Abstract
The human obesity susceptibility gene, FTO, encodes a protein that is homologous to the DNA repair AlkB protein. The AlkB family proteins utilize iron(II), α-ketoglutarate (α-KG) and dioxygen to perform oxidative repair of alkylated nucleobases in DNA and RNA. We demonstrate here the oxidative demethylation of 3-methylthymine (3-meT) in single-stranded DNA (ssDNA) and 3-methyluracil (3-meU) in single-stranded RNA (ssRNA) by recombinant human FTO protein in vitro. Both human and mouse FTO proteins preferentially repair 3-meT in ssDNA over other base lesions tested. They showed negligible activities against 3-meT in double-stranded DNA (dsDNA). In addition, these two proteins can catalyze the demethylation of 3-meU in ssRNA with a slightly higher efficiency over that of 3-meT in ssDNA, suggesting that methylated RNAs are the preferred substrates for FTO.
Keywords: DNA/RNA repair, FTO, oxidative demethylation
1. Introduction
Alkylating agents from the environment or formed inside cells can attack N- or O-atoms of nucleobases, which can lead to formation of alkylation base damages that require prompt repair [1]. Almost all living organisms have evolved various machineries to recognize and process these modifications. Among them, direct removal of alkyl adducts from the damaged bases is one of the most efficient repair strategies [2]. The AlkB protein family performs this type of direct repair by utilizing an oxidative demethylation mechanism (Fig. S1).
The E. coli AlkB protein belongs to a superfamily of α-KG- and Fe2+-dependent dioxygenases [2–9]. Its homologues are found in viruses, bacteria, and eukaryotes. Although eight human homologues (ABH1–8) were identified before 2007 [10,11], only ABH2 and ABH3 were confirmed to have a similar repair function to AlkB [4,5,12]. In particular, ABH2 was found to be primarily responsible for repairing 1-meA base lesions in genomic DNA while the exact role of ABH3 remains unclear [13]. ABH2 prefers dsDNA substrates over ssDNA ones; however, both hABH3 and AlkB are more active with ssDNA and ssRNA substrates [4,14]. Only recently, the mechanisms underlying the substrate preferences of these proteins were elucidated through X-ray structural studies of the AlkB-dsDNA and ABH2-dsDNA complexes [15]. The ABH2 protein adopts a commonly observed base flipping mechanism with a finger residue which intercalates inside the DNA duplex to fill the gap left by the flipped base [15]. The AlkB protein, however, squeezes the DNA duplex to eliminate the gap left by base flipping. This distortion imposed by AlkB on DNA explains its preference to flexible ssDNA over relatively rigid duplex DNA [15].
All three of these proteins exhibited the highest activities against 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) [4–7], but with lower activities they can also process 1-methylguanine (1-meG) and 3-methylthymine (3-meT) [16,17]. In addition, AlkB, ABH2, and ABH3 have been shown to repair exocyclic DNA base lesions: 1,N6-ethenoadenine (εA) and (or) 3,N4-ethenocytosine (εC) [18–20].
