Abstract
Rationale: Superoxide dismutase (SOD) 3 inhibits oxidative fragmentation of lung matrix components collagen I, hyaluronan, and heparan sulfate. Inherited change in SOD3 expression or function could affect lung matrix homeostasis and influence pulmonary function.
Objectives: To identify novel SOD3 polymorphisms that are associated with lung function or chronic obstructive pulmonary disease (COPD).
Methods: Resequencing of 182 individuals identified two novel polymorphisms, E1 (rs8192287) and I1 (rs8192288), in a conserved region of the SOD3 gene of potential relationship to lung function. We next genotyped 9,093 individuals from the Copenhagen City Heart Study for the polymorphisms and recorded spirometry, and admissions and deaths due to COPD during 26-year follow-up. Finally, we validated our findings in a cross-sectional analysis of 35,635 individuals from the Copenhagen General Population Study.
Measurements and Main Results: Genotyping the Copenhagen City Heart Study identified 35 E1/I1 homozygotes, 1,050 heterozygotes, and 8,008 noncarriers (Hardy-Weinberg equilibrium: P = 0.93). Using quadruple lung function measurements, we found that E1/I1 homozygotes had 7% lower FVC % predicted (P = 0.006) and 4% lower FEV1 % predicted (P = 0.12) compared with noncarriers. In the Copenhagen General Population Study, E1/I1 homozygotes also had lower FVC % predicted than noncarriers (P = 0.03), confirming an association between E1/I1 genotype and reduced lung function. E1/I1 homozygotes had adjusted hazard ratios for COPD hospitalization and COPD mortality of 2.5 (95% confidence interval, 1.0–5.9) and 3.7 (95% confidence interval, 0.9–15), respectively; the results were independent of influence from the R213G allele of the SOD3 gene.
Conclusions: We identified two novel polymorphisms in a conserved region of the SOD3 gene and show that individuals that are homozygous for these polymorphisms have reduced FVC % predicted in two large, population-based studies.
Keywords: superoxide dismutase 3, genetics, chronic obstructive pulmonary disease, oxidative stress
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Extracellular superoxide dismutase (EC-SOD) inhibits oxidative fragmentation of lung matrix components collagen I, hyaluronan, and heparan sulfate. Inherited change in EC-SOD expression or function could affect lung matrix homeostasis and influence pulmonary function.
What This Study Adds to the Field
We identified two novel polymorphisms in a conserved region of the EC-SOD gene and show that individuals that are homozygous for these polymorphisms have reduced FVC % predicted in two large, population-based studies.
Oxidative stress is important in pathophysiological events leading to chronic obstructive pulmonary disease (COPD) (1). Uncontained oxidant activity damages lipids, proteins, and extracellular matrix components creating proinflammatory oxidized lipids and chemotactic matrix fragments that contribute to inflammation in the lung (2–6). However, antioxidants are present that can counter the deleterious effects of increased oxidative stress. One such protective molecule is superoxide dismutase (SOD) 3, which catalyses the conversion of superoxide anions into hydrogen peroxide and oxygen (7, 8). SOD3 binds lung matrix components collagen I, hyaluronan, and heparan sulfate, and inhibits their fragmentation in response to oxidative stress (3, 4, 6). As these matrix components are widely distributed throughout the lung interstitium, and because their fragments stimulate inflammatory cell migration, SOD3 could play a central role in lung tissue defenses against oxidative stress.
SOD3 is a major extracellular antioxidant enzyme that is highly expressed in lungs and accounts for the majority of SOD activity in airways and vessels (9, 10). SOD3 is homologous with CuZn SOD within the catalytic region (11), but has a carboxyterminal region with strong heparin binding affinity. A single-nucleotide polymorphism (SNP) within this coding region (rs1799895; i.e., the R213G polymorphism) results in impaired binding to the extracellular matrix and decreased tissue levels (12). In Copenhagen residents, the R213G SNP has been associated with a hazard ratio of only 0.5 for COPD (13). In the current investigation, we perform DNA resequencing of the SOD3 gene to determine whether there are other SOD3 polymorphisms that might be associated with COPD.
In doing so, we completed a polymorphism map of the SOD3 gene using DNA from 66 control subjects and 116 patients with COPD. We identified two novel polymorphisms that occur in the noncoding 5′ untranslated region (E1) and first intron (I1) of the SOD3 gene. Both polymorphisms are situated in a conserved mammalian interspersed repetitive element, and could be functionally relevant. Thus, we hypothesized that the E1/I1 variants were associated with lung function or clinical COPD in a large population study of Danes. Here we show that the genetic E1/I1 variants in SOD3 are tightly coupled and that they associate with reduced levels of FVC % predicted, and with elevated risk of COPD hospitalization during 26 years of follow-up. The reduction in FVC % predicted with E1/I1 was validated in another large cohort, the Copenhagen General Population Study. The data seem to support a role for SOD3 in oxidant-mediated events influencing lung function. Some of the results of these studies have previously been reported in the form of an abstract (14).
