Summary
To maintain phenotypes of cell lineages cells must “remember” which genes were active before mitosis entry and transmit this information to their daughter cells so expression patterns can be faithfully re-established in G1, a phenomenon called gene bookmarking. However, during mitosis transcription ceases, most sequence-specific proteins dissociate from DNA, and the chromatin is tightly compacted, making it difficult to understand how gene activity “memory” is maintained through this stage of the cell cycle1,2. Gene bookmarking is associated with a lack of compaction of promoters of formerly active genes in mitotic cells, but how compaction of these regions is inhibited was unknown3,4. Here we show that during mitosis TATA-binding protein (TBP), which remains bound to DNA during mitosis, recruits PP2A and also interacts with condensin to allow efficient dephosphorylation/inactivation of condensin near these promoters to inhibit their compaction. Further, ChIP-on-chip data show that TBP is bound to many chromosomal sites during mitosis, and is higher in transcribed regions but low in regions containing pseudogenes and genes whose expression is tissue-restricted. These results suggest that TBP is involved not only in gene transcription during interphase but also in preserving the memory of gene activity through mitosis to daughter cells.
The results of a number of studies suggest that the promoters of genes which are active prior to entry into mitosis are marked by the binding of some factor, that this factor remains associated with the promoter throughout this stage of the cell cycle, and that it somehow prevents compaction of these regions so that the transcriptional machinery can be reassembled on them in G1, thereby transmitting memory of gene expression patterns5,6. Because of similarities with the way a bookmark holds a place in a book, this mechanism has been referred to as gene bookmarking, and the putative factors mediating this process termed molecular bookmarks. However, the identity of the factor or factors involved in this mechanism, and how they prevent compaction of these promoter regions, was not known.
One candidate for this active gene bookmarking factor was TBP, based on data showing that TBP remains associated with chromosomal DNA even during mitosis, but only with promoters of genes that had been active prior to entry into mitosis7–9. In a previous study we identified a mechanism for gene-selective bookmarking in which a factor called HSF2 binds the hsp70 promoter during mitosis and prevents its compaction by interacting with condensin and also recruiting the phosphatase PP2A to dephosphorylate and inactivate these nearby condensin complexes10. In this study we sought to test whether TBP functions as a general bookmarking factor for active gene promoters and, if so, whether there is any similarity between the mechanism it employs to prevent promoter compaction and that of the gene-selective HSF2/hsp70 gene bookmarking mechanism.
First, we sought to test for interaction between TBP and condensin, the 5-subunit protein complex involved in compacting DNA during mitosis2, by performing co-immunoprecipitation analyses using extracts of mitotic cells. The results indicate that antibodies against CAP-G subunit of condensin co-immunoprecipitate the TBP protein, indicating that endogenous TBP and condensin exist as a complex in extracts of mitotic cells (Fig. 1a). The reverse experiment, in which antibodies against TBP were used to immunoprecipitate followed by anti-CAP-G Western blot, further support the association between endogenous TBP and condensin in mitotic cell extracts (Fig. 1b). As an independent test of the TBP-condensin interaction, in vitro binding experiments were performed, and the results show that recombinant GST-TBP is able to pull down CAP-G protein from extracts of mitotic cells (Fig. 1c). GST-TBP also interacts with in vitro translated CAP-G (Fig. 1d), suggesting that the association between TBP and condensin in mitotic cells is mediated, at least in part, by direct interaction between TBP and the CAP-G subunit of condensin.
Figure 1.
TBP interacts with condensin subunit CAP-G in mitotic cell extracts. a, Extracts of mitotic HeLa cells were immunoprecipitated using anti-CAP-G antibodies or non-specific IgG and the immunoprecipitates subjected to anti-TBP Western blot. b, Extracts of mitotic HeLa cells were immunoprecipitated using anti-TBP antibodies or non-specific IgG and the immunoprecipitates subjected to anti-CAP-G Western blot. c, Purified GST-TBP bound to glutathioneagarose was incubated with extracts of mitotic HeLa cells, washed, and then bound proteins were analyzed by Western blot using anti-CAP-G antibodies (upper panel) or anti-GST antibodies (lower panel). d, Purified GST-TBP bound to glutathione-agarose was incubated with 35S-labeled in vitro translated CAP-G, washed, and then bound proteins were subjected to SDS-PAGE and autoradiography (upper panel) or to Western blot using anti-GST antibodies (lower panel).