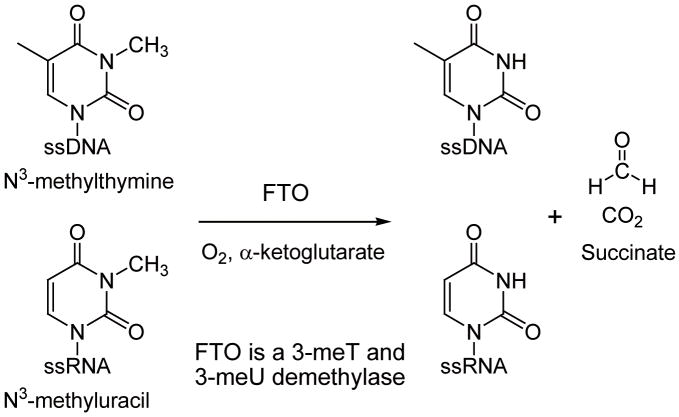
In 2007, it was a great surprise that a human obesity-linked gene, FTO, was found to encode a functional homologue of AlkB [21,22]. The FTO gene is highly expressed in the hypothalamus part of the brain, and a defect of the FTO gene has been linked to an increase of body fat [23–25]. This gene was found to be only present in vertebrates and marine algae [26]. The FTO protein possesses a homologous sequence to the AlkB family proteins, and the purified recombinant mouse FTO protein can oxidatively demethylate 3-meT in ssDNA in the presence of iron(II), α-ketoglutarate (α-KG), and dioxygen [21]. This discovery has assigned FTO as a nucleic acid demethylase that may work on DNA or RNA and also has raised several very interesting questions: i) what is the biochemical activity of the human FTO? ii) is 3-meU in ssRNA a substrate for FTO proteins? iii) how can the DNA/RNA demethylation function of FTO be linked to obesity? Perhaps, the methylation reversed by human FTO is a signal for gene regulation rather than merely for DNA/RNA damage. To help investigate the functional role of FTO, we present here the first study for the biochemical activity of recombinant human FTO protein in vitro. We also include our evaluation of the demethylation of 3-meU in ssRNA mediated by both mouse and human FTO proteins.
2. Experimental procedures
2.1 Construction, expression and purification of mFTO and hFTO
The cDNA sequences encoding full length mFTO (Image ID: 4237261) and hFTO (Genebank Accession No.: NP_001073901.1) were subcloned into pET28a to generate a His-tagged fusion protein. The plasmids were transformed into E. coli BL21 Star (DE3) and bacteria were grown on LB-agar plates containing 50 mg/L of kanamycin. Overnight precultures, which were grown aerobically at 37°C with a shaking speed of 190 rpm, were used to inoculate 1 L LB medium with 50 mg/L kanamycin and grown at 37°C and 250 rpm until OD600 reached ~1.0. Then the bacterial cells were induced by IPTG (0.5 mM) at 15°C and grown overnight at 15°C and 250 rpm. The cells were harvested by centrifugation, frozen by liquid nitrogen, and stored at −80°C. All subsequent steps were performed at 4°C. The cell pellets were resuspended in buffer A (50 mM imidazole, 300 mM NaCl, 50 mM sodium phosphate, pH 8.0), sonicated on ice, and centrifuged at 12,000 rpm for 22 min. The filtered supernatant was purified by Ni-NTA chromatography (GE Healthcare). The fractions collected from the column were further purified with a gel filtration column (GE Healthcare). The fractions were analyzed through denaturing SDS-PAGE.
2.2 Repair of methylated DNA and RNA by mFTO and hFTO
The typical reaction mixture (100 μL) contained DNA/RNA (1 nmol), FTO (0.05–0.5 nmol), (NH4)2Fe2(SO4)2·6H2O (283 μM), α-KG (300 μM), L-ascorbic acid (2 mM), bovine serium albumin (BSA, 50 μg mL−1), and 50 mM MES buffer (pH 6.0). This was incubated for 12 h at 16°C. The reaction was quenched by addition of EDTA to 5 mM.
2.3 Restriction enzyme digestion assay
A previously published procedure [13,15] was followed to evaluate repair of a 49-mer oligonucleotide containing 1-meA, 1-meG, and εA in a DpnII cleavage sequence. The sequence was [5′-TAGACATTGCCATTCTCGATAGG(replaced by 1-meG)A(replaced by 1-meA orεA)TCCGGTCAAACCTAGACGAATTCCA-3′ complementary to 5′-TGGAATTCGTCTAGGTTTGACCGGATCCTATCGAGAATGGCAATGTCTA-3′]. The reactions were run at both 37°C and 16°C for 12 h with 0.2 nmol FTO and 0.2 nmol substrates. For ssDNA, substrates were annealed to the complementary strand for the digestion assay after incubation with the FTO protein and cofactors.
