Abstract
Metridia luciferase is a secreted luciferase from a marine copepod and uses coelenterazine as a substrate to produce a blue bioluminescence (λmax = 480 nm). This luciferase has been successfully applied as a bioluminescent reporter in mammalian cells. The main advantage of secreted luciferase as a reporter is the capability of measuring intracellular events without destroying the cells or tissues and this property is well suited for development of high throughput screening technologies. However because Metridia luciferase is a Cys-rich protein, E. coli expression systems produce an incorrectly folded protein, hindering its biochemical characterization and application for development of in vitro bioluminescent assays. Here we report the successful expression of Metridia luciferase with its signal peptide for secretion, in insect (Sf9) cells using the baculovirus expression system. Functionally active luciferase secreted by insect cells into the culture media has been efficiently purified with a yield of high purity protein of 2–3 mg/L. This Metridia luciferase expressed in the insect cell system is a monomeric protein showing 3.5-fold greater bioluminescence activity than luciferase expressed and purified from E. coli. The near coincidence of the experimental mass of Metridia luciferase purified from insect cells with that calculated from amino acid sequence, indicates that luciferase does not undergo posttranslational modifications such as phosphorylation or glycosylation and also, the cleavage site of the signal peptide for secretion is at VQA-KS, as predicted from sequence analysis.
Although the phenomenon of bioluminescence is widespread most luminous species are marine inhabitants. Many of these marine systems use imidazopyrazinone derivatives as the luciferin and the one most commonly isolated is coelenterazine. Coelenterazine and its derivatives are used by many bioluminescent proteins such as Renilla luciferase [1], the Ca2+-regulated photoproteins [2, 3], luciferase from the scyphozoan medusa Periphylla [4], squid photoproteins [5], etc. The best-characterized examples of such proteins are Ca2+-regulated photoproteins and Renilla luciferase, the ones responsible for bioluminescence of animals belonging to the classes Hydrozoa and Anthozoa respectively. Crystal structures are now available for a number of coelenterazine-dependent proteins, including several structures of different ligand-dependent conformational states of photoproteins [6–10], Renilla luciferase [11], and its natural “substrate” the Ca2+-regulated coelenterazine-binding protein [12]. The spatial structures of the photoproteins for example, have allowed rational engineering using site-directed mutagenesis to produce novel spectral and enzymatic properties, which could find usefulness in imaging and other applications [13–15].
Among coelenterazine-dependent bioluminescent proteins are also found several secreted proteins. These are luciferases from the deep-sea shrimp Oplophorus gracilirostris [16], and the marine copepods Gaussia and Metridia [17, 18]. The bioluminescence of marine copepods originates as a secretion from epidermal glands in response to various stimuli [19]. Metridia luciferase catalyzes an oxygen oxidation of coelenterazine to produce light (~ 480 nm) and requires no other cofactors for the bioluminescent reaction. Metridia luciferase (GenBank accession no. AAR17541) is a small protein (~ 23 kDa) containing an N-terminal signal peptide for secretion (17 amino acids) [18].
The secreted luciferases are very convenient bioluminescent reporters [18, 20–25] because they enable monitoring of intracellular events with high sensitivity, without destroying the cells or tissues. Secretion therefore, is a property well suited for development of high throughput screening technologies. Already the secreted luciferase from Gaussia shows much promise as a bioluminescent label for in vitro hybridization assays [26]. However for in vitro assay development it is important that high purity luciferases need to be available in sufficient amounts.
Most secreted proteins contain Cys residues and disulfide bonds [27] and Metridia luciferase is no exception. The amino acid sequence of Metridia luciferase includes two closely similar motifs of 31 residues containing ten conserved Cys residues, repeating as C-x3-C-x2-C-x5-Cx11-C and C-x2-C-x3-C-x8-C-x11-C in the first and second motifs respectively (Fig. 1B) [18]. Dithiothreitol (DTT)1 addition results in complete loss of activity indicating that active luciferase requires disulfide bonds. The expression of functionally active Cys-rich proteins with numerous S-S bonds in E. coli cells is non-trivial. It has long been noted that proteins containing disulfide bonds are rare in the cytoplasm of any organism, attributable to the reductive environment of the cytoplasm. The E. coli periplasm apparently is more suitable for the expression of recombinant proteins containing the disulfide bonds due to the presence of disulfide oxidoreductase and disulfide isomerase [28]. However the main disadvantage of the E. coli periplasm as a folding compartment is that its enzyme system is not always efficient in catalyzing disulfide bond formation in eukaryotic proteins that contain numerous disulfide bonds.
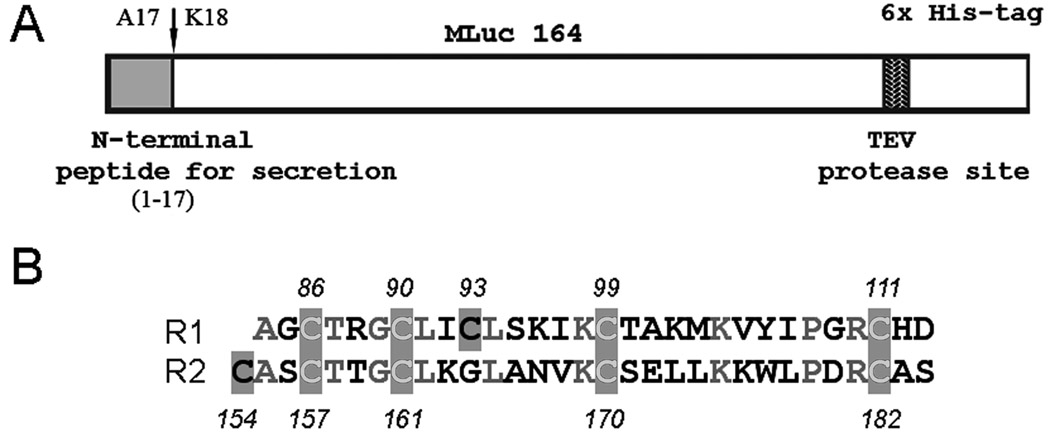
Figure 1.
(A) Schematic representation of the Metridia luciferase gene (MLuc164) and the construct used for its expression. (B) Alignment of two conservative repeats R1 and R2 in Metridia luciferase [18]. The shaded boxes indicate the conserved Cys residues. The residues in gray show the identical amino acids.
Expression in insect cells with the use of a baculovirus expression system does not have these shortcomings and today it is a very powerful tool for expression of recombinant eukaryotic proteins. The eukaryotic insect host cells perform most of the posttranslational modifications of proteins occurring in mammalian cells as well as providing a secretory pathway through the endoplasmic reticulum that allows the correct formation of disulfide bonds. Besides, this expression system provides easy scalability and high levels of protein expression. Also it has been shown that most of the expressed recombinant eukaryotic proteins are as functionally active as their authentic counterparts.