Condensin activity is inhibited by dephosphorylation of three of its subunits, including CAP-G, by the phosphatase PP2A2,11. Therefore, we undertook a set of experiments to test the hypothesis that TBP associates with PP2A as a mechanism for inhibiting compaction of promoters during mitosis. Microcystin is a ligand that binds the catalytic subunits of members of the PPP family of serine/threonine phosphatases, including PP1 and PP2A12,13. As shown in Figure 2a, microcystin-sepharose is able to pull down transfected TBP from cell extracts, suggesting that TBP associates with a member of the PPP phosphatase family. Since PP2A is known to dephosphorylate CAP-G and the other two condensin regulatory subunits CAP-D2 and CAP-H11, we sought to test whether this particular PPP phosphatase interacts with TBP. Immunoprecipitation analysis of extracts of cells transfected with GFP-TBP and then blocked in mitosis support this hypothesis by indicating that PP2A does associate with the transfected TBP protein (Fig. 2b). To test for PP2A association with endogenous TBP protein we immunoprecipitated extracts of mitotic cells using antibodies against the catalytic subunit of PP2A (Fig. 2c) or TBP (Fig. 2d), followed by Western blot using antibodies against TBP or the PP2A catalytic subunit, respectively. The results support the hypothesis that endogenous TBP and PP2A do interact in extracts of mitotic cells. TBP exists in cells as part of the TFIID complex, and thus to discriminate whether PP2A associates with TBP directly vs. some other subunit of TFIID we performed an in vitro binding experiment. The results show that GST-TBP interacts specifically with purified PP2A, indicating that TBP is able to interact directly with this protein phosphatase (Figure 2d).
Figure 2.
TBP and PP2A associate in mitotic cell extracts. a, Extracts of mitotic HeLa cells that had been transfected with a GFP-TBP expression construct were incubated with microcystin-sepharose or protein G-sepharose beads, washed, and then bound proteins were analyzed by anti-GFP Western blot. b PP2A was immunoprecipitated from extracts of mitotic HeLa cells that had been transfected with GFP-TBP expression plasmid using anti-catalytic subunit (PP2Ac) antibodies (with non-specific IgG as negative control), followed by anti-GFP Western blot. c, PP2A was immunoprecipitated from extracts of mitotic HeLa cells using anti-PP2A catalytic subunit (PP2Ac) antibodies (non-specific IgG as negative control), followed by anti-TBP Western blot. d, TBP was immunoprecipitated from extracts of mitotic HeLa cells using anti-TBP antibodies (non-specific IgG as negative control), followed by Western blot using antibodies against the PP2A catalytic subunit. e, Purified GST-TBP or GST bound to glutathione-agarose was incubated with purified PP2A (Upstate Biotech.), washed, and then bound proteins were subjected to SDS-PAGE followed by Western blot using anti-PP2A catalytic subunit antibodies (upper panel) or anti-GST antibodies (lower panel).
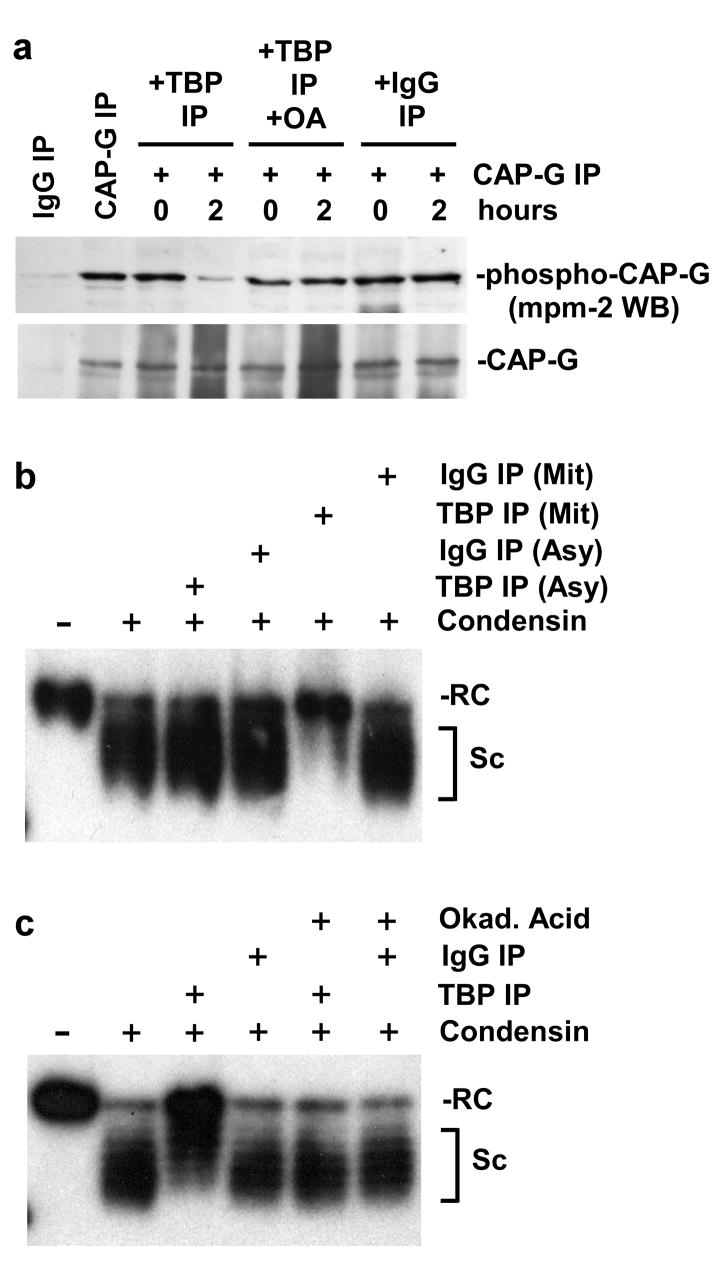