2.4 DNA/RNA digestion and HPLC assay
After the repair reaction, ssDNA/ssRNA was digested into nucleosides with nuclease P1 (Sigma, N8630) and alkaline phosphatase (Sigma, P4252), based on a previous procedure [27]. The digestion solution was analyzed in an isocratic HPLC system equipped with a C18 separation column (150 × 4.6 mm) equilibrated with buffer A (HPLC grade aqueous solution containing 50 mM ammonium acetate) and buffer B (50 mM ammonium acetate, 50% of acetonitrile, 50% water and 0.1% TFA) at 95:5 (v/v) ratio with a flow rate of 1 mL min−1 at room temperature. The detection wavelength was set at 266 nm (for 3-meT) or 261 nm (for 3-meU).
2.5 Kinetics of mFTO and hFTO
To determine Km and Kcat values for the repair reactions, initial rates were obtained by keeping the enzyme concentration constant and varying the substrate concentration. Reactions were adjusted to assure that less than 20% of the substrate was expended. All reactions were performed at 20°C in triplicate and analyzed by Origin 8.0 with the Michaelis-Menten equation.
3. RESULTS AND DISCUSSION
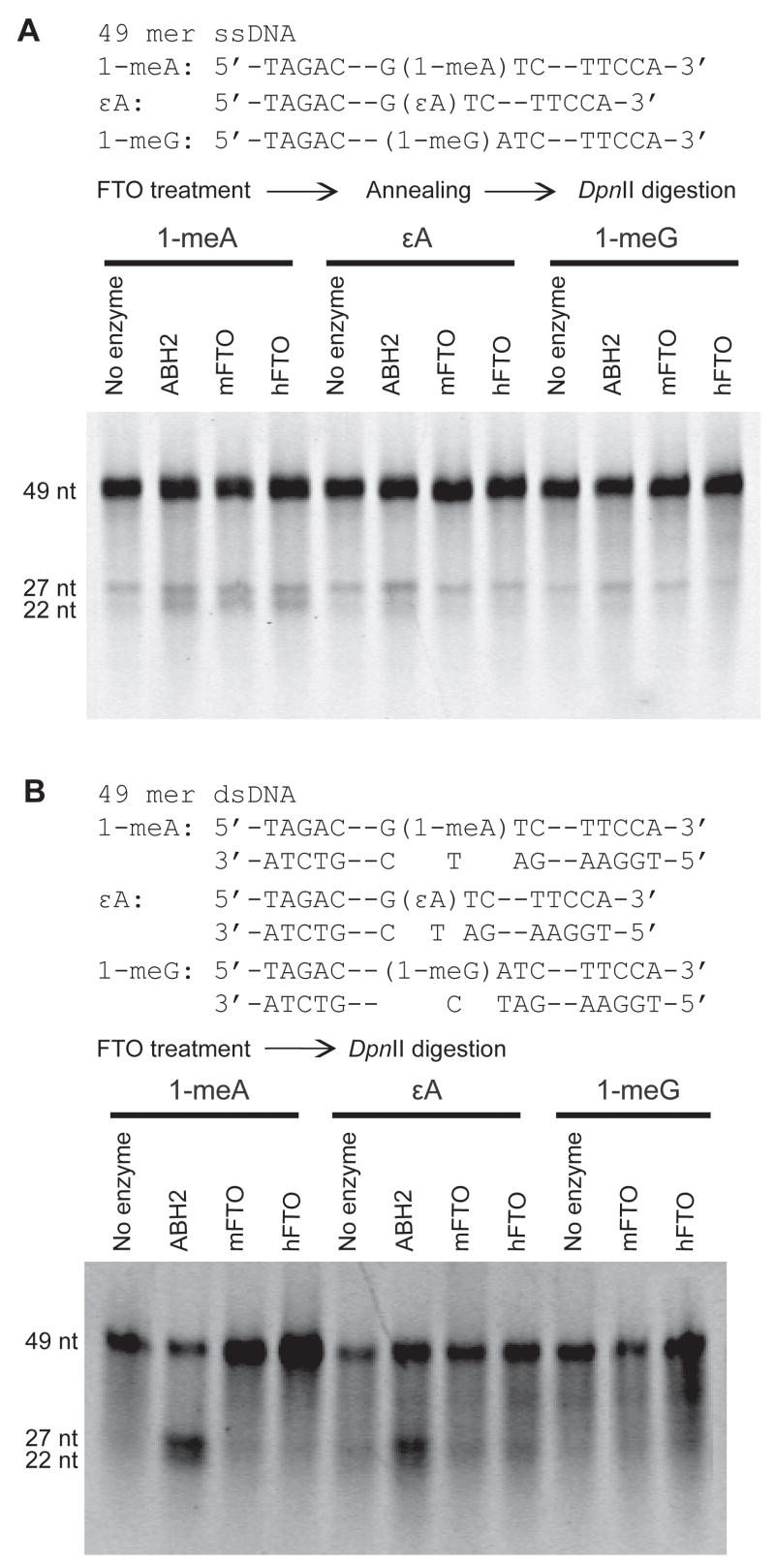
The full length mFTO and hFTO were cloned, expressed, and purified (Fig. S2). A restriction endonuclease digestion assay with the use of DpnII was adopted to evaluate the repair activities of both hFTO and mFTO towards 1-meA, 1-meG and εA in ssDNA or dsDNA [13,15,28]. A modified 49 mer DNA with 1-meA, 1-meG, or εA incorporated into the GATC sequence is resistant to DpnII cleavage. Removal of the base lesion by FTO allows DpnII to cleave the 49 mer DNA probe into two fragments, providing