Summary
Morphogenesis of bacteriophage P22 involves the packaging of double-stranded DNA into a preassembled procapsid. DNA is translocated by a powerful virally-encoded molecular motor called terminase, which comprises large (gp2, 499 residues) and small (gp3, 162 residues) subunits. While gp2 contains the phosphohydrolase and endonuclease activities of terminase, the function of gp3 may be to regulate specific and nonspecific modes of DNA recognition as well as the enzymatic activities of gp2. Electron microscopy shows that wildtype gp3 self-assembles into a stable and monodisperse nonameric ring. A three-dimensional reconstruction at 18 Å resolution provides the first glimpse of P22 terminase architecture and implies two distinct modes of interaction with DNA – involving a central channel of 20 Å diameter and radial spikes separated by 34 Å. Electromobility shift assays indicate that the gp3 ring binds dsDNA nonspecifically in vitro via electrostatic interactions between the positively charged C-terminus of gp3 (residues 143–152) and phosphates of the DNA backbone. Raman spectra show that nonameric rings formed by subunits truncated at residue 142 retain the subunit fold, despite the loss of DNA-binding activity. Difference density maps between gp3 rings containing full-length and C-terminally truncated subunits are consistent with localization of residues 143–152 along the central channel of the nonameric ring. The results suggest a plausible molecular mechanism for gp3 function in DNA recognition and translocation.
Keywords: P22 bacteriophage, terminase structure, DNA binding, electron microscopy, Raman spectroscopy
In the assembly of a double-stranded (ds) DNA bacteriophage the viral genome is packaged into a capsid precursor (procapsid) through the action of an enzyme complex called terminase. The complex is so named because its nucleolytic activity cleaves overlength replicating DNA to establish the termini of the packaged viral genome. Similar packaging mechanisms occur in Herpesviridae and other eukaryotic dsDNA viruses. Typically, the terminase contains two types of subunits, which are responsible for DNA recognition, phosphohydrolysis and translocation.1 For the well-studied bacteriophages P22, SPP1, T3, T4, T7 and λ, the larger of the two terminase subunits exhibits the required phosphohydrolase and translocase activities,2–9 while the smaller subunit is involved in the identification of the dsDNA packaging initiation sites (usually called pac or cos)10–12 and presumably in enzymatic regulation of the large subunit.3–6,10 Small terminase subunits are more variable in amino acid sequence than the large subunits (S. R. Casjens, unpublished results) and it is not known if all are true homologues or have structures with different origins. In the case of P22, the terminase small subunit (gp3, 162 amino acids, 18.6 kDa) and large subunit (gp2, 499 amino acids, 57.6 kDa)13,14 function in concert to package DNA by a “headful” mechanism that is initiated through recognition of a specific DNA packaging signal (pac site).15–17 Similar mechanisms have been proposed for the terminases of phages SPP1 and T4.18,19 A common theme in all of these dsDNA phages is the participation of the terminase large and small subunits in a ternary complex with the appropriate packaging initiation site. To form the functional terminase/pac complex the terminase small subunit of SPP1 (G1P) assembles into an approximately decameric ring,5, while those of T4 (gp16) and T7 (gp18) are reported to assemble into approximately octameric rings.20–22 The terminase small subunit of phageλ, on the other hand, has been proposed to form a dimer alone in solution and bind to specific sequences within the cos site of the λgenome in the form of a hetero-oligomeric ring constructed from a 2:1 ratio of small (gpNu1) and large (gpA) subunits.23,24 While cleavage of λcos is required for packaging, the precise roles of the proposed terminase ring and its heterotrimeric precursor are not known.
A three-dimensional atomic structure has been determined by nuclear magnetic resonance for the N-terminal DNA binding domain of the phage λsmall terminase subunit.25 However, the protein fold has not yet been determined for the full-length small subunit of any terminase. While oligomeric ring formation may be common among the small terminase subunits of many dsDNA phages, no detailed ring structure or uniform oligomer size has yet emerged. Recently, we reported the conformational properties, assembly states and DNA-binding activities of both the small (gp3) and large (gp2) subunits of P22 terminase. Two-dimensional averaging of negatively stained gp3 particles revealed a nonameric ring structure for the wildtype protein and a decameric ring structure for the mutant Ala 112 → Thr.26 Here, we further characterize DNA binding properties of wildtype gp3 and report the first three-dimensional reconstructions of a terminase small subunit assembly by resolving to 18 Å the nonameric ring structure of wildtype gp3. The structure provides new insights into the architecture of the P22 DNA translocation machine and suggests a molecular mechanism for DNA packaging.