To test the ability of the TBP-associated PP2A to function as a condensin phosphatase we performed an experiment in which GFP-TBP immunoprecipitates from mitotic cell extracts were incubated with CAP-G immunoprecipitated from mitotic cells, followed by Western blot assay using the mpm2 antibody that detects cdc2-phosphorylated proteins, including CAP-G14. The results of this experiment indicate that GFP-TBP immunoprecipitates, but not GFP immunoprecipitates, contain a phosphatase activity that dephosphorylates the CAP-G subunit of condensin (Fig. 3a). This experiment also showed that this TBP-associated dephosphorylating activity is blocked by addition of the PP2A inhibitor okadaic acid.
Figure 3.
Mitotic TBP complexes dephosphorylate mitotic CAP-G and inhibit condensin activity. a, Mitotic TBP associates with an okadaic acid-inhibitable phosphatase activity that dephosphorylates mitotic CAP-G. HeLa cells transfected with the GFP-TBP expression construct were blocked in mitosis by nocodazole treatment. The first two lanes show immunoprecipitates pulled down by non-specific IgG or CAP-G antibody and subjected to Western blot with either Mpm-2 antibody (detects phospho-CAP-G) or CAP-G antibody (loading and degradation control). The next lanes show reactions containing CAP-G immunoprecipitates from mitotic cell extracts incubated with either TBP immunoprecipitated (using anti-GFP antibodies) from mitotic extracts, with or without 100 nM okadaic acid, or non-specific IgG immunoprecipitates, for 2 hours or no time. b, Mitotic TBP complexes inhibit condensin activity. TBP-containing complexes immunoprecipitated from extracts of asynchronous or mitotic (nocodazole-treated) HeLa cells, or negative controls immunoprecipitated from these extracts using non-specific IgG, were incubated with condensin immunopurified from mitotic cells and a relaxed circular test plasmid in an in vitro condensation assay. After the incubation, the plasmid was extracted and its supercoiling state analyzed by agarose gel electrophoresis and Southern blot. The labels RC and Sc refer to relaxed circular and supercoiled plasmid DNAs, respectively. c, Inhibition of condensin activity by mitotic TBP complexes is sensitive to okadaic acid. TBP-containing complexes immunoprecipitated from extracts of mitotic (nocodazole-treated) HeLa cells or negative controls immunoprecipitated using non-specific IgG were incubated with condensin immunopurified from mitotic cells and a relaxed circular test plasmid in an in vitro condensation assay, in the presence or absence of 100 nM okadaic acid. The supercoiling state of the plasmid in each sample was analyzed as described in (b).
The results shown above suggest that in mitotic cells TBP, associated with PP2A, could function as a regulator of condensin activity. To test this hypothesis we determined the effect of adding TBP-containing complexes from asynchronous or mitotic cells to an in vitro condensin activity assay15. The results of this experiment show that mitotic TBP immunoprecipitates from extracts of mitotic cells, but not those from extracts of asynchronous cells, are able to inhibit condensin activity (Fig. 3b). This inhibition of condensin activity by TBP complexes from mitotic cells is blocked by the addition of okadaic acid to the reaction, suggesting that PP2A activity is important for this effect (Fig. 3c).