an assay for the repair activity (Fig. 1). As reported previously [21], we found very low activities of both proteins toward repairing 1-meA in 49 mer ssDNA (Fig. 1A). Both proteins failed to repair 1-meA in dsDNA (stoichiometric amount) after a 12 h incubation at either 16°C (Fig. 1B) or 37°C (data not shown); whereas, purified ABH2 exhibited good repair activities for 1-meA and εA in dsDNA under the same conditions (Fig. 1B). In addition, we did not observe any noticeable activity of mFTO or hFTO toward εA and 1-meG in either ssDNA or dsDNA. The 3-meT base lesion, when incorporated into the DpnII cleavage sequence, could not block the enzymatic digestion of the DNA probe. Thus, a different assay was used to evaluate repair of this base lesion.
Fig. 1.
A restriction enzyme digestion assay for repair of 1-meA, εA, and 1-meG by ABH2, mFTO, and hFTO. (A) A 49 mer ssDNA (0.2 nmol) with 1-meA, εA, or 1-meG incorporated into a GATC sequence (can be recognized and cleaved by DpnII) was used for the assay. The modified dsDNA probe is resistant to DpnII cleavage. Repair of the base lesion led to cleavage of the 49 mer dsDNA into two fragments of 27 and 22 base pairs, which was analyzed by a denaturing DNA gel. Both mFTO (0.2 nmol) and hFTO (0.2 nmol) exhibited very low activities toward 1-meA in ssDNA (12 h at 16°C and pH 6.0). Repair of εA and 1-meG by mFTO and hFTO were not observed. (B) No activities toward 1-meA, εA, and 1-meG in dsDNA were observed for either mFTO or hFTO under the same assay conditions. ABH2 showed good activities to repair 1-meA and εA in dsDNA in control experiments.
