Summary
[3H]Epibatidine binds to nAChR subtypes in mouse brain with higher (KD≈0.02 nM) and lower affinity (KD≈7 nM), which can be further subdivided through inhibition by selected agonists and antagonists. These subsets are differentially affected by targeted deletion of α7, β2 or β4 subunits. Most, but not all, higher and lower affinity binding sites require β2 (Marks et al., 2006). Effects of functional α4 gene deletion are reported here. Deletion of α4 virtually eliminated cytisine-sensitive, higher-affinity [3H]epibatidine binding as did β2 deletion, confirming that these sites are α4β2*-nAChR. Cytisine-resistant, higher-affinity [3H]epibatidine binding sites are diverse and some of these sites require α4 expression. Lower affinity [3H]epibatidine binding sites are also heterogeneous and can be subdivided into α-bungarotoxin-sensitive and - resistant components. Deleting α4 did not affect the α-bungarotoxin-sensitive component, but markedly reduced the α-bungarotoxin–resistant component. This effect was similar, but not quite identical, to the effect of β2 deletion. This provides the first evidence that lower-affinity epibatidine binding sites in the brain require expression of α4 subunits. The effects of α4 gene targeting on receptor function were measured using a 86Rb+ efflux assay. Concentration-effect curves for ACh-stimulated 86Rb+ efflux are biphasic (EC50 values = 3.3 µM and 300 µM). Targeting α4 produced substantial gene-dose dependent reductions in both phases in whole brain and in most of the 14 brain regions assayed. These effects are very similar to those following deletion of β2. Thus, α4β2*–nAChRs mediate a significant fraction of both phases of ACh stimulated 86Rb+ efflux.
Keywords: Null mutant mice, Nicotinic acetylcholine receptor, Epibatidine, 86Rb+ efflux, Cytisine, Dihydro-β-erythroidine
Nicotinic cholinergic receptors (nAChRs) are ligand-gated ion channels expressed throughout the body, particularly in skeletal muscle, autonomic ganglia, and the central nervous system. Neuronal nAChRs are pentameric assemblies of homologous subunits, eleven of which have been identified (α2–α7, α9, α10, β2–β4) (Lindstrom, 2000). nAChR influence simple and complex behaviors (Picciotto, 2003), regulate acute and chronic responses to nicotine and similar drugs (Dani and DeBiasi, 2001), and have been implicated in aspects of Alzheimer’s and Parkinson’s diseases as well as schizophrenia, Tourette’s syndrome, and anxiety (Bourin et al., 2003; Leonard et al., 2001; Quik, 2005). Different nAChRs appear to regulate these behaviors and diseases making it important to identify subtype specific function. The potential for nAChR diversity is considerable, but is limited by rules of receptor assembly for and cellular expression of nAChR subunits. Nevertheless, overlapping expression of compatible subunits makes identifying the regional brain compositions of the nAChRs challenging.
Radioligand binding is very useful for the identification of neuronal nAChR subtypes. In particular, epibatidine is an extraordinarily potent nicotinic agonist (Badio and Daly, 1994) that binds to many, if not all, nAChR subtypes in brain (Houghtling et al., 1995; Marks et al., 1998, 2006; Zoli et al., 1998; Perry et al., 2002) and to those heterologously expressed in Xenopus oocytes (Parker et al., 1998; Kuryatov et al., 2000) or HEK cells (Xiao and Kellar, 2004). Immunochemical (Whiting and Lindstrom, 1988; Flores et al., 1992; Gotti et al., 2005; Marritt et al., 2005) or gene deletion (Picciotto et al., 1995; Xu et al., 1999; Zoli et al., 1998; Orr- Urtreger et al., 1997; Marubio et al., 1999; Ross et al., 2000; Whiteaker et al. 2002; Marks et al., 2006) methods confirm the existence of multiple epibatidine binding subtypes.
