Abstract
The F1F0-adenosine triphosphate (ATP) synthase rotational motor synthesizes most of the ATP required for living from adenosine diphosphate, Pi, and a proton electrochemical gradient across energy-transducing membranes of bacteria, chloroplasts, and mitochondria. However, as a reversible nanomotor, it also hydrolyzes ATP during de-energized conditions in all energy-transducing systems. Thus, different subunits and mechanisms have emerged in nature to control the intrinsic rotation of the enzyme to favor the ATP synthase activity over its opposite and commonly wasteful ATPase turnover. Recent advances in the structural analysis of the bacterial and mitochondrial ATP synthases are summarized to review the distribution and mechanism of the subunits that are part of the central rotor and regulate its gyration. In eubacteria, the ε subunit works as a ratchet to favor the rotation of the central stalk in the ATP synthase direction by extending and contracting two α-helixes of its C-terminal side and also by binding ATP with low affinity in thermophilic bacteria. On the other hand, in bovine heart mitochondria, the so-called inhibitor protein (IF1) interferes with the intrinsic rotational mechanism of the central γ subunit and with the opening and closing of the catalytic β-subunits to inhibit its ATPase activity. Besides its inhibitory role, the IF1 protein also promotes the dimerization of the bovine and rat mitochondrial enzymes, albeit it is not essential for dimerization of the yeast F1F0 mitochondrial complex. High-resolution electron microscopy of the dimeric enzyme in its bovine and yeast forms shows a conical shape that is compatible with the role of the ATP synthase dimer in the formation of tubular the cristae membrane of mitochondria after further oligomerization. Dimerization of the mitochondrial ATP synthase diminishes the rotational drag of the central rotor that would decrease the coupling efficiency between rotation of the central stalk and ATP synthesis taking place at the F1 portion. In addition, F1F0 dimerization and its further oligomerization also increase the stability of the enzyme to natural or experimentally induced destabilizing conditions.
Keywords: F1F0 ATPase, F1F0 ATP synthase, IF1, Inhibitor protein, Epsilon, Dimeric, Rotation, Interface, Cristae, Regulation, F1 ATPase
Introduction
The mitochondrial adenosine triphosphate (ATP) synthase is a ubiquitous motor enzyme that provides most of the cellular chemical energy in the form of ATP to fuel all kinds of work in biological nature. This motor functions as a coupling factor between the condensation of adenosine diphosphate (ADP) and Pi that takes place at its catalytic F1-ATPase portion and proton flow through the transmembranous F0-proton channel that consumes energy from electrochemical proton gradients. According to the well-established chemiosmotic theory, this proton gradient is established by oxidative or photosynthetic electron transfer chains of the plasma membrane of bacteria, the inner mitochondrial membrane, and the thylakoid membranes of chloroplasts. Because of thermodynamic and mechanical reversibility, the F1F0-ATP synthase becomes a proton-pumping F1F0-ATPase under conditions of partial or total collapse of the proton gradient; for instance, during anoxia in bacteria where it works as a primary pump to drive secondary transporters, during ischemia in mitochondria, or under dark conditions in chloroplasts. In all these systems, different subunit structures control gyration of the central stalk by favoring rotation in the ATP synthase turnover direction. Chloroplast ATP synthase possesses a unique disulfide bridge in the γ subunit that controls rotation of the central stalk; however, only the structures of the bacterial and the bovine enzymes are reviewed here.
Important new information such as the dimerization of the ATP synthase and the role of the inhibitor protein (IF1) in this process are also reviewed and used to propose a model of the structure of the ATP synthase dimmer that explains the inhibitory and dimerizing roles of IF1. This model also explains how dimerization of the ATP synthase may confer a higher stability and efficiency of the dimeric enzyme to synthesize ATP. This also sheds light on how F1F0 dimerization promotes formation of tubular cristae membrane structures in mitochondria after further polymerization.
Structure and Rotational Mechanism of the ATP Synthase
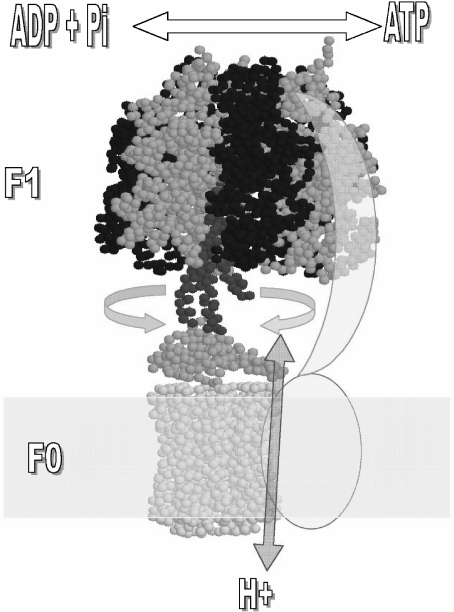
The catalytic part of the enzyme is a water-soluble portion (F1) that can be released in vitro from the membrane, retaining its capacity to hydrolyze ATP (F1-ATPase) [1]. ATP synthesis occurs in the whole F1F0 when the energy derived from proton conduction through the F0 membrane channel is combined with the nucleotide (Mg-ADP) and Pi binding energies [2–5] to drive the release of newly synthesized ATP from each of the three alternating catalytic sites of F1. The coupling between F1 and F0 is critical for efficient ATP synthesis to occur and major progress in the understanding of this coupling mechanism has been achieved. Several approaches at different laboratories showed that a central rotor actually gyrates relative to a stator that holds the catalytic subunits; this rotation induces the alternating binding, catalysis, and product release from three catalytic sites of F1 (for reviews, see [5–9]). These studies also indicated that the γ subunit, together with ε and the ring of 9–15 c subunits of F0, form the rotor in the central part of the enzyme. The more direct evidence demonstrating this rotational movement was the observation by fluorescence microscopy of rotation of a fluorescent actin filament attached to the γ, ε, or c subunits of immobilized F1 and F1F0 complexes [10–13]. These experiments established that the rotor of the enzyme is formed by the central γ–ε–c9 − 15 domain. This core rotor–stator structure is preserved in bacterial and mitochondrial ATP synthases and is shown in Fig. 1 indicating the reversible rotational mechanism of the enzyme.
Fig. 1.
Rotor–stator subunit distribution in the mitochondrial F1F0-ATP synthase. Only the core subunits present in bacteria and mitochondria are shown for simplicity. Rotating subunits are shown in red (γ), orange (ε), and yellow (ring of c subunits) whereas static subunits are in dark blue (β), blue (α), and cyan (subunits a and b). The arrows indicate the reversible rotation of γ–ε–c subunits relative to α and β catalytic subunits of F1 that takes place during ATP synthesis (“clockwise” or right direction) and hydrolysis (“counterclockwise” or left direction). Bidirectional proton flow at the c-ring–sub a interface occurs associated with the gyration of the rotor as indicated by the red arrow. The second-stalk structure is simplified as two cyan subunits (a and b) that work as stator to anchor the catalytic α3β3 to the membranous a subunit. Image is generated in RasMol 2.6 from the mitochondrial F1F0 structure of S. cerevisiae (PDB code 1Q01) and edited as shown
The rotary mechanism of the enzyme implies that, in the central stalk, the γ subunit (together with ε and c9 − 15 subunits) rotates relative to a stator where the catalytic α–β interfaces are held. Therefore, this stator must be somehow anchored to an F0 subunit at the lipid bilayer. A peripheral second stalk was literally “invoked” and found by means of high-resolution electron microscopy studies [14, 15] and by cross-linking of α, δ, b, and a subunits along this peripheral second stalk [7]. The anchoring part of the second stalk with the α subunit of F1 has been solved by nuclear magnetic resonance (NMR) for the Escherichia coli enzyme [16], whereas most of the structure of the second stalk of the bovine mitochondrial enzyme has been resolved by X-ray crystallography [17]. Thus, the whole picture of the simplest ATP synthase of E. coli involves a stator formed by (α–β)3, δ, b2, and a subunits and a central rotor formed by the γ–ε–c9 − 15 domain. This core rotor–stator structure of bacterial ATP synthase becomes more complex with about twice as many different subunits present in chloroplasts and mitochondria. Besides the core subunits and structure of the EF1F0 motor, there are six to eight additional or “supernumerary” subunits that are well described in mitochondrial yeast and bovine ATP synthases. These subunits are d, e, f, g, F6 (h in yeast), A6L (8 in yeast), the inhibitor protein (IF1), and mitochondrial subunit ε which does not have a bacterial counterpart (see below and Table 1). Three additional F0 proteins are also found in the yeast enzyme (see legend of Table 1). The roles of these additional subunits are related to regulation and oligomerization of the ATP synthase as will be described below.
