Abstract
Methanosarcina barkeri inserts pyrrolysine (Pyl) at an in-frame UAG codon in its monomethylamine methyltransferase gene. Pyrrolysyl-tRNA synthetase acylates Pyl onto tRNAPyl, the amber suppressor pyrrolysine tRNA. Here we show that M. barkeri Fusaro tRNAPyl can be misacylated with serine by the M. barkeri bacterial-type seryl-tRNA synthetase in vitro and in vivo in Escherichia coli. Compared to the M. barkeri Fusaro tRNA, the M. barkeri MS tRNAPyl contains two base changes; a G3:U70 pair, the known identity element for E. coli alanyl-tRNA synthetase (AlaRS). While M. barkeri MS tRNAPyl cannot be alanylated by E. coli AlaRS, mutation of the MS tRNAPyl A4:U69 pair into C4:G69 allows aminoacylation by E. coli AlaRS both in vitro and in vivo.
Keywords: tRNAPyl, pyrrolysine, mischarging, Methanosarcina barkeri
1. Introduction
A number of deviations from the universality of the genetic code are known [1]. One of them is the recoding of in-frame UAG codons as pyrrolysine (Pyl) sense codons in the Methanosarcinaceae, a group of methanogenic archaea, and in two evolutionarily unrelated bacteria [2,3]. The co-translational insertion of Pyl is contingent on the presence of the special amber suppressor tRNAPyl (encoded by pylT) [4], and an unusual aminoacyl-tRNA synthetase (aaRS), pyrrolysyl-tRNA synthetase (PylRS, encoded by pylS), specific only for this modified amino acid [4–6]. PylRS specifically recognizes Pyl and tRNAPyl [5,6] and generates Pyl-tRNAPyl both in vitro and in vivo. Transformation of E. coli with the archaeal pylT and pylS genes showed that both gene products are also functional in a bacterial setting [6–9]. Monitoring of UAG read-through efficiency by various reporter systems indicated that the stop codon is suppressed only when tRNAPyl and PylRS are present and the E. coli strain is grown in a culture media containing an exogenous source of Pyl [N-ε-cyclopentyloxycarbonyl-L-lysine (Cyc), a structural analog of Pyl] [6–9].
The molecular basis of PylRS specificity towards tRNAPyl was attributed to the unusual secondary/tertiary structure of the tRNA [8,10] previously seen only in the bovine mitochondrial tRNASer [11,12]. A series of specific interactions mediated by nucleotides located in the acceptor stem and flanking the tRNAPylCUA anticodon were also shown to contribute to the recognition of tRNAPyl by PylRS [8,10]. In contrast to most aaRSs, PylRS does not recognize its tRNA substrate via interaction with nucleotides of the anticodon [8,10,13]. This observation made possible the insertion of Cyc at a UGA codon upon engineering of a complementary UCA anticodon in tRNAPyl [10]. Crystallization of Methanosarcina mazei PylRS in a complex with Pyl and Cyc allowed the visualization of the enzyme’s active site and shed some light on Pyl recognition by archaeal [13,14] and bacterial PylRS [15].
In addition to Pyl and Cyc recognition, recent work describes the engineering of the PylRS active site to accommodate the lysine analogs, N-benzyloxycarbonyl lysine and N-acetyl lysine, so as to allow their incorporation into proteins in human [16] and E. coli [17] cells.
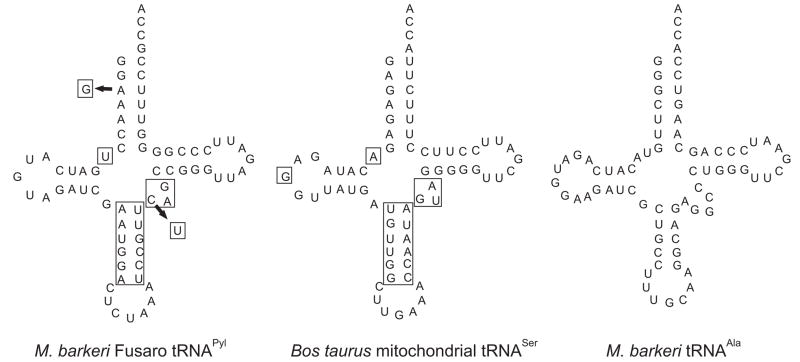
The tRNAPyl sequences of two M. barkeri strains are known; M. barkeri Fusaro and M. barkeri MS tRNAPyl differ in two bases (see Fig. 1), but both share the unique tRNA structure with the one found in the Bos taurus mitochondrial tRNASer species [18]. In these tRNAs the junction between the acceptor stem and D stem is shortened by one nucleotide, the anticodon stem consists of six base pairs instead of the classical five pairs, and the D loop is significantly shorter compared to the canonical cloverleaf structures [5,18]. The M. barkeri MS tRNAPyl has a G3:U70 base pair; this pair was shown to be the primary identity element for tRNAAla recognition by E. coli AlaRS [19,20]. Since the PylRS and tRNAPyl are assumed to be a functional orthogonal pair for genetic code expansion, we wanted to study this further by probing the aminoacylation of tRNAPyl by non-cognate aaRSs.