In this paper we describe the expression of Metridia luciferase in insect (Sf9) cells using the baculovirus expression system. Functionally active luciferase is secreted by the insect cells into the culture media because the Metridia cDNA was cloned with its own signal peptide for secretion and as a result, the luciferase can be efficiently purified as a monomer with a yield of high purity protein of 2–3 mg/L. We also show that this luciferase produced and purified from insect cells, has 3.5-fold greater bioluminescence activity than luciferase expressed and purified from E. coli cells. Using LC–MS we demonstrate also that the experimental mass of Metridia luciferase purified from insect cells is in close agreement with that calculated from its amino acid sequence. This indicates that luciferase does not undergo posttranslational modifications such as phosphorylation or glycosylation and that the cleavage site of signal peptide for secretion is exactly at VQA-KS, as predicted from sequence analysis.
Materials and methods
PCR cloning of the Metridia luciferase cDNA for baculovirus expression system
The oligonucleotide primers were designed to introduce the C-terminal His6-tag followed by a TEV-specific protease site into the MLuc164 coding sequence (Fig. 1A). The cloning strategy also generated a BamHI/XhoI restriction site on the flanking ends of the gene for directional cloning into the baculovirus donor vector. The coding sequence of the full-length MLuc164 cDNA gene containing the N-terminal signal peptide for secretion, was amplified from a cDNA library vector TriplEx2-MLuc164 [18] using two-step PCR. The gene-specific forward S-ML164s (5’-GCCGGGATCCATGGATATAAAGGTTGTCTTTACTCTTGTTTTCTC-3’) and reverse, A-ML164s primers (5’-ATGGTGATGGATGCCCTGAAAATACAGGTTTTCACGATCTCCAGCCATGCCTTT-3’) were used to amplify an approximately 714 bp sequence by polymerase chain reaction using “PCR super-mix” (Invitrogen). PCR was performed for 27 cycles (denaturation: 25 sec at 95°C; annealing: 30 sec at 56°C; extension: 45 sec at 68°C). For the second PCR, the first PCR product was used as a template and the full-size gene was amplified under a similar condition with forward S-ML164s primer and second reverse A2-ML164s primer (5’-GCCGCTCGAGTCAGTGATGGTGATGGTGATGGATGCCCTG-3’). The amplified product was purified using centrifuge spin-columns (Qiagen) and cloned into the pFastBac-1 vector (Invitrogen) after appropriate double digestion with BamHI/XhoI restriction enzymes (Roche). The correct sequence was verified by restriction endonuclease digestion and sequencing.
Transposition
To generate a recombinant bacmid DNA the recombinant plasmid pFastBac-MLuc164 was transformed into Max Efficiency DH10Bac competent cells (Invitrogen) and the gene of interest was transposed into the bacmid through lacZ gene disruption. White clones containing the recombinant bacmid were selected on Luria-agar plates containing 50 mg/ml kanamycin, 7 mg/ml gentamicin, 10 mg/ml tetracycline, 100 mg/ml Bluo-gal, and 40 mg/ml IPTG. After 36 h incubation at 37°C high molecular weight DNA was isolated from the overnight cultures using Qiagen anion exchange resin. The recombinant bacmids were analyzed by performing PCR to verify successful transposition with S-ML164s/A2-ML164s gene-specific primers.
Transfection of Sf9 cells with recombinant bacmid-MLuc164
The minipreparations of recombinant bacmid DNA were transfected into Sf9 nsect cells using CELLFECTIN lipid reagent (Invitrogen). For each transfection 1.0 × 106 cells/ml with viability >97% were seeded in a 35 mm cell culture treated plate and allowed to attach for at least 1 h. The lipid reagent and bacmid DNA (1 µg) were diluted separately into 100 µl of SF-900 II SFM media without antibiotics, and then combined to form lipid-DNA complexes. The lipid-DNA complexes were diluted to 1 ml with SFM and laid over the washed Sf9 cells. The cells were incubated for 5 h at 27°C, rinsed by SFM media, and incubated for another 72 h. Recombinant baculovirus was harvested from the supernatant and amplified in 50-ml cell culture.
Techniques for plaque assay and virus titration were according to the Bac-to-Bac Baculovirus expression system protocol (Invitrogen).
Expression and purification of MLuc164 from E. coli cells
Plasmid pET22b-MLuc164 containing the MLuc164 gene without signal peptide and any purification tags, was used to transform the E. coli expression strain RIPL (DE3) pLysS (Novagen). The signal peptide for secretion was removed from Metridia luciferase sequence [18] for correct comparison of luciferase properties produced in different expression systems. Eukaryotic signal peptides do not allow secretion of proteins in bacteria and therefore, if the complete Metridia luciferase sequence was to be expressed in E. coli cells, it would contain an additional 17 amino acid residue peptide at N-terminus. For protein production, the transformed E. coli was cultivated with vigorous shaking at 37°C in LB medium containing 200 µg/ml ampicillin and induced with 1 mM IPTG when the culture reached an A600 nm of 0.8 – 1.0. After addition of IPTG, the cultivation was continued for 3 h. For purification of luciferase, the cell paste was resuspended in 1 mM EDTA, 20 mM Tris-HCl pH 7.0 (1:20, w/v) and disrupted with ultrasound (1 min × 6) at 0°C. Then the mixture was centrifuged and the supernatant discarded. The pellet was washed sequentially with 0.9% NaCl, 1% Triton X-100, 5 mM EDTA, 20 mM Tris-HCl pH 7.0. All the washing procedures were performed with centrifugation at 4°C (24,000 rpm). The final pellet was resuspended in 6 M urea in 20 mM Tris-HCl pH 8.8, incubated overnight at 4°C, and then centrifuged. The 6 M urea extract was loaded on a DEAE-Sepharose Fast Flow (Amersham Biosciences) column equilibrated with 6 M urea, 20 mM Tris-HCl pH 8.8. The proteins were eluted with a linear salt gradient (0 – 0.5 M NaCl) in the same buffer.
The MLuc164 sample in 6 M urea obtained after chromatography on DEAE Sepharose Fast Flow, was refolded by a 20-fold dilution in 1 mM EDTA, 20 mM Tris-HCI pH 8.8 containing 5 mM GSH and 0.5 mM GSSG. The mixture was incubated overnight at 4°C with stirring and then loaded on an anion exchange HiTrap Q column (Amersham Biosciences). The luciferase sample was eluted by a NaCl linear gradient (0 – 0.5 M) in 1 mM EDTA, 20 mM Tris-HCl pH 8.8. The collected peak was loaded on HiLoad Superdex 75 (Amersham Biosciences) gel filtration column in 0.2 M NaCl, 1 mM EDTA, 20 mM Tris-HCl pH 7.5 and active luciferase monomer fractions were collected.
