1. Summary
Transgenic (Tg) mice expressing HLA class I alleles and lacking murine MHC class I represent a useful model for the pre-clinical evaluation of human vaccines, which focus on induction of CD8+ T-cell responses. We have developed a platform to be used in Tg mice for exploring the immunogenicity of T-cell targets, whose immunologic epitopes have yet to be defined. To test the attributes of the evaluation system in the context of an important human pathogen, we have explored multiple antigens from cytomegalovirus (CMV). A panel of recombinant modified vaccinia Ankara vectors, expressing various CMV proteins (CMV-MVA) was used to immunize HLA-A*0201, B*0702 and A*1101 Tg mice. Immune splenocytes were in vitro stimulated (IVS) either using syngeneic lipo-polysaccharide activated lymphoblasts or Tg HLA-I matched human EBV-transformed B-lymphoblastoid cells (LCL), both loaded with peptide libraries, encompassing the CMV protein under investigation. IVS performed with peptide library loaded lymphoblasts failed to provide a reliable stimulation. In contrast, the usage of LCL as antigen presenting cells of CMV peptide libraries resulted in a consistent and specific amplification of the Tg T-cell response in animals immunized with CMV-MVAs. The LCL IVS method reliably allowed defining the immunogenicity and immunodominant CD8+ T-cell regions of uncharacterized CMV antigens. The combination of CMV-MVA vectors, unbiased pools of CMV-specific peptide libraries presented by Tg HLA-I matched LCL constitutes a valid tool for the pre-clinical evaluation of model candidate vaccines. This convenient method could find application to investigate the immunogenicity profile of cancer antigens or proteins from infectious human pathogens.
Keywords: CMV vaccine candidates, MVA, IVS
2. Introduction
HLA trangenic (Tg) mice have applicability for the study, development and pre-clinical testing of vaccines and immunotherapies[1]. Cellular immune responses to model antigens can be studied in an easily manipulated vertebrate system based on the incorporation of the HLA receptor and antigen processing similarities to humans[2].
HLA class I Tg mice have been extensively used as pre-clinical models to test candidate vaccines for cancer and for many viral pathogens, among which human cytomegalovirus (CMV) is prominent [3–9]. It has been established that cellular immune responses are essential for limiting CMV viraemia during persistent infection, though it remains to be defined the correlation between response to CMV antigens and protective immunity[10–12]. While several studies have focused on the T-cell response to the immunodominant pp65 and IE1 antigens, a limited number of investigations have been carried out for viral proteins whose immunogenicity is less immunodominant[12–15]. Studies of alternative CMV proteins are warranted, since it has been demonstrated that CD8+ T-cell recognition in CMV positive individuals is complex, often very broad, and poorly approximated by responses to any one or two CMV ORFs [12;15].
Highly attenuated modified vaccinia Ankara (MVA) engineered with CMV antigens is an efficient vehicle to present CMV full-length proteins to the immune system, and constitutes an attractive tool to test new candidate CMV antigens, to expand CMV memory T-cells for adoptive immunotherapy, or, as a vaccine, to prime or boost CMV immunity[7;16]. A crucial advantage of this viral vector is its large capacity for foreign DNA, the ease of its construction, and its demonstrated safety in the most susceptible populations[17;18]. Our group has successfully developed several recombinant MVA vectors expressing multiple CMV proteins (CMV-MVA)[7;9;19]. The level of specific activity towards HLA-I restricted CMV pp65 and IE1 CTL nonamer peptide epitopes is used as a screening tool to assess the recombinant vector ability to elicit a primary CMV-specific immune response in the mouse model. In cases in which epitopes are not defined for specific pp65/IE1 HLA types or for newly identified CMV candidate antigens, HLA-allele-specific algorithms based on MHC-binding affinity predictions may be used for epitope discovery[15;20]. While immunogenicity predictions have limits[8], screening tools that take advantage of unbiased pools of overlapping peptide libraries, encompassing the full sequences of antigenic proteins under investigation, greatly facilitate and enhance immune assessments[21]. In fact, libraries can be used without knowledge of specific HLA-restricted T-cell epitopes[22].
We have found that CMV libraries of overlapping 15mer peptides generally provide a much weaker stimulant and unreliable readout than single CTL nonamer peptides, when used to evaluate the primary CMV-specific response in CMV-MVA immunized HLA-I Tg mice. To overcame this limitation we have developed and validated a robust approach to explore the immunogenicity of MVA vectors expressing a panel of poorly defined, but frequently recognized CMV proteins in HLA-A*0201[23], HLA-B*0702[24] and HLA-A*1101[25] Tg mice. This convenient model system can make feasible the immune-profiling of any uncharacterized antigen of clinical interest.
3. Materials and Methods
3.1 Mice and immunizations
HLA-A2.1 HHD II (A2 Tg)[23] and HLA-B*0702 H-2KbDb double knockout (B7 Tg)[24] transgenic mice were obtained from F. Lemonier (Institut Pasteur, France). HLA-A* 1101 H-2KbDb double knockout (A11 Tg) mice were derived at the AAALAC-approved animal care facility at City of Hope (COH) by cross-breeding HLA-A* 1101/Kb[26] with commercially available C57BL/6 KbDb knockout mice (Taconic, model 004215, Hudson, NY)[27]. Correct genotype and Tg expression was confirmed by PCR on tail DNA and blood by FACS analyses using FITC-anti human HLA A, B, C mAb (BD Biosciences, San Jose, CA)[23]. All Tg mice were bred and maintained under pathogen-free conditions at COH AAALAC-approved animal care facility. Ten- to twelve-weeks HLA-I Tg mice were immunized intra-peritoneally (i.p.) for 3 weeks, with 50×106 i.u. of various CMV-MVA vectors (as described in Figure legends).