Preparation and purification of gp3 constructs
The plasmid construct employed for overexpression of full-length wildtype gp3 in E. coli, the methods used for isolation and purification of the recombinant protein, and the demonstration of DNA packaging activity in Salmonella infections have been described.26 The full-length construct contained a twenty amino acid N-terminal addition consisting of a histidine tag (His6) and thrombin cleavage site encoded by the pET-15b expression vector plasmid. The N-terminal addition was removed prior to final purification of the protein.26 The plasmids used for overexpression of truncated variants of gp3 were made as site-directed deletions of the plasmid used for expression of full-length gp3 (QuikChange® Site-Directed Mutagenesis Kit strategy with PFU ULTRA® DNA polymerase, Strategene, La Jolla, CA). Expression and purification of gp3 truncates followed the same protocols used for the full-length protein.
Locus of the nonspecific DNA binding activity of gp3
DNA binding properties of the gp3 nonamer in the presence of a 50-bp DNA target that incorporated either the native 22-bp pac site or a functionally-inactive permuted pac site were assayed previously by native gel electrophoresis.26 Similar gp3 binding affinities were observed for both DNA targets, indicating the formation of nonspecific gp3/DNA complexes. This is consistent with the expectation that genome translocation requires of terminase a nonspecific mode of DNA recognition in addition to pac site recognition. Also consistent with this interpretation is the observation that both of the 50-bp DNA targets are capable of binding several nonameric gp3 rings when excess nonamer is present (data not shown).
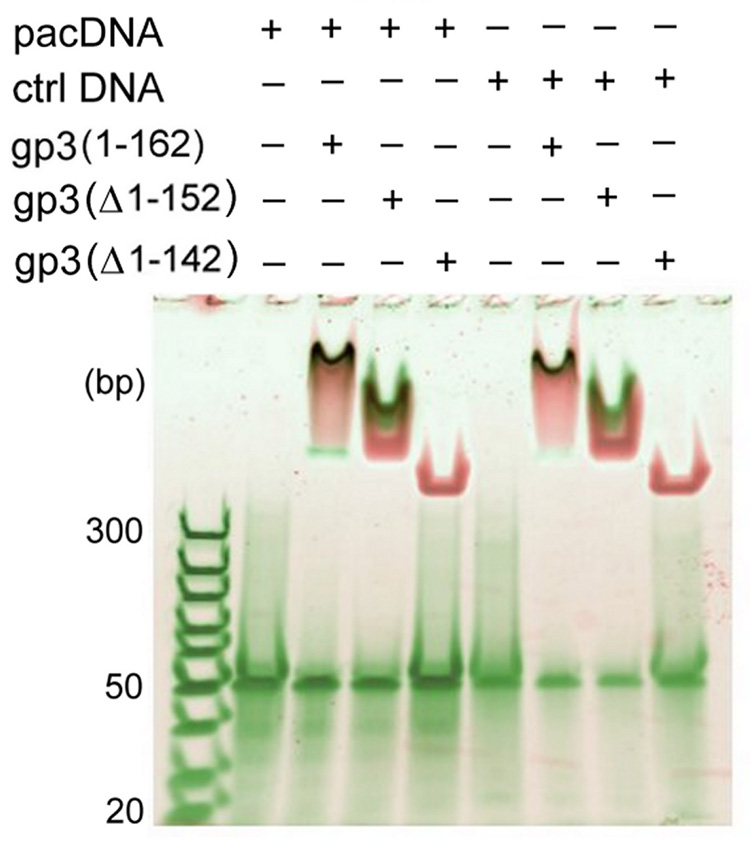
To address the question of whether the highly basic C-terminal region of the 162-residue gp3 subunit14 might be the locus of nonspecific DNA recognition, we constructed plasmids for overexpression of C-terminal truncations of gp3. Two C-terminal truncates, lacking ten and twenty amino acids and designated, respectively, as gp3(Δ1–152) and gp3(Δ1–142), exhibited stable subunit folds (Raman data shown below), the same nonameric oligomer state as the full-length protein (mass spectrometric data not shown), yet distinctly different nonspecific DNA-binding activities. The former contains a net charge of −1 relative to full-length gp3, while the latter contains a net charge of −5 relative to full-length gp3. The similarity in charges of gp3(Δ1–152) and full-length gp3 leads to comparable DNA binding, while the removal of 20 C-terminal amino acids in gp3(Δ1–142) results in the complete loss of DNA binding (Figure 1). Evidently, the highly basic sequence 143RDKRRSRIK that is removed by the deletion of amino acids 143–151 plays an important electrostatic role in nonspecific DNA recognition and binding.