METHODS
Subjects
The Copenhagen City Heart Study and the Copenhagen General Population Study were approved by Herlev Hospital and by Danish ethics committees (KF V.100.2039/91 and KF 01–144/01, Copenhagen and Frederiksberg committee), and were conducted according to the Declaration of Helsinki. Written informed consent was obtained from participants. All participants were white and of Danish descent. Information on subjects and methods in the resequencing study is provided in the online supplement.
The Copenhagen City Heart Study
The Copenhagen City Heart Study is a prospective cardiopulmonary study of the Danish general population initiated in 1976–1978, with follow-up examinations in 1981–1983, 1991–1994, and 2001–2003 (15, 16); DNA was taken in 1991–1994 and 2001–2003. Individuals were randomly selected based on the national Danish Civil Registration System to reflect the Copenhagen general population aged 20 to 80 years and older. All endpoints were recorded in the follow-up period, 1976 through July 2007. Follow-up time was 26 years for COPD hospitalizations (209,395 person-years) and 26 years for COPD mortality (205,246 person-years), and was 100% complete.
Information on diagnoses of COPD (ICD-8 [17] Codes 490–492 and ICD-10 [18] Codes J40–J44) was collected in the national Danish Patient Registry and the national Danish Causes of Death Registry. Pack-years was daily tobacco consumption (in grams) times duration of smoking (in years) divided by 20 (in g/pack).
The Copenhagen General Population Study
The Copenhagen General Population Study is a cross-sectional study of the Danish general population initiated in 2003 and still recruiting (19). At the time of the genotyping for the present study, 35,635 individuals had been included. Individuals were randomly selected based on the national Danish Civil Registration System to reflect the adult Copenhagen general population, aged 20–80 years and older. Information on diagnoses of COPD was ascertained as in the Copenhagen City Heart Study.
Spirometry
FEV1 and FVC were determined using an electronic spirometer (Monoghan N 403; Monaghan, Littleton, CO) at the first and second examinations, and with a dry wedge spirometer (Vitalograph; Maids Moreton, Buckinghamshire, UK) at the third and fourth examinations of the Copenhagen City Heart Study. In the Copenhagen General Population Study, FEV1 and FVC were determined by EasyOne Spirometer (ndd Medizintechnik, Zurich, Switzerland). Algorithms for calculation of FEV1 % predicted and FVC % predicted were made using multiple regression, with age and height as covariates on all individuals for men and women separately. If a subsample of never-smokers was used rather than all individuals, the results were similar to those presented.
Laboratory Analyses
Genotyping was performed on Qiagen isolated DNA using TaqMan assays (ABI Prism 7900HT Sequence Detection System; Applied Biosystems, Foster City, CA). All genotyping was verified by DNA sequencing. Genotype frequencies did not differ between individuals without COPD in the resequencing study (SOD3 E1/I1 noncarriers, heterozygotes, homozygotes: 89, 11, and 0%, respectively), the Copenhagen City Heart Study (88, 12, and 0.4%), and the Copenhagen General Population Study (88, 11, and 0.4%) (χ2: P = 0.97). Plasma SOD3 was determined in a subsample of SOD3 R213G carriers (n = 2456) by ELISA as part of a previously published article (20).
Statistical Analysiss
Statistical analyses were performed using Stata/SE version 10.0 for Windows (StataCorp LP, College Station, TX). We used two-sided Mann-Whitney U test, Pearson's chi-square test, and Fisher's exact test. Because quadruple measures of pulmonary function were not available for all subjects (30% of participants had all four measurements, 39% three measurements, 21% two measurements, and 10% one measurement), lung function was analyzed in a repeated-measures model using the linear mixed modeling option in SPSS version 16.0 (SPSS, Inc., Chicago, IL). The survey number of the Copenhagen City Heart Study specified repeated measurements of lung function. An unstructured covariance type for residuals was used. The unstructured type places no restrictions on the structure and may be preferable to other types. No random effects were specified. The SOD3 genotype and survey number of the Copenhagen City Heart Study were specified as fixed effects, and lung function was the repeated dependent variable. We used one-sided Mann-Whitney U test for validation of the spirometry findings in the Copenhagen General Population Study. When left truncation (i.e., delayed entry) was used with age as the time scale, Cox proportional hazards regression estimated hazard ratios for COPD hospitalization/death with 95% confidence intervals. The crude and adjusted models automatically allowed for age; in addition, the adjusted model included the following covariates, which were forced into all models: sex, smoking habits (never- or ever-smoker), pack-years at study entry, and SOD3 E1/I1 genotype. Linkage disequilibrium coefficient was calculated using the free web tool, CubeX (21).