3.2 CMV-MVA immunogens
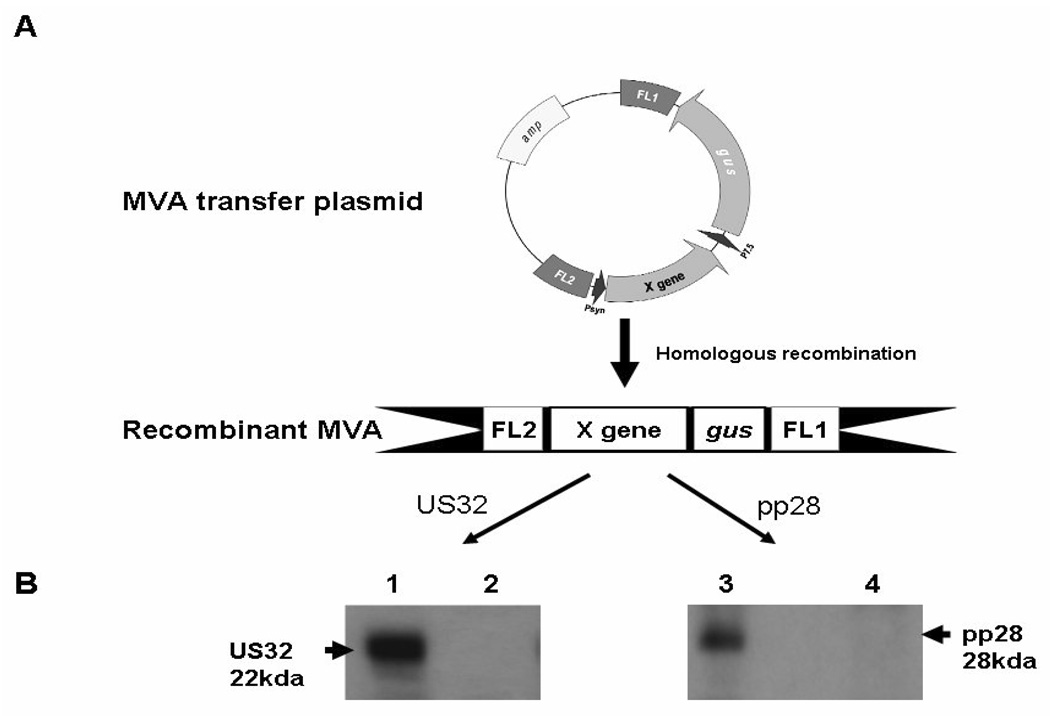
The following CMV-MVA vectors were produced and used as immunogens for the current study: CMV-MVA dually expressing full-length pp65 and IE1 proteins (pp65/IE1 MVA) obtained as described [9]; CMV-MVA dually expressing full-length pp65 and IE1-IE2fusion proteins (pp65/IE1-IE2fusion MVA, unpublished results); CMV-MVA expressing full-length pp28 antigen (pp28 MVA; Figure 1); CMV-MVA expressing full-length US32 antigen (US32 MVA; Figure 1). The CMV pp28 gene was amplified from HCMV genomic DNA, and cloned into MVA transfer plasmid pMCO3 to yield pp28-pMCO3. The CMV US32 gene was also amplified from HCMV genomic DNA, and a 6 x His Tag was added to the C-terminus by PCR approach. The gene product resulted from this PCR was cloned into pMCO3 to yield US32-pMCO3. pp28 and US32 MVA were generated on BHK-21 cells via homologous recombination, followed by eight consecutive rounds of screening[7;9]. The protein expression levels of pp28 and US32 (Figure 1 B) were detected by western blot using ECL chemiluminscence detection kit (Amersham Pharmacia Biotech, Buckinghamshire, UK)[7;9]. After electro-transfer of proteins from the gel onto PVDF membranes (Bio-Rad, Hercules, CA), they were incubated first with purified monoclonal antibody (Ab) 41–18 (kind gift of Bill Britt, University of Alabama) against pp28 or His-Tag for US32 (EMD Chemicals, San Diego), and then followed with HRP-labeled goat anti–mouse Ab according to the manufacturer’s instructions. All CMV-MVA used in this study were expanded and purified by 36% sucrose density untracentrifugation, resuspended in PBS containing 7.5% lactose and frozen at −80 °C.
Figure 1. pp28 MVA and US32 MVA vectors.
(A) Schematic representation of MVA transfer pMCO3 plasmid and recombinant MVA vector. The plasmid has a vaccinia synthetic promoter (Psyn) which drives the expression of pp28 or US32 gene, indicated as X gene, and the synthetic promoter P7.5 for the expression of the gus gene. pMCO3 plasmid targets MVA deletion region III via flanking (FL) regions 1 and 2. pp28 MVA and US32 MVA were generated via homologous recombination by transfecting pMCO3 plasmid into wild type MVA infected BHK-21 cells. gus bacterial marker gene was used for color selection of recombinant MVA. (B) Western blot detection of US32 and pp28 from corresponding recombinant MVA infected BHK-21 cells. Lane 1 and 3: respectively, cell lysates of US32 MVA and pp28 MVA infected BHK-21 cells. In lane 2 and 4: cell lysates of wild type MVA infected BHK-21 cells, as negative controls.
3.3 Peptides and peptide libraries
HLA-A*0201 pp65495–503[13], HLA-B*0702 pp65265–275[28], HLA-A*1101 pp6513–24[28], HLA-A*0201 IE-1316–324[29], HLA-B*0702 IE1307–321[30], HLA-B*0702 IE1308–317[30] restricted CMV peptides were generated as previously described[31]. Overlapping 15 amino acid (AA) peptides (PepMix™) spanning full-length CMV pp65 and IE1 proteins were purchased from JPT Peptide Technologies GmbH (Berlin, Germany). IE2, pp28 and US32 libraries were formulated and generated in house. The 581 AA sequence from the IE2 protein, the 154 AA sequence from the pp28 protein, the 183 AA sequence from the US32 protein were divided into 15mer stretches that overlap successive peptides by four AA, using an online program which excludes impermissible AA at the amino and carboxyl terminus of each 15mer peptide, based on synthetic consideration (http://www.hiv.lanl.gov/content/hiv-db/PEPTGEN/Explanation.html). For the IE2 library, 121 peptides divided into 20 peptides/pool were generated. For the pp28 and US32 libraries, 46 peptides divided into 12 peptides/pool and 48 peptides divided into 12 peptides/pool were respectively produced. Peptide pools (composed on average of 15mers) were subsequently combined into one single super-pool at a concentration of 200 µg/ml, dissolved in 50% DMSO/water.
3.4 In vitro stimulation (IVS) using Tg HLA-I matched LCL as antigen presenting cells (APC)
Three weeks after immunization, spleens were aseptically removed, and splenic suspensions were teased through a sterile nylon mesh[6] using PBS containing 2 mM EDTA (pH 7.4). To remove cell debris, 120 Kunitz units of DNase I (Sigma, St. Louis, MO) per ml of splenic suspension were then added for 15 minutes at 18–25 °C, mixing vigorously. Cell suspensions were further washed with PBS/EDTA and passed once more through a nylon mesh. In cases in which enriched CD8+ T-cell populations were stimulated, a Dynal Mouse CD8 Negative Isolation Kit containing Mouse Depletion Dynabeads® and Antibody Mix with monoclonal antibodies towards CD4, CD14, CD16 (a and b), CD19, CD36, CD56, CDw123, Glycophorin A was used, following the manufacturer procedures. Tg HLA-I matched EBV-lymphoblastoid cell lines (LCL) derived as described[31] from HLA-A*0201, HLA-B*0702 and HLA-A*1101 consented healthy CMV positive volunteers[7] were used as APC to stimulate the splenic suspensions. Briefly, 5×106 LCL in 0.2 ml serum free RPMI medium[6] were loaded either with the relevant CMV-CTL epitope[6] or CMV-peptide library (4µg/ml) or CMV sub-library pools (4µg/ml) for 2½ hours in a 37°C incubator. One million of peptide loaded and irradiated LCL (5000 rads)[6] were co-cultured in a 24-well plate with 3×106 splenic cells in 2 ml of RPMI containing 10% T-Stim Culture Supplement [6] for 8 days. For CD8+ T-cell enriched IVS, 1×106 negatively isolated CD8+ T-cell together with 2×106 feeder syngeneic irradiated (2500 rads) splenocytes were co-cultered with LCL, as described above. Marked cell growth was generally obtained by day 3–4, requiring cell splitting and addition of 10% T-stim/RPMI media.
3.5 IVS using syngeneic lymphoblasts
Splenic single-cell suspensions from immunized mice were also stimulated for 7 days following IVS standard methods[6;7;32] with syngeneic lipo-polysaccharide (LPS) lymphoblasts as APC, loaded either with the relevant CMV-CTL epitope[6] or CMV-peptide library (4µg/ml).
