Abstract
Rationale: Circulating leukocyte RNA transcripts are systemic markers of inflammation, which have not been studied in cystic fibrosis (CF) lung disease. Although the standard assessment of pulmonary treatment response is FEV1, a measure of airflow limitation, the lack of systemic markers to reflect changes in lung inflammation critically limits the testing of proposed therapeutics.
Objectives: We sought to prospectively identify and validate peripheral blood leukocyte genes that could mark resolution of pulmonary infection and inflammation using a model by which RNA transcripts could increase the predictive value of spirometry.
Methods: Peripheral blood mononuclear cells were isolated from 10 patients with CF and acute pulmonary exacerbations before and after therapy. RNA expression profiling revealed that 10 genes significantly changed with treatment when compared with matched non-CF and control subjects with stable CF to establish baseline transcript abundance. Peripheral blood mononuclear cell RNA transcripts were prospectively validated, using real-time polymerase chain reaction amplification, in an independent cohort of acutely ill patients with CF (n = 14). Patients who responded to therapy were analyzed using general estimating equations and multiple logistic regression, such that changes in FEV1% predicted were regressed with transcript changes.
Measurements and Main Results: Three genes, CD64, ADAM9, and CD36, were significant and independent predictors of a therapeutic response beyond that of FEV1 alone (P < 0.05). In both cohorts, receiver operating characteristic analysis revealed greater accuracy when genes were combined with FEV1.
Conclusions: Circulating mononuclear cell transcripts characterize a response to the treatment of pulmonary exacerbations. Even in small patient cohorts, changes in gene expression in conjunction with FEV1 may enhance current outcomes measures for treatment response.
Keywords: cystic fibrosis, peripheral blood mononuclear cells, biomarkers, pulmonary exacerbation
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Although severe airway inflammation is a hallmark of cystic fibrosis lung disease, no systemic marker of inflammation has been validated.
What This Study Adds to the Field
We demonstrate that circulating leukocyte RNA transcripts predict treatment response in cystic fibrosis lung disease and may potentially serve as biomarkers to strengthen current outcomes measures in clinical trials.
Cystic fibrosis (CF) is the most common lethal inherited disease in the Western world, with respiratory failure accounting for more than 80% of deaths from the disease, usually in the third or fourth decade of life. The triad of mucus airway obstruction, chronic Pseudomonas aeruginosa infection, and severe airway inflammation are the major pathogenic factors in CF lung disease (1). There is a critical need for effective antimicrobial and antiinflammatory therapies to mitigate disease in this young population. The design of clinical trials in CF is hampered, in part, by the lack of sensitive measures of treatment response. The most established outcome measure for CF therapies is FEV1, a functional measure of airflow limitation, and a key consideration in the advancement of treatments from phase 2 to phase 3 trials. The process of airway remodeling by inflammatory cells is progressive and may not be detected by FEV1 alone in the timeframe of a typical phase 2 trial. To date, no systemic marker of treatment response has been validated in CF. The gene expression of peripheral leukocytes has never been studied as a predictor of treatment response in CF, yet peripheral blood mononuclear cells (PBMCs) serve as the source of the dense mononuclear infiltrate present at sites of CF airway injury and acquire a unique transcriptional “fingerprint” as a result of repeated passages through the pulmonary vascular bed. Furthermore, circulating leukocytes have demonstrated diagnostic and prognostic utility for multiple pulmonary conditions, including steroid sensitivity in asthma and discrimination between viral and bacterial pulmonary infections (2–6).
A condensed model of deterioration in CF lung disease is represented by episodic pulmonary exacerbations. Primarily a clinical diagnosis, exacerbations are treated with antimicrobials, considered the standard of care to decrease pulmonary bacterial burden and host airway inflammation. The aggressive treatment of typical pulmonary exacerbations results in rapid reduction in pulmonary inflammation due to reduced infection (7–10). Response to treatment is assessed by improvements in spirometry and clinical evaluation (11). Because exacerbation frequency is positively correlated with respiratory decline (12–14), the assessment of the effect of therapies on reducing the incidence of pulmonary exacerbations is a key measure in clinical trials for evaluating the most important drugs used in CF care (15–18).
In two separate cohorts of patients with CF, we identified three PBMC genes whose changes in expression correlate with the onset and resolution of acute pulmonary exacerbations. The genes consistently added independent predictive power to FEV1, and represent a novel tool to quantify CF therapeutic response noninvasively.
METHODS
Patient Recruitment
Subjects older than 18 years with CF (sweat chloride testing and genotype) were enrolled at the time of intravenous antibiotic initiation for a clinically diagnosed pulmonary exacerbation at a Cystic Fibrosis Foundation (CFF)–accredited adult CF clinic. Patients enrolled met CFF Clinical Practice criteria for an acute pulmonary exacerbation and were treated for a minimum of 2 weeks with intravenous antibiotics (19). This observational trial used within-subject comparisons, such that each subject served as his or her own control, after treatment with antibiotics. Blood was drawn at initiation (±2 d) and completion (±1 wk) of antibiotic therapy. At each time point, the following were sampled: (1) blood for PBMC isolation and whole-blood differentials, (2) sputum microbiology, (3) simple spirometry for FEV1, and (4) plasma cytokines. In addition, transcript abundance in study patients was compared with PBMC array data from eight healthy volunteer subjects and six control subjects with stable CF. Next, a validation study was performed on 14 adult patients with CF suffering from acute pulmonary exacerbations. Enrollment and assessment of blood, microbiology, and spirometry occurred as described above.
PBMC Isolation
PBMCs were isolated via density gradient centrifugation followed by RNA isolation, as described in the online supplement. PBMC counts at each isolation were compared using paired t tests.
Microarray Hybridization and Data Analysis
Expression profiling of PBMC RNA (development cohort) identified transcriptional changes before and after antibiotic therapy, using Affymetrix (Foster City, CA) Hu133 Plus 2.0 gene arrays. From known gene sequences, we identified differentially expressed genes with treatment in pairwise comparisons, with a minimum of 1.4-fold change. Ontology analysis for biologic plausibility narrowed the list from 32 to 19 candidate genes. After reverse transcriptase–polymerase chain reaction (RT-PCR) validation, transcript abundance from the 10 remaining genes were compared with PBMC microarray data from matched normal control subjects and control subjects with CF, using analysis of variance (ANOVA). See the online supplement for detailed methods.
Quantitative RT-PCR Validation
Real-time RT-PCR confirmed microarray expression data in the development cohort and measured transcript abundance in the validation cohort, using Sybrgreen indicator on a 7300 Real Time PCR System (Applied Biosystems, Foster City, CA). Details are specified in the online supplement. Log-transformed transcript abundance was compared using paired t tests.
Plasma Cytokine Measurements
Nineteen cytokines and C-reactive protein (CRP) levels were quantified at each blood draw in the development cohort. Details are included in the online supplement. Paired t tests compared log-transformed pre- and postantibiotic values. Spearman correlation coefficients (SAS, Version 9.0; SAS Institute, Cary, NC) assessed relationships between CRP, FEV1, circulating leukocytes, and gene expression pre- and postantibiotic therapy.
