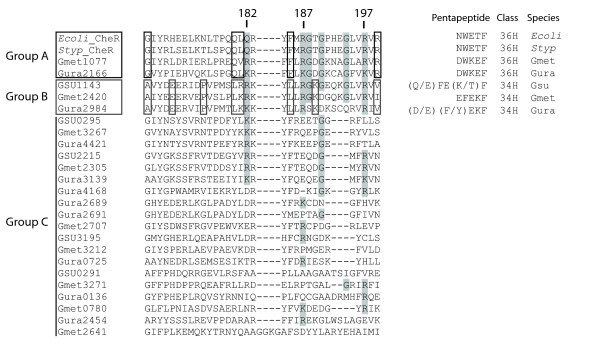
To identify possible class-specific MCP-methyltransferase tethering interactions, we compared the aligned β-subdomain sequences of the Geobacter CheR homologs to the Salmonella and E. coli CheR sequences. The Salmonella CheR structure, co-crystallized with the NWETF pentapeptide has enabled the identification of residues in the β-subdomain that are involved in the peptide-CheR interaction (Q182, G188, R187, G190, G194 and R197, numbered according to Salmonella CheR, PDB# 1bc5) [75,77]. Figure 3 shows aligned sequences from the β-subdomain of all the Geobacter CheR homologs, together with the E. coli and Salmonella sequences (residues 166–199). Using this alignment, we divided the Geobacter CheRs into three groups. Two groups (A and B) displayed significant identity with residues important for binding a pentapeptide; the third and largest group (C) did not (Figure 3). Consequently, we concluded that the CheR homologs in Group C probably do not methylate MCPs by the E. coli mechanism.
Figure 3.
Alignment of the beta-subdomain of Geobacter CheR homologs with E. coli and S. enterica CheR. Based on homology, the Geobacter CheRs were divided into three groups. Two groups (A and B) displayed significant identity with residues important for binding pentapeptide (highlighted in grey) and the third group (C) did not. Gene positioning provides further evidence that the group A and B homologs bind to MCPs containing the C-terminal pentapeptide: these homologs are located in che clusters with pentapeptide-containing MCPs (Figure 1). Group A consists of two CheR homologs that are located near two class-36 H MCPs. The consensus pentapeptide of the MCPs that are cognate to the Group B CheR homologs, EFEKF, is found in class-34 H MCPs.
The colocalization of mcp and CheR genes within the same clusters provided evidence that group A and B CheR homologs bind to MCPs containing a C-terminal pentapeptide; these CheR homologs are located in che gene clusters containing at least one gene that encodes a pentapeptide-containing MCP (Figure 1). The two CheR homologs that comprise group A are located adjacent to class-36 H MCPs (Gmet1078, Gura2167) and have DWKEF as the C-terminal pentapeptide – judged to be more similar to the E. coli consensus (NWETF). By contrast, the consensus pentapeptide coded by mcp genes located in the che gene clusters with group B CheR homologs is EFEKF. All 14 MCPs that contain this consensus pentapeptide belong to class 34 H (Figure 3) [also see Additional file 2], and 10 of these are located in the che gene clusters that contain the group B CheR genes (Figure 1). Differences in the consensus pentapeptide for class 34 versus class 36 MCPs correlate with differences in the β-subdomain amino acid residues in pentapeptide-binding pocket of group A versus B CheR homologs (respectively, Figure 3). Thus, it is plausible that these differences contribute to (and reflect) class-specific MCP-CheR interactions.
By contrast, all group C CheR homologs are either (a) not located in a che gene cluster, (b) located in che clusters that do not contain an mcp gene, or (c) located in che clusters that contain genes encoding class 40 H or 44 H MCPs, These MCPs do not contain recognizable CheR tethering segments (terminating in a pentapeptide) according to criteria defined previously [7]. Thus, it is probable that group C CheRs use a pentapeptide-independent mechanism for receptor methylation, similar to that observed with T. maritima [77]. In addition, we interpret the specific pairings within the che gene clusters, of the CheR groups (A, B, C) and the MCP classes (36 H, 34 H, 40 H/44 H, respectively), as support for the idea of class-specific receptor signaling.