3.6 Immunophenotyping and intracellular cytokine staining (ICS)
Stimulated splenic cell suspensions were phenotyped and evaluated following IVS[7;16] using anti-CD8-FITC (Clone Ly-2) and anti-IFN-γ-APC (Clone XMG1.2) monoclonal antibodies (BD Biosciences, San Jose, CA). Briefly, cell cultures from standard 7-day lymphoblast IVS[6] were harvested and layered over a Ficoll gradient (1:2) to remove cell debris, then 1×106 cells in 1ml of RPMI were stimulated for 10–12 hours with either the relevant CMV-CTL epitope or CMV-peptide library or sub-library peptide pools during ICS assay[7;16;33]. The peptide library diluent was used as control[34]. As for the LCL IVS, 8 day-cultures were starved of 10% T-Stim culture supplement 24 hours prior of ICS stimulation and procedure[7;16]. To assess CMV-specific CD8 T-cell activation state, IVS cultures were incubated for 10–12 hours with either the relevant CMV-CTL epitope, CMV-peptide library, sub-library peptide pools or peptide library diluent [7;16;33]. After re-stimulation, LCL IVS cultures were washed and stained for 30 minutes on ice with CD8-APC[16] antibody. Flow cytometric acquisition was performed on a FACSCanto™ (BD Immunocytometry Systems). Between 0.5 to 1×106 events were acquired for each sample. FACS analysis was performed using FCS Express version 2 software (De Novo, Ontario, Canada). The number of double-positive cells is expressed as a percentage of the CD8+ or CD4+ T-cell population. CD8− T-cells were used as surrogate for CD4+ T-cells, which is an acceptable approximation for this type of study[35].
4. Results
4.1 Comparing CMV-libraries vs. CMV-CTL epitopes
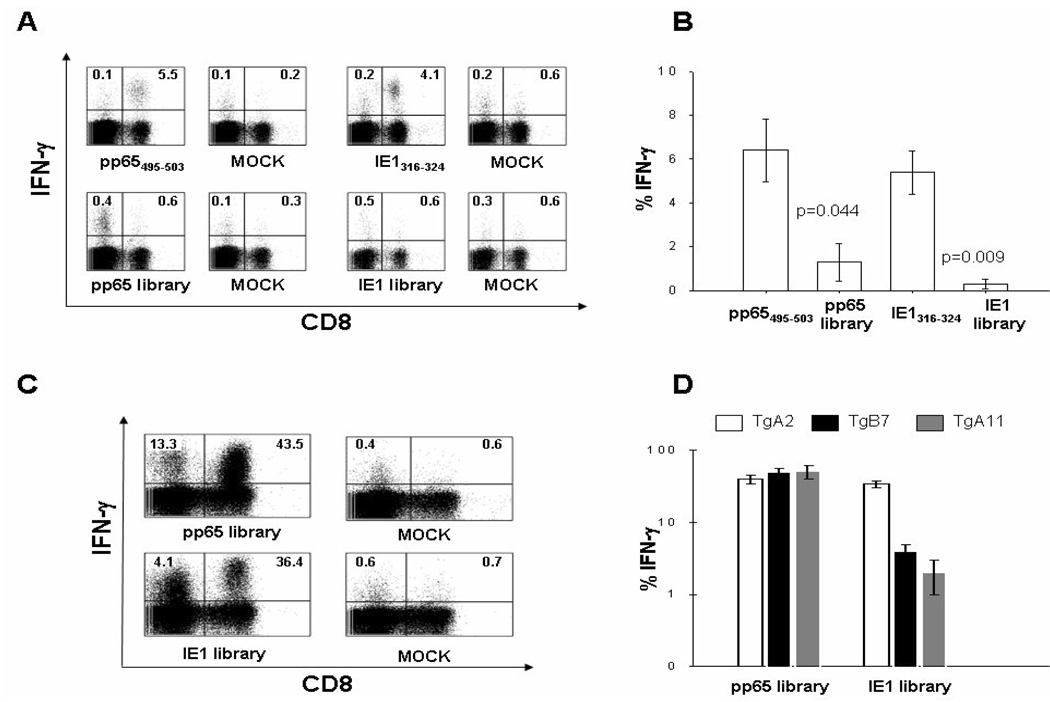
Peptide libraries are indispensable tools to evaluate viral antigens for which HLA-I restricted CTL epitopes are not described[12;34]. In a pilot experiment, we compared the levels of CMV-specific IFN-γ produced by splenic T-cells after in vitro re-stimulations using commercially available pp65 and IE1 Pep-Mix™ libraries with those obtained using the corresponding HLA-A*0201 pp65495–503[13] and HLA-A*0201 IE-1316–324[29] CTL immunodominant epitopes. A2 Tg mice were immunized with a recombinant MVA co-expressing both pp65 and IE1 full-length proteins. In a standard syngeneic LPS blast IVS followed by ICS assay[7], the HLA-A*0201 pp65495–503[13] and HLA-A*0201 IE- 1316–324[29] CTL epitopes[6;7] or the corresponding Pep-Mix™ peptide library spanning the whole pp65 and IE1 proteins were used during the stimulation. As shown in Figure 2 A–B, stimulations by pp65 and IE1 libraries resulted in a significantly lower CMV-specific CD8 T-cell activity than those in which CTL epitopes were used. In particular, stimulations performed with IE1 library produced almost undetectable levels of CMV-specific IFN-γ+ CD8+ T-cells, in mice in which a concomitant strong activity to HLA-A* 0201 IE-1316–324 peptide was detected, when this epitope was used as stimulant. In the case of pp65 library, the significantly reduced CMV-specific CD8 T-cell activity was accompanied by an enhanced murine class II response (Figure 2 A, lower left plot). Both CD4 and CD8 T-cell activation are expected with the usage of libraries composed of 15mer peptides[22]. A similar pattern of diminished CD8 reactivity was found using pp65 library instead of the HLA-B*0702 pp65265–275[28] and HLA-A*1101 pp6513–24[28] epitopes for stimulation in both B7 and A11 Tg mice immunized with pp65/IE1 MVA. Levels of IFN-γ+ CD8+ T-cells were generally undetectable in both B7 and A11 Tg mice, though no comparison could be done with the corresponding epitopes since IE1 HLA-A* 1101 epitope has not been defined, while the HLA-B*0702 IE1307–321[30] and the HLA-B*0702 IE1308–317[30] putative CTL epitopes are minimally recognized in B7 Tg mice (data not shown and [36]). Our results indicated that pp65 and IE1 peptide libraries are less effective and reliable than CTL epitopes as a read-out to assess CMV-specific CD8 T-cell primary response in HLA-I transgenic mice.
Figure 2. LPS lymphoblasts vs. LCL.
(A) IFN-γ production after syngeneic LPS lymphoblast IVS and ICS stimulation using the indicated CTL epitopes (upper plots) or peptide libraries (lower plots) in a representative A2 Tg mouse, immunized i.p. for 3 weeks with 50 M iu of an MVA construct expressing both pp65 and IE1 proteins. (B) Average levels of IFN-γ produced by CD8+ T-cells upon LPS lymphoblast IVS with either the epitope or peptide library as specified on the x axis, in five A2 Tg mice immunized as described in A. Difference significance between groups (p values) was tested by t test. (C) pp65 and IE1 specific IFN-γ production detected by ICS in a representative A2 Tg mouse immunized as in A. IVS was performed using pp65 or IE1 peptide library loaded HLA-A*0201 LCL. (D) Average levels of IFN-γ produced by CD8+ T-cells following Tg HLA-I matched LCL IVS and ICS stimulation with the peptide library specified on the×axis, in five A2 Tg (white histograms), five B7 Tg (black histograms) and five A11 Tg (grey histograms) mice, immunized as in A. The MOCK plots in A and C indicate that no epitope/library was used during the ICS stimulation. Numbers in the plots show percentages of CD8+ (upper right) or CD4+ (as CD8−, upper left) T-cells producing IFN-γ In B and D, IFN-γ production to mock stimulated cells during the ICS procedure was subtracted. Error bars represent the SEM among the immunized mice.