Endpoints
Primary endpoints were lung function (FEV1% predicted) and gene transcript changes after treatment. Patients whose FEV1% predicted increased with therapy and who were not rehospitalized with a diagnosis of acute exacerbation within 7 days were included in the logistic regression model.
Statistical Methods
Using Generalized Estimating Equations software (SAS), multivariate logistic regression models were constructed predicting resolution of acute pulmonary exacerbations as a function of FEV1% predicted, and the discriminative value of combinations of genes identified by the development cohort. To identify the most frugal combination of predictors for resolution of inflammation, we required a P value less than 0.05 for entry into the model, allowing determination of the unique contribution of the genes beyond FEV1. Receiver operating characteristic (ROC) analyses reflected the overall diagnostic value of the gene markers in terms of enhanced sensitivity and specificity over FEV1 alone.
RESULTS
Sequential Data Analysis Strategy
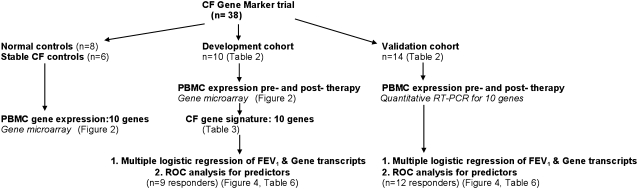
The stepwise execution of this study is shown in Figure 1. Candidate genes, differentially regulated before and after antibiotic therapy in patients with CF and acute lung infection, were identified in 10 patients on the basis of data generated from high-density oligonucleotide array experiments on patients' PBMC samples. Nonparametric univariate tests ranked genes on their ability to discriminate between the two time points. PBMC transcript abundance from patients with stable CF and normal healthy control subjects, matched in age and sex to the CF population and prepared with identical method and array platforms, was included in the analysis. Samples underwent whole-blood differentials as well as cell counts after each isolation event. Although there was a trend toward decreased total white blood cell count after treatment of pulmonary exacerbations (P = 0.052), there were no statistically significant differences in median percentage values of neutrophils, lymphocytes, monocytes, eosinophils, and basophils, pre- and post-therapy, nor were there significant differences between PBMC absolute numbers pre– and post–antibiotic therapy, after isolation. Second, classifier genes were independently confirmed in the same set of samples using quantitative real-time RT-PCR. Of note, 6 of 10 patients in the development cohort had sufficient remaining RNA, after microarray analysis, for transcript measurements of 10 genes. Third, the resulting “CF gene signature,” consisting of genes significantly changed in both microarray and RT-PCR experiments, was then tested for its ability to classify therapeutic response in an independent group of patients with CF undergoing therapy for acute pulmonary infections. Table 1 depicts measurement distributions performed for all 24 samples from patients with CF. Fourth, using FEV1 improvement as a standard measure of clinical response, logistic regression on the change in FEV1% predicted, with gene transcript changes, allowed association between measurements of lung function and genes in the responder groups for both cohorts. Finally, a comparison of the diagnostic value of genes in combination with FEV1 as markers, was depicted using ROC curves.
Figure 1.
Study design and sequence for study groups. Genes identified in the development cohort were tested in the validation cohort. CF = cystic fibrosis; PBMC = peripheral blood mononuclear cell; ROC = receiver operating curve.
TABLE 1.
ANALYSIS PERFORMED ON CYSTIC FIBROSIS SAMPLES IN DEVELOPMENT AND VALIDATION COHORTS
| Patient | PBMC Gene Microarray | Quantitative RT-PCR | Plasma Cytokine Measurements | FEV1 | CRP | Logistic Regression Model |
|---|---|---|---|---|---|---|
| 1 | x | x | x | x | x | |
| 2 | x | x | x | x | x | |
| 3 | x | x | x | x | x | x |
| 4 | x | x | x | x | x | x |
| 5 | x | x | x | x | x | x |
| 6 | x | x | x | x | x | x |
| 7 | x | x | x | x | x | x |
| 8 | x | x | x | x | x | x |
| 9 | x | x | x | x | NR* | |
| 10 | x | x | x | x | x | |
| V11 | x | x | NR | |||
| V12 | x | x | x | |||
| V13 | x | x | x | |||
| V14 | x | x | x | |||
| V15 | x | x | x | |||
| V16 | x | x | x | |||
| V17 | x | x | x | |||
| V18 | x | x | NR | |||
| V19 | x | x | x | |||
| V20 | x | x | x | |||
| V21 | x | x | x | |||
| V22 | x | x | x | |||
| V23 | x | x | x | |||
| V24 | x | x | x |
Definition of abbreviations: CRP = C-reactive protein; NR = nonresponder; PBMC = peripheral blood mononuclear cells; V = validation cohort population.
Nonresponders were not included in logistic regression model.
Baseline Patient Characteristics
PBMCs were isolated from 22 adult patients with CF at the beginning and end of a course of antibiotic treatment for an acute pulmonary exacerbation, representing a total of 24 acute pulmonary exacerbations. The development cohort contained 10 patients, and a subsequent validation cohort consisted of 14 patients. Two patients in the development cohort suffered subsequent exacerbations and were included in the validation cohort 2 years later. All patients met CFF guidelines for an acute pulmonary exacerbation (19). The baseline demographics, severity of airflow limitation, genotype, and sputum microbiology for the development and validation cohorts are shown in Table 2. Both groups had moderate to severe airway disease by American Thoracic Society criteria, on the basis of FEV1% predicted measured at the completion of intravenous antibiotic therapy (20). Ninety percent of patients in both groups grew P. aeruginosa on sputum culture at the time of therapy. Staphylococcus aureus was commonly isolated in both groups, although only one strain was methicillin resistant. Whereas no patients were treated with steroids in the development cohort, over half of the validation cohort received steroids (P = 0.002). The median pretreatment FEV1% was lower in the validation group (P = 0.04); however, post-treatment FEV1% predicted did not significantly differ between groups. All patients exhibited FEV1 increases at the conclusion of therapy except for a single patient in the development cohort, whose FEV1% predicted declined by 26% after therapy. This patient concomitantly suffered a severe flare-up of CF-related arthritis at the end of her antibiotic therapy. Given the drop in FEV1, this patient was considered a nonresponder, and her gene copy changes were not used in the regression analyses for genes with FEV1. Clinical nonresponders for the second cohort are described below.
TABLE 2.