Figure 1.
The binding of full-length and C-terminally truncated gp3 rings to 50-bp DNA targets incorporating either the 21-bp pac site or a permuted (nonfunctional) sequence of the same AT/GC content26 was examined by native gel electrophoretic mobility shift assay. Mixtures of DNA and gp3 rings were incubated at room temperature for 30 min and loaded onto a 4–20% polyacrylamide gel. The total DNA concentration in each lane was 200 nM. DNA and protein were stained with SYBR Green and SYPRO Ruby fluorescent dyes, respectively, and the gel was scanned with a Typhoon 9400 Scanner (Amersham Biosciences, Piscataway, NJ). The DNA-free gp3 ring appears at the top of the gel and protein-free DNA (50 bp) migrates to the middle of the gel. DNA/gp3 complexes are visualized as dark spots. The full-length and truncated gp3 rings progress differently into the gel due to the different net charges of the respective rings. While rings assembled from full-length gp3 and the gp3(Δ1–152) truncate bind DNA comparably, no DNA binding is observed for the gp3(Δ1–142) ring.
The fold of C-terminally truncated gp3
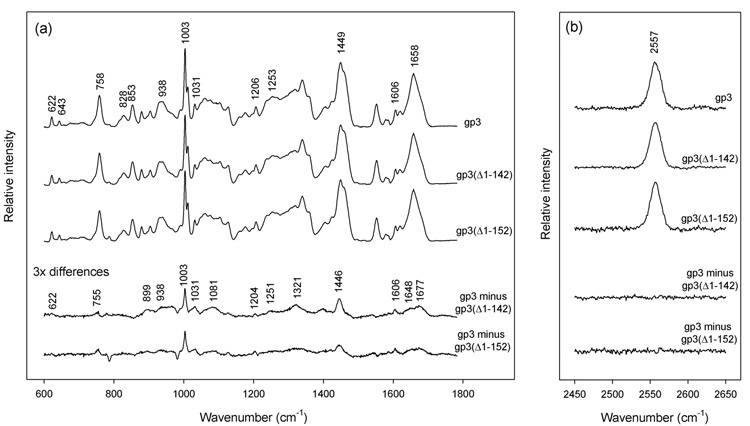
Raman spectra of nonameric assemblies of full-length and C-terminally truncated gp3 (lacking either twenty [gp3(Δ1–142)] or ten [gp3(Δ1–152)] residues) exhibit very similar amide I (1658 cm−1) and amide III (1253 cm−1) markers, as shown in Figure 2a. This suggests that the subunit fold is largely conserved after removal of residues 143–162. Quantitative analysis27 of the amide I band profile of the gp3(Δ1–142) truncate indicates 39 ± 2% α-helix, 21 ± 2% β-strand and 40 ± 3% irregular structure, which differs only marginally from the secondary structure distribution in full-length gp3.26 The weak positive difference peaks, which are centered within the amide I band envelope near 1648 and 1677 cm−1, are consistent with the presence of ordered α-helical and β-strand secondary structures, respectively, in the C-terminal twenty residues of gp3.
Figure 2.
Raman spectroscopy of nonameric ring assemblies of the full-length wildtype gp3 protein and C-terminal truncates gp3(Δ1–142) and gp3(Δ1–152). Solutions of purified rings were concentrated to ~20 mg/mL in 10 mM tris (pH 7.8), 0.1 M NaCl and sealed in 1 mm glass capillaries maintained at 20 °C. Raman spectra were excited at 532 nm (Verdi-5 laser, Coherent, Palo Alto, CA) and collected in the 90° scattering geometry (Spex 500M spectrograph, JY Horiba, Edison, NJ). Standard corrections for contributions of the buffer and glass were applied.26 (a) From top-to-bottom: Spectra in the region 600–1800 cm−1 of rings formed by full-length gp3, C-terminal truncate gp3(Δ1–142), C-terminal truncate gp3(Δ1–152), corresponding difference spectra (amplified 3-fold) between full-length and truncated species, as labeled. The integrated intensity of the tyrosine marker at 643 cm−1 was employed as the internal intensity standard. (b) From top-to-bottom: Spectra in the region 2450–2650 cm−1 of rings formed by full-length gp3, C-terminal truncate gp3(Δ1–142), C-terminal truncate gp3(Δ1–152), and corresponding difference spectra, as labeled. The integrated intensity of the 2557 cm−1 band was employed as the internal intensity standard.