Table 1.
Subunit composition of the E. coli and mitochondrial ATP synthases
| E. coli subunit | Bovine subunit | Yeast subunit | |
|---|---|---|---|
| F1 | α3 | α3 | α3 |
| β3 | β3 | β3 | |
| γ1 | γ1 | γ1 | |
| δ1 | OSCP1 | Sub 51 | |
| ε1 | δ1 | δ1 | |
| – | ε1 | ε1 | |
| – |
IF
|
IF
|
|
| F0 | A1 | Sub. 61 | Sub. 61 |
| B2 | b1 | Sub 41 | |
| C9 − 15 | c9 − 15 | Sub 910 | |
| – | d1 | Sub 71 | |
| – | e1 | e1 | |
| – | f1 | f1 | |
| – | g1 | g1 | |
| – | F61 | h1 | |
| – | A6L1 | Sub 81 |
There are eight and 16 different subunits in the bacterial and bovine enzymes, respectively. Subunits are accommodated according to their corresponding homologs. For example, E. coli δ and ε correspond to bovine OSCP and δ, respectively. Corresponding subunit stoichiometries are indicated as superscripts. The enzyme from yeast (S. cerevisiae) contains at least three additional subunits, namely i, j, and k.
The Central Stalk is Part of the ATP Synthase Rotor
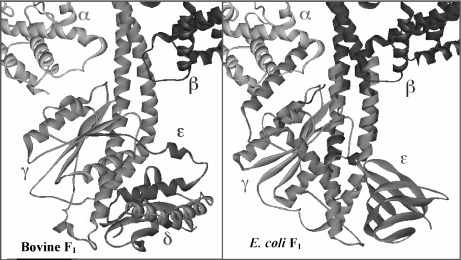
Crystallographic studies have solved most of the central stalk structure in E. coli (EF1) [18] and bovine mitochondrial (MF1) F1-ATPases [19–22]. The tertiary structure and orientation of the globular domain of the γ subunit is very similar in both species (Fig. 2), and it is in agreement with previous cross-linking data obtained with the enzyme from E. coli. However, the ε subunit of E. coli (bovine δ) was found far away from the cross-linking distance to the α or β subunits but closer to the F0 subunit c in the crystals of the yeast [23] and bovine [20–22] enzymes. It was therefore unclear how ε could cross-link with α or β subunits in the native enzyme as found before, until another X-Ray diffraction analysis was made with a soluble γ–ε complex from E. coli [18]. In this study, the structure of soluble ε was very different from the bovine δ subunit F1. The soluble ε subunit associated with γ was found rotated in relation to the vertical axis of the central stalk and extending its two C-terminal helices toward the C-termini of α and β. This position placed the appropriate residues in cross-linking distance [18] (Fig. 2). The conclusion is therefore that ε experiences dramatic changes in conformation that are important for its role as an inhibitor of the ATPase activity of the enzyme, controlling the rate of rotation of the central stalk. Engineered cross-linking in the E. coli F1F0 complex entrapped these two conformations of the ε subunit [24]. Interestingly, when the C-terminus of ε is compacted as an antiparallel α-helix coil with its N-terminal β-sheet domain, the F1F0-ATPase activity is enhanced and the enzyme is coupled during ATP hydrolysis and synthesis. However, when the C-terminus of ε extends toward F1 (as shown in Fig. 2), ATP hydrolysis is inhibited but ATP synthesis remains unaffected [24]. In agreement with this work, the structure of the γ–ε domain in the E. coli F1-ATP synthase [25] was found very similar to that of the isolated subunits [18]. Furthermore, it has been found that the ratchet mechanism of ε can be regulated by ATP binding in some bacteria [9, 26]. When ATP is bound, the closed conformation is stabilized, thus favoring rotation of the central stalk in the ATPase direction; conversely, at low ATP concentrations, ε is unable to bind ATP, and therefore the extended conformation is favored, thus leaving the enzyme prone to rotate into the ATP synthase turnover. This model is supported with the recent crystal structure of the Bacillus PS3 subunit ε with ATP associated to the C-terminus of this subunit [27]. In summary, these studies show that ε works as an ATP sensor in bacteria that posses a novel ATP-binding motif in this subunit [26–29].
Fig. 2.
Comparison of the bovine (left) and E. coli (right) F1F0-ATP synthases at the central stalk domain. Crystallographic structures of MF1 and EF1 central stalks are shown in the same orientation. Homologous subunits are drawn in the same color, γ (blue), ε subunits (green). For clarity, only one α subunit (red) and one β subunit (yellow) are shown. The structure shown on the right is a composite of the E. coli γ–ε structure [18] and the bovine MF1 structure [19], constructed by aligning segments of γ present in both structures. Segments of MF1 γ subunit are shown in darker blue, and those of E. coli γ subunit are shown in lighter blue. This figure was modified from an original courtesy of Dr. Andrew J.W. Rodgers
Besides the control of rotation described so far for the bacterial enzymes by the ε subunit, it is important to introduce a novel inhibitory 11-kDa protein that we recently found in the ATP synthase of the α-proteobacteria Paracoccus denitrificans (Morales-Ríos et al. 2008, submitted). The ATP synthase from P. denitrificans has been only described functionally as the fastest ATP synthase and the slowest ATPase found to date [30]; however, it has never been isolated until we addressed this issue. This novel inhibitory 11-kDa protein is present in F1-ATPase and F1F0-ATPase preparations obtained from P. denitrificans membranes, and it will likely add a novel control and inhibitory mechanism to the α-proteobacteria family where the open reading frame exists. Importantly, the ε subunit of this enzyme does not inhibit the ATPase activity of the F1-ATPase or F1F0-ATPase complexes in P. denitrificans (Morales-Ríos et al. 2008, submitted). Thus, the 11-kDa protein will add a novel control mechanism to the ATP synthases, in addition to the classical inhibitory mechanisms of bacterial, chloroplast, and mitochondrial F1F0 complexes. Unidirectional functioning of ATP synthase turnover has been also described for another bacterial enzyme of the thermoalkaliphilic type, Bacillus sp. TA2.A1 [31]. However, instead of additional regulatory proteins, unique polar interactions at the rotor–stator interface of the F1 subunits allow almost exclusively unidirectional rotation in the ATP synthase direction for this enzyme [31]. To our knowledge, only two other bacterial proteins have been found encoded in the atp operon in addition to the eight core subunits of bacterial F1F0 (α, β, γ, δ, ε, a, b, c); these two proteins are encoded by the unc-I and urf-6 genes that correspond, respectively, to an assembly factor of the c-ring [32] and to majastridin, a cytosolic protein nonassociated with the Rhodospirillum blasticus ATP synthase [33]. In contrast, the gene encoding the 11-kDa protein of P. denitrificans is located upstream to both atp operons (one for F0 and another for F1 subunits) already sequenced on chromosome II of P. denitrificans (see Morales-Ríos et al. 2008, submitted, and the following link: http://genome.jgi-psf.org/finished_microbes/parde/parde.home.htm). Therefore, it seems that the 11-kDa regulatory protein that we found in the F1F0 complex of P. denitrificans is one of the first, if not the first, supernumerary subunit added to bacterial ATP synthases as an exogenous gene of the atp operon (formerly known as the unc operon). This 11-kDa protein therefore emerged during α-proteobacterial evolution and previous to the endosymbiotic event from which mitochondria emerged.