BASELINE CHARACTERISTICS OF STUDY POPULATION
| Characteristic | Development Group (n = 10) | Validation Group (n = 14) |
|---|---|---|
| Age, yr | 26 ± 5.7 | 27 ± 3 |
| Sex | ||
| Male | 3 (30) | 9 (64) |
| Female | 7 (70) | 5 (42) |
| Genotype | ||
| ΔF508/ΔF508 | 8 (80) | 9 (64) |
| Other | 2 (20) | 5 (36) |
| FEV1, % predicted | ||
| Pre-therapy | 44 ± 19 | 31.5 ± 18* |
| Post-therapy | 51 ± 18 | 44 ± 20 |
| Sputum culture | ||
| Pseudomonas aeruginosa | 9 (90) | 13 (93) |
| Staphylococcus aureus (methicillin sensitive) | 7 (70) | 8 (57) |
| Staphylococcus aureus (methicillin resistant) | 1 (10) | 0 |
| Other (Burkholderia cepacia, Alcaligenes xylosoxidans, Aspergillus fumigatus) | 1 (10) | 4 (29) |
| Systemic antibiotic therapy | ||
| APAG + 4th gen Ceph or Carbapenem or Monobactam | 7 (70) | 10 (71) |
| Other combination† | 3 (30) | 4 (29) |
| Systemic steroid use | 0 | 8 (57)* |
| CFRD | 4 (40) | 5 (36) |
Definitions of abbreviations: APAG = antipseudomonal aminoglycoside; CFRD = cystic fibrosis–related diabetes mellitus as diagnosed by Cystic Fibrosis Foundation guidelines; 4th gen Ceph = fourth generation cephalosporin.
Values are n (%) or mean ± SD.
P < 0.05.
Other therapy includes vancomycin, nafcillin, cefazolin, levofloxacin, and carbapenem/monobactam combinations.
A Transcriptional Signature Characterizes the Effect of Antibiotic Therapy in Patients with CF with Acute Pulmonary Exacerbations
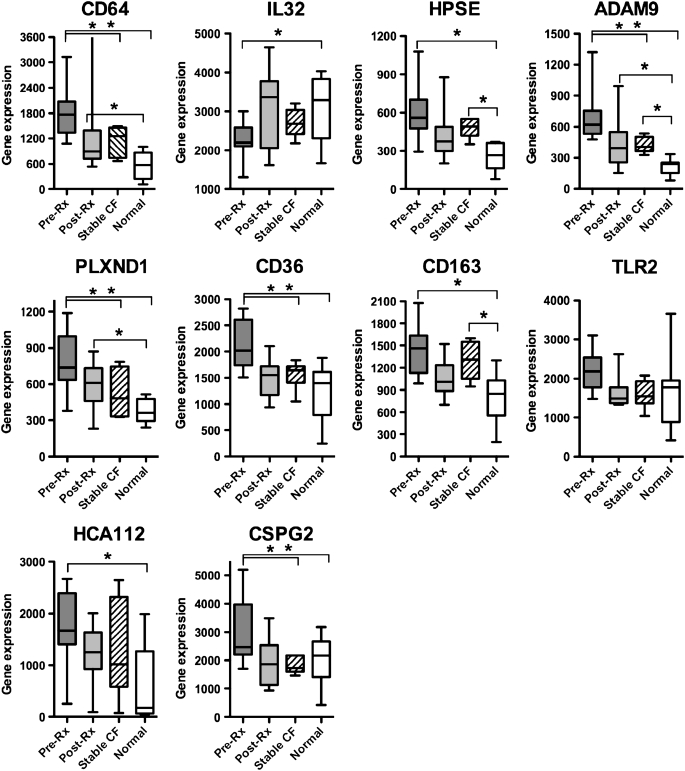
Table 3 lists 10 genes significantly changed (P < 0.05) by both microarray and quantitative real-time RT-PCR analysis in the development cohort of patients with CF. After ontology analysis, 19 candidate genes identified by microarray analysis were evaluated by quantitative PCR in the same samples to validate results. Of 19 genes, 10 were significantly changed when measured by both methods. Of note, the real-time RT-PCR validation occurred in six pairs of RNA samples, because the other samples did not have sufficient remaining RNA after microarray to be evaluated by PCR. We designated these 10 PBMC genes, reflecting treatment of CF pulmonary exacerbations, the “CF therapeutic signature.” The vast majority of genes (9 of 10) were down-regulated after resolution of the acute pulmonary exacerbation. Only IL-32 increased transcription after treatment, suggesting suppression during the acute exacerbation and a return to normal baseline after treatment, as indicated by transcripts in normal control subjects. In Figure 2, we compare pre- and post-antibiotic therapy median values and distribution for CF genes in the development cohort with array data from eight age- and sex-matched normal control subjects, as well as six control subjects with stable CF. Before antibiotic therapy, expression of all genes, except for TLR2, significantly differed from expression in normal subjects (ANOVA, P < 0.05; pairwise comparison with Fisher's protected least significant difference test [PLSD], P < 0.05) and expression of 5 of 10 genes (PLXND1, ADAM9, CSPG2, CD64, and CD36) differed from that in stable subjects with CF (ANOVA, P < 0.05; pairwise comparison with Fisher's PLSD, P < 0.05). After treatment of the acute pulmonary exacerbation, none of the 10 genes differed significantly between post-therapy patients and control subjects with stable CF. The following genes were significantly different between preantibiotic CF and stable CF and unchanged between postantibiotic CF and stable CF: PLXND1, ADAM9, CSPG2, CD64, and CD36 (using ANOVA and pairwise comparison with Fisher's PLSD). In post-therapy patients with CF, expression of three genes, CD64, ADAM9, and PLXND1, remained significantly different in patients with CF compared with normal control subjects (ANOVA, P < 0.05; pairwise comparison with Fisher's PLSD, P < 0.05).
TABLE 3.
CLASSIFICATION OF 10 GENES IN THE CYSTIC FIBROSIS THERAPEUTIC GENE SIGNATURE
| Array*
|
PCR†
|
||||
|---|---|---|---|---|---|
| Gene | Description | Fold Change | P Values | Fold Change | P Value |
| Cell membrane molecules and receptors | |||||
| CD64 | Fcγ receptor IA | −2.4 | 0.004 | −2.1 | 0.007 |
| CD36 | Collagen type 1 receptor/thrombospondin receptor | −1.7 | <0.001 | −1.6 | <0.001 |
| CD163 | Hemoglobin scavenger receptor | −1.6 | 0.003 | −1.7 | 0.024 |
| TLR2 | Toll-like receptor 2 | −1.6 | 0.003 | −1.7 | 0.011 |
| HCA112 | Hepatocellular carcinoma-associated antigen 112 | −1.8 | 0.004 | −1.6 | 0.012 |
| PLXND1 | Plexin D1 | −1.7 | 0.005 | −1.3 | 0.083 |
| Immune response | |||||
| IL32 | Interleukin 32 | 1.4 | 0.033 | 1.4 | 0.001 |
| Matrix degradation/extravasation | |||||
| HPSE | Heparanase | −1.4 | 0.017 | −1.6 | 0.004 |
| ADAM9 | A disintegrin and metalloproteinase, meltrin gamma | −2.5 | 0.004 | −1.7 | 0.016 |
| CSPG2 | Versican, chondroitin sulfate proteoglycan 2 | −1.9 | <0.001 | −1.9 | 0.015 |
Definition of abbreviation: PCR = polymerase chain reaction.
Values are mean fold changes or P values calculated by paired t test after log transformation; n = 6 patient samples pre- and postantibiotic therapy from development cohort.
Pre- and postantibiotic therapy expression values from oligonucleotide arrays.
Pre- and postantibiotic therapy real-time PCR gene expression values as copies per 1,000 copies of housekeeping gene, hypoxanthine-phosphoribosyl transferase (HPRT).
Figure 2.