Predicted Function of E. coli-like Chemotaxis Clusters
Above, we defined Geobacter che clusters operationally as 'E. coli-like' by the presence of one or more mcp genes that encode MCPs with CheR tethering segments. In addition, the Geobacter CheA homologs in these clusters belong to the same phylogenetic grouping as E. coli CheA (Figure 2). These clusters were sorted further into two types. Type 1 clusters – clusters one and two in the G. metallireducens and G. uraniireducens genomes, respectively (Figure 1), have significant resemblances to the E. coli mocha-meche cluster, judged by the gene order and by the percent identities between predicted Geobacter proteins and the E. coli proteins (Figure 2 and Figure S3) [see Additional file 1]. Notably, the Geobacter mcp genes in these two clusters encode for MCPs that belong to class 36 H, the same as E. coli MCPs.
Type 2 clusters are also characterized by significant sequence identity (although lower than Type 1), but the gene positions bear a comparatively small resemblance to the E. coli cluster. Moreover, the Type 2 clusters contain predicted ORFs in significant numbers that have unknown function or assigned functions other than chemotaxis. (See Figure S3 [Additional file 1] for comparisons of gene arrangement and the percent identities of individual gene products.) The Type 2 clusters possess multiple genes coding for MCPs that belong to class 34 H; many of these contain a CheR-tethering segment that terminates in an 'NWETF-like' pentapeptide at the C-terminus.
Three features distinguish the E. coli-like Geobacter che clusters from the E. coli cluster. (1) The E. coli meche operon contains cheZ, but Geobacter genomes do not, so a CheZ-independent signaling mechanism must operate in Geobacter pathways. (2) Multiple CheW genes are found in each cluster (except for G. metallireducens), an observation made previously with other bacteria. Studies of the CheW homologs in R. sphaeroides have led to the suggestion that these homologs do not perform redundant functions, but engender MCP-specific interactions, a proposal based on observed differences in binding affinity [9]. It has also been suggested that multiple CheWs allow additional MCPs to be incorporated within the chemosensory system, since there tends to be more mcp genes than cheW genes [11]. Another interesting hypothesis has been proposed: the different CheWs may recognize MCPs in a class-specific manner, which produces different specific signaling pathways in a che cluster [54]. (3) Finally, cheD and other non-che genes – not found in the E. coli chemotaxis cluster, are present in the E. coli-like Geobacter Type 2 clusters. By analogy to the functions assigned in B. subtilis and T. maritima, the presence of CheD signifies that a different mechanism is in play for deamidating MCPs and regulating CheY~P hydrolysis [78,79]. The presence of genes with unknown function within chemotaxis operons has been reported in various bacteria, and appears to be commonplace in bacteria with more complex chemotaxis pathways [27,80-82].
Based on these observations, we suggest that the Type 1 che clusters functions like the E. coli chemotaxis pathway, albeit with the differences noted above, in which case che cluster 1 of G. metallireducens regulates signaling through a lone class 36 H MCP (Gmet1078) [34]. If cluster 2 of G. uraniireducens serves a similar role, then a lone 36 H MCP (Gura2167) serves to detect the environmental stimuli in this situation as well. According to this reasoning, we do not expect G. sulfurreducens PCA (AE017180.1) to have a flagellar-based chemotaxis pathway that uses this signaling logic, because it lacks both a Type1 E. coli-like che gene cluster and class 36 H MCPs. However, the absence of class 36 H MCPs does not rule out other modes of flagellar-based motility or chemotaxis. For example, the chemotaxis pathway in B. subtilis uses class 44 H MCPs, and the genome of G. sulfurreducens contains several mcp genes that belong to this class. Fewer investigations of Type 2 E. coli-like che clusters have been conducted, yet in their study of a Type 2-like cluster in R. sphaeroides, Armitage and colleagues found that this cluster is essential for flagellar-based motility [83]. All the Geobacter sp. genomes contain at least one Type 2 cluster; these too could potentially participate in flagellar-based chemotaxis. Further work is needed to verify the actual functions and relationships of Type 1 and 2 E. coli-like clusters, which will serve strengthen the confidence of predictions based on percent gene identity, gene cluster organization and mechanistic similarities reflected in protein organization.