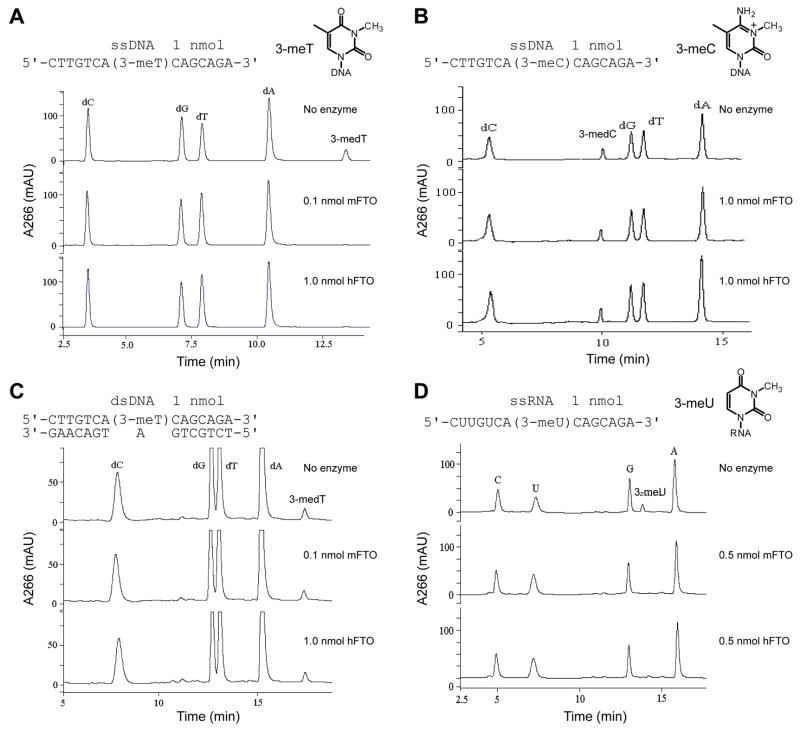
To probe 3-meT demethylation by FTO proteins, a 15 mer ssDNA [5′-CTTGTCA(3-meT)CAGCAGA-3′] with 3-meT incorporated in the middle was synthesized and purified. This DNA was digested into nucleosides by nuclease P1 and alkaline phosphatase, and subsequently analyzed by HPLC. As shown in Fig. 2A, nucleosides dC, dG, dT, dA and 3-medT could be cleanly separated. The change in intensity of the 3-medT peak was monitored with other nucleoside peaks as internal references. The 3-meT-containing ssDNA was incubated with either mFTO or hFTO at 16°C for 12 h. After quenching the reaction, the content of 3-meT was measured by the digestion assay and HPLC analysis. The results indicated complete demethylation of 3-meT in ssDNA by both mFTO and hFTO (Fig. 2A). When a 15 mer 3-meC-containing ssDNA was subjected to the same repair assay, only ~15% of 3-meC was demethylated after 12 h under the same conditions (Fig. 2B), showing strong preference of FTO proteins toward 3-meT over 3-meC.
Fig. 2.
HPLC chromatograms of digested nucleosides from 3-meT-containing DNA (1 nmol), 3-meC-containing DNA (1 nmol) and 3-meU-containing RNA (1 nmol). (A) Complete demethylation of 3-meT in a 15 mer ssDNA by mFTO and hFTO. All reactions were run for 12 h at 16°C and pH 6.0. (B) Both mFTO and hFTO gave ~15% demethylation of 3-meC in 15 mer ssDNA; 100% repair of 3-meT was observed for both enzymes under the same conditions as shown in (A). (C) Negligible repair of 3-meT in dsDNA was observed for both mFTO and hFTO under the same conditions. (D) Complete demethylation of 3-meU in a 15 mer ssRNA by mFTO and hFTO under the same conditions.
However, almost negligible demethylation activities (< 5% after 12 h) were observed for both proteins (Fig. 2C) when a 3-meT-containing dsDNA (obtained by annealing the 15 mer ssDNA with its complementary strand) was subjected to the same repair procedure. Thus, we conclude that recombinant mFTO and hFTO are not likely to catalyze demethylation of dsDNA substrates in vitro and perhaps in vivo.