[3H]Epibatidine binding can be separated into two major classes that differ markedly in affinity (KD ≈.02 nM and KD ≈ 5 nM) (Marks et al., 1999; Whiteaker et al., 2000b). Densities of higher- and lower affinity [3H]epibatidine binding sites in rodent brain are approximately equal. Each of these two classes of binding sites can be subdivided on the basis of differential sensitivity to inhibition by nicotinic agonists and antagonists. Higher affinity [3H]epibatidine binding sites can be separated into cytisine-sensitive and -resistant components. Lower affinity [3H]epibatidine binding sites can be separated into α-bungarotoxin-sensitive and -resistant components (Marks et al., 1998; Whiteaker et al., 2000a; Perry et al., 2002). Recently, we (Marks et al., 2006) reported that deletion of either β2 or β4 markedly reduced many components of both higher and lower affinity [3H]epibatidine binding sites whereas α7 deletion eliminated only the α-bungarotoxin sensitive component of lower affinity [3H]epibatidine binding. Clearly, an α subunit is required to form functional nAChRs. The studies reported here evaluated the effects of α4 gene deletion on diverse [3H]epibatidine binding sites and indicate that α4 is essential to the expression of many [3H]epibatidine binding sites.
Characterization of binding sites provides significant information about nAChR diversity, but characterization of nAChR function provides significant additional information. Electrophysiological methods have been successfully used to demonstrate functional diversity (Alkondon and Albuquerque, 1993) and examine changes in expression following nAChR subunit deletion (Picciotto et al., 1995; Orr-Urtreger et al., 1997; Marubio et al., 1999). The function of nAChRs has also been measured using biochemical methods. Many nAChRs are expressed on presynaptic nerve terminals, and the function of these receptors has been evaluated by measuring neurotransmitter release from synaptosomes or tissue slices (Wonnacott, 1997). Agonist-stimulated 86Rb+ efflux from mouse brain synaptosomes provides a direct biochemical assay for nAChR function (Marks et al., 1994, 1999, 2002). Efflux with pharmacological properties consistent with an α3β4-nAChR is seen in a few brain regions (inferior colliculus and interpeduncular nucleus) (Marks et al., 2002), but β2* nAChRs modulate this response in most brain regions (Marks et al., 1999; 2000). Biphasic agonist dose-response curves have been described for α4β2-nAChR expressed in cell lines or Xenopus oocytes (Zwart and Vijverberg, 1998; Buisson and Bertrand, 2001; Nelson et al., 2003; Zhou et al., 2003). The observation that concentration-effect curves for acetylcholine (Ach) stimulation of agonist-stimulated 86Rb+ efflux are also biphasic suggests that α4β2* nAChRs modulate both of these components of agonist-stimulated 86Rb+ efflux.
The studies reported here evaluated the effects of α4 gene deletion on multiple components of [3H]epibatidine binding as well as ACh-stimulated 86Rb+ efflux. Major findings include the demonstration of a previously undescribed low affinity a4β2* nAChR and confirmation that α4β2* nAChRs modulate both the high and low affinity components of agonist-stimulated 86Rb+ efflux. Portions of this work have been published as an abstract (Marks et al., 2003).
Materials and Methods
Mice
The University of Colorado Animal Care and Utilization Committee approved animal care and experimental procedures. Efforts were made to reduce animal use particularly by dissecting as many brain areas as possible from each individual.
Mice with targeted deletion of the α4 nAChR subunit (Ross et al., 2000) were obtained from the Howard Florey Institute, The University of Melbourne, Victoria, Australia. Animals have subsequently been maintained in the specific pathogen free facility at the University of Colorado, Boulder, CO. Heterozygous α4 nAChR subunit mice had been backcrossed with C57BL/6J mice for 2 generations at the time of the experiments. Homozygous and heterozygous mutant and wild type control mice were generated from heterozygous matings. Mice were weaned at 25 days of age and like-sexed littermates were housed together (2–5 mice per cage). Mice were allowed free access to food (Harlan Teklad, Madison, WI) and water. The vivarium, in which they were housed, was maintained at 22°±1°C. Lights were on from 7 AM to 7 PM.
Genotypes were determined using DNA extracted from tail clippings obtained around postnatal day 40 as described previously (see Salminen et al., 2004 for details). Animals were 60–100 days old at the time of the experiments.
Tissue Preparation
Each mouse was killed by cervical dislocation. The brain was rapidly removed and placed on an ice-cold platform. For some experiments, the whole brain was used, while for others fourteen brain regions were dissected (olfactory bulbs; olfactory tubercles; cerebral cortex; hippocampus; striatum; thalamus; hypothalamus; midbrain; habenula; interpeduncular nucleus; superior colliculus; inferior colliculus; hindbrain; and cerebellum).