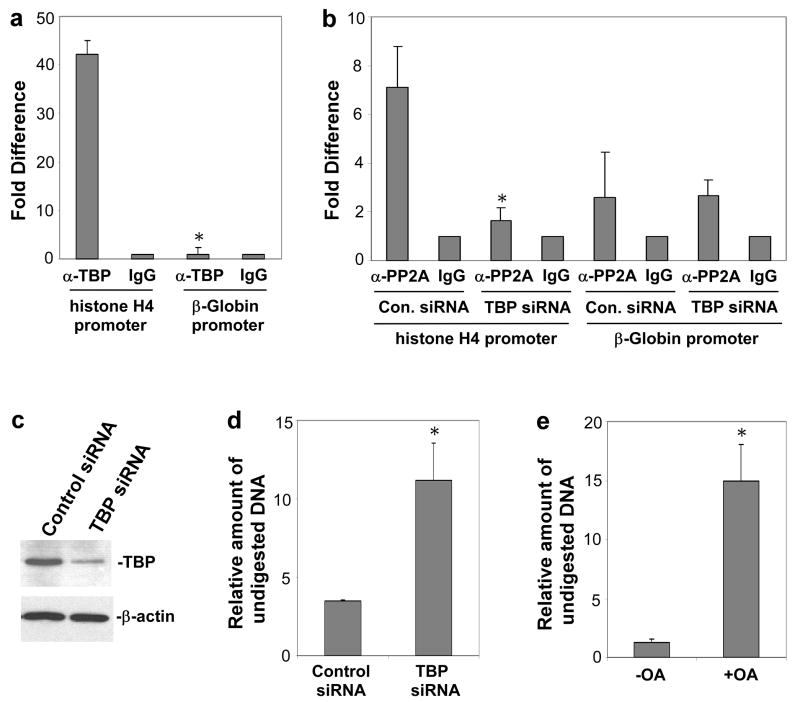
Consistent with the results of previous studies7–9,16, chromatin immunoprecipitation (ChIP) experiments demonstrate that in mitotic cells TBP remains associated with promoters of genes that are active in these cells during interphase, such as the histone H4 gene, but not with the promoter of a gene that is not expressed in these cells (β-globin) (Fig. 4a). As shown in Figure 4b, PP2A also exists within cross-linking distance of the histone H4 gene promoter in mitotic cells but not with the promoter of the β-globin gene. The results in this figure also show that the amount of PP2A associated with this promoter is decreased in cells in which TBP levels are reduced by RNAi (knockdown of TBP protein levels is shown in Figure 4c), consistent with the hypothesis that PP2A is recruited to promoters during mitosis by TBP. To directly test the importance of TBP for preventing compaction of gene promoters during mitosis, we determined whether reduction of cellular TBP levels by RNAi results in decreased accessibility (loss of bookmarking) of the histone H4 gene promoter in mitotic cells. Nuclei of TBP siRNA-transfected (or control siRNA-transfected) mitotic cells were subjected to DNase I accessibility assay, after which DNA isolated from the nuclei was used to perform PCR using a primer pair that amplifies the histone H4 promoter region. The results indicate that treatment with TBP siRNA results in decreased digestion of the histone H4 promoter region, suggesting reduced accessibility of this region (Fig. 4d). To test the importance of PP2A activity for promoter accessibility during mitosis, DNase I accessibility assay was performed using nuclei from mitotic cells that had been incubated (prior to isolation of nuclei) for 3 hours with a concentration of okadaic acid that selectively inhibits PP2A17,18. The results reveal that this treatment, like the TBP siRNA treatment, results in reduced accessibility of the promoter region of the H4 gene (Fig. 4e).
Figure 4.
Analysis of TBP and PP2A promoter binding in mitotic cells, effect of TBP knockdown on mitotic PP2A promoter binding, and effect of TBP RNAi and okadaic acid treatments on mitotic promoter accessibility. a, Chromatin immunoprecipitation (ChIP) assay was performed on mitotic (nocodazole-blocked) Jurkat cells using TBP antibodies or control IgG antibodies, followed by quantitative PCR assay using primer pairs specific to the H4 and β-globin gene promoters. Data are means +/− s.d. (n=3, *P<0.0002). b, PP2A associates specifically with the H4 promoter during mitosis, and this association is decreased by TBP RNAi. HeLa cells transfected with TBP siRNA or negative control siRNA were blocked in mitosis by nocodazole treatment, subjected to ChIP assay using antibodies against the PP2A catalytic subunit, and then analysed by quantitative PCR analysis using H4 and β-globin gene promoter primers. Data are means +/− s.d. (n=3, *P<0.006). c, Western blot assay of extracts of HeLa cells transfected with TBP siRNA or negative control siRNA using anti-TBP or anti- β-actin antibodies. d, Nuclei of HeLa cells transfected with TBP siRNA or negative control siRNA and blocked in mitosis by nocodazole treatment were subjected to DNase I accessibility assay, after which DNA was isolated and analysed by quantitative PCR using histone H4 promoter primers. Data is presented as the ratio of amount of undigested DNA in the DNAse-treated/non-DNAse-treated samples. Data are means +/− s.d. (n=3, *P<0.005). e, HeLa cells were blocked in mitosis by nocodazole treatment and then incubated with 50 nM okadaic acid for 3 hours. Nuclei of these cells were then subjected to DNase I accessibility assay, after which DNA was isolated and analysed by quantitative PCR using histone H4 promoter primers. Data is presented as the ratio of amount of undigested DNA in the DNAse-treated/non-DNAse-treated samples. Data are means +/− s.d. (n=3, *P<0.002).