Expression and purification of MLuc164 from Sf9 cells
Sf9 cells cultured in suspension using HyQ SFX serum-free medium (HyClone), were infected with plaque-pure recombinant virus at a multiplicity of infection (MOI) of 0.2 plaque-forming units (pfu) and harvested at 72 h post-infection. Cells were pelleted at 2000 g for 10 min at 4°C and the supernatant containing Metridia luciferase was kept at 4°C until purification.
Since the recombinant luciferase contained a His6-tag at the N-terminus, the protein was purified using a Ni-NTA resin (5-ml HisTrap column, Amersham Biosciences). Routinely, 3-liter batches of media containing the recombinant Metridia luciferase were concentrated by ammonium sulfate precipitation (60%, m/v). The insoluble particles were spun down (6,000 rpm, 30 min) and dissolved in the Ni-binding buffer (0.2 mM NaCl, 10 mM imidazole, 50 mM Na2HPO4/KH2PO4 buffer pH 7.1). The mixture was passed over a 5-ml His Trap Ni–column (Amersham Biosciences) and eluted with an imidazole gradient (0 – 0.5 M) in 0.2 M NaCl, 50 mM Na2HPO4/KH2PO4 buffer p H 7.1. The luciferase peak was collected and dialyzed against TEV protease cleavage buffer (1 mM EDTA, 20 mM Tris–HCl pH 8.0 containing 2 mM GSH and 0.2 mM GSSG). The His6-tag was cleaved by incubation of the luciferase sample with TEV protease at ratio 50:1 (w/w) overnight at room temperature; then the sample was loaded on a Ni-NTA resin column and the flow-through collected. The collected sample was dialyzed against 1 mM EDTA, 20 mM Tris-HCl pH 8.5 and loaded on an anion exchange HiTrap Q column (Amersham Biosciences). The luciferase was eluted by a NaCl linear gradient (0 – 0.5 M) in 1 mM EDTA, 20 mM Tris-HCl pH 8.5. The eluted peak was passed through a Superdex75 gel filtration column equilibrated by 0.2 M NaCl, 1 mM EDTA, 20 mM Tris-HCl pH 7.5. Typically, at the final purification step the yield of high purity luciferase was 2 mg per liter of insect cell culture.
Bioluminescence assay
The bioluminescence activity of luciferase was measured as the maximum intensity in relative light units (RLU) at room temperature using a Turner luminometer (Turner, USA) on rapid injection of 10 µl of 2 µM coelenterazine in methanol into a cuvette containing 5 µl of the luciferase sample in 0.5 ml buffer (0.3 M NaCl, 20 mM Tris-HCl pH 7.5).
The specific activities of Metridia luciferases obtained with different expression systems were determined as the integrated bioluminescence signal per mg of protein using a Luminoscan Ascent plate luminometer (Thermo Electron Co, Finland) by injection of 50 µl of freshly prepared coelenterazine (7.2 µM in 0.3 M NaCl, 20 mM Tris–HCl pH 7.5) into the wells containing 100 µl luciferase at concentration 5 ng/ml in 0.3 M NaCl, 20 mM Tris–HCl pH 7.5. Coelenterazine solution in buffer was prepared by dilution from stock solution of coelenterazine in methanol with the concentration determined from the extinction coefficient (methanol, ε434 nm = 8900 M−1 cm−1) [1]. The protein concentration was determined by the Dc protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
Spectral measurements
Bioluminescence spectra were measured with an AMINCO spectrofluorimeter (Thermo Spectronic, USA) using a slit width of 8 nm and a scan rate of 16 nm/sec. All spectra were corrected for spectral sensitivity of the instrument with the program supplied by AMINCO and for intensity decay during the scan period. All measurements were carried out at room temperature.
SDS-PAGE and Western blot analysis
The media samples harvested at 72 h post-infection were mixed with SDS sample buffer and heated to 100°C for 5 min. The samples were separated by 12–20% SDS-polyacrylamide gradient gel electrophoresis and then transferred on PVDF membrane. The membrane was blocked overnight at 4–8°C with 3% BSA in PBS containing Tween 20 (0.5%) and incubated at room temperature for 30 min with mouse polyclonal IgG anti-His6 antibodies (Qiagen). The appropriate secondary alkaline phosphatase-conjugated anti-mouse IgG antibody (Pierce) was added and complexes were detected by enhanced color reaction.
Liquid chromatography–mass spectrometry
The precise molecular masses of the pure Metridia luciferase samples were determined using HPLC-Atmospheric Pressure Ionization mass spectrometry (LC-MS) at the Chemical and Biology Science Mass Spectrometry facilities of University of Georgia.
The luciferase samples were analyzed by separation on C-4 HPLC column (Applied Biosystems HPLC system) followed by spraying of the eluted fractions into the mass spectrometer (Perkin Elmer Sciex API I plus quadrapole mass spectrometer). The mass spectrometer was calibrated with polypropylene diol, which produces precisely known fragments up to 2100 a.m.u.