4.2 Tg HLA-I matched LCL loaded with CMV-libraries are potent APC for IVS
Since human LCL are powerful APC for PBMC stimulations[13;37], we loaded HLA-A* 0201 LCL either with pp65 or IE1 peptide libraries to assess whether there would be enhanced recognition of MVA expressing both pp65 and IE1 proteins. Loading of LCL with CMV-peptide libraries greatly amplified in vitro proliferation (>20X) of immune splenocytes, generating a strong HLA class I restricted response to MVA expressing pp65 and IE1, as shown in Figure 2 C–D. A concomitant strong class II murine-specific response was also detected (Figure 2 C, upper left quadrants of left plots)[22]. The applicability of the approach was extended to B7 and A11 Tg mice, using corresponding Tg HLA-I matched LCL as APC, loaded with either the pp65 or IE1 libraries. LCL as APC during the IVS, increased the activity of the pp65 library in all tested Tg mice (Figure 2 D).
4.3 IE2 specific CD8+ T-cells in A2, B7 and A11 Tg mice
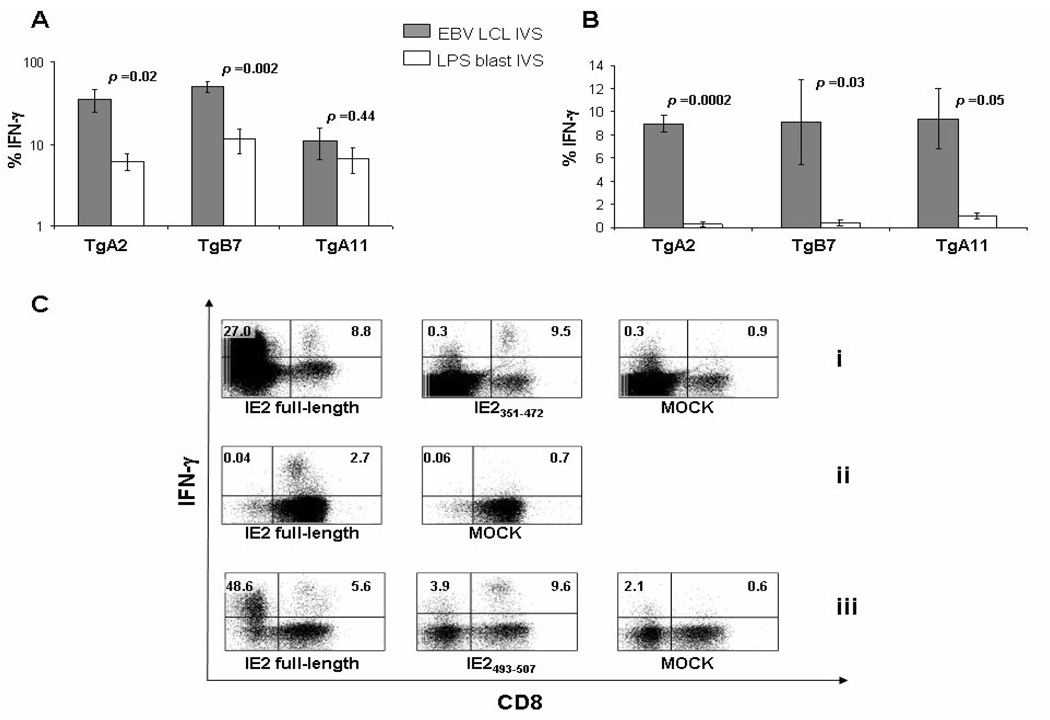
The immediate-early 2 (IE2) regulatory protein stimulates a CD8+ T-cell memory response by a large percentage of asymptomatic CMV-positive adults[12]. IE2 protein has been implicated in the development of coronary restenosis and in the disruption of the transactivation function of p53[38;39]. We investigated the immunogenic property of the IE2 protein incorporated in a MVA co-expressing pp65 and IE1 antigens on the same viral vector (pp65/IE1-IE2fusion MVA). The construct, preliminary tested in A2 Tg mice for response to HLA-A*0201 restricted pp65495–503 and IE-1316–324 epitopes[7], was found to be immunogenic (data not shown). Next, A2 Tg, B7 Tg and A11 Tg mice were injected with pp65/IE1-IE2fusion MVA to compare immune activity upon either LPS blast or LCL IVS. APC were loaded with either an IE2 library synthesized in our lab (see Materials and Methods) or the relative HLA-I restricted pp65 CTL epitope. In all immunized mice there was a strong recognition of the pp65 epitope[7], which was markedly higher when the LCL were used compared to the LPS blast, IVS assay (Figure 3 A). IE2 specific IFN-γ production by CD8+ T-cells was minimal when the IVS was performed using the LPS blast as APC. In contrast, it was always detectable (>5%) when LCL were used as APC during the stimulation (Figure 3 B). The difference in antigen specific IFN-γ stimulation between the two techniques was significant (Figure 3 A–B). We also found that the CD8 activity was concentrated in the overlapping 15mer pool spanning IE2351–472 for both A2 Tg and A11 Tg mice (Table 1 and Figure 3 Ci, middle plot). Using the overlapping 15mers spanning IE2351–472 for ICS stimulation (Figure 3 Ci, middle plot) or performing LCL IVS using a CD8 negative isolation kit (Figure 3 Cii) strongly reduced the murine CD4 response. Attempts to narrow down the CD8+ T-cell response showed that the activity was present in multiple 15mers. In the case of B7 Tg mice, the 15mer IE2493–507 was the best peptide recognized of all components of the IE2 library by the CD8+ T-cells (Table 1 and Figure 3 Ciii, middle plot). These data show that IE2 expressed from rMVA generates a relevant class I primary immune response in Tg mice, which could be consistently detected using the LCL/peptide library stimulation.
Figure 3. pp65/IE1-IE2fusion MVA immunogenic property.
(A–B) Average levels of IFN-γ produced by CD8+ T-cells following either Tg HLA-I matched LCL (grey bars) or syngeneic LPS lymphoblast (white bars) IVS in the Tg mice (5 mice/Tg strain) showed on the x axes, immunized i.p. for 3 weeks with 50 M iu of pp65/IE1-IE2fusion MVA. Error bars represent the SEM among the immunized mice. IFN-γ production to mock stimulated cells during the ICS procedure was subtracted. Difference significance between groups (p values) was tested by t test. In A, the following HLA-I restricted CTL peptide epitopes were used for the cell stimulation: pp65495–503[13] for A2 Tg, pp65265–275[28] for B7 Tg, pp6513–24[28] for A11 Tg mice. In B, the stimulation was performed with the IE2 peptide library for all Tg mice. (C) IFN-γ production in representative Tg mice (i and ii, A2 Tg; iii, B7 Tg) immunized and stimulated as described in B. In ii, a CD8+ enriched cell population (see Materials and Methods) was used during the IVS. The peptide library, sub-library or sequence used during the ICS stimulation is reported under each plot. MOCK plots show IFN-γ backgrounds (no epitope/library was used during the ICS stimulation). Numbers in the plots indicate the percentages of CD8+ (upper right) or CD4+ (as CD8−, upper left) T-cells producing IFN-γ.
Table 1.