Dif-like Clusters May Regulate Extracellular Matrix Formation and Chemotactic Motility
G. sulfurreducens and G. uraniireducens possess clusters comprised of similar genes and gene ordering to the dif cluster of M. xanthus. These clusters contain class 44 H MCPs with two predicted transmembrane segments, but small periplasmic domains (~3 to 10 aa), genes for CheA, CheW, CheY, CheC and CheD, and genes with unidentified function. (Figure S4 lists the gene arrangements and percent identities of the individual gene products [Additional file 1].) The dif signaling system of M. xanthus has been studied most, where it is known to be involved in the regulation of exopolysaccharide formation, an essential component of the Myxococcus social motility apparatus [61,84]. It has been noted that phenomenon of social motility in M. xanthus resembles biofilm formation in other bacteria [85]. In addition, the dif cluster is also involved in sensing of certain lipids [24]. One difference between the M. xanthus dif cluster and the Geobacter dif-like clusters is the presence of CheR in the Geobacter cluster instead of difB. A plausible consequence of this observation is that the Geobacter dif-like pathways are CheR-dependent, whereas the M. xanthus dif system is CheR-independent. The involvement of Geobacter dif-like clusters in the synthesis of extracellular matrix material, which is essential for biofilm formation, is currently under investigation.
Che Clusters with CheA/Y Fusion Proteins
G. metallireducens and G. uraniireducens each have one che cluster with a gene that encodes a CheA-CheY fusion protein (CheA/Y). In R. centenum, M. xanthus, P. aeruginosa, and Synechocystis strain PCC6803, CheA/Y-containing che clusters carry out various functions, including the regulation of flagellar-based motility [86,87], typeIV-pili based motility and/or the biosynthesis of typeIV pili [21,25,26], cell development [17,23], and biofilm formation [16]. The Geobacter che clusters in Figure 1 that encode CheA/Y fusion proteins are most similar to the M. xanthus Frz cluster, cluster 3 of P. aeruginosa, and cluster 3 of R. centenum. This conclusion was reached through comparisons of the gene cluster content, gene order and the percent identity among CheA/Y homologs (Figure S5) [see Additional file 1]. These gene clusters function in developmental cell aggregation [88], biofilm formation [16], and cyst cell development [17], respectively – processes that involve cell-cell interaction. By these same criteria, the Geobacter clusters were least similar to che cluster 1 of R. centenum (chemotactic and photactic responses [86]), M. xanthus cluster 3 (the regulation of fruiting body formation [23]) and Synechocystis cluster 2 [26]. Overall, these findings suggest that the corresponding Geobacter che clusters may also regulate processes involving cell-cell interactions and/or social motility, but this idea is in need of experimental proof.
Che Clusters that are Unique to the Geobacter Species and δ-Proteobacteria
Two groups of che clusters are highly conserved among the Geobacter sp., we refer to these as α and β groups. The clusters belonging to these two groups contain the well-known homologs of chemotaxis genes (cheA, cheW, cheB, and CheR), but no mcp genes. Cluster 1 of G. sulfurreducens and cluster 7 of G. metallireducens and G. uraniireducens belong to the α group; the β group che clusters are 5, 2 and 5, respectively (Figure 1). An extensive search of both completed and draft bacterial genomes led to the finding that group α che gene clusters are present only in the genomes of the Geobacteraceae, including Geobacter bemidjiensis Bem ctg130, Geobacter lovleyi SZ, Pelobacter propionicus DSM 2379, and Geobacter sp. FRC-32.