RESULTS
Resequencing and Identification of Two Novel Polymorphisms in the SOD3 Gene
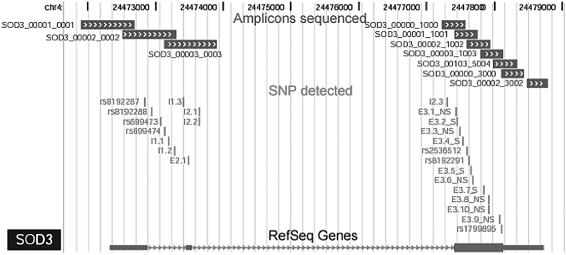
The SOD3 gene has been previously sequenced and is known to contain 3 exons and 2 introns (7); however, there is a paucity of published sequence data from humans. Thus, in a subset of 66 control subjects and 116 subjects with COPD from a U.S. population, we resequenced selected amplicons in the 10-kb genomic span of SOD3 that included all three exons and splice site junctions (Figure 1). Our resequencing detected 24 polymorphisms, 17 of which were novel. The majority of these polymorphisms were rare, and the status of their association with COPD remains unknown; however, we identified two new polymorphisms that were situated in a conserved region with potential functionality. The first polymorphism (rs8192287), which we name E1, is in the noncoding, 5′ untranslated region in exon 1. The second polymorphism (rs8192288), which we name I1, is in the first intron. To determine whether the E1 and I1 polymorphisms altered lung tissue levels, we measured SOD3 lung tissue levels in 27 independent normal subjects from the United States. Both the E1 and I1 polymorphisms were associated with lower, but non–statistically significant levels of lung tissue SOD3 (mean ± SEM: E1 66 ± 18 ng SOD3/mg protein vs. 77 ± 8 ng/ml; P = ns; I1 66 ± 18 ng/ml vs. 80 ± 8 ng/ml; P = ns).
Figure 1.
Positional location of the superoxide dismutase (SOD) 3 polymorphisms. From top to bottom, this schematic shows 7,500 base pairs (7.5 kb) of the human SOD3 gene (chromosome 4): amplicons used for resequencing are shown by boxes, with chevrons with the amplicon name to the left; single-nucleotide polymorphisms that have been identified in this study and dbSNP identifiers are indicated by vertical bars; the last line shows the location of exons 1–3 (boxes), with triangles indicating introns; sequences are read from left to right (5′→3′). SNP = single-nucleotide polymorphism.
To determine the clinical relevance of the polymorphisms, we genotyped 9,093 individuals from a large Danish population sample, the Copenhagen City Heart Study. The r2 was 1.0 between the E1 and I1 alleles of the two loci, indicating complete linkage in the investigated Danish population. Thus, subsequent results will refer to just one genotype (i.e., combined E1/I1 genotype).
The Copenhagen City Heart Study
Of the 9,093 individuals tested in the Copenhagen City Heart Study, 8,008 (88%) were E1/I1 noncarriers, 1,050 (12%) were heterozygotes, and 35 (0.4%) were homozygotes (Table 1). This distribution did not differ from that predicted by the Hardy-Weinberg equilibrium (P = 0.93). None of the baseline characteristics differed between E1/I1 noncarriers, heterozygotes, and homozygotes.
TABLE 1.
CHARACTERISTICS OF PARTICIPANTS AT STUDY ENTRY IN THE COPENHAGEN CITY HEART STUDY
| Characteristics | Noncarriers (n = 8,008) | Heterozygotes (n = 1,050) | Homozygotes (n = 35) |
|---|---|---|---|
| Women/men, n | 4,436/3,572 | 600/450 (P = 0.28) | 14/21 (P = 0.07) |
| Age, yr | 46 ± 23 | 46 ± 23 (P = 0.78) | 46 ± 22 (P = 0.81) |
| Smokers, % | 76 | 75 (P = 0.78) | 74 (P = 0.84) |
| Smoking, pack-years | 14 ± 32 | 14 ± 32 (P = 0.57) | 14 ± 31 (P = 0.98) |
| Body mass index, kg/m2 | 25 ± 7 | 25 ± 8 (P = 0.72) | 25 ± 7 (P = 0.91) |
Values represent number of participants, mean ± 2 SE, or percentages. P values are for comparing heterozygotes or homozygotes with noncarriers by Pearson's chi-square test or Mann-Whitney U test.
Lung Function
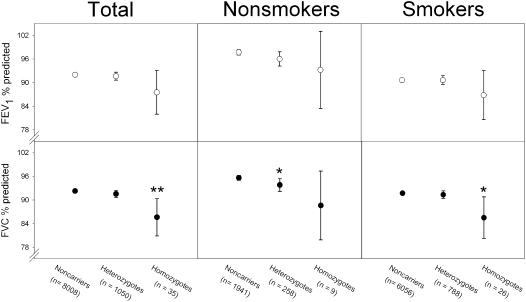
The E1/I1 homozygotes had 7% lower FVC % predicted and 4% lower FEV1 % predicted than noncarriers (P = 0.006 and P = 0.12, respectively; Figure 2). E1/I1 heterozygotes did not differ overall from noncarriers in FVC % predicted and FEV1 % predicted.
Figure 2.
Pulmonary function in SOD3 E1/I1 homozygotes and heterozygotes versus noncarriers in the Copenhagen City Heart Study. Values represent mean ± 2 SE. *P < 0.05 and **P < 0.01 versus noncarriers using repeated measures analysis.
We previously observed that SOD3 R213G heterozygosity protects against COPD in smokers, but not in nonsmokers (13). As such, we also stratified the present analysis for tobacco smoking. Among smokers, homozygotes still had 6% lower FVC % predicted than noncarriers (P = 0.02), whereas, among nonsmokers, homozygotes had 7% lower FVC % predicted (P = 0.12) (Figure 2). Heterozygotes also had slightly lower FVC % predicted among nonsmokers, whereas no such difference was evident among smokers. Smoking did not interact statistically with E1/I1 genotype in predicting FVC % predicted (P = 0.53) or FEV1 % predicted (P = 0.64).