The strong preference for 3-meT in ssDNA by the FTO proteins raised the question of whether 3-meU in ssRNA could also serve as a substrate for these proteins. We synthetically prepared 3-meU-CE phosphoramidite (Fig. S3) and incorporated it into a 15 mer ssRNA via solid state synthesis (Fig. S4). When this RNA substrate was incubated with either mFTO or hFTO, complete removal of the methyl group on 3-meU was observed in both cases (Fig. 2D). Thus, both mFTO and hFTO are capable of repairing 3-meT in ssDNA and 3-meU in ssRNA in vitro.
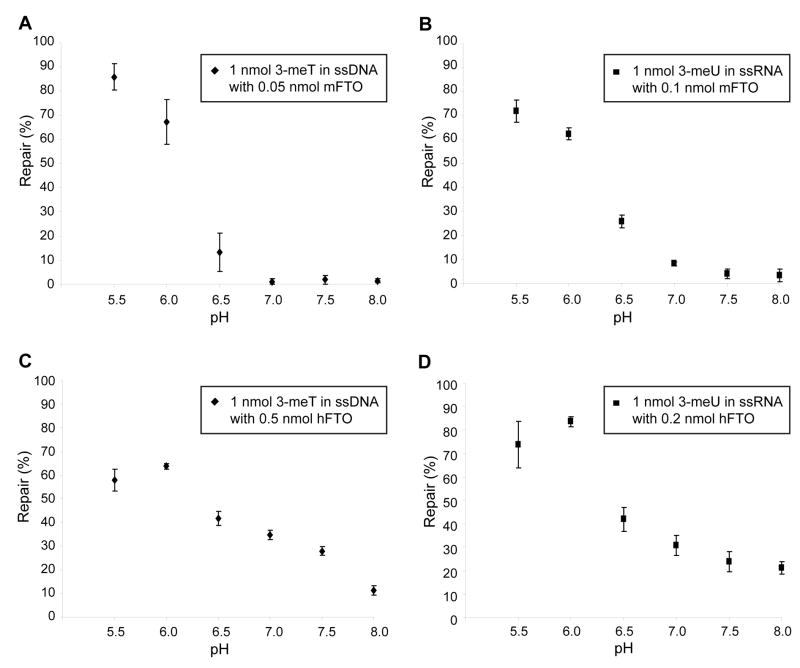
The inhibition effect of α-KG at millimolar concentrations was evaluated as noted previously [29] (Fig. S5), and 300 μM of α-KG was chosen for further studies. Next, the pH-activity profiles were estimated for the demethylation of 3-meT in ssDNA and 3-meU in ssRNA. In the case of mFTO, a decrease of the demethylation activity was observed with increasing pH for both ssDNA and ssRNA substrates (Fig. 3A and 3B). However, hFTO exhibited the highest activity at pH 6.0 for both ssDNA and ssRNA substrates (Fig. 3C and 3D). Thus, we chose pH 6.0 for detailed kinetic analysis.
Fig. 3.
The pH-activity profiles for demethylation reactions of 3-meT in ssDNA and 3-meU in ssRNA by mFTO and hFTO. All reactions were run in triplicate at 16°C for 12 h. (A) Demethylation of 3-meT in the 15 mer ssDNA (1 nmol) by mFTO (0.05 nmol). (B) Demethylation of 3-meU in the 15 mer ssRNA (1 nmol) by mFTO (0.1 nmol). (C) Demethylation of 3-meT in the 15 mer ssDNA (1 nmol) by hFTO (0.5 nmol). (D) Demethylation of 3-meU in the 15 mer ssRNA (1 nmol) by hFTO (0.2 nmol).