Group α clusters have not been found in genomes outside the Geobacteraceae family. (See Figure S6 [Additional file 1] for gene arrangements and percent identities.) Each group α cluster contains a gene encoding a protein with an HD domain – which defines membership in an enzyme superfamily of metal-dependent phosphohydrolases, where the conserved His-Asp (HD) doublet has a role in catalysis [89]. Within a variety of contexts, HD-domain-containing proteins have diverse biochemical functions, including nucleic acid metabolism and signal transduction. The predicted Geobacter homologs contain no other recognizable domains, i.e. they may function as standalone proteins. Standalone HD domain proteins in E. coli have low amino acid identity with each other and to the Geobacter homologs (~10%), yet the E. coli proteins all act on nucleotide substrates [90]. The predicted HD domain proteins located within the group α che clusters are probably regulated by, or participate in, chemotaxis-like signaling pathways of special significance to the cellular physiology of Geobacter.
Group β clusters are conserved among δ-Proteobacteria, and have been detected in the genomes of G. bemidjiensis, G.lovleyi,Geobacter sp. FRC-32, Stigmatella aurantiaca DW4/3-1, Anaeromyxobacter dehalogenans 2CP-C, Plesiocystis pacifica SIR-1, and Myxococcus xanthus DK 1622. (See Figure S6 [Additional file 1] for gene arrangements and percent identities.) In general, the functions of β group clusters are not known. However, we have obtained preliminary results with G. sulfurreducens cluster 5 knockout mutants, which indicate that this β group cluster regulates the expression of extracellular matrix material, and may represent a new way that chemotaxis-like signaling pathways can participate in biofilm formation (HT Tran, DR Lovley and RM Weis, unpublished observations).
Chemotaxis Gene Expression Regulated by Alternative Sigma Factors 28 and 54
The mechanisms for regulating the expression of chemotaxis and flagellar genes are complex, but diverse, and should provide clues to the diversity and purpose of chemotaxis-like signaling systems. Therefore, we conducted a preliminary investigation into the regulation of che gene expression, in particular σ28- and σ54-regulated promoters upstream of che and flagellar gene clusters. In E. coli and Salmonella, the che and late flagellar genes, including fliC (the flagellar filament), are positively regulated by σ28 [91-93]. In other bacteria, especially those with more than one che cluster, expression is also regulated by σ54 [94]. For instance, R. sphaeroides has a σ28-regulated system that shows coupled expression of the chemotaxis proteins and flagella, and a system that regulates flagellar synthesis independently via σ 54 [95].
We searched upstream of the major Geobacter che operons and fliC loci for evidence of σ28-regulated expression. As Figure 4A shows, σ28 binding sites were identified upstream of fliC in G. metallireducens, G. sulfurreducens and G. uraniireducens, but only one major che cluster, the G. sulfurreducens group α cluster (cluster 1, Figure 1), had a recognizable σ28 binding site. Therefore, it seems that the specific mechanisms of regulation for most of the Geobacter che clusters will be different from E. coli (and Salmonella).
Figure 4.
Putative σ28 and σ54 promoter elements. (A) Putative σ28 promoter regulation sites found upstream of G. sulfurreducens che cluster 1 and the fliC genes of G. sulfurreducens (gsu3038), G. metallireducens (gsu0442), and G. uraniireducens (gsu4096) [97]. 125, 160, 127, and 152 nucleotide bases separate the predicted transcription start sites from the start codons, respectively. (B) Putative σ54 promoter elements upstream of the Geobacter major che gene clusters [51]. For G. metallireducens clusters 1 and 6, G. uraniireducens cluster 3, and G. sulfurreducens cluster 6, the predicted transcription start sites are 50, 33, 24 and 16 nucleotide bases upstream of predicted operon ATG start codons, respectively.