COPD
To determine whether E1/I1 homozygotes with decreased FVC % predicted had greater COPD morbidity, we recorded admissions due to COPD during a median follow-up period of 26 years (range, 0.002–27.8 yr). In this period, 774 participants were hospitalized with COPD (incidence rate, 37 events/10,000 person-years). In unadjusted analyses, homozygotes and heterozygotes had hazard ratios for COPD hospitalization of 1.8 (95% confidence interval [CI], 0.7–4.3) and 0.9 (95% CI, 0.7–1.2) compared with noncarriers (Table 2). After adjustment for age, sex, smoking habits, and pack-years at study entry, equivalent hazard ratios for COPD hospitalization were 2.5 (95% CI, 1.0–5.9) and 0.9 (95% CI, 0.7–1.2). In an analysis stratified for smoking habits, homozygotes had an increased hazard ratio for COPD hospitalization after adjustment among smokers only. No nonsmoking homozygotes were hospitalized due to COPD during the observation period. Consistent with the lung function data, nonsmoking heterozygotes had an increased hazard ratio of 2.0 (95% CI, 1.0–4.0) for COPD hospitalization in an unadjusted model, but this result did not reach statistical significance after adjustment.
TABLE 2.
RISK OF CHRONIC OBSTRUCTIVE PULMONARY DISEASE IN SOD3 E1/I1 HOMOZYGOTES AND HETEROZYGOTES VERSUS NONCARRIERS
| Heterozygotes
|
Homozygotes
|
|||||
|---|---|---|---|---|---|---|
| Crude HR (95% CI) | Adjusted HR (95% CI)* | P Value | Crude HR (95% CI) | Adjusted HR (95% CI)* | P Value | |
| COPD hospitalization | ||||||
| All (n = 9,093) | 0.9 (0.7–1.2) | 0.9 (0.7–1.2) | 0.46 | 1.8 (0.7–4.3) | 2.5 (1.0–5.9) | < 0.05 |
| Nonsmokers (n = 2,208) | 2.0 (1.0–4.0) | 1.9 (0.9–3.9) | 0.10 | |||
| Smokers (n = 6,870) | 0.9 (0.7–1.1) | 0.8 (0.6–1.1) | 0.22 | 1.8 (0.7–4.3) | 2.7 (1.1–6.4) | 0.03 |
| COPD mortality | ||||||
| All (n = 9,093) | 1.1 (0.7–1.8) | 1.2 (0.7–2.0) | 0.44 | 3.6 (0.9–15) | 3.7 (0.9–15) | 0.07 |
| Nonsmokers (n = 2,208) | 1.2 (0.1–11) | 1.3 (0.1–11) | 0.82 | |||
| Smokers (n = 6,870) | 1.2 (0.7–1.9) | 1.2 (0.7–2.0) | 0.47 | 3.1 (0.8–13) | 3.8 (0.9–15) | 0.07 |
Definition of abbreviations: CI = confidence interval; COPD = chronic obstructive pulmonary disease; HR = hazard ratio.
P values are for comparing time to COPD event in heterozygotes or homozygotes with noncarriers by adjusted Cox regression.
Allowed for age, sex, smoking status, pack-years at study entry, and SOD3 E1/I1 genotype.
To determine whether E1/I1 homozygotes with decreased FVC % predicted had greater COPD mortality, we recorded deaths due to COPD during a median follow-up period of 26 years (range, 0.04–28.0 yr). In this period, 166 participants died due to COPD (incidence rate, 8.1 events/10,000 person-years). In unadjusted analyses, homozygotes and heterozygotes had hazard ratios for COPD mortality of 3.6 (95% CI, 0.9–15) and 1.1 (95% CI, 0.7–1.8) compared with noncarriers (Table 2). After adjustment for age, sex, smoking habits, and pack-years at study entry, equivalent hazard ratios for COPD mortality were 3.7 (95% CI, 0.9–15) and 1.2 (95% CI, 0.7–2.0). In an analysis stratified for smoking habits, homozygotes had a hazard ratio for COPD mortality of 3.8 (95% CI, 0.9–15) after adjustment among smokers. No nonsmoking homozygotes died due to COPD during the observation period.