Comparison of median expression values of the cystic fibrosis (CF) gene signature in preantibiotic (Pre-Rx) and postantibiotic (Post-Rx) peripheral blood mononuclear cell samples versus age- and sex-matched normal control subjects and patients with stable CF. Whiskers on box plots represent range of expression values between patient samples for each individual gene. Median is represented within each box whose boundaries represent the 25th–75th quartiles. Differences in transcript abundance between pre- and post-therapy patients with CF were significant (P < 0.05) for all genes, except for HCA112. *P < 0.05 using analysis of variance with Fisher's protected least significant difference test for difference between bracketed groups.
The CF Therapeutic Signature and Associations with Common Clinical Outcome Measures
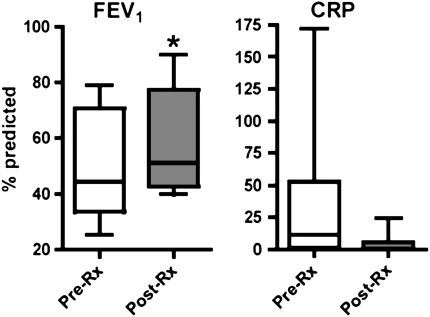
Measurements of CRP and FEV1 were performed simultaneously with PBMC isolation in the development cohort. As expected, improvement in FEV1% predicted was statistically significant after treatment, (P = 0.02 by paired t test; 95% confidence intervals, −1.8 to −15.78) (Figure 3). The change in plasma CRP levels between pairs did not reach statistical significance (P = 0.09). The correlations between gene expression values and both FEV1 and CRP levels were evaluated with Spearman's rank order correlation coefficient (rs) in Table 4. A significant correlation exists between the development cohort expression values and CRP values for 7 of the 10 genes: TLR2, CD64, CSPG2, HPSE, IL-32, CD163, and ADAM9. No correlation was noted between FEV1 change and the 10-gene CF signature. Furthermore, we conducted correlation analyses between genes and circulating cellular markers of inflammation, namely, neutrophil counts and total white blood cell counts (Table 5). Four genes were highly correlated (P < 0.05) to circulating neutrophil counts before and after therapy: ADAM9, CSPG2, IL32, and CD163.
Figure 3.
Clinical outcomes as represented by changes in FEV1% predicted and C-reactive protein (CRP) pre– and post–antibiotic therapy in the development cohort. The left panel represents changes in FEV1% predicted at initiation (Pre-Rx) and termination (Post-Rx) of antibiotics. Box plot boundaries represent 25th–75th quartiles of values, with median line within the box, and whiskers representing FEV1% predicted ranges. CRP values in mg/dl are also depicted in pre- and post-therapy groups, with normal standard range. *P = 0.02 for differences between FEV1% predicted pre- and post-therapy by paired t test. P = 0.09 for differences between pre- and post-therapy CRP values by paired t test.
TABLE 4.
SPEARMAN'S RANK CORRELATION COEFFICIENTS (rS) FOR GENES, FEV1, AND C-REACTIVE PROTEIN PRE– AND POST–ANTIBIOTIC THERAPY
| FEV1
|
CRP
|
|||
|---|---|---|---|---|
| Variable | rs | P Value | rs | P Value |
| FEV1 | 1.00 | −0.45 | 0.05* | |
| CRP | −0.44 | 0.05* | 1.00 | |
| PLXND1 | −0.38 | 0.10 | 0.42 | 0.07 |
| HCA112 | 0.42 | 0.07 | 0.21 | 0.38 |
| ADAM9 | −0.35 | 0.13 | 0.57 | 0.01* |
| HPSE | 0.08 | 0.72 | 0.54 | 0.01* |
| CSPG2 | −0.31 | 0.18 | 0.45 | 0.05* |
| IL32 | 0.09 | 0.71 | −0.47 | 0.04* |
| CD64 | 0.03 | 0.88 | 0.49 | 0.03* |
| CD36 | −0.25 | 0.30 | 0.31 | 0.18 |
| CD163 | −0.42 | 0.06 | 0.48 | 0.03* |
| TLR2 | −0.18 | 0.44 | 0.49 | 0.02* |
Definition of abbreviation: CRP = C-reactive protein.
P values were calculated by paired t test on log-transformed values.
TABLE 5.
SPEARMAN'S RANK CORRELATION COEFFICIENTS (rS) FOR GENES, CIRCULATING BLOOD NEUTROPHILS, AND TOTAL WHITE BLOOD CELLS PRE– AND POST–ANTIBIOTIC THERAPY
| PMNS
|
WBC
|
|||
|---|---|---|---|---|
| Variable | rs | P Value | rs | P Value |
| PMNS | 1.00 | 0.87 | <0.01* | |
| WBC | 0.87 | <0.01* | 1.00 | |
| PLNX | 0.42 | 0.07 | 0.39 | 0.09 |
| HCA112 | −0.04 | 0.89 | −0.25 | 0.29 |
| ADAM9 | 0.47 | 0.04* | 0.28 | 0.23 |
| HPSE | 0.25 | 0.31 | −0.02 | 0.93 |
| CSPG2 | 0.54 | 0.02* | 0.31 | 0.19 |
| IL32 | −0.60 | <0.01* | −0.31 | 0.18 |
| CD64 | 0.13 | 0.61 | −0.08 | 0.75 |
| CD36 | 0.42 | 0.07 | 0.21 | 0.37 |
| CD163 | 0.50 | 0.03* | 0.36 | 0.12 |
| TLR2 | 0.24 | 0.32 | 0.21 | 0.37 |
Definition of abbreviations: PMNS = polymorphonuclear leukocytes; WBC = white blood cells.
P values were calculated by paired t test on log-transformed values.
Plasma Cytokine Changes in the Resolution of the Acute Pulmonary Exacerbations in CF
Multiple analytes were measured in aliquots of plasma with the Luminex system (Luminex Corp., Austin, TX), taken from the same blood specimens from which PBMCs were isolated before and after antibiotic therapy. Table 6 depicts mean cytokine measurements at both time points and their associated P values. After the Bonferroni's correction for multiple tests, the change in granulocyte colony–stimulating factor fails to reach significance at the P ≤ 0.05 level.
TABLE 6.