Fig. 1.
Cloverleaf representation of M. barkeri Fusaro tRNAPyl, Bos taurus mitochondrial tRNASer, M. barkeri MS tRNAPyl and M. barkeri tRNAAla UGC. Differences in M. barkeri MS tRNAPyl are indicated by arrows. The boxes indicate the unusual structure shared by the tRNAs.
2. Materials and Methods
2.1 General
Oligonucleotide synthesis, DNA sequencing and Edman degradation were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. [14C]Serine (155 mCi/mmol) and [14C]alanine (150 mCi/mmol) were from Amersham Biosciences (Uppsala, Sweden).
2.2 Construction of tRNA, minihelix, and enzyme constructs
M. barkeri strain Fusaro and strain MS tRNAPyl were produced and purified as described in [5]. Minihelices were constructed based on the sequences of M. barkeri MS tRNAAla (tRNAAla), M. barkeri MS tRNAPyl (tRNAPyl), and an M. barkeri MS tRNAPyl mutant (A4:U69→C4:G69) (tRNAPyl C4:G69). The minihelices are shown in Fig. 4B with their designated names (indicated in parentheses in the previous sentence). The pGF-1b plasmid containing the E. coli suppressor tRNALys gene was obtained from W. McClain (University of Wisconsin, Madison). The pET-15b plasmids containing the M. barkeri Fusaro bacterial and methanogenic SerRS genes were as described [21]. The pET15 E. coli AlaRS construct was a gift from M. Frugier (IBMC, Strasbourg, France).
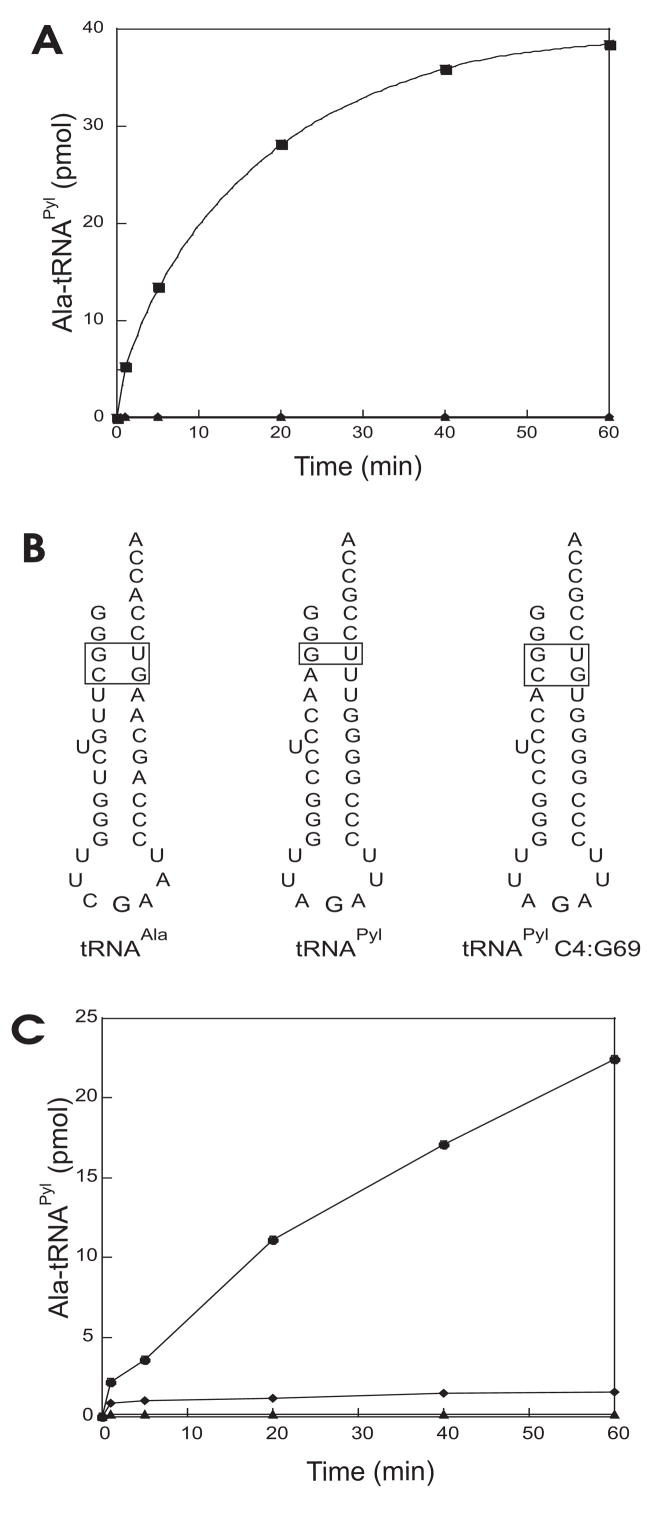
Fig. 4.
Misacylation of M. barkeri MS tRNAPyl with alanine in vitro. (A) In vitro aminoacylation of wild type M. barkeri MS tRNAPyl. Total E. coli tRNA with E. coli AlaRS (■), M. barkeri MS tRNAPyl with E. coli AlaRS (▲), no enzyme (◆). (B) Minihelices based on the sequences of, from left to right, M. barkeri MS tRNAAla (tRNAAla), M. barkeri MS tRNAPyl (tRNAPyl), M. barkeri MS C4:G69 tRNAPyl mutant (tRNAPyl C4:G69). (C) In vitro aminoacylation of minihelices. MS tRNAAla minihelix with E. coli AlaRS (●), MS tRNAPyl minihelix with the E. coli AlaRS (▲), MS tRNAPyl C4:G69 mutant minihelix with E. coli AlaRS (◆).
2.3 Aminoacylation Assays