R213G Polymorphism
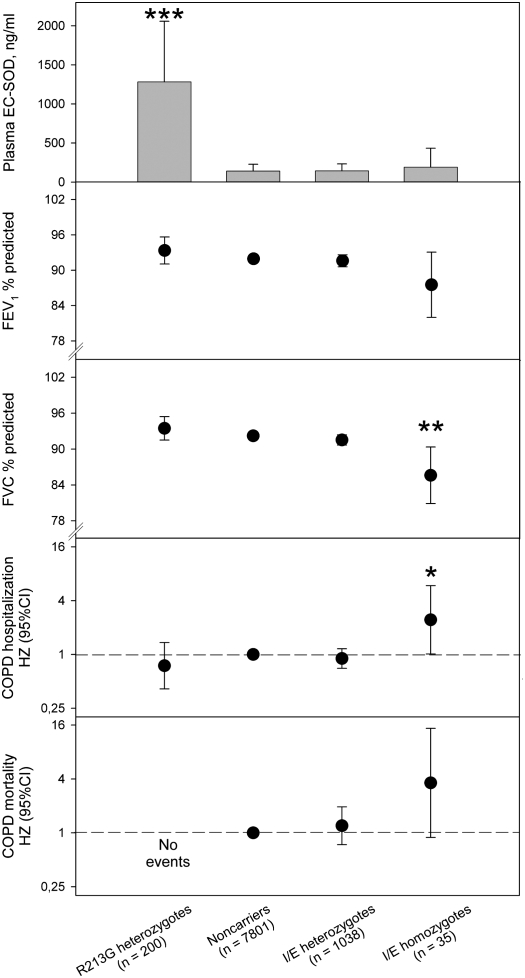
The R213G polymorphism in the SOD3 gene causes ninefold or higher levels of SOD3 in plasma (8, 20). To adjust our analyses for the R213G polymorphism in the SOD3 gene, we also stratified or adjusted the analyses for this variant. As expected, R213G heterozygotes had ninefold higher levels of plasma SOD3 compared with noncarriers (Figure 3) (13). However, E1/I1 noncarriers, heterozygotes, and homozygotes did not differ in level of plasma SOD3 after excluding heterozygotes (n = 200) and homozygotes (n = 2) of the R213G polymorphism from analysis. After this exclusion, E1/I1 homozygotes had 7% lower FVC % predicted and 4% lower FEV1 % predicted than noncarriers (P = 0.006 and P = 0.12, respectively; Figure 3). After adjustment for R213G genotype, age, sex, smoking habits, and pack-years at study entry, E1/I1 homozygotes and heterozygotes had hazard ratios for COPD hospitalization of 2.4 (95% CI, 1.0–5.9; P < 0.05) and 0.9 (95% CI, 0.7–1.2; P = 0.42) compared with noncarriers. Equivalent hazard ratios for COPD mortality were 3.6 (95% CI, 0.9–15; P = 0.07) and 1.2 (95% CI, 0.7–1.9; P = 0.46). The R213G and E1/I1 polymorphisms were in minimal linkage disequilibrium (D′= −0.811; r2=0.0005) (see Table E1 in the online supplement).
Figure 3.
Plasma SOD3, pulmonary function, and risk of chronic obstructive pulmonary disease (COPD) according to SOD3 genotype. Values represent mean ± 2 SE or hazard ratio (HZ) and 95% confidence interval (CI). Plasma SOD3 was measured in a subset of 194 R213G carriers, 1,953 noncarriers, 281 E1/I1 heterozygotes, and 9 E1/I1 homozygotes. The Cox regression models allowed for age, sex, smoking, packyears at entry, and SOD3 E1/I1 genotype. *P < 0.05, **P < 0.01, and ***P < 0.001 versus noncarriers using Mann-Whitney U test (for plasma SOD3), repeated measures analysis (for FEV1 % predicted and FVC % predicted), or Wald's test (for COPD hospitalization/mortality).
The Copenhagen General Population Study
We also genotyped the Copenhagen General Population Study for the E1/I1 polymorphisms. Of the 35,635 individuals tested, 31,422 (88%) were E1/I1 noncarriers, 4,079 (11%) were heterozygotes, and 134 (0.4%) were homozygotes. This distribution did not differ from that predicted by the Hardy-Weinberg equilibrium (P = 0.90). Sex, age, smoking habits, pack-years, and body mass index did not differ between E1/I1 noncarriers, heterozygotes, and homozygotes in the Copenhagen General Population Study (Table E2).
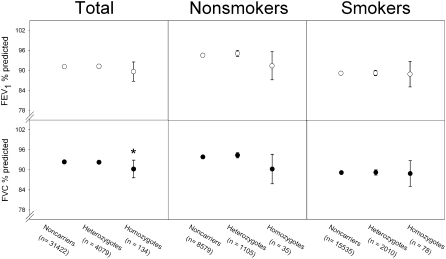
Using a cross-sectional design, we found that E1/I1 homozygotes had 2% lower FVC % predicted and 1% lower FEV1 % predicted compared with noncarriers (P = 0.03 and P = 0.14, respectively, Figure 4). Smoking did not interact statistically with E1/I1 genotype in predicting FVC % predicted (P = 0.28) or FEV1 % predicted (P = 0.54).
Figure 4.
Pulmonary function in SOD3 E1/I1 homozygotes and heterozygotes versus noncarriers in the Copenhagen General Population Study. Values represent mean ± 2 SE. *P < 0.05 versus noncarriers using one-sided Mann-Whitney U test.
Prevalence of COPD hospitalization was 2% in homozygotes, 4% in heterozygotes, and 4% in noncarriers (χ2: P = 0.41). The Copenhagen General Population Study is currently recruiting subjects with lower daily tobacco consumption and prevalence of COPD hospitalization than the Copenhagen City Heart Study, which may explain the less pronounced effect in this study compared with the Copenhagen City Heart Study.