Tissue Preparation for Ligand Binding Assays
To prepare tissue for [3H]epibatidine binding assays, dissected regions were placed in 10 volumes (w/v) of ice-cold hypotonic buffer (NaCl, 14 mM; KCl, 0.15 mM; CaCl2, 0.2 mM; MgSO4, 0.1 mM; HEPES, 2.5 mM; pH=7.5) and homogenized using a motor-driven pestle. Homogenized samples were centrifuged at 12,000 × g for 20 min. The pellet was resuspended in hypotonic buffer and again centrifuged. The resuspension/centrifugation cycle was repeated two more times. The resulting pellet was stored frozen under fresh hypotonic buffer until assayed.
On the assay day, the sample was thawed, resuspended in the overlying buffer and centrifuged at 12,000 × g. The resulting pellet was resuspended in water for use in the [3H]epibatidine binding assays.
[3H]Epibatidine Binding to Whole Brain Samples
Tissue prepared from the whole brains of mice of each genotype [wild-type (+/+), heterozygous (+/−) and homozygous (−/−)] was assayed for [3H]epibatidine binding (Dupont NEN, Boston, MA; 55.5 Ci/mmol). All binding experiments were performed in a buffer of the following composition: NaCl, 140 mM; KCl, 1.5 mM; CaCl2, 2 mM; MgSO4, 1 mM; HEPES, 25 mM; pH=7.5. Following the 2 hr incubation, particulate protein was collected under vacuum using an Inotech Cell Harvester (Inotech Biosystems, Rockville, MD) onto glass fiber filters that had been soaked in 0.5% polyethylenimine (top filter; type GB, Micro Filtration Systems, Dublin, CA; bottom filter Type A/E, Gelman, Ann Arbor, MI). Samples were subsequently washed six times with cold (4° C) buffer. Following filtration and wash, filters were transferred to 5 ml scintillation vials to which 1 ml of Budget Solve scintillation fluid (RPI, Mt. Prospect, IL) was added. Radioactivity was determined using a Packard Tricarb 1600 Liquid Scintillation Analyzer. Counting efficiency was 45%.
[3H]Epibatidine saturation binding was determined using ligand concentrations from 0.01 nM to 32 nM. Incubation volume for the eight lower concentrations (0.01 nM – 1.28 nM) was 500 µL, while the incubation volume for the eight higher concentrations (0.25 nM – 32 nM) was 65 µL. All samples were arranged in a standard 96 well format. Incubations in the lower concentration range were conducted in 1.2 ml polypropylene tubes, while those in the higher concentration range were conducted in polystyrene plates. Concentrations were chosen such that the three higher concentrations from the lower concentration range (0.32, 0.64 and 1.28 nM) were similar to the three lower concentrations from the higher concentration range (0.25, 0.5 and 1.0 nM) in order to ensure that binding under the two conditions was comparable. Results for the overlapping samples were averaged yielding a saturation curve with thirteen concentrations of [3H]epibatidine. Aliquots of each ligand concentration were counted to determine initial concentration. Free ligand concentrations were estimated by adjusting the initial ligand concentrations for the amount of bound ligand thereby correcting for ligand depletion.
Inhibition of [3H]epibatidine binding by cytisine was measured using a ligand concentration of approximately 0.4 nM, while inhibition by d-tubocurarine was determined using approximately 8 nM [3H]epibatidine. Actual concentrations in each experiment were determined by counting aliquots of the appropriate [3H]epibatidine solution.
An incubation volume of 500 µL was used with 0.4 nM [3H]epibatidine. Ten concentrations of cytisine (0.1. nM, 0.3 nM, 1 nM, 3 nM, 10 nM, 30 nM, 100 nM, 300 nM, 1 µM, and 3 µM) were used to construct the concentration-effect curve. Blanks were established by measuring binding in the presence 100 µM nicotine.
An incubation volume of 65 µL was used with 8 nM [3H]epibatidine. Nine concentrations of d-tubocurarine (0.3 µM, 1 µM, 3 µM, 10 µM, 30 µM, 100 µM, 300 µM, 1 mM, and 3 mM) Blanks were established using 1 mM nicotine.