Aminoacylation was performed at 37° C in 100 mM Hepes-KOH pH 7.2, 20 mM MgCl2, 5 mM KCl, 5 mM dithiotreitol, 5 mM ATP, 300 μM [14C]Ser or [14C]Ala, 2 μM tRNA transcript, and 1 μM enzyme. 20 μL aliquots were taken at 1, 5, 20, 40, and 60 min and spotted onto Whatman filters (3MM). The filters were washed twice with 10% TCA for 15 min each, and once with 5% TCA for 15 min. The filters were then rinsed with ethanol and allowed to dry. Radioactivity was measured with a Beckman Coulter LS 6500 scintillation counter.
2.4 TrpA mutant auxotrophy
To determine charging of a tRNAPyl mutant with alanine in vivo, an E. coli trpA94 strain (amber mutation in position 94 of trpA) was used. Active TrpA protein can only be made by the insertion of Ala or Gly into this position [22]. Cells were transformed with pGF-1b plasmid carrying the tRNAPyl C4:G69 mutant and plated on M9 minimal medium supplemented with ampicillin, either with or without Trp and grown at 37°C.
2.5 Suppression efficiency
Read-through efficiency using the dual luciferase/β-galactosidase reporter system was performed as indicated earlier [9].
2.6 Suppression of a UAG codon in an E. coli folA reporter gene
A mutant E. coli folA gene (encoding dihydrofolate reductase, DHFR) containing a UAG codon in place of codon 3 was amplified from genomic DNA, cloned into the pCR 2.1-TOPO plasmid vector (Invitrogen, Carlsbad, CA), sequenced, and subcloned into pRSFDuet-1. E. coli BL21(DE3) competent cells were cotransformed with wild-type M. barkeri Fusaro tRNAPyl (cloned in pTECH), the folA reporter construct (cloned in pRSF) and the M. barkeri bacterial type SerRS in pET15b. The resulting strains were grown in Luria broth at 37° C. Production of the recombinant DHFR was induced with 1 mM isopropyl-β-D-thiogalactoside when cells reached an A600 of 0.6. Cells were harvested after 17 h. DHFR was purified using a Ni-NTA affinity column and then blotted onto a PVDF membrane. The first eight amino acids were identified by Edman degradation. Background was determined in the same conditions but in the absence of any overexpressed proteins.