DISCUSSION
We identified two new polymorphisms, E1 and I1, in a conserved region of the SOD3 gene. Screening the Copenhagen City Heart Study for these variants, we found that E1/I1 homozygotes had lower FVC % predicted than noncarriers. Using a similarly sized sample of pulmonary function results from another large Danish general population sample, we confirmed a relationship between the SOD3 E1/I1 genotype and reduced FVC % predicted. In support, E1/I1 homozygotes had greater risk of clinical COPD in terms of hospitalizations from COPD; however, this finding could not be confirmed in the Copenhagen General Population Study. Collectively, the data seem to support a role for SOD3 in oxidant-mediated events influencing lung function.
The E1 and I1 polymorphisms reside in a sequence that is identified as a mammalian interspersed repetitive element, and which is conserved within the mouse genome, suggesting that even though this region is non–protein coding, it is still likely functionally relevant, either in translational or transcriptional control or mRNA stability. The E1 polymorphism occurs in a putative binding site for 5′TG3′ interacting factor protein and the I1 SNP occurs in the middle of a putative c-Myc/Max heterodimer E12 binding site (Figure E1). 5′TG3′ interacting factor is a homeobox gene that belongs to the three–amino acid loop extension superclass of homeodomains (22). The c-Myc/Max belongs to a family of heterodimeric transcription factors that have diverse functions in proliferation, differentiation, and tumorigenesis (23). Whether these sites have functional significance in SOD3 biology remains to be proven. Furthermore, our analyses do not preclude the possibility that these polymorphisms are linked to another part of the SOD3 gene that we have not yet resequenced (e.g., the introns). Additional fine mapping, resequencing, and functional studies will be necessary to characterize how the E1/I1 polymorphism modulates SOD3 expression and function.
Earlier studies support SOD3 or nearby variation could be related to lung function, as was observed for the E1/I1 variant in the present study. Wilk and coworkers (24) demonstrated that a polymorphism nearby the SOD3 gene exhibited association to percent predicted phenotypes in humans in a recent genomewide association study. The polymorphism that they identified lay within a hypothetical protein 3′ of the SOD3 gene. Another study using linkage analysis data and expression profiling in different mouse strains also identified SOD3 as a possible determinant of lung function (25). Findings with gene-targeted mice subsequently suggested that SOD3 was a contributing factor defining the complex trait of conducting airway volume. In addition, we and others have shown that the R213G variant of the SOD3 gene associates with lower risk of COPD, indicating that this variant may protect from COPD development (13, 26).
An association between SOD3 E1/I1 polymorphisms and reduced lung function could also be biologically plausible, as SOD3 protects lung tissue matrix collagen I, hyaluronan, and heparin sulfate from oxidative fragmentation (3, 4, 6). The fragments created from collagen I, hyaluronan, and heparin sulfate during an increased oxidative burden are chemotactic and contribute to inflammatory cell recruitment in vitro or in vivo (3, 4, 6). Similarly, SOD3 binds fibulin-5, and could also provide antioxidative protection of this lung matrix protein, which normally associates with elastin in the lung (27, 28). Thus, changes in SOD3 expression or function could cause less protection of lung matrix components against oxidants and lead to increased inflammatory cell recruitment and injury. This could cause reduced lung function and COPD over time.
A second mechanism by which SOD3 may play a beneficial role in host defenses against oxidative stress is by attenuating inflammation through decreasing the release of proinflammatory cytokines from phagocytes. In an animal model of LPS-induced lung injury, Bowler and coworkers (29) showed that SOD3 attenuates the inflammatory response to LPS in vivo, and that SOD3 inhibited LPS-induced cytokine release from macrophages. This potential protective capability may not be trivial, as SOD3 is released in large amounts from inflammatory cells into the airways during inflammation (30, 31). Moreover, studies of SOD3-deficient and SOD3-overexpressing mice suggest that SOD3 may protect the lung from injury in response to oxidant inhalation (32–35), fibronegic agents (36, 37), and radiation (38, 39).
Figures 2 and 4 may indicate different effects of smoking on the lung function–genotype associations; however, smoking did not interact with genotype in predicting FVC or FEV1 in either figure (P values ≥ 0.28). Thus, smoking had no effect on the lung function–genotype associations in either of the two studies. Although our results could suggest that FVC was reduced more than FEV1, statistically there was no significant difference between the reductions in FVC and FEV1 in either of the two studies (mean reduction ± 2 SE: FEV1 % predicted, 4 ± 6 vs. FVC % predicted, 7 ± 5 in the Copenhagen City Heart Study; FEV1 % predicted, 1 ± 2 vs. FVC % predicted, 2 ± 1 in the Copenhagen General Population Study) (40). Further research into this area will be required to examine whether E1/I1 could also be associated with restriction.