CYSTIC FIBROSIS PLASMA CYTOKINE LEVELS PRE– AND POST–ANTIBIOTIC THERAPY IN DEVELOPMENT GROUP
| Cytokine | Pre-RX | Post-RX | P Value* |
|---|---|---|---|
| IL-1α | 0.15 ±0.15 | <0.01 | 0.30 |
| IL-1β | 1.09 ± 0.41 | 0.88 ± 0.50 | 0.60 |
| IL-1RA | 2,608 ± 1412 | 1,637 ± 667 | 0.20 |
| IL-2 | 0.95 ± 0.34 | 0.45 ± 0.29 | 0.30 |
| IL-4 | <0.01 | 0.78 ± 0.40 | 0.08 |
| IL-5 | 0.93 ± 0.64 | 0.20 ± 0.11 | 0.08 |
| IL-6 | 7.34 ± 3.39 | 3.57 ± 2.03 | 0.10 |
| IL-8 | 5.37 ± 2.25 | 2.56 ± 1.31 | 0.10 |
| IL-10 | 3.24 ± 2.74 | 1.50 ± 0.55 | 0.90 |
| IL-13 | 1,118 ± 388 | 1,539 ± 495 | 0.30 |
| IL-17 | 2.38 ± 1.36 | 0.15 ± 0.15 | 0.08 |
| TNF-α | 2.99 ± 0.83 | 3.04 ± 0.68 | 0.80 |
| IFN-γ | 0.04 ± 0.04 | 0.37 ± 0.20 | 0.80 |
| G-CSF | 29.50 ± 10.30 | 12.27 ± 4.96 | 0.02 |
| GM-CSF | 0.14 ± 0.08 | 0.45 ± 0.33 | 0.06 |
| MIP-1β | 39.10 ± 13.50 | 31.51 ± 8.83 | 0.20 |
| VEGF | 10.75 ± 2.82 | 10.57 ± 2.76 | 0.50 |
| RANTES | 3216 ± 433 | 4,268 ± 776 | 0.30 |
Definition of abbreviations: G-CSF = granulocyte colony–stimulating factor ; GM-CSF = granulocyte-macrophage colony–stimulating factor ; MIP-1β = macrophage inflammatory protein-1β; Pre-RX = before antibiotic therapy; Post-RX = after antibiotic therapy; RANTES = regulated upon activation, normal T-cell expressed and secreted; TNF-α = tumor necrosis factor-α; VEGF = vascular endothelial growth factor.
Values are mean ± SEM.
P values were calculated by paired t test on log-transformed values.
Prospective Evaluation of the CF Therapeutic Signature in a Validation Cohort of Patients with CF
Expression of the 10 identified classifier genes was evaluated in an independent population of patients with CF and acute pulmonary exacerbations via real-time RT-PCR (n = 14). PBMC isolation and FEV1 measurements were performed on adult patients with CF at the initiation and termination of antibiotic therapy for acute pulmonary exacerbations using identical methods as for the development cohort. This validation group of patients was heterogeneous in terms of sputum microbiology and variable use of systemic steroids and was ultimately more representative of a realistic clinical setting (Table 2). Although many patients were treated with steroids, as opposed to none in the initial study group, the presence of steroids did not significantly alter gene expression between steroid-treated and steroid-naive patients for 9 of 10 genes (by multi-way ANOVA). Only transcript changes in HPSE significantly differed between steroid-treated and steroid-naive patients (P = 0.03). FEV1 improved significantly between pre- and post-measurement in this cohort (P = 0.003 by paired t test). All patients in the validation cohort had a higher FEV1% predicted at the second measurement. However, two patients in this cohort with severe airway limitation (FEV1 <25% predicted at both measurements) manifested an improvement of 100 ml or less in FEV1% predicted and were considered nonresponders based on clinical parameters. Both patients had poor outcomes: one was readmitted with recurrent pulmonary symptoms in 6 days and the second was never discharged due to a persistently poor clinical response to antibiotics and underwent lung transplant 43 days later. These nonresponders were not included in the logistic regression analysis of the validation cohort, because the model was predicated on identifying genes associated with therapeutic response. In a univariate analysis of individual gene changes within the validation cohort, five genes were significantly changed in responders after antibiotic therapy, based on log-transformed gene expression values measured by real-time RT-PCR: CD36, P = 0.002; CD64, P = 0.007; PLXND1, P = 0.01; CSPG2, P = 0.002; and TLR2, P = 0.05.
Diagnostic Value of the CF Therapeutic Signature in Association with Percentage of Change in FEV1
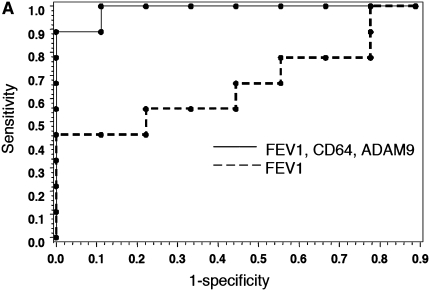
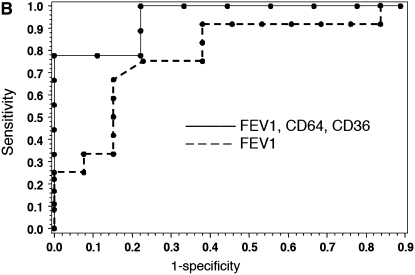
Using multivariate analysis, we evaluated the combined explanatory power of FEV1 in combination with gene expression values. The addition of two gene measurements to the regression substantially increased the explanatory power of the model. ROC curves, shown in Figures 4A and 4B, demonstrate the sensitivity and specificity of gene expression values for assessing treatment response. Juxtaposition of ROC curves demonstrates the additional discriminatory capacity of gene measurements, compared with FEV1 alone, to diagnose resolution of airway inflammation. In the development cohort, four different pairs of genes were combined with FEV1 (c-statistic = 0.75, 0.85, and 0.88, respectively) to give an overall better diagnostic performance than FEV1 alone (c-statistic = 0.58). In the validation cohort, four different pairs of genes combined with FEV1 (c-statistic ranging from 0.73 to 0.80) demonstrated a more robust performance than did FEV1 alone (c-statistic = 0.69). Table 7 demonstrates diagnostic values (area under ROC curves) for pairs of PBMC gene markers and their independent association with improvement in FEV1, in both development and validation groups. As demonstrated by P values less than 0.05 for each gene in the model, the genes contributed meaningful diagnostic information not available from FEV1 alone. The use of two gene markers combined with FEV1 was an optimal pairing, as less than two lost significance and greater than two did not improve significance by logistic function. From the 10-gene signature, 7 genes were strong independent predictors for treatment response in the regression model for the two groups. Three genes significantly improved diagnostic value (P ≤ 0.05) in both cohorts. In the first cohort, the gene pair with the highest predictive accuracy, based on c-statistic, as well as statistical significance for each gene in the model, consisted of CD64 and ADAM9 (c = 0.88). In the validation cohort, the best predictive pair, in terms of c-statistic and significance in all genes, was represented by CD64 and CD36 (c = 0.80). The independent, significant explanatory power contributed by these genes to both patient groups demonstrates that gene expression values from the CF therapeutic signature enhance the predictive discriminating value of FEV1 alone.
Figure 4.
Receiver operating characteristic (ROC) curves of gene combinations and FEV1. ROC curves depict the fraction of true-positive (sensitivity) and false-positive (1 − specificity) values plotted for RNA transcripts and FEV1% predicted. A perfect test is indicated by an area under curve = 1. A test with no discriminatory value has an area under curve = 0.50. (A) ROC curves for the development cohort depicting discriminatory capacity of FEV1% predicted alone versus FEV1 with CD64 and ADAM9 transcripts (c-statistic = 0.88). (B) ROC curves for validation group comparing FEV1% predicted alone with FEV1 plus CD64 and CD36 transcripts (c-statistic = 0.80).
TABLE 7.