BL21(DE3) competent cells were alsocotransformed with wild-type M. barkeri Fusaro tRNAPyl (cloned in pTECH), the folA reporter construct (cloned in pRSF) and M. barkeri PylRS in pET15b. Cells were grown in the presence of Cyc, recombinant DHFR was purified and sequenced by Edman degradation as indicated above.
3. Results and Discussion
3.1 Mischarging of M. barkeri Fusaro tRNAPyl with serine in vitro
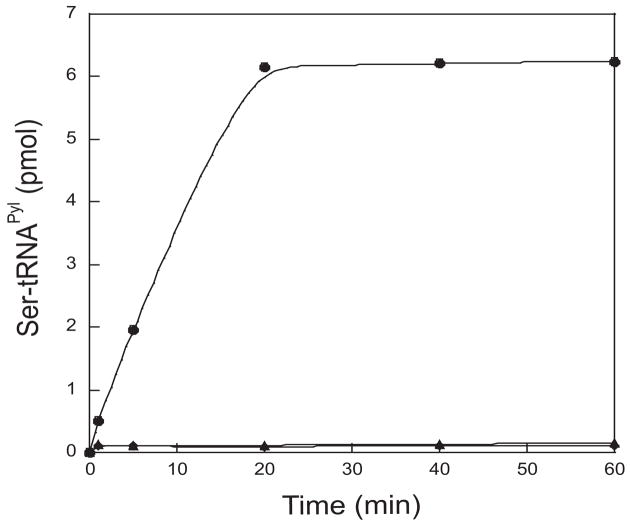
The unique secondary structure of M. barkeri Fusaro tRNAPyl is only shared by mammalian mitochondrial tRNASerUGA [23] (Fig. 1). Therefore, we investigated whether tRNAPyl could be a substrate for the two M. barkeri SerRSs. M. barkeri contains two serS genes that encode two different types of SerRSs: a bacterial-type SerRS present in most organisms, and a highly divergent SerRS only found in some methanogenic archaea [24]. The methanogenic and bacterial-type SerRSs use overlapping identity elements to recognize the same set of tRNASer species [21]. In vitro aminoacylation reactions showed that the bacterial-type SerRS was able to acylate up to 15% of the available M. barkeri tRNAPyl transcript with serine after 60 min (Fig. 2). In contrast, the methanogenic SerRS showed no detectable charging of tRNAPyl (Fig. 2).
Fig. 2.
Misacylation of M. barkeri Fusaro tRNAPyl with serine in vitro. tRNAPyl and bacterial-type SerRS (●), tRNAPyl and methanogenic type SerRS (▲), no enzyme (◆).
3.2 Mischarging of M. barkeri Fusaro tRNAPyl with serine in vivo
We then determined whether the mischarging of tRNAPyl with serine observed in vitro also occurs in vivo. To do so, we measured the level of UAG read-through in E. coli in the presence of the two M. barkeri SerRSs using a plasmid-based reporter system that contains in frame the genes encoding β-galactosidase (lacZ) and luciferase (luc). The two genes are transcribed as a single mRNA unit; an in frame UAG codon is located between the two open reading frames. In the absence of read-through only the β-galactosidase is produced. Upon read-through a β-galactosidase-luciferase fusion protein is synthesized. Luciferase enzymatic activity directly correlates to the frequency of read-through, and when normalized to β-galactosidase activity allows sample-to-sample comparison. Read-through efficiency is expressed as the ratio of luciferase to β-galactosidase enzymatic activities observed with the in frame UAG-containing reporter plasmid over the same ratio obtained when the stop codon is a standard lysine codon. This reporter system, previously used to study internal ribosome reentry sites [25], permits to reliably measure read-through even when it occurs at very low level.