Supernumerary Subunits and Their Role in the Regulation and Dimerization of the Mitochondrial ATP Synthase
Most of the supernumerary subunits in the mitochondrial enzyme correspond to membrane proteins associated with the F0 proton channel. These additional subunits are d, e, f, g, F6, and A6L. Subunits d and F6 are part of the second stalk, and A6L is a membrane protein of F0 that is essential for the assembly of subunit 6, the one that forms the proton-conducting interface with the c9 − 15 ring. On the other hand, the roles of some of these subunits were recently unveiled by studies in yeast showing that subunits e and g are needed to form F1F0 dimers in situ [34, 35]. However, an unexpected result was the finding that genetic removal of these e and g subunits deformed the inner mitochondrial membrane and the classical cristae transformed into concentric membrane layers inside enlarged mitochondria [35]. This demonstrated that dimerization of the ATP synthase is not an artifact of detergent extraction as suspected but a natural and important biological process that improves the ATP synthase activity and the stability of the enzyme. Thus, besides dimerizing to improve somehow the ATP synthesis reaction, the dimeric enzyme also promotes mitochondrial cristae formation, thus optimizing the overall process of oxidative phosphorylation.
Two supernumerary subunits are part of the mitochondrial F1, namely ε and the so-called inhibitor protein (IF1). Bovine ε is different with its bacterial homonym; it is a 5.7-kDa protein, whereas that of E. coli is 15 kDa in size (Table 1 and Fig. 2). Bovine ε stabilizes the structure of the central stalk by interacting with the globular part of γ (Figs. 2, 3, and 4). Closely interacting with ε, bovine subunit δ has a similar structure to that of its homologous E. coli ε. However, neither bovine δ or ε subunits inhibit the ATPase activity of bovine F1F0; together, they form a compact and noninhibitory structure at the central stalk, in contrast to the flexible structure of E. coli ε (Fig. 2) [20]. The bovine and E. coli δ subunits also correspond to different proteins. E. coli δ does not form part of the central stalk as the bovine δ does (Table 1, Fig. 2). E. coli δ is the connection between the “tip” of the F1 subunit α and the “top” of the peripheral second stalk (reviewed in [7]). It is homologous to bovine oligomycin sensitivity conferring protein (OSCP) which also interacts with subunits of the second stalk (see Fig. 4, and [17, 36–38]). As mentioned above, the connection between E. coli δ and α subunits has been resolved by NMR [16].
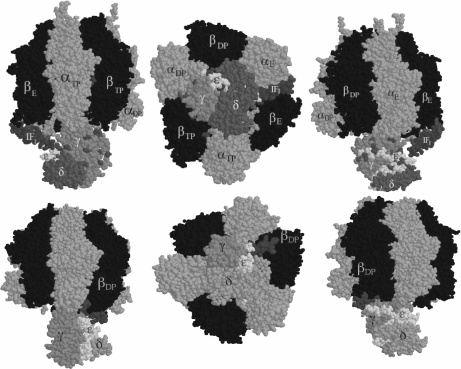
Fig. 3.
Model and crystal structures of the F1–IF1 complex from bovine heart mitochondria. Top three panels, our model: we positioned the IF1 N-terminal domain at an entrance-binding site (αE–βE interface) at about 12-Å cross-linking distance from γ and ε subunits as we found [44]. From the side (left and right) and “bottom” (center) views, it was clearly shown and proposed for the first time that IF1 is close enough to the rotor of the enzyme to block gyration of the central stalk as part of its inhibitory mechanism [44]. Bottom panels, the crystal structure from the F1–IF1 crystal with a nondimerizing fragment of IF1 [21]: the same IF1 N-terminal side was resolved and observed actually bound to the γ subunit at an αDP–βDP interface [21, 22]. The top structure depicts the entrance site of IF1, whereas the bottom structure shows the final inhibited structure where IF1 is locked into the same αE–βE interface that became αDP–βDP after two counterclockwise 120° gyration steps (shift from top to bottom panels). IF1 therefore inhibits rotation of the central stalk and the opening and closing conformational changes of a single catalytic interface
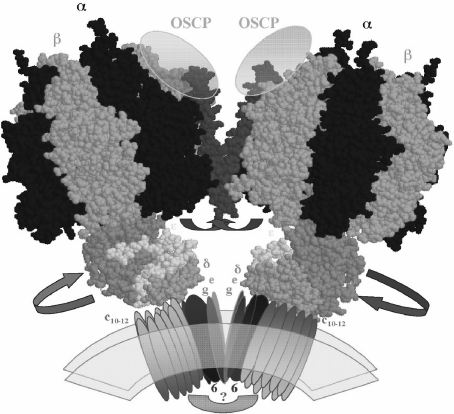
Fig. 4.
Model of the dimeric-mitochondrial ATP synthase: possible localization of the IF1 protein and its movements to allow rotation of the central stalk during ATP synthesis. The model depicts the overall shape of the dimeric ATP synthase molecule that we observed for the bovine mitochondrial enzyme [54]. The dimeric interface involves F0 subunits (e and g) and two protein bridges, one at the F0–F0 side of unknown composition (question mark) and another at the F1–F1 interface where the second stalks (not shown for clarity) and the IF1 protein (red) are likely to be located. The C-terminal side of the IF1 molecule is assumed to cross the dimer interface and to stabilize the dimer by interacting with subunits OSCP [65] and possibly subunits of the second stalk. The N-terminal inhibitory domain that in the absence of the proton gradient blocks rotation of the central stalk by entering at an α–β–γ interface (Fig. 2) is removed from this position and exposed into the media after establishment of a transmembrane proton gradient, thus allowing rotation of the central stalk during ATP synthesis. The F1 structures were constructed from the bovine F1-DCCD coordinates available (PDF code 1E79)
The Inhibitor Protein (IF1) and its Inhibitory and Dimerizing Roles on the Mitochondrial ATP Synthase
A key regulatory subunit absent in bacterial or chloroplast F1F0 is the mitochondrial inhibitor protein (IF1). Since its first isolation in 1963 by Pullman and Monroy [39], this protein was shown to inhibit the ATPase activity of the catalytic F1 part. This protein is therefore crucial to preventing the hydrolysis of newly synthesized ATP in conditions of low membrane potential in mitochondria. Upon membrane energization, IF1 is believed to be relocated from its inhibitory site into an unknown position within [40, 41] or outside the F1F0 complex [42, 43], therefore allowing ATP synthesis to occur. In de-energized or uncoupled conditions, such as ischemia, the bovine IF1 is productively associated with the enzyme, inhibiting the ATPase turnover of the F1I or F1F0I complexes. However, this protein allows the rotational ATP synthesis turnover during energization of mitochondrial membranes. Therefore, IF1 is an important physiological regulator of the functioning of the ATP synthase.