Results
Metridia luciferase expressed in E. coli cells
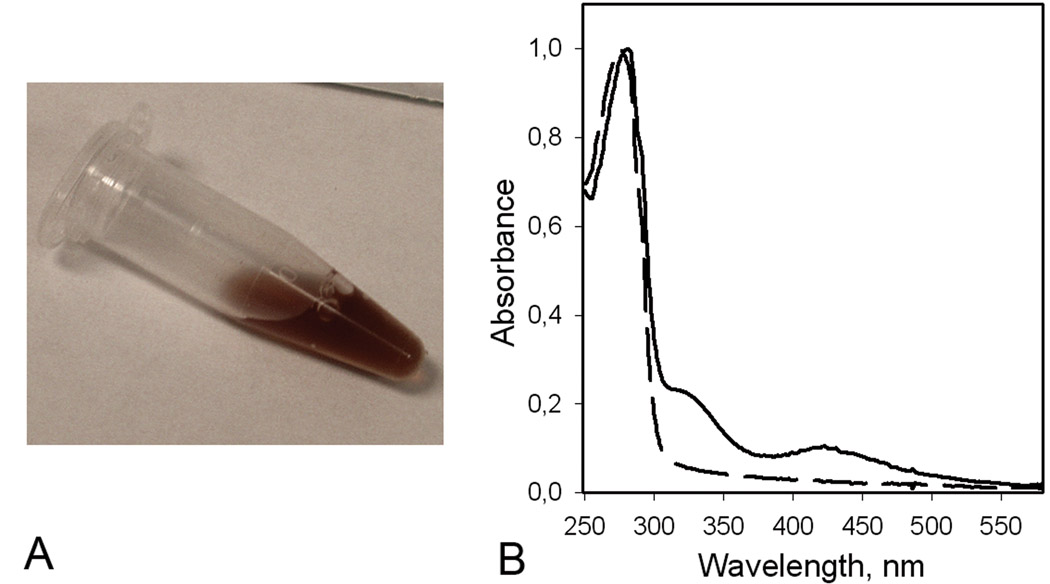
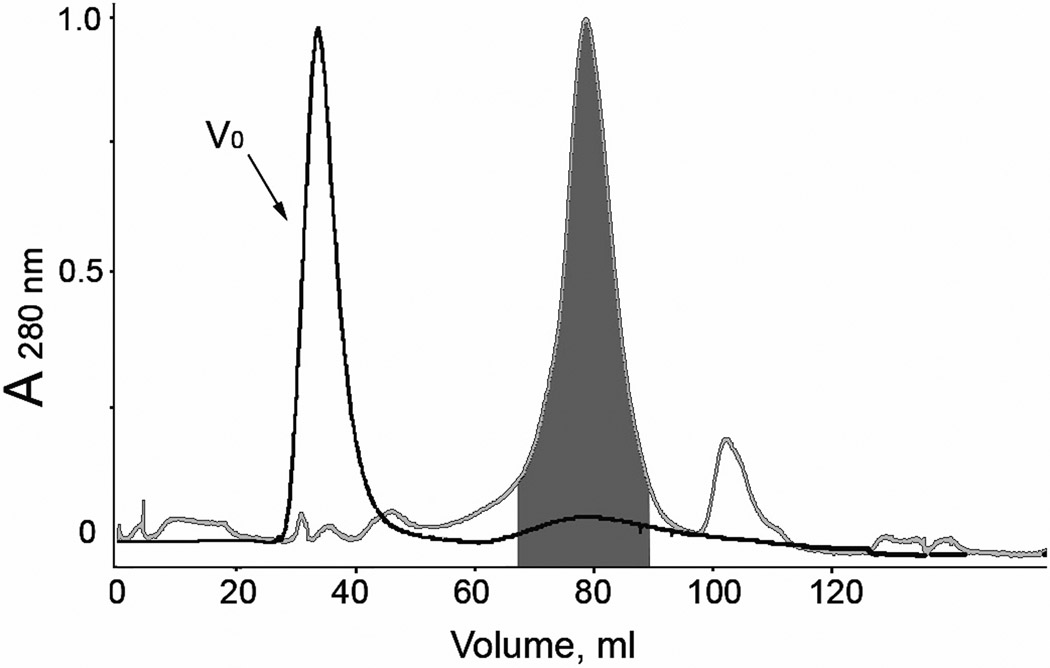
Previous studies have shown that on expression in E. coli, the luciferase lacking the secretion sequence accumulates in insoluble inclusion bodies and therefore a refolding step is required to recover the functionally active protein [18]. It was found that the best conditions for refolding from inclusion bodies and forming disulfide bonds are those that mimic the endoplasmic reticulum of eukaryotic cells [29–31], i.e., a mix of reduced and oxidized thiols (5 mM GSH, 0.5 mM GSSH) in 1 mM EDTA, 20 mM Tris-HCl pH 8.8. Although adding of the mix of reduced and oxidized thiols in refolding buffer results in 25-fold increase of luciferase bioluminescence activity, nevertheless only a minor part of the total protein attempted to be refolded could be purified as functionally active monomeric luciferase. Besides the inability of E. coli cells to provide properly folded protein and correct formation of disulfide bonds, expression MLuc164 in E. coli results in nonspecific binding of metal ions by the Cys residues of luciferase, evidenced by a surprisingly brown color of the inclusion bodies. After solubilization of inclusion bodies in 6 M urea, purification by anion exchange chromatography on DEAE-Sepharose Fast Flow column, and refolding, the MLuc164 retains the red-brown color (Fig. 2A). The absorbance spectrum showed maxima at 412 nm and 320 nm characteristic of proteins with an Fe-S cluster (Fig. 2B) [32]. The elemental chemical analysis of MLuc164 revealed the presence of iron at a ratio of 1:0.6 (protein/iron) and some zinc traces. In addition, we found that adding Fe3+ caused an irreversible aggregation of luciferase indicated by gel filtration where an iron-enriched luciferase fraction eluted in the void volume (Fig. 3). The aggregated luciferase contained only 5–10% bioluminescence activity of the monomeric fraction. Inclusion of chelation agents such as EDTA or an iron specific chelator (deferoxamine mesylate) to the protein sample buffers and during the washing step of the inclusion bodies, did not eliminate the contamination of luciferase by metals. Incubation of the luciferase sample (1 mg/ml) in 6 M guanidine-HCl with slowly added HCl to a final concentration 0.5 M at 4°C, merely resulted in partial decoloration of the sample but did not completely remove iron, indicated by the remaining absorbance bands at 320 nm and 412 nm, though with lower intensity than the sample before treatment.
Figure 2.
(A) MLuc164 purified from E. coli cells at protein concentration 42 mg/ml. (B) Normalized absorption spectra of MLuc164 purified from insect Sf9 (dashed line) and E. coli (solid line) cells respectively. The peak at 412 nm and shoulder at 320 nm are characteristic for proteins with an iron-sulfur cluster [32].
Figure 3.
The elution profile of Metridia luciferases purified from E. coli (black line) and insect Sf9 (gray line) cells on a HiLoad Superdex 75 column. The area highlighted in the dark gray color shows the collected fractions containing the active monomers of MLuc164. V0, void volume of the column.
Several different approaches have been tested to improve MLuc164 expression in E. coli cells. We attempted to increase the Mluc164 solubility by fusing the luciferase with MBP (maltose-binding protein) tag-protein. This increases the luciferase solubility but does not decrease its aggregation. The use of the special E. coli strain Rosetta-gami (Novagen) with reduced thioredoxin reductase activity designed for proper disulfide bond formation in the E. coli cytoplasm, yielded the luciferase in inclusion bodies, which could be solubilized and refolded but was completely inactive. The expression with secretion of Mluc164 into the E. coli periplasm gave extremely low protein yield.
Apparently, Metridia luciferase expressed in E. coli cells must aggregate through intermolecular disulfide bond formation. This process might be accelerated by binding the metal ions because the aggregated luciferase fraction contains iron. It is reasonable to assume that instead of intramolecular bridging the free Cys residues into S-S bonds, there are some specific (or unspecific) mechanisms in E. coli cells, which promote binding the iron to Cys residues but which apparently are not required for Mluc164 bioluminescence, causing only its aggregation and misfolding. For instance, the first Cys-rich repeat (C-x3-C-x2-C-x8-C-x11-C) of Metridia luciferase sequence has some characteristic features peculiar to the C-x3-C-x2-C conserved binding motif of the iron-sulfur cluster of the radical-SAM enzyme superfamily [33].