DIAGNOSTIC VALUE OF CYSTIC FIBROSIS THERAPEUTIC SIGNATURE FOR RESOLUTION OF AIRWAY INFLAMMATION
| Area under ROC Curve |
P Values (logistic regression)
|
|||
|---|---|---|---|---|
| Markers | Gene 1 | Gene 2 | FEV1 | |
| Development cohort | ||||
| CD64 ADAM9 | 0.88 | 0.0003 | 0.03 | 0.89 |
| CD64 PLXND1 | 0.85 | 0.003 | 0.01 | 0.19 |
| CSPG2 ADAM9 | 0.85 | 0.01 | 0.005 | 0.85 |
| CSPG2 CD163 | 0.75 | 0.008 | 0.03 | 0.64 |
| FEV1 alone | 0.58 | 0.004 | ||
| Validation cohort | ||||
| CD64 CD36 | 0.8 | 0.02 | 0.03 | 0.5 |
| CD64 CSPG2 | 0.77 | 0.02 | 0.004 | 0.6 |
| CD64 IL32 | 0.76 | 0.03 | 0.02 | 0.5 |
| CD36 ADAM9 | 0.73 | 0.02 | 0.03 | 0.4 |
| FEV1 alone | 0.69 | 0.25 | ||
Definition of abbreviations: ROC = receiver operating characteristic.
DISCUSSION
In CF lung tissue, mononuclear cells that originate from the peripheral blood are abundantly present at sites of airway injury. Histologic studies of CF airways highlight mononuclear cells as the predominant cell population in areas of cartilaginous destruction and identify lymphocytes as the majority cell type infiltrating airway submucosa (21). (22). The consistent change in expression of a small group of PBMC genes among patients with CF with heterogeneous genotypes, lung function, sputum microbiology, and antibiotic regimens creates a strong basis on which to study the participation of these cells in CF lung disease pathogenesis.
This study is unique in several aspects. It is the first study of its kind to use circulating leukocyte transcripts to assess response to therapy in CF lung disease. By correlation with known inflammatory markers, CRP and circulating neutrophil counts, the gene predictors are plausible markers of inflammation. Second, the gene transcript changes demonstrate reproducibility across two patient groups and discriminatory capacity to differentiate between acutely ill and subsequently treated patients. Third, our regression model demonstrates that mRNA changes in circulating leukocytes add meaningful diagnostic information to FEV1 in assessing treatment response in acute pulmonary exacerbations.
Genes identified in this study may serve to generate additional hypotheses concerning disease pathogenesis, inflammatory regulation, and treatment response in the CF airway. Seven of the 10 genes (and the proteins they encode) featured in this article (IL32, HPSE, ADAM9, PLXND1, HCA112, CSPG2, and CD163) have not previously been linked to CF lung disease. It is important to note that these genes are not specific to CF and have varying roles in other conditions characterized by pathologic pulmonary inflammation, including asthma and pneumonia (23, 24). The fact that these genes encode for proteins implicated in inflammatory processes lends biologic plausibility to naming them as potential markers of the resolution of CF airway infection and inflammation. As a group, these genes represent functions of immune recognition and response, phagocytosis, and matrix degradation. TLR2 represents a central pattern recognition receptor for the innate response against bacterial infection. IL-32 is a newly described tumor necrosis factor–inducible intracellular cytokine (25). An inducer of proinflammatory cytokines, IL-32 induces blood monocyte differentiation to macrophages, with subsequent phagocytic activity for live bacteria (26). The expression of IL-32 by inflamed luminal epithelia may facilitate differentiation of blood monocytes infiltrating infected lung (26). Three surface-receptor genes participate in phagocytosis. CD64, or FcγRIA, mediates receptor-mediated endocytosis of IgG-antigen complexes in macrophages (27). CD36, a scavenger receptor, mediates macrophage uptake of oxidized low-density lipoprotein, as well as serving as a surface receptor for thrombospondin-1 (28–30). CD163 serves as a macrophage cell surface hemoglobin scavenger receptor and was recently shown to be highly predictive of mortality in pneumococcal bacteremia (24, 31). Degradative enzymes, including heparanase, ADAM9, and versican, facilitate extravasation of leukocytes to inflamed tissues. In persistent airway inflammation, this process may culminate in marked and irreversible structural injury to lung parenchyma by modification of extracellular matrix architecture (32).
This study was designed to measure markers that change with aggressive treatment of a pulmonary exacerbation, the current best therapy for reduction of acute increases in airway infection and inflammation in CF lung disease. This design has several advantages. First, our design closely parallels a clinical trial sequence in which a treatment would be tested for its effect on decreasing inflammation. Second, our gene signature is not pathogen limited. Our study population suffered from infection with a representative variety of bacterial pathogens, typical of the CF population as a whole. When patients with CF were treated for pulmonary exacerbations, expression of most of the candidate genes more closely resembled the normal control subjects (Figure 2), which supports the biologic roles of these genes as markers of decreased infection and inflammation. Furthermore, one-half of the genes were specific for exacerbation among patients with CF, meaning that, when compared with patients with stable CF, values differed significantly before treatment but not after treatment. This includes the three genes (CD64, ADAM9, and CD36) that were most highly diagnostic of therapeutic response. One of the genes that added significant explanatory power to the regression model in both patient groups, ADAM9, is highly representative of immediate postexacerbation transcriptional changes and is the only gene whose postexacerbation expression remained significantly different from both normal PBMC and control subjects with stable CF.
Simultaneous measurements of respiratory physiology and plasma markers allowed for statistical comparisons of multiple outcomes measures. The significant correlation of more than one-half of the genes in the CF therapeutic signature with changes in CRP, a well-characterized serum marker of inflammation, in addition to correlations to neutrophil counts, further strengthens the association of these genes with inflammatory processes. The concomitant evaluation of a relatively comprehensive panel of plasma cytokines in the development cohort did not demonstrate statistical significance. Post-treatment cytokine measurements were lower than pretreatment measurements; however, the specific cytokines reduced differed from patient to patient, making none of them broadly applicable as a marker for exacerbation resolution in a cohort of this size. Although this study was not powered to detect a significant difference in plasma cytokine concentrations, our group and others are currently conducting larger-scale longitudinal studies of blood, sputum, and plasma markers in the resolution of acute exacerbations, which will allow simultaneous analysis of markers reflecting compartmental and circulating inflammation.
The complexity of CF airway inflammation is such that local events within airways may be heterogeneous and compartmentalized across lung segments. Although PBMCs are easily accessible, they may not completely reflect inflammatory processes occurring in all pulmonary compartments. It would be of great interest to compare the response of PBMC RNA transcripts to direct measures of airway inflammation.
Although many markers of airway inflammation can be detected in the sputum, multiple previous studies (particularly of this size) have not been able to detect changes in sputum concentrations of inflammatory markers associated with the treatment of an acute pulmonary exacerbation (33–38). Thus, to directly compare identified changes in PBMC expression with specific inflammatory markers in the sputum, sputum markers of inflammation were not analyzed as a part of this study.. The compartmental quality of local pulmonary events may also reflect, in part, the relative steroid independence of the gene expression change in the steroid-exposed validation cohort. In animal models, glucocorticoids typically suppress local blood flow at sites of inflammation, reduce vascular permeability, and reduce leukocyte migration. These responses would likely have greater effect on local transcription profiles at the site of inflammation rather than on circulating expression (39–41). In addition, whereas corticosteroid therapy did not affect expression of 9 of 10 genes, the study was not powered to detect significant steroid-modulated differences in gene expression.