Although the location of the endogenous IF1 in the whole F1F0I-ATP synthase remains unknown, on the basis of the available structural studies, we proposed a model of the binding site for the IF1 that would explain the inhibitory role of IF1 [44]. It is well known that the inhibitory domain of IF1 lies on the N-terminal side of the molecule [45–47]; thus, according to our cross-linking data showing for the first time a relatively short distance (12 Å) between IF1 and the γ and ε subunits, we placed the N-terminal inhibitory side in a cross-linking position of about 12 Å from the γ and ε subunits [44]. Figure 3 (top panels) shows this position of the N-terminal side on a cleft formed by the βE catalytic subunit and the γ–ε part of the central rotor; lysine residues that are at 12-Å cross-linking distance are shown in purple. This β E–γ–ε cleft was the wider binding site available for entrance of the IF1 N-terminal side into the rotor–stator interface. It is clearly demonstrated from different perspectives that the binding of the N-terminal side of IF1 in this position interferes not only with the conformational changes of the β subunits, as proposed before [19, 48], but also with the intrinsic rotation of the central stalk. Thus, we supported and proposed a novel mechanism of action for this protein that was later confirmed by the elegant crystal structure of the reconstituted dimeric F1–IF1 complex [22]. In the latter complex, the central coiled α-helixes of the γ subunit that extend along the central pseudosymmetry axis of the F1-ATPase particle were found not only in proximity but in actual close contact with the its N-terminal side of IF1 [21, 22] (see Fig. 3, bottom panels). The IF1 was found locked into a βDP–γ cleft rather than in a βE–γ–ε interface, as we originally proposed [44]. This shows that what we found by cross-linking and model building was the entrance site of IF1 into the F1-ATPase particle and that two further angular movements of the γ subunit of 120° lock the IF1 into the βDP form of the catalytic subunit that previously received the IF1 in its open βE conformation (see Fig. 3 and [21]). Thus, the mechanism of action of IF1 as inhibitor involves blocking the rotation of the central stalk and inhibiting the opening–closing conformational changes of the catalytic β subunit that leads to substrate binding, catalysis, and product release from F1.
Besides showing the close-up view of the IF1–γ interaction, the isolation and resolution of the F1–IF1 crystal structure also showed that reconstitution of recombinant IF1 induces dimerization of the soluble F1-ATPase particles in the expected 1:1 IF1–F1 stoichiometry [22, 49]. Earlier blue native polyacrylamide gel electrophoresis analyses of mitochondria showed a dimeric ATP synthase species that appears after mitochondrial solubilization with several detergents [50] and that some F0 subunits such as e and g are essential for ATP synthase dimerization [34, 35]. Thus, the question emerged of whether the IF1 participates in the dimerization of the whole F1F0-ATP synthase in mitochondria, besides dimerizing the soluble F1-ATPase in vitro [49]. Initially, several groups found that genetic or physical removal of the yeast or bovine IF1, respectively, did not prevent F1F0 dimerization; thus, it was concluded that IF1 does not participate in the homodimerization of the whole F1F0 [51, 52]. However, because the yeast IF1 protein lacks most of the C-terminal dimerizing domain and it is much less prone to dimerize [53], it was conceivable that the role of IF1 in dimerization of the ATP synthase might be excluded from the yeast enzyme but present in the bovine and rat mitochondrial enzymes. Besides, the results where IF1 removal did not change the dimer to monomer ratio of the bovine ATP synthase were obtained in the presence of triton X-110 where the F1F0-ATPase is inactive [51]. Therefore, we reassessed the role of IF1 as a dimerizing factor of the bovine and rat mitochondrial enzymes in digitonin-extraction conditions where the dimeric and monomeric forms of the F1F0 complex are functional [54]. Besides, instead of looking to the decrease in the dimeric ATP synthase after IF1 removal, we looked for the recovery or promotion of the dimeric species after reconstitution of increasing amounts of IF1 into submitochondrial particles. With this approach, we demonstrated that removal of IF1 dissociated the whole ATP synthase into monomers of high ATPase activity, and the reconstitution of IF1 into SMP brought a partial recovery of the dimer content of the SMP extract accompanied by an overall inhibition of the ATPase activity [55]. Interestingly, the larger ATP synthase oligomers were also partially recovered by IF1 reconstitution [55], suggesting that IF1 participates in the formation of aggregation states of the ATP synthase larger than the ATP synthase dimer. Thus, it seems that yeast IF1 is not essential for F1F0 dimerization in Saccharomyces cerevisiae simply because it is much less prone to dimerize since it lacks most of the C-terminal coiled-coil dimerizing domain [53]. The latter seems therefore essential for bovine and rat IF1 to promote and/or stabilize the dimer and higher oligomer structures of the mitochondrial ATP synthase. This is consistent with previous findings where it has been shown that IF1 confers structural stability to the F1F0I and F1I complexes during high-pressure denaturation that leads to dissociation of oligomeric species [56]. In line with the dimerizing role of IF1, it has been recently shown that, among other metabolic effects, the overexpression of IF1 increases the amount of mitochondrial cristae, and its downregulation decreases the number of cristae in mitochondria of cultured cells [57]. Taken together, these studies confirm that, besides the inhibitory role of IF1, it is also an important factor that stabilizes dimerization and further oligomerization of the mitochondrial ATP synthase, thus promoting formation of mitochondrial cristae as detailed below.
Structure of the Dimeric-Mitochondrial ATP Synthase: Improving Rotational Catalysis, Adding Stability, and Giving Shape to Mitochondrial Cristae
In Fig. 4, the possible quaternary structure of the dimeric bovine F0F1I complex is depicted according to the structural data available from crystallographic [20], genetic [58–60], subunit association [36, 37], cross-linking [38, 44, 61–65], and protease accessibility [38, 66, 67] evidence. How does this model accommodate the inhibitory and dimerizing functions of IF1 in the F1F0 dimer? We assumed a crossed IF1 structure at the dimer interface, given that we also resolved by high-resolution electron microscopy the dimeric F1F0 and found a conical homodimeric molecule containing a protein bridge at the F1–F1 interface [54]. In this model, the IF1 N-terminal side is located at the rotor–stator interface in inhibitory position, whereas the C-terminal side of IF1 crosses the dimer interface and interacts with the opposite monomer probably through the OSCP subunit at the top of the side stalk as found by cross-linking evidence [65]. This model explains both the inhibitory and dimerizing roles of IF1; however, both functions of IF1 would require some further distortion from the fully extended helix observed in the isolated IF1 to a bent or random coil conformation. This distortion is necessary to introduce the N-terminal side of IF1 into the βDP–γ interface as shown by the crystal structures [21, 22]. In the F1F0 dimer model, we used a crystal IF1 conformer that is bent in the middle of the IF1 protein, and this fits better at this interface than the extended IF1 dimer conformers [68]. Similar crossed IF1 dimeric structures have been observed in the IF1 crystal [68]; this arrangement would be different from the observed antiparallel coiled-coil dimer of isolated IF1 [68]. It was necessary to invoke this crossed structure because the distance between the N-terminal inhibitory domains in the IF1–IF1 extended dimer is about 60 Å [68], whereas the F1–F1 distance observed in the soluble F1–IF1 dimer [22] or in the (F1F0I)2 dimer is ≤10 Å [54]. This implies that the IF1 dimer must bend or cross somehow to be accommodated at the F1–F1 interface of the ATP synthase dimer that had an angle of about 40° which gives its conical shape.