The tyrosines (Tyr 35, Tyr 52, Tyr 86, Tyr 104) and tryptophans (Trp 10, Trp 29, Trp 38, Trp 45, Trp 83) of gp3 are located in the region of the sequence that is present in all three variants, and their Raman markers are potentially informative of any tertiary structural changes that might result from the C-terminal truncations. Figure 2a shows that the diagnostic Raman doublets of the tyrosines (828/853 cm−1)28,29 and tryptophans (1340/1360 cm−1)30 of gp3(Δ1–142) are similar to those of full-length gp3. This supports the conclusion that the tertiary structure within residues 1–142 is largely invariant to the C-terminal truncations. The small spectral differences between gp3(Δ1–142) and full-length gp3, which are revealed as positive difference bands in the 3x-amplified difference spectrum of Figure 2a, can be assigned confidently to the excised side chains of the 143–162 sequence.31 Notable among these are the difference peaks at 622, 755, 1003, 1031, 1081, 1204 and 1606 cm−1, all of which are attributed to the excised Phe 154 residue, and the difference peaks near 885, 936 and 1321 cm−1, which are due primarily to arginines and lysines of the excised C-terminus.
Each cysteine (Cys 32, Cys 33) of gp3 is also expected to generate a Raman marker that is informative of the local environment of the side-chain sulfhydryl (S–H) group.32 The single symmetrical band observed at 2557 cm−1 (Figure 2b) in the full-length gp3 assembly indicates that the S–H donors of Cys 32 and Cys 33 are not exposed to the aqueous solvent but participate in robust S–H⋯X interactions with other protein acceptor groups. These interactions are unaffected by C-terminal truncations. Strong S–H⋯X bonding by the Cys 32 and Cys 33 side chains suggests that this region of the gp3 chain is buried either within the subunit fold or is protected from solvent exposure by the intersubunit interface. The results are also consistent with very little change in the subunit fold after removal of residues 143–162, as noted above.
The spectral differences observed between the gp3(Δ1–152) truncate and full-length gp3 (Figure 2) are consistent with the above results. In combination with the gel binding assays of Figure 1, the Raman spectra thus show that: (i) the region of the gp3 sequence responsible for nonspecific DNA recognition (residues 143–162) adopts a stable α/βfold, and (ii) excision of the putative DNA binding segment does not appreciably perturb either the secondary or tertiary structure of the remaining gp3 sequence (i.e. residues 1–142).
Three-dimensional structure of the gp3 assembly
Recently, we reported that the terminase small subunit of P22 assembles into a highly stable and symmetric ring.26 Two-dimensional averaging of the gp3 assemblies identified in electron micrographs of negatively stained particles revealed a central hole of approximately 20 Å in diameter with electron density emerging radially from the central annulus. Here, we have further analyzed the structure of the gp3 ring assembly by three-dimensional single particle analysis of negatively stained gp3 ring particles.
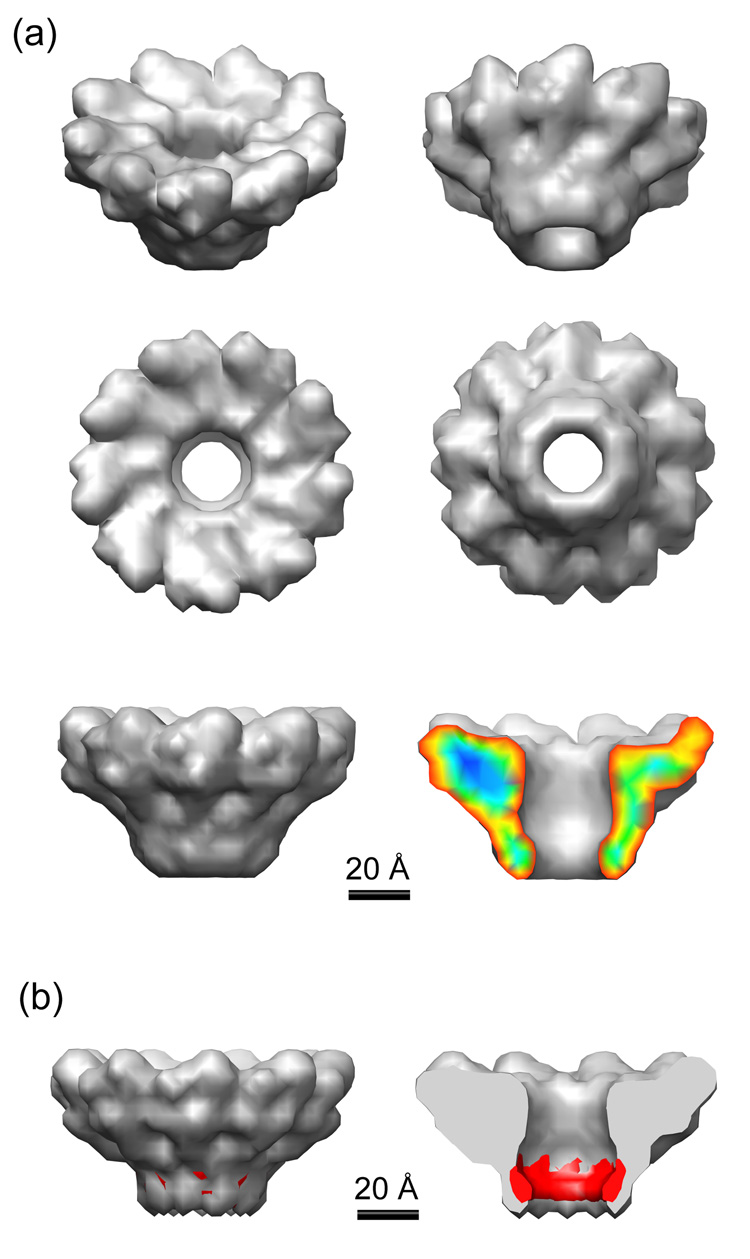