A systemic marker of lung inflammation has many advantages, because blood can be obtained from subjects of any age and disease severity, and may reflect the status of inflammation throughout the lung, rather than one segment, as is assessed by bronchoalveolar lavage. The current study establishes that the use of PBMC expression permits the assessment of leukocyte activity, concomitantly with the functional information represented by FEV1, to reflect treatment of an acute pulmonary exacerbation. This analysis is sensitive, inexpensive, and obtained from tissue that is easily accessible in pediatric and adult populations, and has the potential to be performed in a clinical laboratory.
The identification of relevant biomarkers could have more immediate clinical implications for several large subpopulations of patients with CF. In children, airway infection and inflammation can occur as early as 4 weeks of age (42). Computed tomography radiographic studies have demonstrated considerable bronchiectasis and parenchymal abnormalities in children with normal lung function (43). Sensitive markers could allow for a personalized strategy of antiinfectious and antiinflammatory treatment in young children, with the ability to rapidly monitor outcomes from these interventions. Conversely, in patients with severe lung destruction and multiple antibiotic drug–resistant organisms, assessment of response to a particular treatment is often difficult, given day-to-day variability in disease and the degree of irreversibility in airway damage. A sensitive measure of leukocyte activities could be used to gauge response to therapeutics when clinical response lags far behind.
It is important to acknowledge the small size of our study. In addition, two patients provided samples in both cohorts, defining two separate pulmonary exacerbations for each individual. A larger trial is underway by our group to define the optimal combination of genes to characterize a successful therapeutic response. Validation will also require determination of intra- and interindividual variability, responders versus nonresponders, and corticosteroid effect. Herein, we have identified potential gene biomarkers, from which various gene combinations with FEV1 serve as accurate predictors of resolution of pulmonary exacerbations, with greater sensitivity, specificity, and discriminatory capacity than FEV1 alone. Future separate studies will be necessary to determine whether genes have applicability as outcomes measures in clinical trials for antiinflammatory drugs. The utilization of leukocyte gene expression to bolster standard physiologic outcome variables has major implications in terms of the conduct of small trials in rare human diseases.
Supplementary Material
Supported by National Institutes of Health grants K08HL74512 (M.T.S.), U01 HL081335 (F.J.A.), HL090991 (J.A.N.), the Max and Yetta Karasik Foundation (J.A.N.), the Rebecca Runyon Bryan Chair for Cystic Fibrosis (J.A.N.), and the Cystic Fibrosis Foundation (M.T.S. and J.A.N.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200803-387OC on August 21, 2008
Conflict of Interest Statement: M.T.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.J.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.A.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.M.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.D.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.W.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.D.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. F.J.A. received $2,500 from Medpro, Inc., for teaching a course. J.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.A.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Konstan MW. Therapies aimed at airway inflammation in cystic fibrosis. Clin Chest Med 1998;19:505–513. [DOI] [PubMed] [Google Scholar]
- 2.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007;109:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakonarson H, Bjornsdottir US, Halapi E, Bradfield J, Zink F, Mouy M, Helgadottir H, Gudmundsdottir AS, Andrason H, Adalsteinsdottir AE, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci USA 2005;102:14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutherford RM, Staedtler F, Kehren J, Chibout SD, Joos L, Tamm M, Gilmartin JJ, Brutsche MH. Functional genomics and prognosis in sarcoidosis: the critical role of antigen presentation. Sarcoidosis Vasc Diffuse Lung Dis 2004;21:10–18. [PubMed] [Google Scholar]
- 5.Shlobin OA, West EE, Lechtzin N, Miller SM, Borja M, Orens JB, Dropulic LK, McDyer JF. Persistent cytomegalovirus-specific memory responses in the lung allograft and blood following primary infection in lung transplant recipients. J Immunol 2006;176:2625–2634. [DOI] [PubMed] [Google Scholar]
- 6.Bull TM, Coldren CD, Moore M, Sotto-Santiago SM, Pham DV, Nana-Sinkam SP, Voelkel NF, Geraci MW. Gene microarray analysis of peripheral blood cells in pulmonary arterial hypertension. Am J Respir Crit Care Med 2004;170:911–919. [DOI] [PubMed] [Google Scholar]
- 7.Padoan R, Brienza A, Crossignani RM, Lodi G, Giunta A, Assael BM, Granata F, Passarella E, Vallaperta PA, Xerri L. Ceftazidime in treatment of acute pulmonary exacerbations in patients with cystic fibrosis. J Pediatr 1983;103:320–324. [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin FJ, Matthews WJ Jr, Strieder DJ, Sullivan B, Taneja A, Murphy P, Goldmann DA. Clinical and bacteriological responses to three antibiotic regimens for acute exacerbations of cystic fibrosis: ticarcillin-tobramycin, azlocillin-tobramycin, and azlocillin-placebo. J Infect Dis 1983;147:559–567. [DOI] [PubMed] [Google Scholar]
- 9.Richard DA, Nousia-Arvanitakis S, Sollich V, Hampel BJ, Sommerauer B, Schaad UB. Oral ciprofloxacin vs. intravenous ceftazidime plus tobramycin in pediatric cystic fibrosis patients: comparison of antipseudomonas efficacy and assessment of safety with ultrasonography and magnetic resonance imaging. Cystic Fibrosis Study Group. Pediatr Infect Dis J 1997;16:572–578. [DOI] [PubMed] [Google Scholar]
- 10.Ordonez CL, Henig NR, Mayer-Hamblett N, Accurso FJ, Burns JL, Chmiel JF, Daines CL, Gibson RL, McNamara S, Retsch-Bogart GZ, et al. Inflammatory and microbiologic markers in induced sputum after intravenous antibiotics in cystic fibrosis. Am J Respir Crit Care Med 2003;168:1471–1475. [DOI] [PubMed] [Google Scholar]
- 11.Ferkol T, Rosenfeld M, Milla CE. Cystic fibrosis pulmonary exacerbations. J Pediatr 2006;148:259–264. [DOI] [PubMed] [Google Scholar]