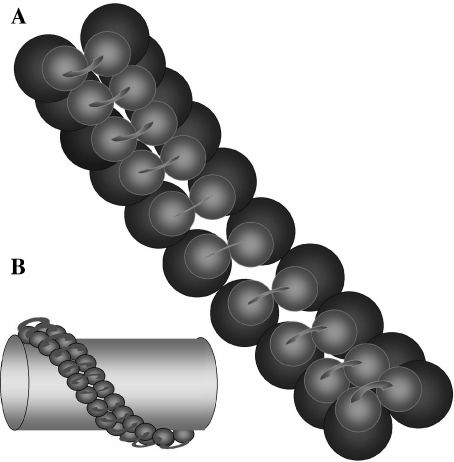
On the other hand, it is also noted that, besides the bovine dimeric ATP synthase [54], other similar dimeric structures have been subsequently observed by electron microscopy in S. cerevisiae and Polytomella sp mitochondria. The latter species has a unique second-stalk composition and is therefore nonrepresentative of other mitochondrial ATP synthases [69]; however, in both cases, the dimeric structure adopted two angles of about 40° and 70° [69]. Dudkina and colleagues [70] named their open (70°) structure as the “true dimer”, and our compact (40°) structure as a “pseudo-dimer”; furthermore, they also suggest that, in line with other reports, only their open “true” dimer actually participates in cristae formation [71, 72]. However, their dimer structures have several drawbacks: (1) their image averages are collected not by hand but automatically by image analysis software; in consequence, a large proportion of their dimer particles lack one or both of the F1-portions, showing that their preparation is largely unstable compared to our preparation, which contains mostly complete F1F0 structures. (2) The larger detergent concentration used to isolate the enriched open dimers [69, 70] decreases the dimer yield and stability, and, importantly, it also decreases the functional coupling between F1 and F0; in contrast, our dimer enriched at lower detergent concentrations preserves essentially full oligomycin sensitivity, i.e., F1F0 functional coupling (Minauro-Sanmiguel and García-Trejo, unpublished results). This parameter has not been reported in the preparations enriched with the open (70°) and unstable dimer; it would not be surprising to find there a decreased F0 inhibition. (3) There is emerging evidence from others [73] and from our recent studies with the yeast F1F0 dimer (not shown) indicating that both structures (open and closed) coexist with a wide distribution of dimers showing different angles after detergent extraction, but there is no clear evidence indicating which protein or factor is controlling the opening or closing of the dimer angle. Although IF1 is not essential for IF1 dimerization in yeast [52], the possibility remains that the shift from an extended to a compact conformation of the IF1 dimer could participate in determining the angle of dimeric F1F0. Therefore, we conclude that there is no reason to name arbitrarily the open or closed conformations as “pseudo” or “true” dimers; instead, we propose to refer to them just as “open” (≅70°) and “closed” (≅40°) dimers, with the understanding that the dimer population actually spreads through all angles between these values. Regardless of the observed angle values after detergent extraction, two major dimeric species correlate well with two distinct dimeric interfaces at the F0 side that have been found in yeast F1F0 [74]; these two interfaces would build a helical polymer of dimers that wraps and gives shape to the tubular cristae of mitochondria [75], as it is currently proposed (Fig. 5).
Fig. 5.
Possible arrangement of the ATP synthase helical polymer that wraps and gives shape to the mitochondrial tubular cristae. a Inner view of the polymer from the interior of the cristae (membrane is transparent); the green spheres represent the F0 channels connected by the protein bridge we observed [54]; the blue spheres are the F1 heads. b The ATP synthase polymer is depicted from the outer surface of a single cristae (yellow). The F1 particles are in blue and connected with a protein bridge that could be composed by the IF1 and second-stalk subunits. The original model is from Allen et al. [75]
In summary, the dimeric structure of the F1F0 ATP synthase is stabilized by the so-called inhibitor protein (IF1) in the mitochondria of complex organisms such as rat or cow. Literally, on the other hand, the conserved N-terminal side of IF1 inhibits the F1F0-ATPase activity by entering through the open catalytic αE–βE interface in a cleft formed by β–γ–ε subunits. With the IF1 bound at this interface, the F1-ATPase carries out two 120° gyrations of the central stalk and the N-terminal side of IF1 locks at the βDP–αDP–γ interface, completely blocking rotation of the central stalk and the opening and closing of the catalytic sites. A further question that emerges is, how this deep inhibitory interaction of IF1 with the rotor–stator interface of F1 is reversed in the presence of the mitochondrial electrochemical proton gradient to allow ATP synthesis turnover? We are currently addressing this question by limited proteolysis experiments; interestingly, we observed that the N-terminal side of IF1 becomes exposed to the media upon membrane energization, whereas the C-terminal side of IF1 becomes shielded to proteolysis, indicating that it hides behind another F1F0 subunit (García-Trejo et al., unpublished). We propose here how this might happen in the dimeric F1F0 structure of bovine heart mitochondria. Upon membrane energization, the C-terminal side of IF1 might become occluded between OSCP or second-stalk subunits at the dimer interface, whereas the N-terminal inhibitory domain is released from the αDP–βDP–γ cleft where it is bound, thus restoring rotation of the central stalk and the opening–closing conformational changes of the β subunits that are essential for F1 catalysis. In this model, second-stalk subunits are not depicted for clarity, but they should contribute significantly to the dimer interface, as shown for the yeast H subunit (bovine subunit F6, see [75]). Once formed, the dimer structure seems more stable and in better shape to resist the rotational drag of the continuous gyration of the central stalk than its dimeric form (Fig. 4). In other words, the monomeric enzyme could lose coupling energy by rotating as a rigid body following the angular drag of the rotor; this would hardly occur in a dimerized or oligomerized ATP synthase. Indeed, it has been proposed that the rotational drag of each monomer promotes closer F0–F0 interactions in the dimer as observed by atomic force microscopy in the dimeric enzyme [71]. It can also be questioned whether dimerization actually increases the coupling efficiency of the enzyme, given that the monomeric bacterial enzyme is already highly efficient as a coupling factor; indeed, the most efficient and practically unidirectional ATP synthases described so far are those of P. denitrificans [30] and of a thermoalkaliphilic bacterium [31]. However, it is also recalled that, in α-proteobacteria and even in eubacteria such as E. coli, it has been described that the rotary turnover of the F1 portion undergoes slippage from the proton conduction through F0 under conditions of low ADP and Pi concentrations [76, 77]. This slipping has not been observed for the mitochondrial enzyme, probably because the rotor and stator interfaces of each monomer interact more efficiently in the dimeric or oligomeric forms of the enzyme. In this line, we are currently collecting evidence to respond to the question of whether the dimeric enzyme possesses a higher stability and better efficiency as ATP synthase in comparison with its monomeric species; preliminary results indicate that it is actually the case. Together with its role in formation of the mitochondrial cristae, these studies and models shed light on the mechanisms by which the F1F0-ATP synthase becomes not only the most efficient nanomotor in nature by its regulation in bacteria and by its dimerization in mitochondria, but also becomes a dimeric building block of a hypothetical helical polymer that wraps and gives shape to the mitochondrial cristae (Fig. 5).
Acknowledgements
This work and our work reviewed here were supported by the CONACyT grants No. I32903-N, J34744-N, and V43814-M.