The G. sulfurreducens genome was searched for σ54 recognition sites to determine the number of che gene-related sites relative to all the sites that may exhibit σ54 regulation. Of the 110 sites identified genome-wide, nine were located in noncoding regions upstream of che, mcp or flagellar operons (Figure S7 lists positions and sequences of the chemotaxis-related promoter sites [Additional file 1]) – one of these was the Dif-like cluster (cluster 6, Figure 1). Focused searches upstream of the major che clusters in the other two species identified possible σ54-regulated promoter sites before cluster 3 in G. uraniireducens, and clusters 1 and 6 in G. metallireducens (Figure 4B). No correlation was apparent between the identity of these clusters and their mode of regulation, i.e. G. metallireducens cluster 3 is classified as an E. coli-like cluster, and the other two do not belong to any identified class. Consequently, little specific insight can be gleaned from these early findings. Nonetheless, the results may presage a diversity of mechanisms for regulating expression. Indeed, we can expect that once the che-gene-specific regulatory elements are known (which is significant in itself), it will be a challenge to determine how these systems map onto the global patterns of gene expression; this pattern should reflect how Geobacter adapts to the complex environment it inhabits.
Conclusion
The comparative analysis of che gene clusters and regulatory sequences among Geobacter sp. and to other bacteria has provided valuable insight into the functions of the various Geobacter chemotaxis-like pathways. The genomes of Geobacter sp. have multiple copies of chemotaxis genes – more than 60 per genome. Their arrangement in six to seven major clusters reflects both greater complexity and diversity in comparison to the single cluster on the E. coli chromosome. This diversity is also reflected in the presence of both σ54- and σ28-dependent regulatory sequences. The presence of multiple chemotaxis-like clusters and mechanisms of regulation both suggest that the pathways in Geobacter are not redundant, but instead each fills a specific cellular need.
Geobacter species have several clusters in addition to a che cluster that is similar in organization to the chemotaxis operons of E. coli and S. enterica. These clusters are similar to known clusters in other bacteria that regulate functions other than flagellar-dependent motility. From our analysis, it seems probable that Geobacter sp. use chemotaxis-like signaling pathways for a variety of functions, which probably include type IV pili-based motility, regulation of motility apparatus expression (flagellar, pili, and extracellular matrix), and biofilm formation. Interestingly, the Geobacter sp. have che clusters that – at the present time – appear to be unique, which may plausibly mean these pathways regulate physiological functions that are unique to the Geobacters. Sensory inputs to the chemotaxis-like pathways are likely to be diverse, because the Geobacter genomes contain a large number of chemoreceptor (mcp) genes, which display a diversity of sensing domain architecture. The presence of this large number of proteins – receptors and Che proteins – undoubtedly reflects a greater need for the Geobacters to respond to a variety of environmental conditions, which allows them to thrive in subsurface environments. The presence of MCPs that belong to different MA domain classes in one genome – i.e. express MCPs in the same cell membrane with MA domains of different lengths, may contribute to the segregation of receptors into class-specific clusters with their cognate Che signaling proteins. We postulate that this mechanism will generate pathway specificity and diminish unwanted cross-talk. Such a mechanism can be general for bacteria with multiple chemotaxis-like pathways.
Abbreviations
MA: Methyl-accepting; MCP: Methyl-accepting chemotaxis protein.
Authors' contributions
HT and RMW developed the main concepts and drafted the manuscript. HT carried out the sequence alignments, and homology and phylogenetic tree analyses. JK directed computational analyses of operon predictions and σ54 promoter regulation. FA participated in the computational analysis of the MCPs. RMW, JK, and DRL read and provided feedback on the manuscript.
Supplementary Material
The file contains the additional information figures and tables.
The file contains the multiple sequences alignment of Geobacter mcp genes.
Acknowledgments
Acknowledgements
We thank Marko Puljic and Yanhua Qu (The University of Tennessee, Memphis) for computational assistance with promoter predictions and operon comparisons, and Roger Alexander and Igor Zhulin (ORNL and University of Tennessee, Knoxville) for helpful discussions about Geobacter MCPs classes. We thank John R. Kirby (University of Iowa) for comments on an early draft of the manuscript and the anonymous reviewers for constructive feedback. JK thanks Victor Solovyev (Softberry, Inc.) for helpful discussions of the algorithms in FGENESB. HT gives special thanks to Ned Young (University of Massachusetts, Amherst) for advice on the use of Phylip. The Geobacter sp. sequence data used in this study were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/. This research was supported by the U.S. Department of Energy Office of Science (BER) under the Cooperative Agreement No. DE-FC02-02ER63446.