- 12.Liou TG, Adler FR, Cahill BC, FitzSimmons SC, Huang D, Hibbs JR, Marshall BC. Survival effect of lung transplantation among patients with cystic fibrosis. JAMA 2001;286:2683–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liou TG, Adler FR, Fitzsimmons SC, Cahill BC, Hibbs JR, Marshall BC. Predictive 5-year survivorship model of cystic fibrosis. Am J Epidemiol 2001;153:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer-Hamblett N, Rosenfeld M, Emerson J, Goss CH, Aitken ML. Developing cystic fibrosis lung transplant referral criteria using predictors of 2-year mortality. Am J Respir Crit Care Med 2002;166:1550–1555. [DOI] [PubMed] [Google Scholar]
- 15.Elkins MR, Robinson M, Rose BR, Harbour C, Moriarty CP, Marks GB, Belousova EG, Xuan W, Bye PT. A controlled trial of long-term inhaled hypertonic saline in patients with cystic fibrosis. N Engl J Med 2006;354:229–240. [DOI] [PubMed] [Google Scholar]
- 16.Burns JL, Van Dalfsen JM, Shawar RM, Otto KL, Garber RL, Quan JM, Montgomery AB, Albers GM, Ramsey BW, Smith AL. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J Infect Dis 1999;179:1190–1196. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW, Rosenstein BJ, Smith AL, Wohl ME. Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. The Pulmozyme Study Group. N Engl J Med 1994;331:637–642. [DOI] [PubMed] [Google Scholar]
- 18.Saiman L, Marshall BC, Mayer-Hamblett N, Burns JL, Quittner AL, Cibene DA, Coquillette S, Fieberg AY, Accurso FJ, Campbell PW III. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 2003;290:1749–1756. [DOI] [PubMed] [Google Scholar]
- 19.Yankaskas JR, Marshall BC, Sufian B, Simon RH, Rodman D. Cystic fibrosis adult care: consensus conference report. Chest 2004;125:1S–39S. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrino R, Viegi G, Brusasco V, Crapo RO, Burgos F, Casaburi R, Coates A, van der Grinten CP, Gustafsson P, Hankinson J, et al. Interpretative strategies for lung function tests. Eur Respir J 2005;26:948–968. [DOI] [PubMed] [Google Scholar]
- 21.Hubeau C, Lorenzato M, Couetil JP, Hubert D, Dusser D, Puchelle E, Gaillard D. Quantitative analysis of inflammatory cells infiltrating the cystic fibrosis airway mucosa. Clin Exp Immunol 2001;124:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durieu I, Peyrol S, Gindre D, Bellon G, Durand DV, Pacheco Y. Subepithelial fibrosis and degradation of the bronchial extracellular matrix in cystic fibrosis. Am J Respir Crit Care Med 1998;158:580–588. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q, Martin RJ, Lafasto S, Efaw BJ, Rino JG, Harbeck RJ, Chu HW. Toll-like receptor 2 down-regulation in established mouse allergic lungs contributes to decreased mycoplasma clearance. Am J Respir Crit Care Med 2008;177:720–729. [DOI] [PubMed] [Google Scholar]
- 24.Moller HJ, Moestrup SK, Weis N, Wejse C, Nielsen H, Pedersen SS, Attermann J, Nexo E, Kronborg G. Macrophage serum markers in pneumococcal bacteremia: prediction of survival by soluble CD163. Crit Care Med 2006;34:2561–2566. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity 2005;22:131–142. [DOI] [PubMed] [Google Scholar]
- 26.Netea MG, Lewis EC, Azam T, Joosten LA, Jaekal J, Bae SY, Dinarello CA, Kim SH. Interleukin-32 induces the differentiation of monocytes into macrophage-like cells. Proc Natl Acad Sci USA 2008;105:3515–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigo WW, Jin X, Blackley SD, Rose RC, Schlesinger JJ. Differential enhancement of dengue virus immune complex infectivity mediated by signaling-competent and signaling-incompetent human Fcgamma RIA (CD64) or FcgammaRIIA (CD32). J Virol 2006;80:10128–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok CF, Juan CC, Ho LT. Endothelin-1 decreases CD36 protein expression in vascular smooth muscle cells. Am J Physiol Endocrinol Metab 2007;292:E648–E652. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira V, van Dijk KW, Groen AK, Vos RM, van der Kaa J, Gijbels MJ, Havekes LM, Pannekoek H. Macrophage-specific inhibition of NF-kappaB activation reduces foam-cell formation. Atherosclerosis 2007;192:283–290. [DOI] [PubMed] [Google Scholar]
- 30.Doyen V, Rubio M, Braun D, Nakajima T, Abe J, Saito H, Delespesse G, Sarfati M. Thrombospondin 1 is an autocrine negative regulator of human dendritic cell activation. J Exp Med 2003;198:1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss M, Schneider EM. Soluble CD163: an age-dependent, anti-inflammatory biomarker predicting outcome in sepsis. Crit Care Med 2006;34:2682–2683. [DOI] [PubMed] [Google Scholar]
- 32.Vaday GG, Lider O. Extracellular matrix moieties, cytokines, and enzymes: dynamic effects on immune cell behavior and inflammation. J Leukoc Biol 2000;67:149–159. [DOI] [PubMed] [Google Scholar]
- 33.Downey DG, Brockbank S, Martin SL, Ennis M, Elborn JS. The effect of treatment of cystic fibrosis pulmonary exacerbations on airways and systemic inflammation. Pediatr Pulmonol 2007;42:729–735. [DOI] [PubMed] [Google Scholar]
- 34.Wolter JM, Rodwell RL, Bowler SD, McCormack JG. Cytokines and inflammatory mediators do not indicate acute infection in cystic fibrosis. Clin Diagn Lab Immunol 1999;6:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham S, McColm JR, Mallinson A, Boyd I, Marshall TG. Duration of effect of intravenous antibiotics on spirometry and sputum cytokines in children with cystic fibrosis. Pediatr Pulmonol 2003;36:43–48. [DOI] [PubMed] [Google Scholar]
- 36.Karpati F, Hjelte FL, Wretlind B. TNF-alpha and IL-8 in consecutive sputum samples from cystic fibrosis patients during antibiotic treatment. Scand J Infect Dis 2000;32:75–79. [DOI] [PubMed] [Google Scholar]
- 37.Reix P, Bellon G, Bienvenu J, Pavirani A, Levrey-Hadden H. Cytokine pattern in cystic fibrosis patients during antibiotic therapy and gene therapy using adenoviral vector. Eur Cytokine Netw 2002;13:324–330. [PubMed] [Google Scholar]
- 38.Nixon LS, Yung B, Bell SC, Elborn JS, Shale DJ. Circulating immunoreactive interleukin-6 in cystic fibrosis. Am J Respir Crit Care Med 1998;157:1764–1769. [DOI] [PubMed] [Google Scholar]
- 39.Perretti M, Ahluwalia A. The microcirculation and inflammation: site of action for glucocorticoids. Microcirculation 2000;7:147–161. [PubMed] [Google Scholar]
- 40.Oda T, Katori M. Inhibition site of dexamethasone on extravasation of polymorphonuclear leukocytes in the hamster cheek pouch microcirculation. J Leukoc Biol 1992;52:337–342. [DOI] [PubMed] [Google Scholar]
- 41.Wahl SM, Feldman GM, McCarthy JB. Regulation of leukocyte adhesion and signaling in inflammation and disease. J Leukoc Biol 1996;59:789–796. [DOI] [PubMed] [Google Scholar]
- 42.Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW. Early pulmonary inflammation in infants with cystic fibrosis. Am J Respir Crit Care Med 1995;151:1075–1082. [DOI] [PubMed] [Google Scholar]
- 43.de Jong PA, Nakano Y, Lequin MH, Mayo JR, Woods R, Pare PD, Tiddens HA. Progressive damage on high resolution computed tomography despite stable lung function in cystic fibrosis. Eur Respir J 2004;23:93–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.