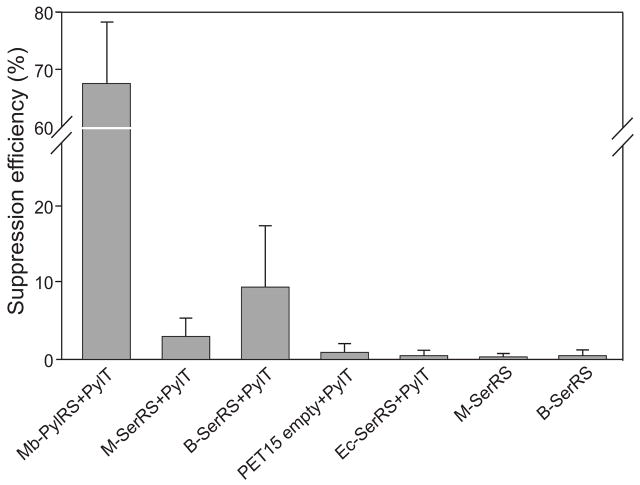
We quantified UAG suppression efficiency in the presence of tRNAPyl and each one of the two M. barkeri SerRSs and compared these values to those obtained with the appropriate negative and positive controls. When tRNAPyl was co-expressed with the bacterial type SerRS, read-through to up to 8.6% was detected and only to about 3.3% with the methanogenic SerRS (Fig. 3). In the absence of any overexpressed enzyme, background read-through was measured as 1.3% (Fig. 3). These results indicated that when expressed in E. coli the two M. barkeri SerRSs are able to misacylate M. barkeri tRNAPyl with serine at a low but detectable level. To gauge the significance of such read-through we compared it with UAG suppression observed in the presence of PylRS, tRNAPyl and Cyc, a structural analog of Pyl (Fig. 3). Read-through in the presence of the bacterial-type SerRS accounted for only 11% of that observed with PylRS and Cyc. The in vivo observations are in line with our above in vitro results that showed the ability of the M. barkeri bacterial-like SerRS to serylate tRNAPyl. More surprising was the fact that we could detect read-through activity with the methanogenic enzyme while no aminoacylation could be detected in vitro. We attribute this apparent discrepancy to the higher sensitivity of the in vivo assay for very low mischarging activities. Neither the methanogen-type M. barkeri SerRS nor E. coli SerRS formed significant amounts of Ser-tRNAPyl, and suppression was entirely dependent on the presence of tRNAPyl (Fig. 3).
Fig. 3.
Incorporation of serine and Cyc in a β-galactosidase/luciferase fusion reporter protein. Bars represent UAG read-through efficiency: M. barkeri tRNAPyl and either M. barkeri bacterial like SerRS (B-SerRS), M. barkeri methanogenic SerRSs (M-SerRS), E. coli SerRS (Ec-SerRS) or an empty expression vector (pET15 empty). As positive control, M. barkeri tRNAPyl and M. barkeri PylRS (Mb-PylRS) in the presence of Cyc was measured. Negative controls include measurements with either M-SerRS or B-SerRS without PylT.
3.3 Direct evidence for serine incorporation into protein
Next we sought to confirm the direct insertion of serine into a reporter protein in E. coli. We utilized an E. coli folA (encoding DHFR) reporter gene containing an in-frame UAG codon at position 3 and a six-histidine codon repeat at the 3′ end. E. coli cells were co-transformed with tRNAPyl, the folA gene reporter and either M. barkeri bacterial-type serS, or an empty plasmid for background determination. For each condition, a DHFR-His6 was purified by Ni-NTA chromatography and SDS gel electrophoresis. N-terminal sequencing of the purified DHFR-His6 from cells co-transformed with the bacterial-type serS revealed the presence of Ser at position 3 in the protein. Pro, Gln and Val were also detected at the third position albeit in much smaller amounts. Pro was found to be at position 3 of the protein sample extracted from the cells lacking tRNAPyl (negative control).
To ensure that our reporter system was functioning properly we measured the incorporation of Cyc at position 3 of the recombinant DHFR-His6 in cells grown on Cyc and containing the tRNAPyl:PylRS pair. Expression of the DHFR-His6 was significantly higher than in the previous experiments and allowed purification of the protein to homogeneity. Amino acid 3 of the purified DHFR-His6 was identified as Cyc, in line with earlier mass spectroscopy data [17]. No other amino acid was detected in position 3 (data not shown).