The three-dimensional reconstruction of the gp3 assembly at 18 Å resolution, as reported by RMEASURE,33 reveals a morphology that is roughly funnel-like in shape, being much wider at one end than the other (Figure 3a). Despite the relatively low resolution of this structure, certain features and dimensions can be clearly distinguished. Protruding structures approximately 27 Å in height surround the wider end or mouth of the funnel, extending a distance of 38 Å from the inner edge at the narrowest opening of the central channel. Although these “spikes” have a definite skewed appearance relative to the central channel, the handedness of the reconstruction cannot be determined without further experiments. The spike domain of the structure accounts for nearly 80% of the total mass of the ring assembly, and due to its complexity, is likely to be involved in multiple recognition events (see below). The annulus region, which may be important for intersubunit association and DNA passage, may also provide surfaces for binding of gp2 or other proteins involved in the formation of an active translocation complex. Given that the electron density of proteins correspond on average to 0.8 Da/Å3, the reconstructed density was depicted at a contour level corresponding to 167.4 kDa (the mass of nonameric gp3). At this contour level salient features of the structure emerge, including an overall height of 57 Å and a maximum outer diameter of 99 Å in the spike domain that narrows to a diameter of 49 Å at the annulus. At the funnel mouth (top surface as oriented in the bottom images of Figure 3a), the central channel entrance has a diameter of ~25 Å. This opening widens slightly to ~28 Å before narrowing to ~20 Å in diameter at the neck of the annulus domain. The channel appears to be sufficient to accommodate the passage of B-form DNA. The height of the channel opening between the neck and mouth of the funnel is roughly 42 Å. (See also Table 1.)
Figure 3.
(a) 3D reconstruction of the gp3 ring assembly from negatively stained particles. The map was rotated 30° about the horizontal axis in the top 2 images, and viewed from above and below in the central images. The funnel-like morphology is evident when viewed from the side in the lowest two images, and a cutaway view on the right is colored by density strength. The ring-like structure of gp3 is clearly divided into two domains – the wider spike domain, in which 9 protrusions can be seen reaching out from the central channel at a skewed orientation, and the annulus domain, which forms a thin-walled channel extending away from the spike domain. The central channel accommodates the passage of double-stranded (B form) DNA, as shown in Figure 4a, with a slightly smaller opening at the annulus than at the spike domain. Dimensions are listed in Table 1. The 9-fold symmetry of the wild type gp3 particles discerned in previous negative stain work was confirmed by performing a reference-free classification of selected top-view particles. The resulting class averages presented only 9-fold views, without the enforcement of symmetry. To rule out the possibility of symmetrization bias, we performed separate 3D reconstructions enforcing either 8-fold (C8), 9-fold (C9) or 10-fold (C10) symmetries, each with a single radially averaged starting model. While the C8 and C10 reconstructions resulted in smooth featureless ring-like densities, the C9 reconstruction showed structural details for which the top-view projections matched those of the reference-free classification. Further discussion of the assembly architecture and its relevance to terminase function are discussed in the text. (b) 3D reconstruction (side and cutaway views) of the nonameric truncated gp3(Δ1–127) ring, along with the difference density shown in red. This difference, which corresponds primarily to the C-terminal 35 amino acids (see text), suggests that the C-terminal residues involved in nonspecific DNA recognition are localized mainly along the inner surface of the central channel.