Metridia luciferase expressed in insect cells
As a consequence of the problem of producing functionally active Metridia luciferase from E. coli expression systems, we turned to the Bac-to-Bac system for expression of Metridia luciferase along with its secretion sequence in the Spodoptera frugiperda (Sf9) cell line. Exploiting the fact that the luciferase is a naturally secreted protein, we cloned the full-length luciferase cDNA including the signal sequence for secretion, to the pFastBac-1 vector. The amount of Metridia luciferase being expressed by the Sf9 cells was monitored by the bioluminescence activity of a sample of the media, which should directly correspond to the amounts of protein expressed. For this purpose, samples were taken at 0, 24, 48, 72 and 96 h post-infection and the bioluminescent activity of the supernatant separated from the cell pellet was measured. The appearance of MLuc164 activity in the culture media indicated that the natural signal peptide successfully provides the luciferase secretion from these insect cells themselves. The highest bioluminescence activity with the lowest cell breakdown, was found in the supernatant of cells infected with virus at an MOI of 0.2 pfu for 72 h. These optimal conditions were selected for large-scale luciferase expression and its following purification. The purification procedure comprised concentration of the luciferase from the cell culture media by ammonium sulfate precipitation, Ni-affinity chromatography, anion exchange chromatography, and gel-filtration (Table 1). Because MLuc164 contains disulfide bonds, all procedures were carried out under non-reducing conditions, i.e. without including DTT or β-mercaptoethanol.
Table 1.
Purification of Metridia luciferase
| Total protein (mg) |
Total activity (RLU† × 106) |
% activity recovered |
Specific activity (RLU × 106/mg) |
|
|---|---|---|---|---|
| Culture media (sf9 cells) | ||||
| Media | 2300.0 | 27500.0 | 100 | 11.9 |
| Precipitation with (NH4)2SO4 | 1610.0 | 26125.0 | 95 | 16.2 |
| His-Trap column | 9.8 | 20993.8 | 75.6 | 2142.2 |
| TEV digested, His-Trap column flow-through | 5.4 | 12335.8 | 44.5 | 2284.4 |
| HiTrap Q | 4.2 | 9644.1 | 34.8 | 2296.0 |
| Superdex 75 | 3.4 | 8261.3 | 29.3 | 2429.8 |
| Inclusion bodies (E. coli cells) | ||||
| 6 M urea extract of inclusion bodies | 51.6 | – | – | – |
| Sepharose Fast Flow (6 M urea) | 7.4 | – | – | – |
| Refolding | 6.1 | 456.9 | 100 | 74.9 |
| HiTrap Q | 3.4 | 401.2 | 87.8 | 118 |
| Superdex 75 | 0.6 | 332.3 | 72.8 | 553.8 |
RLU – at maximum bioluminescent intensity
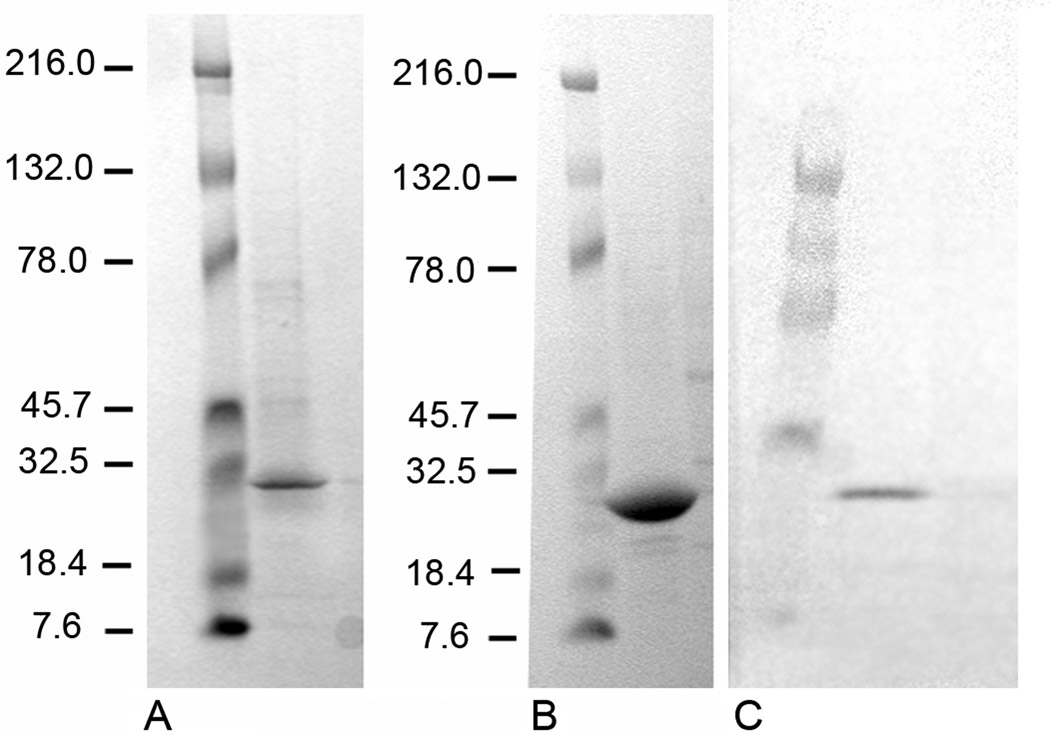
According to SDS-PAGE (Fig. 4A,B), the luciferase obtained is of high purity. Western-blot analysis using anti-His antibody indicates that this is actually the required protein with molecular mass ~ 24 kDa (Fig. 4C). Large-scale baculovirus expression of MLuc164 routinely yielded approximately 2–3 mg luciferase of high purity per liter of culture media.
Figure 4.
SDS-PAGE and Western-blot analysis of Metridia luciferase expressed in and purified from Sf9 cells. (A) MLuc164 after purification on the Ni-HisTrap column (line 2); (B) MLuc164 after gel filtration on HiLoad Superdex 75 column (line 2); (C) Western-blot analysis of MLuc164 (sample of 72 h post-transfected culture media) (line 2); negative control (line 3). Molecular mass markers (Kaleidoscope prestained standards, Bio-Rad) are shown always in line 1.
Metridia luciferase expressed in insect cells in contrast to the luciferase expressed in E. coli cells, is a fully active monomeric protein with no evidence of aggregation as confirmed by gel filtration (Fig. 3). The absorption spectrum of MLuc164 expressed in Sf9 cells shows only one maximum at 280 nm without the maxima at 412 nm and 320 nm observed in the absorption spectrum of luciferase purified from E. coli (Fig. 2). Also elemental chemical analysis revealed no traces of iron or zinc in the MLuc164 purified from insect cells.