Abbreviations
- EF1, EF1F0
Escherichia coli F1 and F1F0 complexes
- EM
electron microscopy
- F1F0
the whole ATP synthase complex with its catalytic (F1) and proton channel (F0) parts
- F1F0I
the whole ATP synthase containing its physiological inhibitor protein (IF1)
- IF1
the intrinsic inhibitor protein of the mitochondrial ATP synthase
- MF1, MF1F0
bovine heart mitochondrial F1 and F1F0 complexes, respectively
- NMR
nuclear magnetic resonance spectroscopy
References
- 1.Penefsky, H.S., Pullman, M.E., Datta, A., Racker, E.: Partial resolution of the enzymes catalyzing oxidative phosphorylation. II. Participation of a soluble adenosine triphosphatase in oxidative phosphorylation. J. Biol. Chem. 235, 3330–3336 (1960) [PubMed]
- 2.Souid, A.K., Penefsky, H.S.: Mechanism of ATP synthesis by mitochondrial ATP synthase from beef heart. J. Bioenerg. Biomembranes 26, 627–630 (1994). doi:10.1007/BF00831537 [DOI] [PubMed]
- 3.García, J.J., Gómez-Puyou, A., Maldonado, E., Tuena de Gómez-Puyou, M.: Acceleration of unisite catalysis of mitochondrial F1-adenosinetriphosphatase by ATP, ADP and pyrophosphate—hydrolysis and release of the previously bound [gamma-32P]ATP. Eur. J. Biochem. 249(2), 622–229 (1997). doi:10.1111/j.1432-1033.1997.00622.x [DOI] [PubMed]
- 4.García, J.J., Capaldi, R.A.: Unisite catalysis without rotation of the gamma-epsilon domain in Escherichia coli F1-ATPase. J. Biol. Chem. 273(26), 15940–15945 (1998). doi:10.1074/jbc.273.26.15940 [DOI] [PubMed]
- 5.García, J.J.: The F0F1-ATP synthase: binding energy, coupling and rotational catalysis. In: Pandalai, S.G. (ed.) Recent Research Developments in Bioenergetics, p. 41. Transworld Research Network, Trivandrum (2000)
- 6.Boyer, P.D.: The ATP synthase a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997). doi:10.1146/annurev.biochem.66.1.717 [DOI] [PubMed]
- 7.Capaldi, R.A., Schulenberg, B., Murray, J., Aggeler, R.: Cross-linking and electron microscopy studies of the structure and functioning of the Escherichia coli ATP synthase. J. Exp. Biol. 203, 29–33 (2000) [DOI] [PubMed]
- 8.Iino, R., Rondelez, Y., Yoshida, M., Noji, H.: Chemomechanical coupling in single-molecule F-type ATP synthase. J. Bioenerg. Biomembranes 37, 451–454 (2005). doi:10.1007/s10863-005-9489-5 [DOI] [PubMed]
- 9.Feniouk, B.A., Yoshida, M.: Regulatory mechanisms of proton-translocating F(O)F (1)-ATP synthase. Results Probl. Cell Differ. 45, 279–308 (2008). doi:10.1007/400_2007_043 [DOI] [PubMed]
- 10.Noji, H., Yasuda, R., Yoshida, M., Kinosita, K., Jr.: Direct observation of the rotation of F1-ATPase. Nature 386, (6622), 299–302 (1997). doi:10.1038/386299a0 [DOI] [PubMed]
- 11.Kato-Yamada, Y., Noji, H., Yasuda, R., Kinosita, K., Jr., Yoshida, M.: Direct observation of the rotation of epsilon subunit in F1-ATPase. J. Biol. Chem. 273(31), 19375–19377 (1998). doi:10.1074/jbc.273.31.19375 [DOI] [PubMed]
- 12.Omote, H., Iwamoto-Kihara, A., Ueda, T., Yanagida, Y., Wada, M., Futai, M.: Mechanical rotation of the c subunit oligomer in ATP synthase (F0F1): direct observation. Science 286(5445), 1722–1724 (1999). doi:10.1126/science.286.5445.1722 [DOI] [PubMed]
- 13.Tsunoda, S.P., Aggeler, R., Yoshida, M., Capaldi, R.A.: Rotation of the c subunit oligomer in fully functional F1Fo ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 98(3), 898–902 (2001). doi:10.1073/pnas.031564198 [DOI] [PMC free article] [PubMed]
- 14.Wilkens, S., Capaldi, R.: Electron microscopic evidence of two stalks linking the F1 and F0 parts of the Escherichia coli ATP synthase. Biochim. Biophys. Acta 1365(1–2), 93–97 (1998). doi:10.1016/S0005-2728(98)00048-6 [DOI] [PubMed]
- 15.Karrasch, S., Walker, J.E.: Novel features in the structure of bovine ATP synthase. J. Mol. Biol. 290(2), 379–384 (1999). doi:10.1006/jmbi.1999.2897 [DOI] [PubMed]
- 16.Wilkens, S., Borchardt, D., Weber, J., Senior, A.: Structural characterization of the interaction of the $\updelta$ and $\upalpha$ subunits of the Escherichia coli F1F0-ATP synthase by NMR spectroscopy. Biochemistry 44(35), 11786–11794 (2005) [DOI] [PubMed]
- 17.Dickson, V.K., Silvester, J.A., Fearnley, I.M., Leslie, A.G., Walker, J.E.: On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 25(12), 2911–2918 (2006). doi:10.1038/sj.emboj.7601177 [DOI] [PMC free article] [PubMed]
- 18.Rodgers, A.J., Wilce, M.C.: Structure of the gamma-epsilon complex of ATP synthase. Nat. Struct. Biol. 7(11), 1051–1054 (2000). doi:10.1038/80975 [DOI] [PubMed]
- 19.Abrahams, J.P., Leslie, A.G., Lutter, R., Walker, J.E.: Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370(6491), 621–628 (1994) [DOI] [PubMed]
- 20.Gibbons, C., Montgomery, M.G., Leslie, A.G.W., Walker, J.E.: The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat. Struct. Biol. 7(11), 1055–1061 (2000). doi:10.1038/80981 [DOI] [PubMed]
- 21.Gledhill, J.R., Montgomery, M.G., Leslie, A.G., Walker, J.E.: How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U. S. A. 104(40), 15671–15676 (2007). doi:10.1073/pnas.0707326104 [DOI] [PMC free article] [PubMed]
- 22.Cabezón, E., Montgomery, M.G., Leslie, A.G., Walker, J.E.: The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 10(9), 744–750 (2003). doi:10.1038/nsb966 [DOI] [PubMed]
- 23.Stock, D., Leslie, A.G.W., Walker, J.E.: Molecular architecture of the rotary motor in ATP synthase. Science 286(5445), 1700–1705 (1999). doi:10.1126/science.286.5445.1700 [DOI] [PubMed]
- 24.Tsunoda, S., Rodgers, A.J.W., Aggeler, R., Wilce, M.C.J., Yoshida, M., Capaldi, R.A.: Large conformational changes of the epsilon subunit in the bacterial F1F0 ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. U. S. A. 98(12), 6560–6564 (2001). doi:10.1073/pnas.111128098 [DOI] [PMC free article] [PubMed]
- 25.Hausrath, A.C., Capaldi, R.A., Matthews, B.W.: The conformation of the epsilon- and gamma-subunits within the Escherichia coli F(1) ATPase. J. Biol. Chem. 276(50), 47227–47232 (2001). doi:10.1074/jbc.M107536200 [DOI] [PubMed]
- 26.Iino, R., Murakami, T., Iizuka, S., Kato-Yamada, Y., Suzuki, T., Yoshida, M.: Real-time monitoring of conformational dynamics of the epsilon subunit in F1-ATPase. J. Biol. Chem. 280(48), 40130–40134 (2005). doi:10.1074/jbc.M506160200 [DOI] [PubMed]