CD8+ T-cell recognition of CMV IE2, pp28 and US32 libraries in immunized Tg mice
| CMV antigen | |||
|---|---|---|---|
| Tg mouse strain | IE2 | pp28 | US32 |
| A2 | 351–472 ND IFN-γ average: 11.4% |
134–148: LDEEDTSIYLSPPPV IFN-γ average: 18.5% |
NONE |
| B7 | 493–507: MIIHAATPVDLLGAL IFN-γ average: 2.9% |
153–166: VVAKRLPRPDTPRT IFN-γ average: 7.1% |
48–62: RAERRAANWRRQMRR IFN-γ average: 70.1% |
| A11 | 351–472 ND IFN-γ average: 1.4% |
NONE | 109–123: TVRAFSRAYHHRINR IFN-γ average: 17.1% |
Tg mice were immunized with MVA vectors expressing the CMV antigen indicated. Following LCL IVS using the corresponding peptide library, mice were evaluated by ICS for CMV antigen-specific IFN-γ+ production (see Material and Methods). In the 3 row cells, first line indicates the highest recognized sub-library or 15mer from the corresponding CMV antigen peptide library; 2nd one shows the AA sequence of the 15mer (bold character) when identified (ND=not defined); the 3rd one reports the average of IFN-γ + CD8+ T-cells detected upon stimulation with either the CMV sublibrary or the 15mer indicated. NONE: minimal IFN-γ production (<0.1%). Backgrounds IFN-γ levels to mock stimulated cells during the ICS procedure were subtracted
4.4 CMV-MVA pp28 and US32 elicit a primary immune response in Tg mice
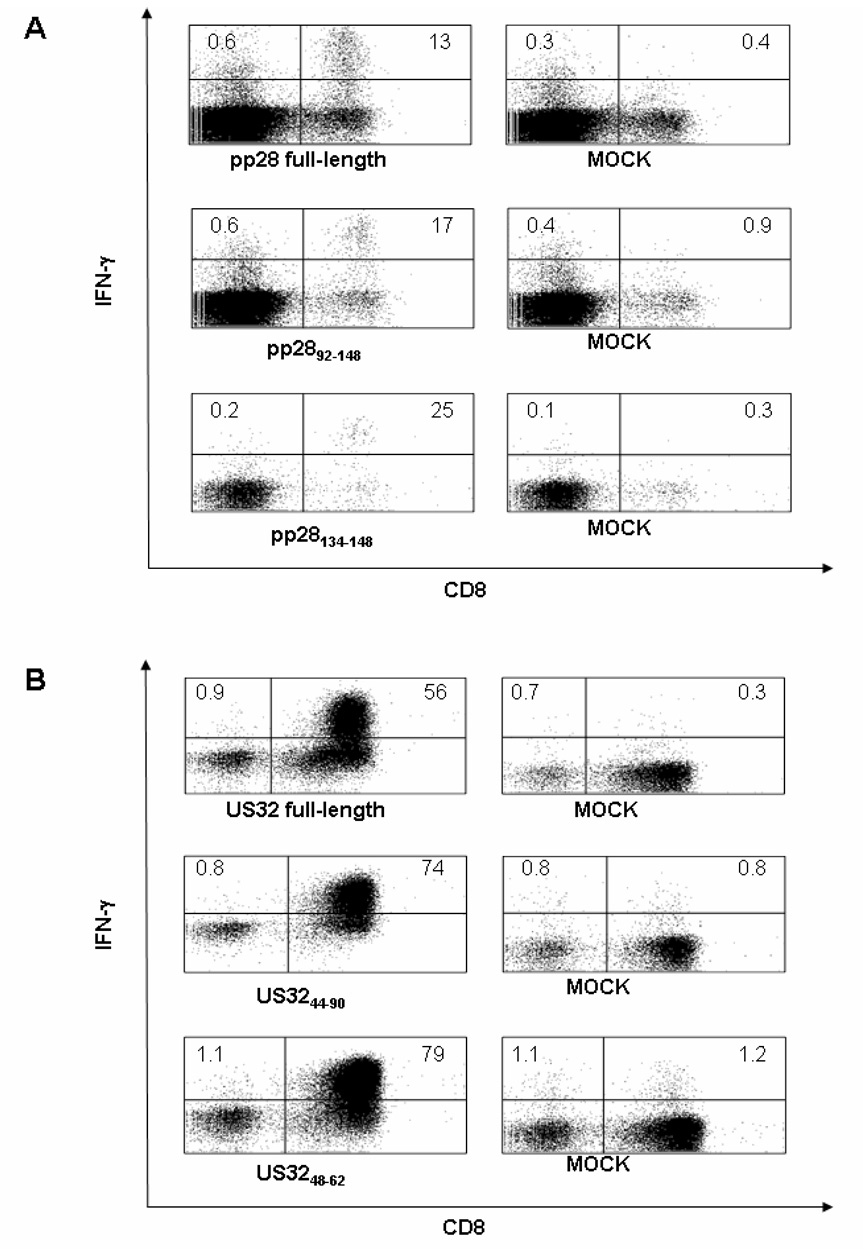
pp28 and US32 are two additional CMV antigens that are frequently recognized by healthy, CMV positive individuals[12], and whose immunogenicity have not yet explored. pp28, encoded by a CMV genome sequence called unique long region 99 (UL99), is a structural phosphoprotein essential for assembly of infectious virus[40]. Encoded by a cluster of CMV genes called unique short region 32 (US32), US32 was found to be dispensable for viral growth and replication, however its functional profile has not been assessed[41]. We produced MVA either expressing full-length pp28 or full-length US32 (Figure 1) and synthesized peptide libraries of overlapping 15mers, respectively encompassing the whole pp28 and US32 antigens. No specific activity was detected in Tg mice immunized with either pp28 or US32 MVA when the murine splenocytes were stimulated respectively with pp28 or US32 pulsed syngeneic LPS lymphoblasts (data not shown). In contrast, pp28 specific IFN-γ was detected in cell cultures from A2 and B7 Tg mice immunized with either pp28 MVA and stimulated with the respective library loaded Tg HLA-I matched LCL (Table 1 and Figure 4 A). As for US32 MVA immunized and peptide loaded LCL stimulated mice, the vector induced marked US32-specific IFN-γ response in B7 and A11 Tg mice, while minimal IFN-γ production was elicited in A2 Tg mice (Table 1 and Figure 4 B). By using pp28 or US32 sub-library peptide pools we narrowed the responsive region to subpools composed of overlapping 15-mers for each HLA Tg mouse strain evaluated (Table 1 and Figure 4). Our data are the first evidence that the pp28 and US32 antigens can be immunogenic in a pre-clinical model for CMV vaccine studies.
Figure 4. Response to pp28 and US32 CMV antigens.
IFN-γ levels produced following IVS using either (A) pp28 full-length library loaded HLA-A*0201 LCL in a A2 Tg mouse immunized i.p. for 3 weeks with 50 M iu of pp28 MVA or (B) US32 full-length library loaded HLA-B*0702 LCL in a B7 Tg mouse immunized i.p. for 3 weeks with 50 M iu of US32 MVA. Numbers in the plots indicate the percentages of IFN-γ+CD8+ (upper right) or CD4+ (as CD8−, upper left) T-cells. The peptide library, sub-library or sequence used during the ICS stimulation is reported under each plot. MOCK plots (no epitope/library used during the ICS stimulation) show IFN-γ backgrounds.