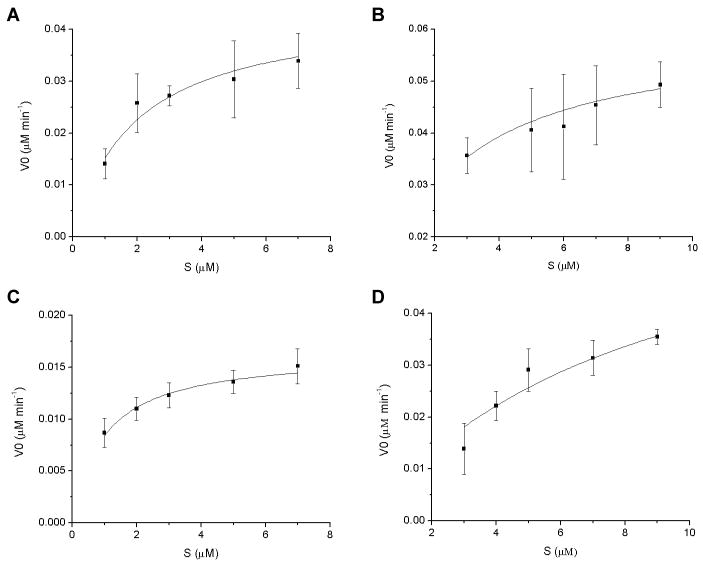
The kinetic studies were performed at 20°C with varying concentrations of ssDNA or ssRNA substrates. The results are shown in Fig. 4 and summarized in Table 1. The mFTO protein showed higher activities toward both ssDNA and ssRNA substrates than hFTO. It should be noted that this result may not reflect the in vivo activities of these proteins because the purified recombinant mFTO appears to be more stable than the purified recombinant hFTO in vitro. The kinetic studies revealed that both mFTO and hFTO exhibit a 2-fold preference for 3-meU in ssRNA as the substrate over 3-meT in ssDNA. The experiments were repeated in triplicate with similar results obtained.
Fig. 4.
Kinetics of demethylation reactions catalyzed by mFTO and hFTO. All reactions were run in triplicate at 20°C and pH 6.0. (A) Demethylation of 3-meT in the 15 mer ssDNA by mFTO (0.5 μM). (B) Demethylation of 3-meU in the 15 mer ssRNA by mFTO (0.3 μM). (C) Demethylation of 3-meT in the 15 mer ssDNA by hFTO (2.5 μM). (D) Demethylation of 3-meU in the 15 mer ssRNA by hFTO (0.4 μM).
Table 1.
Kinetic constants for 3-meT and 3-meU demethylation by mFTO and hFTO at 20°C and pH 6.0
| enzyme | substrate | Km (μM) | kcat (min−1) | kcat/Km (min−1μM−1) |
|---|---|---|---|---|
| mFTO | ssDNA (3-meT) | 1.92 ± 0.43 | 0.089 ± 0.008 | 0.046 ± 0.019 |
| ssRNA (3-meU) | 2.08 ± 0.31 | 0.199 ± 0.009 | 0.096 ± 0.029 | |
| hFTO | ssDNA (3-meT) | 0.95 ± 0.12 | 0.007 ± 0.0002 | 0.007 ± 0.002 |
| ssRNA (3-meU) | 8.51 ± 3.13 | 0.115 ± 0.022 | 0.014 ± 0.007 |
In summary, this study has established the DNA/RNA demethylation activity of the recombinant human FTO for the first time (Fig. 5). Both mFTO and hFTO showed no observable activity towards εA and 1-meG in ssDNA. A very low demethylation activity of 1-meA in ssDNA was observed for these two proteins. They could also catalyze demethylation of 3-meC in ssDNA, but with a much lower efficiency as compared to 3-meT in ssDNA. Both mFTO and hFTO failed to repair base lesions in dsDNA, strongly suggesting that they are involved in ssDNA or ssRNA processing.
Fig. 5.
FTO can demethylate both 3-meT from ssDNA and 3-meU from ssRNA.