If correction for four multiple comparisons was performed, only the reduced FVC % predicted would be of statistical significance. Replication is the gold standard in genetic epidemiological studies, and the replicated lung function data, we believe, is a robust finding. Although these data were confirmed in two ethnically homogenous and independent studies, it would of course be desirable that the finding is replicated in further independent studies in the future. The COPD finding in the longitudinal Copenhagen City Heart Study could not be confirmed in the cross-sectional Copenhagen General Population Study, and replication in a well characterized, independent COPD population would be necessary to conclusively determine whether the E1/I1 genotype is associated with COPD. This would also allow the possible association with some COPD subphenotypes.
In conclusion, our studies show that individuals that are homozygous for the E1/I1 polymorphisms in the SOD3 gene have reduced FVC % predicted in two large, population-based studies. We believe that the data support a role for SOD3 in oxidant-mediated events influencing lung function. Recombinant SODs are available, and studies of intervention therapy with SOD3 in animal models with reduced lung function seem feasible and warranted (41, 42).
Supplementary Material
Acknowledgments
The authors thank Hanne Damm, Jessica Orcutt, Christina Wheeler, Michael Nicks, Dana Busam, Karen Beeson, and Timothy Stockwell for their invaluable assistance in the laboratory.
Supported by the Danish Heart Foundation, the Danish Lung Association, the Copenhagen County Foundation, Flight Attendant Medical Research Institute (R.P.B.), National Institutes of Health General Clinical Research Center (M01 RR00051), and the Monfort Foundation.
This article has an online supplement, which is available from the issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200804-549OC on August 14, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.MacNee W, Rahman I. Is oxidative stress central to the pathogenesis of chronic obstructive pulmonary disease? Trends Mol Med 2001;7:55–62. [DOI] [PubMed] [Google Scholar]
- 2.Dahl M, Bauer AK, Arredouani M, Soininen R, Tryggvason K, Kleeberger SR, Kobzik L. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. J Clin Invest 2007;117:757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao F, Koenitzer JR, Tobolewski JM, Jiang D, Liang J, Noble PW, Oury TD. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J Biol Chem 2008;283:6058–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kliment CR, Tobolewski JM, Manni ML, Tan RJ, Enghild J, Oury TD. Extracellular superoxide dismutase protects against matrix degradation of heparan sulfate in the lung. Antioxid Redox Signal 2008;10:261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mudway IS, Kelly FJ. Ozone and the lung: a sensitive issue. Mol Aspects Med 2000;21:1–48. [DOI] [PubMed] [Google Scholar]
- 6.Petersen SV, Oury TD, Ostergaard L, Valnickova Z, Wegrzyn J, Thogersen IB, Jacobsen C, Bowler RP, Fattman CL, Crapo JD, et al. Extracellular superoxide dismutase (EC-SOD) binds to type I collagen and protects against oxidative fragmentation. J Biol Chem 2004;279:13705–13710. [DOI] [PubMed] [Google Scholar]
- 7.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics 1994;22:162–171. [DOI] [PubMed] [Google Scholar]
- 8.Kinnula VL, Crapo JD. Superoxide dismutases in the lung and human lung diseases. Am J Respir Crit Care Med 2003;167:1600–1619. [DOI] [PubMed] [Google Scholar]
- 9.Oury TD, Chang LY, Marklund SL, Day BJ, Crapo JD. Immunocytochemical localization of extracellular superoxide dismutase in human lung. Lab Invest 1994;70:889–898. [PubMed] [Google Scholar]
- 10.Oury TD, Day BJ, Crapo JD. Extracellular superoxide dismutase in vessels and airways of humans and baboons. Free Radic Biol Med 1996;20:957–965. [DOI] [PubMed] [Google Scholar]
- 11.Hjalmarsson K, Marklund SL, Engstrom A, Edlund T. Isolation and sequence of complementary DNA encoding human extracellular superoxide dismutase. Proc Natl Acad Sci USA 1987;84:6340–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu Y, Alwahdani A, Iida S, Lund DD, Faraci FM, Heistad DD. Vascular effects of the human extracellular superoxide dismutase R213G variant. Circulation 2005;112:1047–1053. [DOI] [PubMed] [Google Scholar]
- 13.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:858–864. [DOI] [PubMed] [Google Scholar]
- 14.Dahl M, Juul K, Bowler R, Crapo J, Nordestgaard BG. Extracellular superoxide dismutase polymorphisms, lung function, and chronic obstructive pulmonary disease. Nihon Kokyuki Gakkai Zasshi 2008;46:92–95. In Japanese.18318249 [Google Scholar]
- 15.The Copenhagen City Heart Study. Osterbroundersogelsen: a book of tables with data from the first examination (1976–78) and a five year follow-up (1981–83). The Copenhagen City Heart Study Group. Scand J Soc Med Suppl 1989;41:1–160. [PubMed] [Google Scholar]
- 16.Dahl M, Vestbo J, Lange P, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. C-reactive protein as a predictor of prognosis in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:250–255. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization. Manual of the international statistical classification of diseases, injuries, and causes of death, 8th revision. Geneva: the Organization, 1965.
- 18.World Health Organization. International statistical classification of diseases and related health problems, 10th revision. Geneva: the Organization, 1992.