5. Discussion
Tg mice represents a useful model for preclinical discovery and quality control testing of vaccines designed to induce cellular immune responses in humans[1;4;5].We have described a reliable approach to be used in HLA-I Tg animals for delineating the immune properties of viral proteins for which immunogenicity profiles and/or HLA-I restricted CTL epitopes have not yet been defined[12], or whose immunodominance requires further investigation[15;30;36].
We exclusively used H-2 class I knock out Tg mice, since they can direct a more focused CD8+ T-cell repertoire than classical Tg animal[27], and consequently have an improved ability to utilize the HLA class I molecule as a restriction element[24].
We chose CMV as a prominent pathogen model to explore and validate our experimental platform. CMV is a large double-stranded DNA virus encoding 213 open reading frames of which 70% are immunogenic for T-cells[12]. In healthy individuals, CMV establishes a life-long asymptomatic infection, however it can cause severe disease in the setting of cellular immune deficiency or immaturity, including transplant recipients, individuals with late-stage HIV infection, and congenitally infected neonates[42]. Developing a vaccine to both prevent CMV infection and control its intensity has been the object of investigation for over three decades, no licensed product has resulted[43;44]
A panel of commercially available or in house synthesized CMV-specific 15-mer overlapping peptide libraries was used to evaluate the immune response of Tg mice injected with recombinant MVAs expressing CMV proteins pp65, IE1, IE2, pp28 or US32[6;7;9;19;45]. While for pp65 immunodominant epitopes have been defined for several HLA types[28], epitope knowledge for the IE1 antigen is mainly restricted to the HLA A*0201 type, while immunodominance in other HLA types remain controversial [29;30;36]. In the case of IE2, pp28 and US32 proteins[12], no putative T-cell domains or HLA-I restricted CTL epitopes have been described. Using overlapping peptides spanning an entire protein sequence, CD8+ T-cell responses can be detected to multiple epitopes, regardless of HLA type[22]. Peptide libraries corresponding to specific antigens are increasingly been used to identify T-cell immune responses to vaccines or pathogens since there is mounting evidence that the correlation of particular restricting HLA alleles and epitope immunodominance is either not absolute or not adequately represented by the response to a single epitope, and that individual HLA I alleles do not predict an immunodominant response restricted by that allele [12;21;22;46–48]. Therefore, reactivity to single immunodominant epitopes may not be representative or predictive of the total response to the entire antigen[46]. In addition, predicting immunodominant CD8+ T-cell responses in humans is complex, given the high diversity of MHC class I haplotypes found within the general population and the likely similar heterogeneity among non-MHC class I genes influencing this process[49]. These results call into question the validity of using the response to a single viral epitope as a surrogate measure of the total virus-specific CD8+ T-cell response. Thus, an appreciation of the overall immune response to a viral pathogen and its relationship to parameters of viral infection it is supposed to be best achieved by examining the response to every potential viral epitope in the absence of assumptions of immunodominance[50].
The classical approach of using peptide-pulsed LPS induced, syngeneic lymphoblasts[6;7;9;23;24;51–53] showed inconsistent results and significantly reduced antigen-specific response when overlapping 15mer peptide pp65 or IE1 libraries were used, instead of the corresponding HLA-I restricted CTL-epitopes in pp65 or IE1 immunized A2 Tg mice (Figure 2 A–B). In the case of A2, B7 and A11 Tg mice, immunized with MVA expressing either IE2 (Figure 3 B), pp28 or US32 (data not shown) CMV proteins, minimal to undetectable activity was found when LPS lymphoblasts, used as APC in IVS, were sensitized with the corresponding 15mer library. Our results showed a general inability of the 15mer library sensitized LPS lymphoblasts to generate a consistent antigen-specific response. In contrast, unbiased pools of peptide libraries covering full-length CMV proteins have been successfully used for studying both CMV-specific CD4+ and CD8+ T-cell response ex-vivo and after IVS procedure with human PBMC [12;16;21;22;34]. It has been shown that species-specific generation of peptide epitopes may elicit a differential presentation of antigenic epitope by murine and human cells[54]. Differently from 8–11 AA CTL epitopes that can directly bind to their restricted HLA class I, it is likely that 15-mers undergo internalization and processing [13;55]. Cytosolic proteases digest antigens into fragments that contain a variable number of N-terminal flanking residues[56]. The extra N-terminal residues are trimmed in the ER to generate the peptide repertoire which will bind to the MHC I[57]. The protease for this function is called ERAAP, the endoplasmic reticulum (ER) aminopeptidase associated with antigen processing in the mouse[58], or ERAP1 in the human[59]. The two enzymes share 86% identity[56]. Differences between these proteases may be partly responsible for this variation between the human and Tg murine system.
Human LCL have been extensively used as professional APC in in vitro systems[7;16]. Human cell lines, such as Jurkat and TAP deficient T2 cells have been previously used as APC for Tg murine IVS procedures[60–63]. However, human LCL have never been described for stimulation of Tg HLA-I splenocytes. LCL are professional APC and powerful stimulators[7;37]. To evaluate their presentation properties in the Tg murine context, we used HLA-A*0201, HLA-B*0702, HLA-A*1101 LCL, derived from CMV-positive volunteers, as APC for CMV–peptide libraries during the IVS of splenocyte cultures from cognate Tg mouse strains, MVA immunized. Both CMV epitope or library pulsed Tg HLA-I matched LCL induced much higher CD8+ T-cell activity than syngeneic LPS lymphoblast loaded with the same epitope or library in the CMV MVA immunized mice (Figure 2 C–D, Figure 3, Figure 4). The usage of human cells for processing in the context of the HLA class I complex could facilitate the recognition by the HLA-Tg T-cells. In fact, it has been reported that LCL pulsed with long overlapping peptides, or polypeptides (typically 15–30 AA) are able to present short (8–11 AA) epitopes in the context of HLA class I molecules[64]. The marked post-IVS cell expansions (>20X) mirrors what found using CMV-MVA in PBMC from healthy CMV positive adults[7;37]. It is interesting to note that the high backgrounds generated by the EBV proteins[7;16], which are usually detected when LCL stimulation are performed with human PBMC, were not found in the case of LCL stimulated immune-splenocytes (Figure 2 C and Figure 3 C). These results further increase the attractiveness of using LCL as APC in the Tg murine IVS system.
LCL IVS system allowed showing for the first time the immunogenicity of undefined antigens of possible clinical interest such IE2, pp28 and US32[12;41;65]. CD8+ T-cell reactive domains identified in the Tg mice analyzed (Table 1) could be of potential interest for further evaluation/monitoring in humans. However, it has to be taken in consideration that our observations of CD8+ T-cell immunodominance are made in inbred Tg mice and transposing these finding to humans requires caution. In fact, in this controlled rodent model, contrary to the human population, heterogeneity in the Tg HLA and non-MHC genes influencing T-cell receptor repertoire is at a minimum. Relative dominance of HLA-B*07 restricted CD8+ T-cell responses to pp65 in persons sharing HLA-A*02 and HLA-B*07 alleles has been reported [48]. Others have shown that Tg mice appear unable to respond to some known HLA-I restricted epitopes[2;5]. In a different study, some epitopes in the CMV IE1 protein were preferentially recognized in Tg mice than in humans[66].
Using libraries composed of 15mer peptides both CD4+ and CD8+ T-cell activation are expected to be found[22]. While the transgenic mice are a valuable tool to evaluate HLA Class I restricted CD8 T cell responses, they have an intact full complement of murine MHC Class II genes and cannot be directly compared to humans who possess a different repertoire of Class II MHC. Our data showed that parallelisms between the mouse and the human class II responses were not always possible for all the CMV-libraries tested. In fact sequences, such as IE1 and IE2, that do not stimulate a human class II response [12;34;67] were optimal for murine class II (Figure 2 C and Figure 3 C). Consequently, the murine T-helper evaluation was strictly a means to assess the potential immunogenicity of the construct in a mammalian system rather than direct comparison to humans.