Table 1.
Dimensions of the reconstructed gp3 ring
| outer diameter (spike domain) | 99 Å |
| outer diameter (annulus) | 49 Å |
| inner diameter (spike domain) | 25 Å |
| inner diameter (annulus) | 20 Å |
| maximum channel diameter | 28 Å |
| channel height | 42 Å |
| overall height | 57 Å |
The elongated subunit shape in the gp3 assembly (Figure 3a, bottom right) implies a multidomain structure, in which at least one domain (the central annulus) plays a role in subunit/subunit recognition and one or more domains interact with DNA, with the terminase large subunit (gp2) and possibly with the DNA entry channel of the procapsid (portal protein, gp1). Details of these structure-function relationships remain to be determined. Our reconstruction shows, however, that the subunit electron density is significantly elongated in a direction close to perpendicular to the plane of the ring, thus providing a channel with large surface area for recognition and/or passage of agents of appropriate cross-sectional diameter (~20 Å).
Location of the gp3/DNA interface
Figure 1–Figure 3 show that the excision of gp3 residues 143–162 eliminates nonspecific DNA binding, but does not impact either the native subunit fold of residues 1–142 or the overall nonameric ring structure that is visualized by negative stain EM. We attempted to further localize the nonspecific DNA-binding interface of gp3, within the context of the ring assembly, by computing a protein density difference map between rings consisting of full-length gp3 and the truncate gp3(Δ1–142). However, the result was inconclusive (data not shown), owing to the relatively small mass difference between rings consisting of the full-length and truncated subunits (<22 kDa). We therefore examined the difference between reconstructed rings of full-length gp3 and the more radically truncated variant gp3(Δ1–127). The greater mass differential in this case (~37 kDa) does allow visualization of the truncated C-terminus in the computed difference map, as shown in Figure 3b. The external and cutaway side views shown in Figure 3b indicate that 35 amino acids of the gp3 C-terminus (128KEQSQVEDVTPDKGDRDKRRSRIKELFNRGTGRDS), which includes the interface for nonspecific DNA recognition, is localized principally along the inner surface of the central channel and proximal to the neck opening.
Function of gp3 in the DNA packaging machine
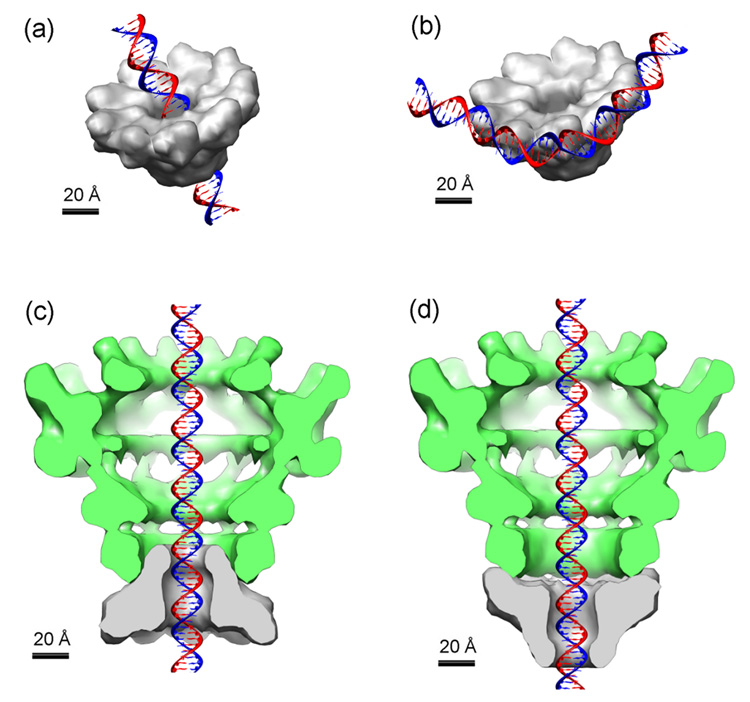
The overall dimensions of the reconstruction of Figure 3a are given in Table 1. These parameters suggest intriguing possibilities for DNA binding, pac site recognition and genome translocation. For example, the central hole of the gp3 ring exhibits a diameter that is appropriate to the passage of dsDNA. It could thus provide a DNA-translocating channel that is dimensionally compatible with the translocating channel of the procapsid portal. The likely location of gp3 sites of nonspecific DNA binding activity (residues 142–162) along this channel (Figure 3b) is consistent with a DNA translocation model like that shown in Figure 4a.