Contributor Information
Hoa T Tran, Email: htt@chem.umass.edu.
Julia Krushkal, Email: jkrushka@utmem.edu.
Frances M Antommattei, Email: antommattei.fm@pg.com.
Derek R Lovley, Email: dlovley@microbio.umass.edu.
Robert M Weis, Email: rmweis@chem.umass.edu.
References
- Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Antommattei MF, Weis RM. Reversible methylation of glutamate residues in the receptor proteins of bacterial sensory systems. In: Tamanoi F, Clarke S, editor. The Enzymes. XXIV. San Diego: Elsevier; 2006. pp. 325–382. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Wadhams GH, Armitage JP. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol Microbiol. 2001;40:1261–1272. doi: 10.1046/j.1365-2958.2001.02468.x. [DOI] [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, et al. Genome of Geobacter sulfurreducens : metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2004;52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Bauer CE. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol Microbiol. 2005;56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- Berleman JE, Bauer CE. A che-like signal transduction cascade involved in controlling flagella biosynthesis in Rhodospirillum centenum. Mol Microbiol. 2005;55:1390–1402. doi: 10.1111/j.1365-2958.2005.04489.x. [DOI] [PubMed] [Google Scholar]
- Berleman JE, Hasselbring BM, Bauer CE. Hypercyst mutants in Rhodospirillum centenum identify regulatory loci involved in cyst cell differentiation. J Bacteriol. 2004;186:5834–5841. doi: 10.1128/JB.186.17.5834-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Micro. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- Vlamakis HC, Kirby JR, Zusman DR. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol Microbiol. 2004;52:1799–1811. doi: 10.1111/j.1365-2958.2004.04098.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JR, Zusman DR. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc Natl Acad Sci USA. 2003;100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner PJ, Xu Q, Black WP, Li Z, Yang Z, Shimkets LJ. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol Microbiol. 2005;57:1499–1508. doi: 10.1111/j.1365-2958.2005.04785.x. [DOI] [PubMed] [Google Scholar]
- Blackhart BD, Zusman DR. Frizzy genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Takahashi A, Grossman AR. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci USA. 2001;98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. 2005;3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR. Bug juice: harvesting electricity with microorganisms. Nat Rev Microbiol. 2006;4:497–508. doi: 10.1038/nrmicro1442. [DOI] [PubMed] [Google Scholar]
- Lovley DR. Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- Newman DK, Kolter R. A role for excreted quinones in extracellular electron transfer. Nature. 2000;405:94–97. doi: 10.1038/35011098. [DOI] [PubMed] [Google Scholar]
- Nevin KPL, D R. ASM 101st General Meeting 588 (ASM, Washington DC, 2001)
- Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- Childers SE, Ciufo S, Lovley DR. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature. 2002;416:767–769. doi: 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G, Pollina RB, Nicoll JS, Lovley DR. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J Bacteriol. 2007;189:2125–2127. doi: 10.1128/JB.01284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol. 2006;72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Stoffel KHW. TMbase – A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:116. [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Felsenstein J. PHYLIP, programs for inferring phylogenies. University of Washington; [Google Scholar]
- Krushkal J, Yan B, DiDonato LN, Puljic M, Nevin KP, Woodard TL, Adkins RM, Methe BA, Lovley DR. Genome-wide expression profiling in Geobacter sulfurreducens : identification of Fur and RpoS transcription regulatory sites in a relGsu mutant. Funct Integr Genomics. 2007;7:229–255. doi: 10.1007/s10142-007-0048-5. [DOI] [PubMed] [Google Scholar]
- Krushkal J, Puljic M, Yan B, Barbe JF, Mahadevan R, Postier B, O'Neil RA, Reguera G, Leang C, DiDonato LN, et al. Genome regions involved in multiple regulatory pathways identified using GSEL, a genome-wide database of regulatory sequence elements of Geobacter sulfurreducens. In: Peng Y, Zhang Y, editor. BMEI2008 Biomedical engineering and informatics: new developments and the future Proceedings the First International Conference on Biomedical Engineering and Informatics. IEEE Computer Society, Las Alamitos, CA; 2008. pp. 423–431. [Google Scholar]