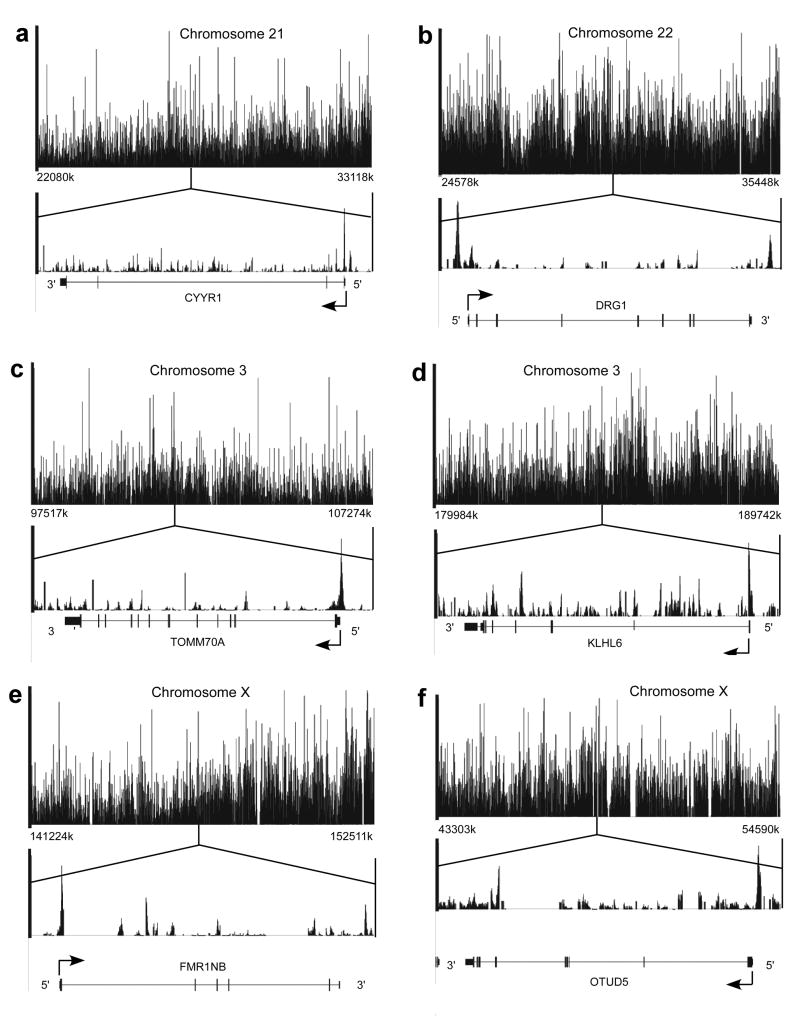
Finally, we performed a “ChIP on Chip” experiment to examine the binding of TBP during mitosis over large regions of chromosomal DNA, using arrays that include chromosomes 3, 21, 22, and X. The results of this analysis, shown in Figure 5, indicate that TBP binds a very large number of different sites in these chromosomes. Analysis of TBP binding to individual genes within these chromosomal regions, shown in the bottom part of each panel, indicates the association of TBP with the promoter regions of these genes. In agreement with a previous study that examined TBP binding to chromosomal regions of interphase cells, we also observed mitotic TBP binding to intronic sequences and intergenic sequences that have not yet been associated with a gene or mRNA annotation19. We also found, similar to the results of another study of TBP binding in interphase cells20, that mitotic TBP binding was only observed for a fraction of the presumed total number of binding sites. This may be due, at least in part, to technical limitations of ChIP-on-chip experiments, including the inability to efficiently cross-link all chromosomal bound TBP protein and/or to immunoprecipitate the DNA-linked TBP. Analysis of the ChIP-on-chip data revealed that more TBP binding is found in transcribed regions of the chromosomes (Supplementary Figure 1), and that TBP binding signals are low in chromosomal sequences containing pseudogenes, genes whose expression is tissue-restricted and not transcribed in the cell type used for this ChIP-on-Chip experiment (Jurkat), and regions that are devoid of annotated genes (Supplementary Figure 2).