Figure 4.
Models for interaction of the gp3 ring assembly of Figure 3 with double-stranded B DNA and the P22 portal. The dsDNA, terminase ring and portal vertex are plotted on the same scale. (a) Passage of dsDNA through the central hole of the gp3 ring assembly. (b) Binding of dsDNA to the periphery of the gp3 ring assembly. Docking of the funnel-shaped gp3 ring at the portal neck is depicted in neck-to-neck (c) and mouth-to-neck (d) orientations.
The recruitment of concatemeric DNA and the specific recognition of the pac site by nonameric gp3 might be initiated by DNA contact along the spikes of the gp3 nonamer at high ring radius. Such a model for pac site recognition would appear to be favored over monomer-initiated binding because of the very rapid self-assembly kinetics of gp3.26 This hypothesis for nonamer/pac site recognition is particularly attractive given the apparent circumferential spacing between adjacent spikes of protein density (~34 Å), which is close to the contour length of one turn of the DNA helix (~33 Å pitch).34 The model depicted in Figure 4b would allow interaction of adjacent subunits of the ring with phosphate binding sites along a single face of DNA. Bending of the 21 bp pac site by ~40° would be sufficient to facilitate cooperative binding of three neighboring subunits of the gp3 nonamer, thus suggesting a plausible structural basis for pac site selection.
Studies of phages λ, T3, SPP1 and T4 provide strong evidence that the translocation of DNA into the procapsid is powered by the ATPase activity of the terminase large subunit. However, little is known about the molecular structures of the terminase motor components or the three-dimensional organization of its parts. To generate a force for DNA packaging, a terminase complex must be bound to the procapsid. In the phages λ and T4 current evidence indicates that the terminase large subunit binds directly to the portal protein of the procapsid during DNA packaging.35–37 These findings do not preclude additional contacts to the procapsid involving the terminase small subunit. In fact, for phages SPP1 and T3 experimental evidence has been obtained for direct procapsid contact by both the small and large subunits.2,5,38 A phylogenetic analysis of terminase large subunits of many phages suggests also that the terminase of P22 is most closely related to that of SPP1.39 While it is not known if terminases of different phages bind to their respective procapsids via different or similar mechanisms, the possibility exists that both the large and small subunits of P22 terminase may make contact with the procapsid. Docking of the terminase ring at the portal vertex could thus provide an effective translocating channel, as noted above. Using the present and previous40 EM reconstructions, we illustrate two feasible coaxial arrangements for such a DNA translocating channel in Figures 4c and 4d. In both arrangements, the conical shape of the gp3 ring elongates the stalk of the portal that emerges from the procapsid. While the models depicted in Figures 4c and 4d allow for significant contact surfaces, it is not known whether a rigid or flexible interface would be required to accommodate the obvious symmetry mismatch between the two rings. It is conceivable, however, that a trio of subunits of the nine-fold gp3 ring could interact with a quartet of subunits of the twelve-fold portal ring, since both have 120° radial spacing. In Figures 4c and 4d, the end of the gp3 funnel that protrudes outwardly from the portal ring would presumably serve as the locus for contact with the terminase large subunit during ATPase and DNase directed activities. Because a lobe of portal subunit (gp1) density would extend beyond the gp3 ring in either Figure 4c or 4d, both allow the terminase large subunit to simultaneously contact gp3 and gp1 subunits. Experiments in progress, which are designed to identify the gp3/gp2 and gp3/gp1 interaction surfaces, are expected to shed further light on the mode of portal docking.