3.4 Misacylation of M. barkeri MS tRNAPyl with alanine in vitro
M. barkeri MS tRNAPyl differs from M. barkeri Fusaro tRNAPyl at two positions: the first nucleotide of the variable loop is U44 instead of C44, and in the acceptor stem MS tRNAPyl contains a G3 instead of A3 (Fig. 1). The G3A deviation results in the formation of the wobble base pair G3:U70. A G3:U70 pair is also the primary identity element for tRNAAla recognition by AlaRS [19,20].
We then tested whether M. barkeri MS tRNAPyl could be a substrate for E. coli AlaRS. As seen in Fig. 4A, the M. barkeri MS tRNAPyl is not misacylated with alanine by E. coli AlaRS in vitro, possibly the result of a tRNA anti-determinant whose presence prevents AlaRS recognition. A good candidate for an anti-determinant in wild-type MS tRNAPyl is the A4:U69 base pair, as insertion of A4:U69 into tRNAAla disrupts the acceptor stem helix and reduces recognition by AlaRS [19]. We tested whether this particular base pair plays a role in precluding MS tRNAPyl from being misacylated with alanine by E. coli AlaRS. Inspection of the M. barkeri MS tRNAAla sequence revealed a C4:G69 base pair underneath the G3:U70 main identity element. We decided to replace M. barkeri MS tRNAPyl A4:U69 with a C4:G69 pair. AlaRS is one of the few aaRSs shown to be able to charge stem-loop helices derived from its cognate tRNAAla [26]. We then constructed three minihelices (Fig. 4B) based on (i) M. barkeri MS tRNAAla (tRNAAla), (ii) acceptor and T-stem sequences of M. barkeri MS tRNAPyl (tRNAPyl), and (iii) a C4:G69 M. barkeri MS tRNAPyl mutant (tRNAPyl C4:G69). The M. barkeri MS tRNAPyl minihelix, like the full length tRNAPyl is not misacylated by E. coli AlaRS. The control M. barkeri MS wild-type tRNAAla stem-loop helix was charged close to 50%. The C4:G69 mutant minihelix was acylated with Ala but remained a poor substrate for AlaRS since it was charged to only 2% (Fig. 4C).
3.5 Misacylation of M. barkeri MS tRNAPyl with alanine in vivo
To determine whether M. barkeri MS tRNAPyl and its C4:G69 mutant can be misacylated with Ala in vivo, we made use of the E. coli trpA94 amber mutant strain that will only grow in the absence of exogenously added Trp is the amber codon is suppressed by either Gly or Ala [22]. We transformed this strain with plasmids containing either the M. barkeri MS wild type tRNAPyl, the C4:G69 tRNAPyl mutant, an E. coli tRNAAla amber suppressor, or an empty vector. The transformed strains were plated on M9 minimal medium with or without Trp (Fig. 5).
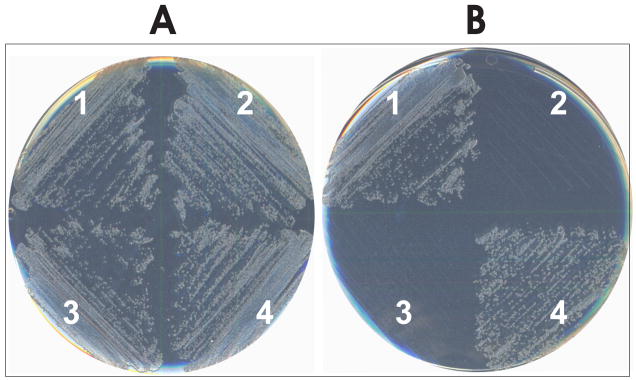
Fig. 5.
Suppression of an E. coli strain trpA94 with the M. barkeri MS tRNAPyl C4:G69 mutant. E. coli transformants were streaked on M9 medium (A) with Trp (B) without Trp. E. coli transformed with 1, C4:G69 M. barkeri MS tRNAPyl mutant; 2, M. barkeri MS tRNAPyl wild type; 3, empty plasmid; 4, tRNAAla amber suppressor.
After incubation at 37 C for 72 h, the E. coli strain trpA94 transformed with the tRNAPyl C4:G69 mutant showed growth on plates without Trp, albeit at a slower rate (72 h) compared to the E. coli tRNAAla amber suppressor (24–48 h) (Fig. 5). This result indicated the misacylation of the mutant tRNAPyl with Ala, and the production of a sufficient amount of active TrpA enzyme, permitting cell survival. The cells transformed with M. barkeri MS wild-type tRNAPyl or with the empty plasmid did not show any growth after 72 h (Fig. 5), confirming our in vitro results. Lastly, as expected, all transformants grew on minimal plates supplemented with Trp (Fig. 5).