Molecular masses of MLuc164 purified from Sf9 and E. coli cells
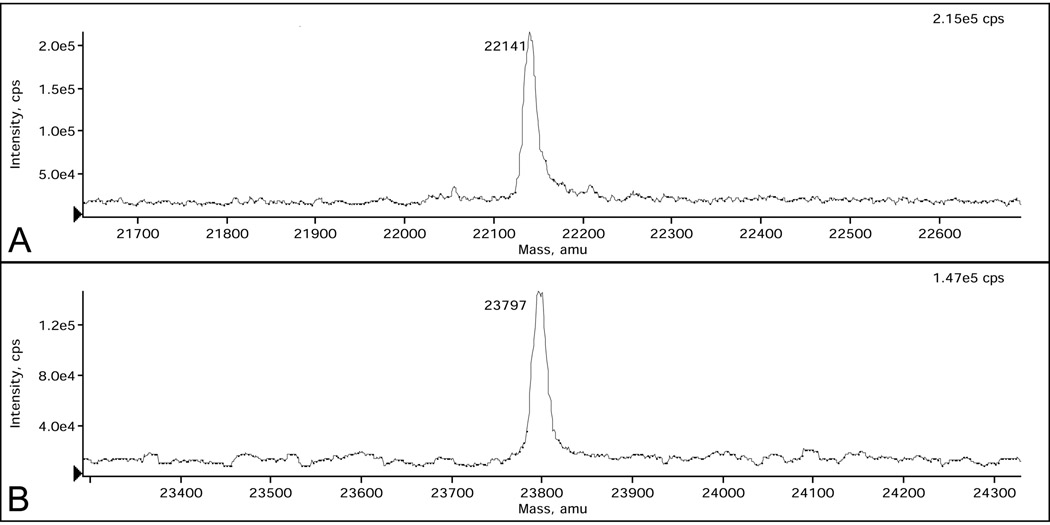
The molecular mass of MLuc164 purified from Sf9 cells determined by the LC–MS method is 23,797 (± 2.4) Da, 10 a.m.u. less than calculated (Table 2, Fig. 5B). The near coincidence of experimental mass with that calculated from amino acid sequence indicates that luciferase does not undergo posttranslational modifications such as phosphorylation or glycosylation. The discrepancy of 10 a.m.u. can be accounted for by the formation of several S-S bonds (4 or 5) accompanied by removal of the corresponding hydrogens. Moreover, the coincidence of molecular masses unambiguously shows that the cleavage site of signal peptide for secretion is at VQA-KS, i.e. between Ala17 and Lys18, as predicted from the amino acid sequence analysis of Metridia luciferase [18].
Table 2.
Molecular masses of the pure monomeric Metridia luciferase obtained with different expression systems
| Protein expression system |
Molecular mass: experimental (calculated‡) |
|---|---|
| E. coli cells | 22,141 ± 2.0 (22,021) |
| Δ+120 | |
| Sf9 cells | 23,797 ± 2.4 (23,807)† |
| Δ−10 |
Molecular mass for MLuc164 with His6-tag
Average molecular mass
Figure 5.
LC–MS spectra of pure Metridia luciferase samples purified from E. coli (A) and Sf9 insect (B) cells. Each peak is labeled with its mass in Daltons.
In contrast, the molecular mass of MLuc164 obtained from E. coli cells is higher by 120 a.m.u. than the calculated mass (Table 2, Fig. 5A). Since luciferase purified from E. coli is contaminated by iron (as well as some zinc traces revealed in the protein sample by elemental chemical analysis), it would be reasonable to assume that higher molecular mass of this luciferase sample is a result of the presence of this metal ion.
Bioluminescence properties of MLuc164 purified from Sf9 and E. coli cells
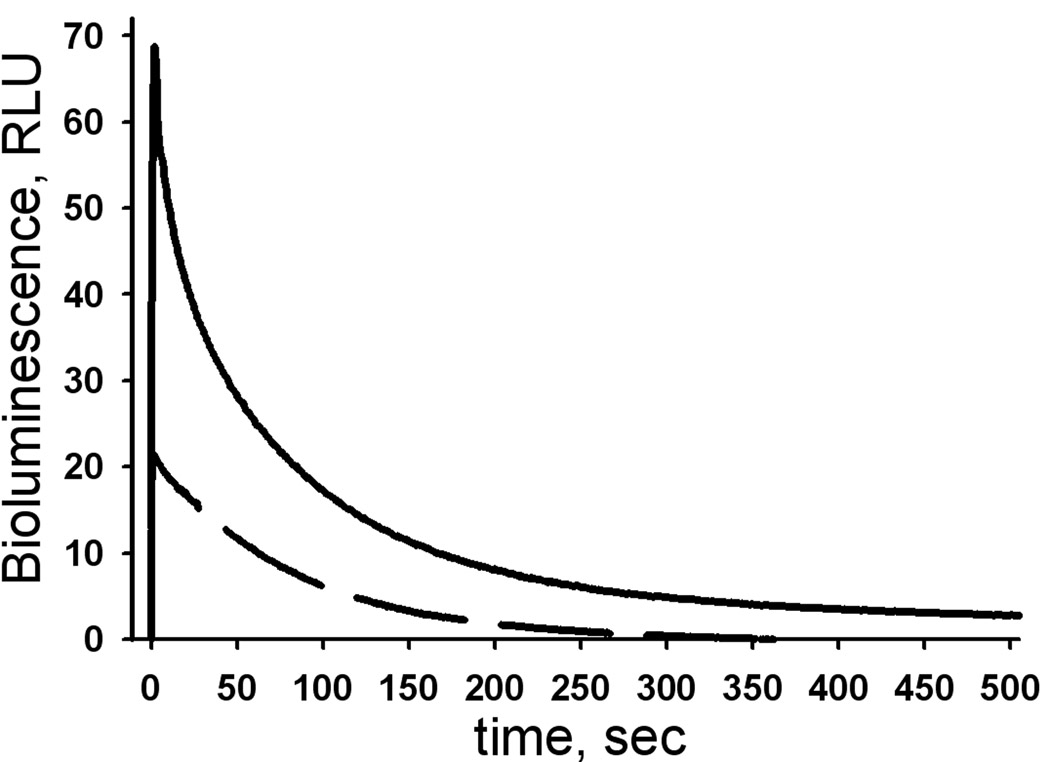
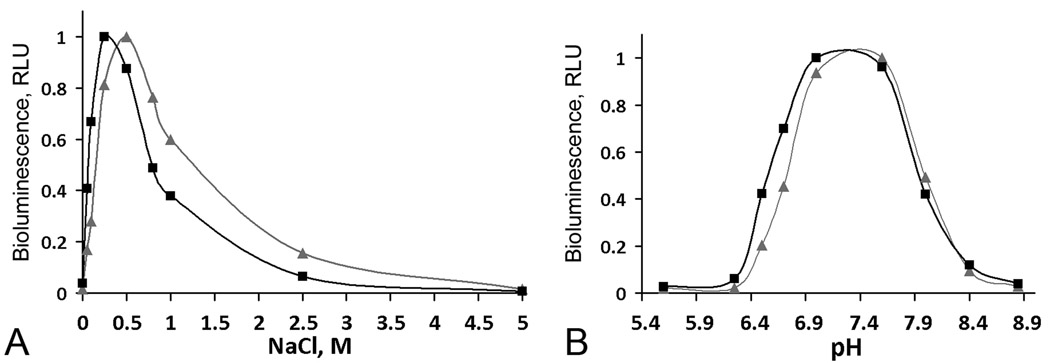
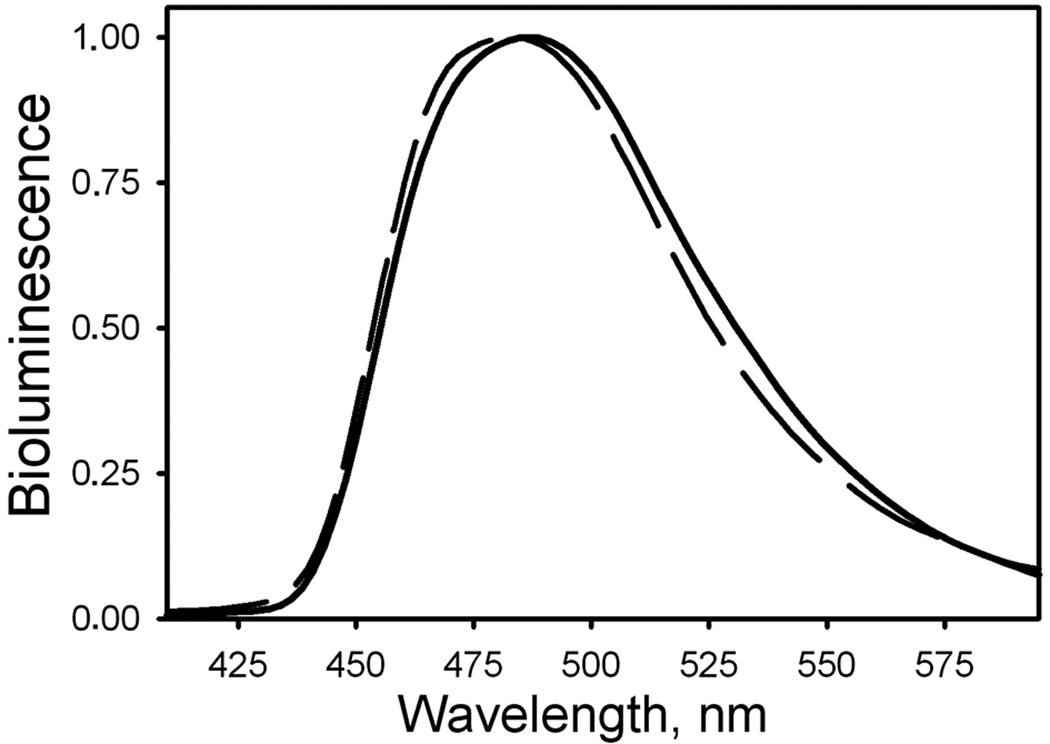
Metridia luciferase expressed in Sf9 cells reveals 3.5 times higher specific bioluminescence activity compared to monomeric MLuc164 expressed and purified from E. coli cells (Fig. 6). The bioluminescence of both luciferases is also activated more than 100-fold in the presence of NaCl but with a slightly shifted optimum for NaCl concentration for luciferase obtained from insect cells (0.5 M) than for MLuc164 purified from E. coli (0.3 M) (Fig. 7). Besides, MLuc164 expressed in insect cells displays higher tolerance towards ionic strength (1 – 2 M of NaCl) than does luciferase from E. coli, which might indicate a more rigid and stable protein structure. It is likely that the effect of NaCl on bioluminescence activity of MLuc164 results from chloride ion interaction as has been reported for Gaussia luciferase [34], not unexpected from the high identity of amino acid sequence of the two proteins [18]. Activation of Metridia luciferase purified both from insect and E. coli cells by bivalent cations such as Mg2+ or Ca2+ is not so evident. At the same time, both Metridia luciferases reveal a similar pH optimum for bioluminescence, around 