Figure 5.
TBP is bound to many chromosomal loci during mitosis. DNA fragments immunoprecipitated by TBP antibodies by ChIP assay using mitotic cells, with input genomic DNA as control, were hybridized to the Affymetrix Genechip human tiling array 2.0c, which includes genomic DNA arrays spanning human chromosomes 3, 21, 22, and X. Shown at the top of each panel are the locations of TBP binding in regions representing 10 million base-pair segments of the indicated chromosome. In the lower part of each panel are close-up views of TBP binding to a gene within that region, indicating the association of TBP with the promoter regions of each gene. The genes are CYRR1 (panel a), DRG1 (panel b), TOMM70A (panel c), KLHL6 (panel d), FMR1NB (panel e), and UTUD5 (panel f).
The results of this study suggest that TBP plays an important role in the transmission of active gene memory to daughter cells by interacting with and inactivating condensin in the vicinity of these promoters via its associated PP2A, thereby inhibiting compaction of these regions. This role for TBP in bookmarking active genes makes sense because its function as an essential basal transcription factor at promoters transcribed by all three RNA polymerases not only provides a straightforward mechanism for identifying many genes that are active in a cell, and therefore should be bookmarked, but also for efficiently reassembling the transcriptional machinery on these promoters upon entry into G121,22. These results also reveal that some elements of this mechanism, for example the recruitment of PP2A to dephosphorylate/inactivate condensin, are also found in the gene family-specific HSF2-mediated bookmarking of the hsp70i promoter. This presumably reflects the important role of PP2A as a negative regulator of condensin activity11. In addition to increasing understanding of how the memory of gene activity is transmitted through the transcription block of mitosis, knowledge of the gene bookmarking mechanism could lead to advances in generating stem cells from adult cells and cloning animals via somatic cell nuclear transfer, because failure to properly reprogram gene bookmarking is believed to be a key barrier limiting the success of both of these processes23–25.
Methods
See Supplementary Information for detailed Methods.
Generation of hTBP and hPP2Ac antibodies
Affinity purified goat polyclonal antibodies to hTBP and hPP2Ac were prepared by Bethyl Labs against peptides (see SI for sequences) corresponding to N-terminal and C-terminal sequences of hTBP and hPP2Ac polypeptides, respectively.
Cell culture, preparation of M-phase populations, and transfection
Culture of HeLa cells and enrichment of M-phase populations using nocodazole treatment (200ng/ml for 16 hours) were performed as described previously10. HeLa cells were transfected with pEGFP-TBP9 using Effectene reagent according to the manufacturer’s instructions (Qiagen).
Expression/purification of GST-TBP and in vitro binding assay
GST-hTBP (plasmid from Dr. Arnold Berk) or GST alone (pGEX-2T plasmid) were expressed and purified from Escherichia coli BL21 (DE3) cells as described previously26. Pull-down assays between GST-TBP (or GST alone as negative control) on glutathione-agarose beads and either in vitro translated CAP-G or endogenous CAP-G-containing complexes in mitotic HeLa cell extracts were performed as described previously26. Detection was either by autoradiography to detect the presence of [35S] methionine-labeled hCAP-G, Western blot with anti-hCAP-G polyclonal antibodies/ECL, or anti-GST Western blot to ensure similar levels of GST-TBP and GST.
Immunoprecipitations and western blot analysis
The antibodies used for the immunoprecipitation/western blot experiments were the anti-hCAP-G rabbit polyclonal antibodies10 (1:200 IP, 1:5,000 WB), anti-hTBP goat polyclonal antibodies (1:200 IP, 1:5,000 WB), and anti-hPP2Ac goat polyclonal antibodies (described above) (1:200 IP, 1:2,000 WB), all made by Bethyl Laboratories (Montgomery, TX), anti-PP2A catalytic subunit mouse monoclonal antibodies (Upstate Biotechnology) (1:200 IP, 1:1,000 WB), anti-GFP mouse monoclonal and rabbit polyclonal antibodies (Clontech) (1:5,000 WB), anti-GST goat polyclonal antibodies (Amersham Pharmacia Biotech Inc.) (1:5,000 WB), anti-mpm2 mouse monoclonal antibody (Upstate Biotechnology) (1:1,000 WB) and control IgG (Sigma) (same final concentration as experimental antibodies). Immunoprecipitation assays using HeLa cells were performed as described previously10.