In summary, the novel system described here for the identification of CMV vaccine candidates permits to define the immune profile of any antigenic protein. Its application may result instrumental for studies of human class I presentation in murine models of human diseases.
Acknowledgments
We thank Angelo Mandarino and Tahmineh Akbarnejad for early experiments related to the generation of pp65-IE1IE2fusion MVA and US32 MVA constructs; and Heang Ly, Joy Martinez and Zhongqi Li for excellent technical assistance. We acknowledge the administrative assistance of Denise Marsano and secretarial support of Donna Packer. These studies were partially supported by grants from the National Cancer Institute (NCI; RO1-CA77544 and PO1-CA30206, Project III) to DJD, and by The Edwin and Bea Wolfe Charitable Foundation to the Laboratory of Vaccine Research. The City of Hope (COH) Comprehensive Cancer Center and General Clinical Research Center (GCRC; satellite of University of Southern California GCRC) are supported respectively by the NCI (CA33572) and the NIH (MO1-RR0043-38).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lemonnier FA. The utility of H-2 class I knockout mice. Virus Res. 2002;82:87–90. doi: 10.1016/s0168-1702(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 2.Wentworth PA, Vitiello A, Sidney J, Keogh E, Chesnut RW, Grey H, et al. Differences and similarities in the A2.1-restricted cytotoxic T cell repertoire in humans and human leukocyte antigen-transgenic mice. Eur J Immunol. 1996;26:97–101. doi: 10.1002/eji.1830260115. [DOI] [PubMed] [Google Scholar]
- 3.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human- mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firat H, Garcia-Pons F, Tourdot S, Pascolo S, Scardino A, Garcia Z, et al. H-2 class I knockout, HLA-A2.1-transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur J Immunol. 1999;29:3112–3121. doi: 10.1002/(SICI)1521-4141(199910)29:10<3112::AID-IMMU3112>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 5.Mateo L, Gardner J, Chen Q, Schmidt C, Down M, Elliott SL, et al. An HLA-A2 polyepitope vaccine for melanoma immunotherapy. J Immunol. 1999;163:4058–4063. [PubMed] [Google Scholar]
- 6.La Rosa C, Wang Z, Brewer JC, Lacey SF, Villacres MC, Sharan R, et al. Preclinical development of an adjuvant-free peptide vaccine with activity against CMV pp65 in HLA transgenic mice. Blood. 2002;100:3681–3689. doi: 10.1182/blood-2002-03-0926. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, La Rosa C, Mekhoubad S, Lacey SF, Villacres MC, Markel S, et al. Attenuated poxviruses generate clinically relevant frequencies of CMV-specific T cells. Blood. 2004;104:847–856. doi: 10.1182/blood-2003-10-3469. [DOI] [PubMed] [Google Scholar]
- 8.Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, et al. HLA-A* 0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol. 2005;175:5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, La Rosa C, Li Z, Ly H, Krishnan A, Martinez J, et al. Vaccine properties of a novel marker gene-free recombinant modified vaccinia Ankara expressing immunodominant CMV antigens pp65 and IE1. Vaccine. 2007;25:1132–1141. doi: 10.1016/j.vaccine.2006.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg PD, Riddell SR. Deficient cellular immunity--finding and fixing the defects. Science. 1999;285:546–551. doi: 10.1126/science.285.5427.546. [DOI] [PubMed] [Google Scholar]
- 11.Reusser P, Riddell SR, Meyers JD, Greenberg PD. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 12.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond DJ, York J, Sun J, Wright CL, Forman SJ. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- 14.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 15.Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, et al. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol. 2003;77:5226–5240. doi: 10.1128/JVI.77.9.5226-5240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Rosa C, Wang Z, Lacey SF, Lalimarmo MM, Krishnan A, Longmate J, et al. In vitro expansion of polyclonal T-cell subsets for adoptive immunotherapy by recombinant modified vaccinia Ankara. Exp Hematol. 2006;34:497–507. doi: 10.1016/j.exphem.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Mayr A, Danner K. Vaccination against pox diseases under immunosuppressive conditions. Dev Biol Stand. 1978;41:225–234. [PubMed] [Google Scholar]
- 18.Meyer H, Sutter G, Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen Virol. 1991;72:1031–1038. doi: 10.1099/0022-1317-72-5-1031. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, et al. Recombinant MVA expressing a soluble form of glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of CMV. J Virol. 2004;78:847–856. doi: 10.1128/JVI.78.8.3965-3976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kern F, Faulhaber N, Frommel C, Khatamzas E, Prosch S, Schonemann C, et al. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur J Immunol. 2000;30:1676–1682. doi: 10.1002/1521-4141(200006)30:6<1676::AID-IMMU1676>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 22.Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 23.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8+ T lymphocytes from beta2 microglobulin (β2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohrlich PS, Cardinaud S, Firat H, Lamari M, Briand P, Escriou N, et al. HLA-B*0702 transgenic, H-2KbDb double-knockout mice: phenotypical and functional characterization in response to influenza virus. Int Immunol. 2003;15:765–772. doi: 10.1093/intimm/dxg073. [DOI] [PubMed] [Google Scholar]
- 25.Alexander J, Oseroff C, Sidney J, Wentworth P, Keogh E, Hermanson G, et al. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol. 1997;159:4753–4761. [PubMed] [Google Scholar]
- 26.Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 27.Ureta-Vidal A, Firat H, Perarnau B, Lemonnier FA. Phenotypical and functional characterization of the CD8+ T cell repertoire of HLA-A2.1 transgenic, H-2KbnullDbnull double knockout mice. J Immunol. 1999;163:2555–2560. [PubMed] [Google Scholar]
- 28.Longmate J, York J, La Rosa C, Krishnan R, Zhang M, Senitzer D, et al. Population coverage by HLA class-I restricted cytotoxic T-lymphocyte epitopes. Immunogenetics. 2001;52:165–173. doi: 10.1007/s002510000271. [DOI] [PubMed] [Google Scholar]
- 29.Khan N, Cobbold M, Keenan R, Moss PA. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J Infect Dis. 2002;185:1025–1034. doi: 10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 30.Kern F, Surel IP, Faulhaber N, Frommel C, Schneider-Mergener J, Schonemann C, et al. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73:8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Rosa C, Krishnan R, Markel S, Schneck JP, Houghten R, Pinilla C, et al. Enhanced immune activity of cytotoxic T-lymphocyte epitope analogs derived from positional scanning synthetic combinatorial libraries. Blood. 2001;97:1776–1786. doi: 10.1182/blood.v97.6.1776. [DOI] [PubMed] [Google Scholar]
- 32.Vitiello A, Sette A, Yuan L, Farness P, Southwood S, Sidney J, et al. Comparison of cytotoxic T lymphocyte responses induced by peptide or DNA immunization: implications on immunogenicity and immunodominance. Eur J Immunol. 1997;27:671–678. doi: 10.1002/eji.1830270315. [DOI] [PubMed] [Google Scholar]