7.3–7.4 (Fig. 7) and bioluminescence emission maxima (Fig. 8) that are very close to those of natural Metridia luciferase [18]. It should be noted that while the pH optima for bioluminescence of Metridia luciferases correspond to those of Gaussia luciferase [33], the light emission maxima (483 and 486 nm for luciferases purified from E. coli and insect cells respectively) are red-shifted from the maximum of Gaussia luciferase (473 nm) [34]. This spectral shift could be a result of differences in the amino acid residue composition of the active sites of Metridia and Gaussia luciferases, as also found for the two Ca2+-regulated photoproteins, aequorin and obelin, where one residue difference in the coelenterazine-binding cavity appears to result in a more than 10 nm difference in their bioluminescence maxima [14].
Figure 6.
Bioluminescent activity of Metridia luciferase purified from E. coli (dashed line) and Sf9 insect (solid line) cells at equal protein concentrations of (5 ng/ml). The coelenterazine concentration was 2.4 µM in both experiments. The comparison was carried out in 0.3 M NaCl, 20 mM Tris–HCl pH 7.5, a condition optimizing the bioluminescence activity of Metridia luciferase purified from E. coli.
Figure 7.
Bioluminescence of Metridia luciferase purified from E. coli (▪) and insect (▲) cells depending on NaCl concentration (A) and pH (B). RLU, the maximum light intensity.
Figure 8.
Normalized bioluminescence spectra of Metridia luciferase purified from E. coli (dashed line) and Sf9 insect (solid line) cells.
Conclusion
The work described here shows that secreted Metridia luciferase expressed in a baculovirus insect cell system is functionally active and has a higher specific bioluminescent activity as compared with monomeric MLuc164 expressed in E. coli cells. In addition, we found that Metridia luciferase from insect cells is a stable monomeric protein with molecular mass closely corresponding to that calculated from sequence, thereby indicating the lack of any post-translation modifications. In addition, the Metridia luciferase purified from insect cells has no tendency for aggregation even at high protein concentration and shows no contamination by metal ions, in contrast to luciferase expressed in E. coli. Also we show that the baculovirus expression system is convenient for both production and purification of the large amounts of Metridia luciferase necessary for crystallization and spatial structure determination, biochemical characterization, or development of in vitro analytical assays.
Acknowledgments
This work was supported by the National Institutes of Health (Grant 1P50 GM62407), University of Georgia Research Foundation and Georgia Research Alliance, the Russian Foundation for Basic Research and Taiwan National Science Council (Grant 06-04-89502), and the program for “Molecular and Cellular Biology” of Russian Academy of Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: MLuc164, Metridia luciferase; GSSH, oxidized glutathione; GSH, reduced glutathione; DTT, dithiothreitol; a.m.u., atom mass unit; RLU, relative light units; pfu, plaque-forming unit; MOI, multiplicity of infection.