Microcystin-sepharose chromatography
Extracts prepared from HeLa cells transfected with pEGFP-TBP or pEGFP-C1 in NP-40 lysis buffer were incubated with microcystin-Sepharose (Upstate Biotechnology) for 2 hours at 4°C with gentle inversion mixing. After collecting by centrifugation, protein complexes were washed three times with lysis buffer. The samples were analyzed by SDS-PAGE and Western blot using anti-GFP monoclonal antibody (BD Clontech) (1:5,000).
Chromatin immunoprecipitation assay/Quantitative PCR analysis
Chromatin immunoprecipitations (ChIP) were performed as previously described10. The antibodies used were goat polyclonal TBP antibodies (described above) (1:200), PP2A monoclonal catalytic subunit antibodies (Upstate Biotechnology) (1:200), and control goat or mouse IgGs (Sigma) (same final concentration as experimental antibodies), and the DNAs were analyzed by Quantitative PCR assay as detailed in Supplementary Information (SI). The sequences of the H4 and β-globin primers used are provided in SI.
Dephosphorylation of mitotic condensin by endogenous TBP-associated phosphatase
This assay was performed as described previously10, except here using GFP antibodies (Clontech) (1:200) to immunoprecipitate GFP-TBP from transfected cells.
Purification of condensin complexes and supercoiling assay
Purification of HeLa condensin was performed essentially as described15, with minor modifications detailed in Supplementary Information. Purified mitotic condensin was incubated with anti-TBP (1:200) or control IgG immunocomplexes in 30kl PP2A buffer (catalog number V2460, Promega) for 30 minutes at 30ºC in the absence or presence of 100 nM okadaic acid. The samples were then subjected to supercoiling assay15.
siRNA treatment
TBP-specific siRNA or non-specific negative control siRNA (negative control siRNA #1) were obtained from Ambion and transfected into HeLa cells using GeneSilencer transfection reagent from Gene Therapy Systems.
DNase I accessibility assay
This was performed as described27. In one experiment HeLa cells were transfected with TBP siRNA or negative control siRNA as described above before blocking in mitosis with nocodazole treatment (200 ng/ml for 16 hours), while in another untransfected HeLa cells were nocodazole-blocked and then incubated with 50 nM okadaic acid for 3 hours. Nuclei prepared from these cells were subjected to digestion at 37ºC with different concentrations of DNase I (Gibco), after which DNA was extracted and analyzed by Quantitative PCR using primers that amplify the histone H4 promoter region.
Statistical analysis
The graphical data shown in Figure 4, each representing the results of three independent experiments, are presented as means ± s.d.. Statistical significance was determined using the Student t-test.
ChIP on Chip analysis of chromosomal TBP binding during mitosis
ChIP assays using anti-TBP antibodies (1:200) were performed using Jurkat cells as previously described10. The resulting precipitated DNA, along with genomic input DNA (control), was amplified and then these DNAs were labeled and hybridized to Affymetrix Genechip Human tiling array 2.0 c (following the manufacturer’s instructions), which includes genomic DNA spanning human chromosomes 3, 21, 22, and X. The results (from three TBP IP DNA-probed chips and three genomic input DNA-probed chips) were quantile-normalized within treatment/control replicate groups, and analyzed as described in the detailed methods provided in the SI section. The NCBI GEO accession number for these data is GSE12256.
Supplementary Material
Correlation between TBP binding and Transcriptome. The top parts of panels a and b show the data for TBP binding to a 56 million bp region of chromosome 3 (vertical lines represent locations of TBP binding), along with a depiction of the RefSeq genes and Transcriptome dataS1 for this region. In the bottom parts of panels a and b are shown the same information for two different sub-portions of this 56 million bp region, one that is approximately 9 million bp long and the other 6 million bp in length.
S1. Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316,1484–1488 (2007).
Lack of strong mitotic TBP binding in a chromosomal region containing pseudogenes, genes expressed in a tissue-restricted manner, and large regions lacking annotated genes. Panel a shows the data for TBP binding to the KLHL6 gene from Figure 5d, but compressed so that its size is displayed relative to the region shown in panel b. The asterisk indicates the TBP peak associated with this gene’s promoter region, and the dotted line relates the height of this TBP signal to the TBP signals in panel b. In panel b is shown a region of chromosome 21 that contains a number of pseudogenes, genes whose expression is tissue-restricted (C21orf99: testis and muscle; ANKRD21: testis; LOC441956: mammary gland), as well as regions that are devoid of any annotated genes. The position of each pseudogene is indicate by the vertical bar next to its identifier. The sizes of the two regions in panels a and b are displayed on the same relative scale, as are the magnitudes of the TBP binding signals.