Supplementary Material
Acknowledgments
Support of this research by National Institutes of Health grants GM50776 (GJT) and AI074825 (SRC) is gratefully acknowledged. Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through a P41 program grant (RR17573) from the National Center for Research Resources. We are grateful to Drs. Bridget Carragher and Clint Potter for their interest and helpful discussions during EM analysis. We also thank Prof. Peter E. Prevelige (University of Alabama at Birmingham) for providing resources for mass spectrometry of gp3 assemblies and contributing valuable discussions throughout the course of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Catalano CE. In: Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. Georgetown, TX and New York: Landes Bioscience / Eurekah.com and Kluwer Academic / Plenum Publishers; 2005. [Google Scholar]
- 2.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 3.Gual A, Camacho AG, Alonso JC. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 2000;275:35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 4.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 5.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J. Biol. Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 6.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 7.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 2006;281:21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 8.Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 1992;227:1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- 11.Shinder G, Gold M. The Nul subunit of bacteriophage λ terminase binds to specific sites in cos DNA. J. Virol. 1988;62:387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 13.Backhaus H. DNA packaging initiation of Salmonella bacteriophage P22: determination of cut sites within the DNA sequence coding for gene 3. J. Virol. 1985;55:458–465. doi: 10.1128/jvi.55.2.458-465.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 15.Tye BK, Chan RK, Botstein D. Packaging of an oversize transducing genome by Salmonella phage P22. J. Mol. Biol. 1974;85:485–500. doi: 10.1016/0022-2836(74)90311-8. [DOI] [PubMed] [Google Scholar]
- 16.Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 18.Streisinger G, Emrich J, Stahl MM. Chromosome structure in phage T4, III. Terminal redundancy and length determination. Proc. Natl. Acad. Sci. USA. 1967;57:292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavares P, Lurz R, Stiege A, Ruckert B, Trautner TA. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 1996;264:954–967. doi: 10.1006/jmbi.1996.0689. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 21.Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: insights into DNA packaging initiation and assembly of packaging motor. J. Mol. Biol. 2006;358:67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 22.White JH, Richardson CC. Gene 18 protein of bacteriophage T7. Overproduction, purification, and characterization. J. Biol. Chem. 1987;262:8845–8850. [PubMed] [Google Scholar]
- 23.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 24.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage λ terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 25.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 26.Nemecek D, Gilcrease EB, Kang S, Prevelige PE, Jr, Casjens S, Thomas GJ., Jr Subunit conformations and assembly states of a DNA-translocating motor: the terminase of bacteriophage P22. J. Mol. Biol. 2007;374:817–836. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berjot M, Marx J, Alix AJP. Determination of the secondary structure of proteins from the Raman amide I band: the reference intensity profiles method. J. Raman Spectrosc. 1987;18:289–300. [Google Scholar]
- 28.Siamwiza MN, Lord RC, Chen MC, Takamatsu T, Harada I, Matsuura H, Shimanouchi T. Interpretation of the doublet at 850 and 830 cm−1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry. 1975;14:4870–4876. doi: 10.1021/bi00693a014. [DOI] [PubMed] [Google Scholar]
- 29.Arp Z, Autrey D, Laane J, Overman SA, Thomas GJ., Jr Tyrosine Raman signatures of the filamentous virus Ff are diagnostic of non-hydrogen-bonded phenoxyls: demonstration by Raman and infrared spectroscopy of p-cresol vapor. Biochemistry. 2001;40:2522–2529. doi: 10.1021/bi0023753. [DOI] [PubMed] [Google Scholar]
- 30.Miura T, Takeuchi H, Harada I. Tryptophan Raman bands sensitive to hydrogen bonding and side-chain conformation. J. Raman Spectrosc. 1989;20:667–671. [Google Scholar]
- 31.Overman SA, Thomas GJ., Jr Raman markers of nonaromatic side chains in an alpha-helix assembly: Ala, Asp, Glu, Gly, Ile, Leu, Lys, Ser, and Val residues of phage fd subunits. Biochemistry. 1999;38:4018–4027. doi: 10.1021/bi982901e. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Thomas GJ. Cysteine conformation and sulfhydryl interactions in proteins and viruses. 1. Correlation of the Raman S-H band with hydrogen-bonding and intramolecular geometry in model compounds. J. Am. Chem. Soc. 1991;113:456–462. [Google Scholar]
- 33.Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J. Struct. Biol. 2007;157:201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. The anatomy of A-, B-, and Z-DNA. Science. 1982;216:475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 35.Frackman S, Siegele DA, Feiss M. A functional domain of bacteriophage λ terminase for prohead binding. J. Mol. Biol. 1984;180:283–300. doi: 10.1016/s0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- 36.Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 1995;245:141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 37.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 38.Fujisawa H, Shibata H, Kato H. Analysis of interactions among factors involved in the bacteriophage T3 DNA packaging reaction in a defined in vitro system. Virology. 1991;185:788–794. doi: 10.1016/0042-6822(91)90550-u. [DOI] [PubMed] [Google Scholar]
- 39.Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005;187:1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.