- 27.Yagi, H., Kajiwara, N., Tanaka, H., Tsukihara, T., Kato-Yamada, Y., Yoshida, M., Akutsu, H.: Structures of the thermophilic F1-ATPase epsilon subunit suggesting ATP-regulated arm motion of its C-terminal domain in F1. Proc. Natl. Acad. Sci. U. S. A. 104(27), 11233–11238 (2007). doi:10.1073/pnas.0701045104 [DOI] [PMC free article] [PubMed]
- 28.Kato-Yamada, Y.: Isolated epsilon subunit of Bacillus subtilis F1-ATPase binds ATP. FEBS Lett. 579(30), 6875–6878 (2005). doi:10.1016/j.febslet.2005.11.036 [DOI] [PubMed]
- 29.Yoshida, M., Kato-Yamada, Y.: Role of the epsilon subunit of thermophilic F1-ATPase as a sensor for ATP. J. Biol. Chem. 282(52), 37618–37623 (2007). doi:10.1074/jbc.M707509200 [DOI] [PubMed]
- 30.Zharova, T.V., Vinogradov, A.D.: Proton-translocating ATP-synthase of Paracoccus denitrificans: ATP-hydrolytic activity. Biochemistry (Mosc.) 68(10), 1101–1108 (2003). doi:10.1023/A:1026306611821 [DOI] [PubMed]
- 31.Stocker, A., Keis, S., Vonck, J., Cook, G.M., Dimroth, P.: The structural basis for unidirectional rotation of thermoalkaliphilic F1-ATPase. Structure 15(8), 904–914 (2007). doi:10.1016/j.str.2007.06.009 [DOI] [PubMed]
- 32.Suzuki, T., Ozaki, Y., Sone, N., Feniouk, B.A., Yoshida, M.: The product of uncI gene in F1F0-ATP synthase operon plays a chaperone-like role to assist c ring assembly. Proc. Natl. Acad. Sci. U. S. A. 104(52), 20776–20781 (2007). doi:10.1073/pnas.0708075105 [DOI] [PMC free article] [PubMed]
- 33.Brosché, M., Kalbina, I., Arnfelt, M., Benito, G., Karlsson, B.G., Strid, Å.: Occurrence, overexpression and partial purification of the protein (majastridin) corresponding to the URF6 gene of the Rhodobacter blasticus atp operon. Eur. J. Biochem. 255(1), 87–92 (1998). doi:10.1046/j.1432-1327.1998.2550087.x [DOI] [PubMed]
- 34.Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R.A., Schagger, H.: Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17(24), 7170–7178 (1998). doi:10.1093/emboj/17.24.7170 [DOI] [PMC free article] [PubMed]
- 35.Paumard, P., Vaillier, J., Coulary, B., Schaeffer, J., Soubannier, V., Mueller, D.M., Brèthes, D., di Rago, J.P., Velours, J.: The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21(3), 221–230 (2002). doi:10.1093/emboj/21.3.221 [DOI] [PMC free article] [PubMed]
- 36.Collinson, I.R., Van Raaij, M.J., Runswick, M.J., Fearnley, I.M., Skehel, J.M., Orriss, G.L., Miroux, B., Walker, J.E.: ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J. Mol. Biol. 242(4), 408–421 (1994) [DOI] [PubMed]
- 37.Collinson, I.R., Skehel, J.M., Fearnley, I.M., Runswick, M.J., Walker, J.E.: The F1F0-ATPase complex from bovine heart mitochondria: the molar ratio of the subunits in the stalk region linking the F1 and F0 domains. Biochemistry 35(38), 12640–12646 (1996). doi:10.1021/bi960969t [DOI] [PubMed]
- 38.Xu, T., Zanotti, F., Gaballo, A., Raho, G., Papa, S.: F1 and F0 connections in the bovine mitochondrial ATP synthase: the role of the of alpha subunit N-terminus, oligomycin-sensitivity conferring protein (OCSP) and subunit d. Eur. J. Biochem. 267(14), 4445–4455 (2000). doi:10.1046/j.1432-1327.2000.01492.x [DOI] [PubMed]
- 39.Pullman, M.E., Monroy, G.C.: A naturally occurring inhibitor of mitochondrial adenosine triphosphatase. J. Biol. Chem. 238, 3762–3769 (1963) [PubMed]
- 40.Dreyfus, G., Gómez-Puyou, A., Tuena de Gómez-Puyou, M.: Electrochemical gradient induced displacement of the natural ATPase inhibitor protein from mitochondrial ATPase as directed by antibodies against the inhibitor protein. Biochem. Biophys. Res. Commun. 100(1), 400–406 (1981). doi:10.1016/S0006-291X(81)80110-6 [DOI] [PubMed]
- 41.Sánchez-Bustamante, V.J., Darszon, A., Gómez-Puyou, A.: On the function of the natural ATPase inhibitor protein in intact mitochondria. Eur. J. Biochem. 126(3), 611–616 (1982). doi:10.1111/j.1432-1033.1982.tb06824.x [DOI] [PubMed]
- 42.Schwerzmann, K., Pedersen, P.L.: Proton adenosine triphosphatase complex of rat liver mitochondria: effect of energy state on its interaction with the adenosine triphosphatase inhibitory peptide. Biochemistry 20(22), 6305–6311 (1981). doi:10.1021/bi00525a004 [DOI] [PubMed]
- 43.Power, J., Cross, R.L., Harris, D.A.: Interaction of F1-ATPase, from ox heart mitochondria with its naturally occurring inhibitor protein. Studies using radio-iodinated inhibitor protein. Biochim. Biophys. Acta 724(1), 128–141 (1983). doi:10.1016/0005-2728(83)90034-8 [DOI] [PubMed]
- 44.Minauro-Sanmiguel, F., Bravo, C., García, J.J.: Cross-linking of the endogenous inhibitor protein (IF1) with rotor (gamma, epsilon) and stator (alpha) subunits of the mitochondrial ATP synthase. J. Bioenerg. Biomembranes 34(6), 433–443 (2002). doi:10.1023/A:1022514008462 [DOI] [PubMed]
- 45.Papa, S., Zanotti, F., Cocco, T., Perrucci, C., Candita, C., Minuto, M.: Identification of functional domains and critical residues in the adenosine triphosphatase inhibitor protein of mitochondrial F0F1 ATP synthase. Eur. J. Biochem. 240(2), 461–467 (1996). doi:10.1111/j.1432-1033.1996.0461h.x [DOI] [PubMed]
- 46.Harris, D.A.: Functional regions of the H(+)-ATPase inhibitory protein from ox heart mitochondria. Biochim. Biophys. Acta 13208–16 (1997). doi:10.1016/S0005-2728(97)00003-0 [DOI] [PubMed]
- 47.van Raaij, M.J., Orriss, G.L., Montgomery, M.G., Runswick, M.J., Fearnley, I.M., Skehel, J.M., Walker, J.E.: The ATPase inhibitor protein from bovine heart mitochondria: the minimal inhibitory sequence. Biochemistry 35(49), 15618–15625 (1996). doi:10.1021/bi960628f [DOI] [PubMed]
- 48.Jackson, P.J., Harris, D.A.: The mitochondrial ATP synthase inhibitor protein binds near the C-terminus of the F1 beta-subunit. FEBS Lett. 229(1), 224–228 (1988). doi:10.1016/0014-5793(88)80832-9 [DOI] [PubMed]
- 49.Cabezón, E., Arechaga, I., Jonathan, P., Butler, G., Walker, J.E.: Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J. Biol. Chem. 275(37), 28353–28355 (2000). doi:10.1074/jbc.C000427200 [DOI] [PubMed]
- 50.Schägger, H., Pfeiffer, K.: Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19(8), 1777–1783 (2000). doi:10.1093/emboj/19.8.1777 [DOI] [PMC free article] [PubMed]
- 51.Tomasetig, L., Di Pancrazio, F., Harris, D.A., Mavelli, I., Lippe, G.: Dimerization of F0F1ATP synthase from bovine heart is independent from the binding of the inhibitor protein IF1. Biochim. Biophys. Acta. 1556(2–3), 133–141 (2002). doi:10.1016/S0005-2728(02)00344-4 [DOI] [PubMed]