Acknowledgments
We are very grateful to Dr. Sui Huang and Dr. Arnold Berk for their generous gifts of GFP-TBP and GST-TBP plasmids, respectively, and to lab members for insightful discussions. This research was supported by NIH grants GM61053 and GM64606 to K.D.S.
Footnotes
Author Information The authors declare no competing financial interests.
Author Contributions The study was directed and funded by K.D.S. N.L.V performed the in vitro binding experiments between GST-TBP and in vitro translated CAP-G, H.X. performed all other experiments, and the paper was written by K.D.S.
References
- 1.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 2.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 3.Larsen A, Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;29:609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 4.Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 5.John S, Workman JL. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–610. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 8.Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing H, et al. Mechanism of hsp70i bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 12.Gulledge BM, Aggen JB, Huang HB, Nairn AC, Chamberlin AR. The microcystins and nodularins: cyclic polypeptide inhibitors of PP1 and PP2A. Curr Med Chem. 2002;9:1991–2003. doi: 10.2174/0929867023368845. [DOI] [PubMed] [Google Scholar]
- 13.Kloeker S, et al. Parallel purification of three catalytic subunits of the protein serine/threonine phosphatase 2A family (PP2A(C), PP4(C), and PP6(C)) and analysis of the interaction of PP2A(C) with alpha4 protein. Protein Expr Purif. 2003;31:19–33. doi: 10.1016/s1046-5928(03)00141-4. [DOI] [PubMed] [Google Scholar]
- 14.Takemoto A, Kimura K, Yokoyama S, Hanaoka F. Cell cycle-dependent phosphorylation, nuclear localization, and activation of human condensin. J Biol Chem. 2004;279:4551–4559. doi: 10.1074/jbc.M310925200. [DOI] [PubMed] [Google Scholar]
- 15.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 16.Fairley JA, Scott PH, White RJ. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003;22:5841–5850. doi: 10.1093/emboj/cdg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favre B, Turowski P, Hemmings BA. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- 18.Honkanen RE, Golden T. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr Med Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- 19.Denissov S, et al. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 22.Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong L, Lako M, Dean W, Stojkovic M. Epigenetic Modification is Central to Genome Reprogramming in Somatic Cell Nuclear Transfer. Stem Cells. 2006;24:805–814. doi: 10.1634/stemcells.2005-0350. [DOI] [PubMed] [Google Scholar]
- 24.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 25.Allegrucci C, Denning C, Priddle H, Young L. Stem-cell consequences of embryo epigenetic defects. Lancet. 2004;364:206–208. doi: 10.1016/S0140-6736(04)16636-1. [DOI] [PubMed] [Google Scholar]
- 26.Schubart DB, et al. Gene structure and characterization of the murine homologue of the B cell-specific transcriptional coactivator OBF-1. Nucleic Acids Res. 1996;24:1913–1920. doi: 10.1093/nar/24.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The Forkhead Transcription Factor FoxI1 Remains Bound to Condensed Mitotic Chromosomes and Stably Remodels Chromatin Structure. Mol and Cell Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between TBP binding and Transcriptome. The top parts of panels a and b show the data for TBP binding to a 56 million bp region of chromosome 3 (vertical lines represent locations of TBP binding), along with a depiction of the RefSeq genes and Transcriptome dataS1 for this region. In the bottom parts of panels a and b are shown the same information for two different sub-portions of this 56 million bp region, one that is approximately 9 million bp long and the other 6 million bp in length.
S1. Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316,1484–1488 (2007).
Lack of strong mitotic TBP binding in a chromosomal region containing pseudogenes, genes expressed in a tissue-restricted manner, and large regions lacking annotated genes. Panel a shows the data for TBP binding to the KLHL6 gene from Figure 5d, but compressed so that its size is displayed relative to the region shown in panel b. The asterisk indicates the TBP peak associated with this gene’s promoter region, and the dotted line relates the height of this TBP signal to the TBP signals in panel b. In panel b is shown a region of chromosome 21 that contains a number of pseudogenes, genes whose expression is tissue-restricted (C21orf99: testis and muscle; ANKRD21: testis; LOC441956: mammary gland), as well as regions that are devoid of any annotated genes. The position of each pseudogene is indicate by the vertical bar next to its identifier. The sizes of the two regions in panels a and b are displayed on the same relative scale, as are the magnitudes of the TBP binding signals.