4. Outlook
The current study demonstrates two instances of tRNAPyl misacylation, the formation of Ser-tRNAPyl and Ala-tRNAPyl. M. barkeri bacterial-type SerRS recognizes its three tRNASer isoacceptors via interaction of amino acids in the long α-helix domain with nucleotides of the extended variable loop of the tRNASer; this specificity mechanism is shared with E. coli SerRS [21]. Bovine mitochondrial tRNASer lacks such an extended variable loop and presents a non-canonical structure; a mitochondrial SerRS:tRNASer complex requires an additional N-terminal enzyme domain interacting with nucleotides of the D-and T-loops [23]. Future studies will establish details of the interaction between SerRS and tRNAPyl, and should also provide more insight into possible uses of misacylated suppressor tRNAPyl protein synthesis.
Acknowledgments
We thank O. Namy for the reporter system, and E.J. Murgola for the E. coli trpA94 mutant strain. This work was supported by grants from the National Institute for General Medical Sciences and from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 2.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Herring S, Ambrogelly A, Gundllapalli S, O’Donoghue P, Polycarpo CR, Söll D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 5.Polycarpo C, Ambrogelly A, Bérubé A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 7.Polycarpo C, Herring S, Bérubé A, Wood JL, Söll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Söll D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci USA. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namy O, Zhou Y, Gundllapalli S, Polycarpo CR, Denise A, Rousset JP, Söll D, Ambrogelly A. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581:5282–5288. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Herring S, Ambrogelly A, Polycarpo CR, Söll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35:1270–1278. doi: 10.1093/nar/gkl1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polycarpo C, Ambrogelly A, Ruan B, Tumbula-Hansen D, Ataide SF, Ishitani R, Yokoyama S, Nureki O, Ibba M, Söll D. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol Cell. 2003;12:287–294. doi: 10.1016/s1097-2765(03)00280-6. [DOI] [PubMed] [Google Scholar]
- 12.Théobald-Dietrich A, Frugier M, Giegé R, Rudinger-Thirion J. Atypical archaeal tRNA pyrrolysine transcript behaves toward EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004;32:1091–1096. doi: 10.1093/nar/gkh266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 14.Kavran JM, Gundllapalli S, O’Donoghue P, Englert M, Söll D, Steitz TA. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MM, Jiang R, Jain R, Larue RC, Krzycki J, Chan MK. Structure of Desulfitobacterium hafniense PylSc, a pyrrolysyl-tRNA synthetase. Biochem Biophys Res Commun. 2008;374:470–474. doi: 10.1016/j.bbrc.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 17.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N-ε-acetylly-sine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y, Kawai G, Yokogawa T, Hayashi N, Kumazawa Y, Ueda T, Nishikawa K, Hirao I, Miura K, Watanabe K. Higher-order structure of bovine mitochondrial tRNASerUGA: chemical modification and computer modeling. Nucleic Acids Res. 1994;22:5378–5384. doi: 10.1093/nar/22.24.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 20.McClain WH, Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 21.Korencic D, Polycarpo C, Weygand-Durasevic I, Söll D. Differential modes of transfer RNASer recognition in Methanosarcina barkeri. J Biol Chem. 2004;279:48780–48786. doi: 10.1074/jbc.M408753200. [DOI] [PubMed] [Google Scholar]
- 22.Murgola EJ. tRNA, suppression, and the code. Ann Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- 23.Chimnaronk S, Jeppesen MG, Suzuki T, Nyborg J, Watanabe K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HS, Vothknecht U, Hedderich R, Celic I, Söll D. Sequence divergence of seryl-tRNA synthetases in archaea. J Bacteriol. 1998;180:6446–6449. doi: 10.1128/jb.180.24.6446-6449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 26.Hou YM, Francklyn C, Schimmel P. Molecular dissection of a transfer RNA and the basis for its identity. Trends Biochem Sci. 1989;14:233–237. doi: 10.1016/0968-0004(89)90033-9. [DOI] [PubMed] [Google Scholar]