- 52.Dienhart, M., Pfeiffer, K., Schagger, H., Stuart, R.A.: Formation of the yeast F1F0-ATP synthase dimeric complex does not require the ATPase inhibitor protein, Inh1. J. Biol. Chem. 277(42), 39289–39295 (2002). doi:10.1074/jbc.M205720200 [DOI] [PubMed]
- 53.Cabezón, E., Butler, P.J., Runswick, M.J., Carbajo, R.J., Walker, J.E.: Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J. Biol. Chem. 277(44), 41334–41341 (2002). doi:10.1074/jbc.M207169200 [DOI] [PubMed]
- 54.Minauro-Sanmiguel, F., Wilkens, S., García, J.J.: Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc. Natl. Acad. Sci. U. S. A. 102(35), 12356–12358 (2005). doi:10.1073/pnas.0503893102 [DOI] [PMC free article] [PubMed]
- 55.García, J.J., Morales-Ríos, E., Cortés-Hernandez, P., Rodríguez-Zavala, J.S.: The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry 45(42), 12695–12703 (2006). doi:10.1021/bi060339j [DOI] [PubMed]
- 56.Fornells, L.A., Guimaraes-Motta, H., Nehme, J.S., Martins, O.B., Silva, J.L.: Pressure effects on the interaction between natural inhibitor protein and mitochondrial F1-ATPase. Arch. Biochem. Biophys. 349(2), 304–312 (1998). doi:10.1006/abbi.1997.0454 [DOI] [PubMed]
- 57.Campanella, M., Casswell, E., Chong, S., Farah, Z., Wieckowski, M.R., Abramov, A.Y., Tinker, A., Duchen, M.R.: Regulation of mitochondrial structure and function by the F1F0-ATPase inhibitor protein, IF1. Cell Metab. 8(1), 13–25 (2008). doi:10.1016/j.cmet.2008.06.001 [DOI] [PubMed]
- 58.Fillingame, R.H., Jiang, W., Dmitriev, O.Y., Jones, P.C.: Structural interpretations of F(0) rotary function in the Escherichia coli F(1)F(0) ATP synthase. Biochim. Biophys. Acta 1458(2–3), 387–403 (2000). doi:10.1016/S0005-2728(00)00089-X [DOI] [PubMed]
- 59.Vik, S.B., Patterson, A.R., Antonio, B.J.: Insertion scanning mutagenesis of subunit a of the F1F0 ATP synthase near His245 and implications on gating of the proton channel. J. Biol. Chem. 273(26), 16229–16234 (1998). doi:10.1074/jbc.273.26.16229 [DOI] [PubMed]
- 60.Devenish, R.J., Prescott, M., Roucou, X., Nagley, P.: Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim. Biophys. Acta 458(2–3), 428–442 (2000) [DOI] [PubMed]
- 61.Dupuis, A., Lunardi, J., Issartel, J.P., Vignais, P.V.: Interactions between the oligomycin sensitivity conferring protein (OSCP) and beef heart mitochondrial F1-ATPase. 2. Identification of the interacting F1 subunits by cross-linking. Biochemistry 24(3), 734–739 (1985) [DOI] [PubMed]
- 62.Joshi, S., Burrows, R.: ATP synthase complex from bovine heart mitochondria. Subunit arrangement as revealed by nearest neighbor analysis and susceptibility to trypsin. J. Biol. Chem. 265(24), 14518–14525 (1990) [PubMed]
- 63.Belogrudov, G.I., Tomich, J.M., Hatefi, Y.: Membrane topography and near-neighbor relationships of the mitochondrial ATP synthase subunits e, f, and g. J. Biol. Chem. 271(34), 20340–20345 (1996). doi:10.1074/jbc.271.34.20340 [DOI] [PubMed]
- 64.Velours, J., Paumard, P., Soubannier, V., Spannagel, C., Vaillier, J., Arselin, G., Graves, P.V.: Organisation of the yeast ATP synthase F(0): a study based on cysteine mutants, thiol modification and cross-linking reagents. Biochim. Biophys. Acta. 1458(2–3), 443–456 (2000). doi:10.1016/S0005-2728(00)00093-1 [DOI] [PubMed]
- 65.Zanotti, F., Raho, G., Gaballo, A., Papa, S.: Inhibitory and anchoring domains in the ATPase inhibitor protein IF1 of bovine heart mitochondrial ATP synthase. J. Bioenerg. Biomembranes 36(5), 447–457 (2004). doi:10.1023/B:JOBB.0000047327.68173.9b [DOI] [PubMed]
- 66.Pan, W., Ko, Y.H., Pedersen, P.L.: Delta subunit of rat liver mitochondrial ATP synthase: molecular description and novel insights into the nature of its association with the F1-moiety. Biochemistry 37(19), 6911–6923 (1998). doi:10.1021/bi9800698 [DOI] [PubMed]
- 67.Ko, Y.H., Hullihen, J., Hong, S., Pedersen, P.L.: Mitochondrial F(0)F(1) ATP synthase. Subunit regions on the F1 motor shielded by F(0), Functional significance, and evidence for an involvement of the unique F(0) subunit F(6). J. Biol. Chem. 275(42), 32931–32939 (2000). doi:10.1074/jbc.M004453200 [DOI] [PubMed]
- 68.Cabezón, E., Runswick, M.J., Leslie, A.G., Walker, J.E.: The structure of bovine IF(1), the regulatory subunit of mitochondrial F-ATPase. EMBO J. 20(24), 6990–6996 (2001). doi:10.1093/emboj/20.24.6990 [DOI] [PMC free article] [PubMed]
- 69.Dudkina, N.V., Heinemeyer, J., Keegstra, W., Boekema, E.J., Braun, H.P.: Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 579(25), 5769–5772 (2005) [DOI] [PubMed]
- 70.Dudkina, N.V., Sunderhaus, S., Braun, H.P., Boekema, E.J.: Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 580(14), 3427–3432 (2006). doi:10.1016/j.febslet.2006.04.097 [DOI] [PubMed]
- 71.Strauss, M., Hofhaus, G., Schröder, R.R., Kühlbrandt, W.: Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27(7), 1154–1160 (2008). doi:10.1038/emboj.2008.35 [DOI] [PMC free article] [PubMed]
- 72.Vonck, J., Schäfer, E.: Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim. Biophys. Acta; Epub ahead of print (2008). doi:1016/j.bbamcr.2008.05.019 [DOI] [PubMed]
- 73.Thomas, D., Bron, P., Weimann, T., Dautant, A., Giraud, M.F., Paumard, P., Salin, B., Cavalier, A., Velours, J., Brèthes, D.: Supramolecular organization of the yeast F1Fo-ATP synthase. Biol. Cell; Epub ahead of print (2008). doi:10.1042/BC20080022 [DOI] [PubMed]
- 74.Fronzes, R., Weimann, T., Vaillier, J., Velours, J., Brèthes, D.: The peripheral stalk participates in the yeast ATP synthase dimerization independently of e and g subunits. Biochemistry 45(21), 6715–6723 (2006). doi:10.1021/bi0601407 [DOI] [PubMed]
- 75.Allen, R.D., Schroeder, C.C., Fok, A.K.: An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J. Cell Biol. 108(6), 2233–2240 (1989). doi:10.1083/jcb.108.6.2233 [DOI] [PMC free article] [PubMed]
- 76.Turina, P., Rebecchi, A., D’Alessandro, M., Anefors, S., Melandri, B.A.: Modulation of proton pumping efficiency in bacterial ATP synthases. Biochim. Biophys. Acta 1757(5–6), 320–325 (2006). doi:10.1016/j.bbabio.2006.04.018 [DOI] [PubMed]
- 77.Feniouk, B.A., Mulkidjanian, A.Y., Junge, W.: Proton slip in the ATP synthase of Rhodobacter capsulatus: induction, proton conduction, and nucleotide dependence. Biochim. Biophys. Acta. 1706(1–2), 184–194 (2005). doi:10.1016/j.bbabio.2004.10.010 [DOI] [PubMed]