- 19.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA 2007;298:299–308. [DOI] [PubMed] [Google Scholar]
- 20.Juul K, Tybjaerg-Hansen A, Marklund S, Heegaard NH, Steffensen R, Sillesen H, Jensen G, Nordestgaard BG. Genetically reduced antioxidative protection and increased ischemic heart disease risk: The Copenhagen City Heart Study. Circulation 2004;109:59–65. [DOI] [PubMed] [Google Scholar]
- 21.Gaunt TR, Rodriguez S, Day IN. Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool ‘CubeX’. BMC Bioinformatics 2007;8:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem 1995;270:31178–31188. [DOI] [PubMed] [Google Scholar]
- 23.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000;16:653–699. [DOI] [PubMed] [Google Scholar]
- 24.Wilk JB, Walter RE, Laramie JM, Gottlieb DJ, O'Connor GT. Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet 2007;8:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguly K, Stoeger T, Wesselkamper SC, Reinhard C, Sartor MA, Medvedovic M, Tomlinson CR, Bolle I, Mason JM, Leikauf GD, et al. Candidate genes controlling pulmonary function in mice: transcript profiling and predicted protein structure. Physiol Genomics 2007;31:410–421. [DOI] [PubMed] [Google Scholar]
- 26.Young RP, Hopkins R, Black PN, Eddy C, Wu L, Gamble GD, Mills GD, Garrett JE, Eaton TE, Rees MI. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax 2006;61:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang PP, Joyce-Brady M, Zhang XH, Jean JC, Goldstein RH. Fibulin-5 gene expression in human lung fibroblasts is regulated by TGF-beta and phosphatidylinositol 3-kinase activity. Am J Physiol Cell Physiol 2006;291:C1412–C1421. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen AD, Itoh S, Jeney V, Yanagisawa H, Fujimoto M, Ushio-Fukai M, Fukai T. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circ Res 2004;95:1067–1074. [DOI] [PubMed] [Google Scholar]
- 29.Bowler RP, Nicks M, Tran K, Tanner G, Chang LY, Young SK, Worthen GS. Extracellular superoxide dismutase attenuates lipopolysaccharide-induced neutrophilic inflammation. Am J Respir Cell Mol Biol 2004;31:432–439. [DOI] [PubMed] [Google Scholar]
- 30.Su WY, Folz R, Chen JS, Crapo JD, Chang LY. Extracellular superoxide dismutase mRNA expressions in the human lung by in situ hybridization. Am J Respir Cell Mol Biol 1997;16:162–170. [DOI] [PubMed] [Google Scholar]
- 31.Tan RJ, Lee JS, Manni ML, Fattman CL, Tobolewski JM, Zheng M, Kolls JK, Martin TR, Oury TD. Inflammatory cells as a source of airspace extracellular superoxide dismutase after pulmonary injury. Am J Respir Cell Mol Biol 2006;34:226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlsson LM, Jonsson J, Edlund T, Marklund SL. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc Natl Acad Sci USA 1995;92:6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fakhrzadeh L, Laskin JD, Gardner CR, Laskin DL. Superoxide dismutase-overexpressing mice are resistant to ozone-induced tissue injury and increases in nitric oxide and tumor necrosis factor-alpha. Am J Respir Cell Mol Biol 2004;30:280–287. [DOI] [PubMed] [Google Scholar]
- 34.Folz RJ, Abushamaa AM, Suliman HB. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J Clin Invest 1999;103:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghio AJ, Suliman HB, Carter JD, Abushamaa AM, Folz RJ. Overexpression of extracellular superoxide dismutase decreases lung injury after exposure to oil fly ash. Am J Physiol Lung Cell Mol Physiol 2002;283:L211–L218. [DOI] [PubMed] [Google Scholar]
- 36.Bowler RP, Nicks M, Warnick K, Crapo JD. Role of extracellular superoxide dismutase in bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2002;282:L719–L726. [DOI] [PubMed] [Google Scholar]
- 37.Fattman CL, Tan RJ, Tobolewski JM, Oury TD. Increased sensitivity to asbestos-induced lung injury in mice lacking extracellular superoxide dismutase. Free Radic Biol Med 2006;40:601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang SK, Rabbani ZN, Folz RJ, Golson ML, Huang H, Yu D, Samulski TS, Dewhirst MW, Anscher MS, Vujaskovic Z. Overexpression of extracellular superoxide dismutase protects mice from radiation-induced lung injury. Int J Radiat Oncol Biol Phys 2003;57:1056–1066. [DOI] [PubMed] [Google Scholar]
- 39.Rabbani ZN, Anscher MS, Folz RJ, Archer E, Huang H, Chen L, Golson ML, Samulski TS, Dewhirst MW, Vujaskovic Z. Overexpression of extracellular superoxide dismutase reduces acute radiation induced lung toxicity. BMC Cancer 2005;5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosner B. Fundamentals of biostatistics, 4th ed. Belmont, CA: Duxbury Press; 1995.
- 41.Bowler RP, Barnes PJ, Crapo JD. The role of oxidative stress in chronic obstructive pulmonary disease. COPD 2004;1:255–277. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez-Saavedra D, Zhou H, McCord JM. Anti-inflammatory properties of a chimeric recombinant superoxide dismutase: SOD2/3. Biomed Pharmacother 2005;59:204–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.