References
- 1.Hori K, Charbonneau H, Hart RC, Cormier MJ. Structure of native Renilla reniformis luciferin. Proc. Natl. Acad. Sci. USA. 1977;74:4285–4287. doi: 10.1073/pnas.74.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vysotski ES, Lee J. Ca2+-regulated photoproteins: Structural insight into the bioluminescence mechanism. Acc. Chem. Res. 2004;37:405–415. doi: 10.1021/ar0400037. [DOI] [PubMed] [Google Scholar]
- 3.Shimomura O. The discovery of aequorin and green fluorescent protein. J. Microsc. 2005;217:1–15. doi: 10.1111/j.0022-2720.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- 4.Shimomura O, Flood PR. Luciferase of the scyphozoan medusa Periphylla periphylla. Biol. Bull. 1998;194:244–252. doi: 10.2307/1543094. [DOI] [PubMed] [Google Scholar]
- 5.Isobe M, Kuse M, Yasuda Y, Takahashi H. Synthesis of 13C-dehydrocoelenterazine and model studies on Symplectoteuthis squid bioluminescence. Bioorg. Med. Chem. Lett. 1998;8:2919–2924. doi: 10.1016/s0960-894x(98)00525-3. [DOI] [PubMed] [Google Scholar]
- 6.Head JF, Inouye S, Teranishi K, Shimomura O. The crystal structure of the photoprotein aequorin at 2.3 Å resolution. Nature. 2000;405:372–376. doi: 10.1038/35012659. [DOI] [PubMed] [Google Scholar]
- 7.Liu ZJ, Vysotski ES, Chen CJ, Rose J, Lee J, Wang BC. Structure of the Ca2+-regulated photoprotein obelin at 1.7 Å resolution determined directly from its sulfur substructure. Protein Sci. 2000;9:2085–2093. doi: 10.1110/ps.9.11.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Markova SV, Vysotski ES, Liu ZJ, Lee J, Rose J, Wang BC. Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence. J. Biol. Chem. 2004;279:33647–33652. doi: 10.1074/jbc.M402427200. [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Vysotski ES, Markova SV, Liu ZJ, Lee J, Rose J, Wang BC. All three Ca2+-binding loops of photoproteins bind calcium ions: the crystal structures of calcium-loaded apo-aequorin and apo-obelin. Protein Sci. 2005;14:663–675. doi: 10.1110/ps.041142905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ZJ, Stepanyuk GA, Vysotski ES, Lee J, Markova SV, Malikova NP, Wang BC. Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state. Proc. Natl. Acad. Sci. USA. 2006;103:2570–2575. doi: 10.1073/pnas.0511142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loening AM, Fenn TD, Gambhir SS. Crystal structures of the luciferase and green fluorescent protein from Renilla reniformis. J. Mol. Biol. 2007;374:1017–1028. doi: 10.1016/j.jmb.2007.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepanyuk GA, Liu ZJ, Lee J, Markova SV, Frank LA, Vysotski ES, Wang BC. Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.72 Å: Why it is not a calcium-regulated photoprotein. Photochem. Photobiol. Sci. 2008;7:442–447. doi: 10.1039/b716535h. [DOI] [PubMed] [Google Scholar]
- 13.Malikova NP, Stepanyuk GA, Frank LA, Markova SV, Vysotski ES, Lee J. Spectral tuning of obelin bioluminescence by mutations of Trp92. FEBS Lett. 2003;544:184–188. doi: 10.1016/s0014-5793(03)01166-9. [DOI] [PubMed] [Google Scholar]
- 14.Stepanyuk GA, Golz S, Markova SV, Frank LA, Lee J, Vysotski ES. Interchange of aequorin and obelin bioluminescence color is determined by substitution of one active site residue of each photoprotein. FEBS Lett. 2005;579:1008–1014. doi: 10.1016/j.febslet.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat. Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 16.Inouye S, Watanabe K, Nakamura H, Shimomura O. Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase. FEBS Lett. 2000;481:19–25. doi: 10.1016/s0014-5793(00)01963-3. [DOI] [PubMed] [Google Scholar]
- 17.Bryan BJ, Sent-Gyorgyi CS. Luciferases, fluorescent proteins, nucleic acids encoding the luciferases and fluorescent proteins and the use thereof in diagnostics, high throughput screening and novelty items. International Pub. 1999 No. WO 99/49019. [Google Scholar]
- 18.Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES. Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa. J. Biol. Chem. 2004;279:3212–3217. doi: 10.1074/jbc.M309639200. [DOI] [PubMed] [Google Scholar]
- 19.Evstigneev PV, Bitukov EP. In: Bioluminescence of Marine Copepods. Greze BN, editor. Kiev, Ukraine: Naukova Dumka; 1990. [Google Scholar]
- 20.Thompson EM, Nagata S, Tsuji FI. Vargula hilgendorfii luciferase: a secreted reporter enzyme for monitoring gene expression in mammalian cells. Gene. 1990;96:257–262. [PubMed] [Google Scholar]
- 21.Inouye S, Ohmiya Y, Toya Y, Tsuji FI. Imaging of luciferase secretion from transformed Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA. 1992;89:9584–9587. doi: 10.1073/pnas.89.20.9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanahashi A, Ohmiya Y, Honma Y, Katsuno S, Ohta Y, Nakamura H, Honma K. Continuous measurement of targeted promoter activity by a secreted bioluminescence reporter Vargula hilgendorfii luciferase. Anal. Biochem. 2001;289:260–266. doi: 10.1006/abio.2000.4932. [DOI] [PubMed] [Google Scholar]
- 23.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codonoptimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Usuda S, Ichinose H, Inouye S. Real-time bioluminescence imaging of a protein secretory pathway in living mammalian cells using Gaussia luciferase. FEBS Lett. 2007;581:4551–4556. doi: 10.1016/j.febslet.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 25.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, Tannous BA. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat. Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhaegen M, Christopoulos T. Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal. Chem. 2002;74:4378–4385. doi: 10.1021/ac025742k. [DOI] [PubMed] [Google Scholar]
- 27.Fahey RC, Hunt JS, Windham CC. On the cysteine and cystine content of proteins. Differences between intracellular and extracellular proteins. Mol. Evol. 1977;10:155–163. doi: 10.1007/BF01751808. [DOI] [PubMed] [Google Scholar]
- 28.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 29.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 30.Wetlaufer DB, Branca PA, Chen G-X. The oxidative folding of proteins by disulfide plus thiol does not correlate with redox potential. Protein Eng. 1987;2:141–146. doi: 10.1093/protein/1.2.141. [DOI] [PubMed] [Google Scholar]
- 31.Rudolph R, Lilie H. In vitro folding of inclusion body proteins. FASEB J. 1996;10:49–56. [PubMed] [Google Scholar]
- 32.Cosper MM, Cosper NJ, Hong W, Shokes JE, Broderick WE, Broderick JB, Johnson MK, Scott RA. Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme. Protein Sci. 2003;12:1573–1577. doi: 10.1110/ps.0302203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inouye S, Sahara Y. Identification of two catalytic domains in a luciferase secreted by the copepod Gaussia princes. Biochem. Biophys. Res. Commun. 2008;365:96–101. doi: 10.1016/j.bbrc.2007.10.152. [DOI] [PubMed] [Google Scholar]