- 33.Lacey SF, La Rosa C, Zhou W, Sharma MC, Martinez J, Krishnan A, et al. Functional comparison of T cells recognizing cytomegalovirus pp65 and intermediate-early antigen polypeptides in hematopoietic stem-cell transplant and solid organ transplant recipients. J Infect Dis. 2006;194:1410–1421. doi: 10.1086/508495. [DOI] [PubMed] [Google Scholar]
- 34.La Rosa C, Limaye AP, Krishnan A, Longmate J, Diamond DJ. Longitudinal assessment of cytomegalovirus (CMV)-specific immune responses in liver transplant recipients at high risk for late CMV disease. J Infect Dis. 2007;195:633–644. doi: 10.1086/511307. [DOI] [PubMed] [Google Scholar]
- 35.Kern F, Bunde T, Faulhaber N, Kiecker F, Khatamzas E, Rudawski IM, et al. Cytomegalovirus (CMV) Phosphoprotein 65 Makes a Large Contribution to Shaping the T Cell Repertoire in CMV-Exposed Individuals. J Infect Dis. 2002;185:1709–1716. doi: 10.1086/340637. [DOI] [PubMed] [Google Scholar]
- 36.Rohrlich PS, Cardinaud S, Lule J, Montero-Julian FA, Prodhomme V, Firat H, et al. Use of a lentiviral vector encoding a HCMV-chimeric IE1-pp65 protein for epitope identification in HLA-Transgenic mice and for ex vivo stimulation and expansion of CD8+ cytotoxic T cells from human peripheral blood cells. Hum Immunol. 2004;65:514–522. doi: 10.1016/j.humimm.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 37.La Rosa C, Wang Z, Lacey SF, Markel SF, Sharma MC, Martinez J, et al. Characterization of Host Immunity to cytomegalovirus pp150 (UL32) Hum Immunol. 2005;66:116–126. doi: 10.1016/j.humimm.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, et al. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 39.Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271:3534–3540. [PubMed] [Google Scholar]
- 40.Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci USA. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khanna R, Diamond DJ. Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol Med. 2006;12:26–33. doi: 10.1016/j.molmed.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Stagno S, Reynolds DW, Huang ES, Thames SD, Smith RJ, Alford CA. Congenital cytomegalovirus infection. N Engl J Med. 1977;296:1254–1258. doi: 10.1056/NEJM197706022962203. [DOI] [PubMed] [Google Scholar]
- 44.Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R. Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clin Infect Dis. 2004;39:233–239. doi: 10.1086/421999. [DOI] [PubMed] [Google Scholar]
- 45.Song GY, Gibson G, Haq W, Huang EC, Srivasta T, Hollstein M, et al. An MVA vaccine overcomes tolerance to human p53 in mice and humans. Cancer Immunol Immunother. 2007;56:1193–1205. doi: 10.1007/s00262-006-0270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betts MR, Casazza JP, Patterson BA, Waldrop S, Trigona W, Fu TM, et al. Putative immunodominant human immunodeficiency virus-specific CD8+ T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol. 2000;74:9144–9151. doi: 10.1128/jvi.74.19.9144-9151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lacey SF, Villacres MC, La Rosa C, Wang Z, Longmate J, Martinez J, et al. Relative dominance of HLA-B*07 restricted CD8+ T-Lymphocyte immune responses to human cytomegalovirus pp65 in persons sharing HLA-A*02 and HLA-B*07 alleles. Hum Immunol. 2003;64:440–452. doi: 10.1016/s0198-8859(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 49.Hoffmeister B, Kiecker F, Tesfa L, Volk HD, Picker LJ, Kern F. Mapping T cell epitopes by flow cytometry. Methods. 2003;29:270–281. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 50.Roederer M, Koup RA. Optimized determination of T cell epitope responses. J Immunol Methods. 2003;274:221–228. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 51.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 52.Perarnau B, Saron MF, San Martin BR, Bervas N, Ong H, Soloski MJ, et al. Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur J Immunol. 1999;29:1243–1252. doi: 10.1002/(SICI)1521-4141(199904)29:04<1243::AID-IMMU1243>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, et al. Recombinant modified vaccinia virus Ankara expressing a soluble form of glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of human cytomegalovirus. J Virol. 2004;78:3965–3976. doi: 10.1128/JVI.78.8.3965-3976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braud VM, McMichael AJ, Cerundolo V. Differential processing of influenza nucleoprotein in human and mouse cells. Eur J Immunol. 1998;28:625–635. doi: 10.1002/(SICI)1521-4141(199802)28:02<625::AID-IMMU625>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 55.Rammensee HG, Falk K, Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 56.Blanchard N, Shastri N. Coping with loss of perfection in the MHC class I peptide repertoire. Curr Opin Immunol. 2008 doi: 10.1016/j.coi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, et al. Characterizing the N-terminal processing motif of MHC class I ligands. J Immunol. 2008;180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 58.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 59.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 60.Yu Z, Liu X, McCarty TM, Diamond DJ, Ellenhorn JD. The use of transgenic mice to generate p53 specific and HLA restricted cytolytic T cells. Journal of Surgical Research. 1997;69:337–343. doi: 10.1006/jsre.1997.5058. [DOI] [PubMed] [Google Scholar]
- 61.Zhou H, Luo Y, Mizutani M, Mizutani N, Becker JC, Primus FJ, et al. A novel transgenic mouse model for immunological evaluation of carcinoembryonic antigen-based DNA minigene vaccines. J Clin Invest. 2004;113:1792–1798. doi: 10.1172/JCI21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takaki T, Marron MP, Mathews CE, Guttmann ST, Bottino R, Trucco M, et al. HLA-A*0201-restricted T cells from humanized NOD mice recognize autoantigens of potential clinical relevance to type 1 diabetes. J Immunol. 2006;176:3257–3265. doi: 10.4049/jimmunol.176.5.3257. [DOI] [PubMed] [Google Scholar]
- 63.Santomasso BD, Roberts WK, Thomas A, Williams T, Blachere NE, Dudley ME, et al. A T cell receptor associated with naturally occurring human tumor immunity. Proc Natl Acad Sci USA. 2007;104:19073–19078. doi: 10.1073/pnas.0704336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gnjatic S, Atanackovic D, Matsuo M, Jager E, Lee SY, Valmori D, et al. Cross-presentation of HLA class I epitopes from exogenous NY-ESO-1 polypeptides by nonprofessional APCs. J Immunol. 2003;170:1191–1196. doi: 10.4049/jimmunol.170.3.1191. [DOI] [PubMed] [Google Scholar]
- 65.Gyulai Z, Endresz V, Burian K, Pincus S, Toldy J, Cox WI, et al. Cytotoxic T Lymphocyte (CTL) Responses to Human Cytomegalovirus pp65, IE1-Exon4, gB, pp150, and pp28 in Healthy Individuals: Reevaluation of Prevalence of IE1-Specific CTLs. J Infect Dis. 2000;181:1537–1546. doi: 10.1086/315445. [DOI] [PubMed] [Google Scholar]
- 66.Gallez-Hawkins G, Villacres MC, Li X, Sanborn MC, Lomeli NA, Zaia JA. Use of transgenic HLA A*0201/Kb and HHD II mice to evaluate frequency of cytomegalovirus IE1-derived peptide usage in eliciting human CD8 cytokine response. J Virol. 2003;77:4457–4462. doi: 10.1128/JVI.77.7.4457-4462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bunde T, Kirchner A, Hoffmeister B, Habedank D, Hetzer R, Cherepnev G, et al. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J Exp Med. 2005;201:1031–1036. doi: 10.1084/jem.20042384. [DOI] [PMC free article] [PubMed] [Google Scholar]