- Studholme DJ, Buck M, Nixon T. Identification of potential sigma(N)-dependent promoters in bacterial genomes. Microbiology. 2000;146:3021–3023. doi: 10.1099/00221287-146-12-3021. [DOI] [PubMed] [Google Scholar]
- Barrios H, Valderrama B, Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Reese MG, Harris NL, Eeckman FH. Biocomputing: Proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co, Singapore; 1996. Large Scale Sequencing Specific Neural Networks for Promoter and Splice Site Recognition. [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Hagen KD, Cohen MF, Summers ML, Meeks JC. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1996;178:2037–2043. doi: 10.1128/jb.178.7.2037-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis : how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort EN, Goy MF, Larsen SH, Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci USA. 1975;72:3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moual H, Koshland DE., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- Pokkuluri PR, Pessanha M, Londer YY, Wood SJ, Duke NEC, Wilton R, Catarino T, Salgueiro CA, Schiffer M. Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction. J Mol Biol. 2008;377:1498–1517. doi: 10.1016/j.jmb.2008.01.087. [DOI] [PubMed] [Google Scholar]
- Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Warren AV, Martin AC, Armitage JP. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol Microbiol. 2003;50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- Le Moual H, Quang T, Koshland DE., Jr Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Weis RM. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Location of the Receptor-interaction Site on CheB, the Methylesterase Response Regulator of Bacterial Chemotaxis. J Biol Chem. 2001;276:32984–32989. doi: 10.1074/jbc.M105925200. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- Antommattei FM, Munzner JB, Weis RM. Ligand-specific activation of Escherichia coli chemoreceptor transmethylation. J Bacteriol. 2004;186:7556–7563. doi: 10.1128/JB.186.22.7556-7563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E, Stock AM. Characterization of the Thermotoga maritima chemotaxis methylation system that lacks pentapeptide-dependent methyltransferase CheR:MCP tethering. Mol Microbiol. 2007;63:363–378. doi: 10.1111/j.1365-2958.2006.05518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario MM, Ordal GW. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol Microbiol. 1996;21:511–518. doi: 10.1111/j.1365-2958.1996.tb02560.x. [DOI] [PubMed] [Google Scholar]
- Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Marchant J, Wren B, Ketley J. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 2002;10:155–159. doi: 10.1016/s0966-842x(02)02323-5. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Porter SL, Warren AV, Martin AC, Armitage JP. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol Microbiol. 2002;46:1081–1094. doi: 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Jiang ZY, Gest H, Bauer CE. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J Bacteriol. 1997;179:5720–5727. doi: 10.1128/jb.179.18.5720-5727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZY, Bauer CE. Analysis of a chemotaxis operon from Rhodospirillum centenum. J Bacteriol. 1997;179:5712–5719. doi: 10.1128/jb.179.18.5712-5719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bustamante VH, Lux R, Zusman D, Shi W. Divergent regulatory pathways control A and S motility in Myxococcus xanthus through FrzE, a CheA-CheY fusion protein. J Bacteriol. 2005;187:1716–1723. doi: 10.1128/JB.187.5.1716-1723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli. J Mol Biol. 2008;378:215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Macnab R. In: Flagella and motility, in Escherichia coli and Salmonella. Neidhardt FC, editor. 1996. pp. 123–145. [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Martin AC, Gould M, Byles E, Roberts MA, Armitage JP. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54. J Bacteriol. 2006;188:7932–7940. doi: 10.1128/JB.00964-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The file contains the additional information figures and tables.
The file contains the multiple sequences alignment of Geobacter mcp genes.