Importantly, we showed that both recombinant hFTO and mFTO can demethylate 3-meU in ssRNA in vitro (Fig. 5). The FTO protein’s RNA demethylation activity is slightly more efficient than the demethylation of 3-meT in ssDNA mediated by the same proteins. Considering the negligible repair of dsDNA substrates by the FTO proteins and their slight preferences for 3-meU in ssRNA over 3-meT in ssDNA, it is attractive to suggest FTO as a RNA demethylase [21]. Perhaps it catalyzes the reverse reaction of a previously unrecognized RNA methylation and exerts gene regulation function at the RNA level. Disruption of this regulatory role of FTO may lead to the obesity phenotype linked to this protein. Of course, detailed in vivo experiments are required to further test this hypothesis. This current work serves as the first comprehensive evaluation of the activities of both mFTO and hFTO in vitro, and provides a foundation for further inquiry into the role of this very interesting nucleic acid demethylase.
Acknowledgments
This work was supported by grants from the National Institute of Health (GM071440), the W. M. Keck Foundation, the Camille & Henry Dreyfus Foundation, and the Research Corporation.
Abbreviations
- mFTO
mouse FTO
- hFTO
human FTO
- 3-meT
3-methylthymine
- 3-meU
3-methyluracil
- 1-meA
1-methyladenine
- 1-meG
1-methylguanine
- 3-meC
3-methylcytosine
- εA
1,N6-ethenoadenine
- εC
3,N4-ethenocytosine
- IPTG
isopropyl-β-D-thiogalactopyranoside
- α-KG
α-ketoglutarate
- BSA
bovine serium albumin
- MES
2-(N-morpholino)ethanesulfonic acid
- EDTA
ethylene diamine tetraacetic acid
- TFA
trifluoroacetic acid
- TLC
thin layer chromatography
- NMR
nuclear magnetic resonance
- MALDI-MS
matrix-assisted laser desorption-ionisation mass spectrometry
- HPLC
high performance liquid chromatography
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version, at doi:XXXX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Mishina Y, Duguid ME, He C. Direct reversal of DNA alkylation damage. Chem Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu B, Edstrom EC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 4.Aas PA, Otterlei M, Falnes PØ, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 5.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 7.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 8.Mishina Y, He C. Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins. J Inorg Biochem. 2006;100:670–678. doi: 10.1016/j.jinorgbio.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welford RWD, Kirkpatrick JM, McNeill L, Puri M, Oldham NJ, Schofield CJ. Incorporation of oxygen into the succinate co-product of iron(II) and 2-oxoglutarate dependent oxygenases from bacteria, plants and humans. FEBS Lett. 2005;579:5170–5174. doi: 10.1016/j.febslet.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 10.Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T, Tsuchiya M, Makino Y, Furukawa T, Konishi N, Yamamoto H. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundheim O, Vågbø CB, Bjørås M, Sousa MML, Talstad V, Aas PA, Drabløs F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ringvoll J, Nordstrand LM, Vågbø CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjørk A, Doughty RW, Falnes PØ, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falnes PØ, Bjørås M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of the DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Eshcherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falnes PØ. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 19.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PØ, Klungland A. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res. 2008;68:4142–4149. doi: 10.1158/0008-5472.CAN-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerken T, Girard CA, Tung YL, Webby CJ, Saudek V, Hewitson KS, Yeo GSH, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O’Rahilly S, Schofield CJ. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi: 10.1186/1471-2091-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Stunff CL, Bougnères P, Kovacs P, Marre M, Balkau B, Cauchi S, Chèvre J, Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 24.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch A, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin M, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJF, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CNA, Doney ASF, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbens S, Rouzé P, Cock JM, Spring J, Worden AZ, de Peer YV. The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J Mol Evol. 2008;66:80–84. doi: 10.1007/s00239-007-9059-z. [DOI] [PubMed] [Google Scholar]
- 27.Crain PC. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods in Enzymology. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 28.Lee D, Jin S, Cai S, Chen Y, Pfeifer GP, O’Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 29.Welford RWD, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ. The selectivity and inhibition of AlkB. J Biol Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]