[3H]Epibatidine Binding to Brain Regions
The fourteen brain regions were dissected, homogenized and particulate fractions prepared as described above. Differential inhibition of [3H]epibatidine (0.4 nM) binding by 100 nM cytisine was measured to estimate the cytisine-sensitive and cytisine-resistant binding sites (Marks et al., 1998: Whiteaker et al., 2000b; Perry et al., 2002). In addition, the effect of 100 nM αBgt and 300 µM d-tubocurarine on binding measured with 15 nM [3H]epibatidine was determined in order to estimate αBgt-sensitive and αBgt-resistant, lower-affinity [3H]epibatidine binding. These inhibitor concentrations were selected following experiments that measured inhibition of [3H]epibatidine binding in whole brain. Tissue samples were incubated with αBgt and αBgt plus d-tubocurarine for 1 hr before addition of [3H]epibatidine to allow toxin to associate with its binding sites. Incubation volumes and times, sample filtration and wash, and liquid scintillation counting were as described above.
Crude Synaptosomal Tissue Preparation for 86Rb+ Efflux
To prepare samples for assay of ACh-stimulated 86Rb+ efflux, dissected regions were placed in 10 volumes of cold (4°C) isotonic 0.32 M sucrose buffered to pH=7.5 with 5 mM HEPES and homogenized by hand using a Teflon/glass tissue grinder. Samples were centrifuged at 12,000 × g for 20 min. The resulting pellet was resuspended in isotonic uptake buffer (60 µl to 800 µl, depending on brain region). Composition of the uptake buffer was: NaCl, 140 mM; KCl, 1.5 mM; CaCl2, 2 mM; MgSO4, 1 mM; HEPES hemisodium, 25 mM; glucose, 20 mM; pH=7.5.
86Rb+ Uptake
25µl aliquots of crude synaptosomal suspension were added to 10 µl of uptake buffer containing 4 µCi of 86Rb+. Following a 30 min incubation, 5 µl of 80 µM diisopropylfluorophosphate was added to each sample (final concentration = 10 µM) and the incubation was continued for 5 min. Uptake was terminated by filtration onto a 6-mm diameter Gelman A/E glass fiber filter disc under gentle vacuum (0.8 atm) and the sample was washed once with 0.5 ml of uptake buffer.
Sample Perfusion and ACh Stimulation
Filters containing the synaptosomes loaded with 86Rb+ were transferred to an open-air platform and superfused with buffer (NaCl, 135 mM; CsCl, 5 mM; KCl, 1.5 mM; CaCl2, 2 mM; MgSO4, 1 mM; glucose, 20 mM; tetrodotoxin, 50 nM; atropine 1 µM; HEPES hemisodium, 25 mM; bovine serum albumin, 0.1%; pH=7.5). A Gilson Minipuls 3 (Gilson, Middleton, WI) was used to apply the buffer to the top of the filter containing the synaptosomes at a flow rate of 2.5 ml/min. Buffer was actively removed from the bottom of the filter platform with a second Gilson Minipuls 3 pump set a flow rate of 3.2 ml/min and pumped through a 200 ul Cherenkov cell in a β-Ram HPLC detector (IN/US Systems, Tampa, FL) to monitor continuously radioactivity in the effluent.
Samples were superfused for 5 min before beginning data collection to allow baseline efflux to stabilize. Concentration-effect curves for whole brain samples were constructed by monitoring the effect of a single 5-sec stimulation with various concentrations of ACh. ACh stimulation of 86Rb+ efflux from the fourteen brain regions was determined by measuring response of each sample to stimulation by three concentrations of ACh. Total superfusion time was 9 min with 5-sec ACh stimulations at 1, 4 and 7 min (in order: 3 µM, 10 µM and 30 µM ACh with 0 µM DHβE or 30 µM, 300 µM and 3000 µM ACh with 2 µM DHβE). Preliminary studies established that under these conditions there was little desensitization. When DHβE-resistant responses were measured, 2 µM DHβE was included in the buffer and in the ACh solutions throughout the experiment. Agonist stimulations were achieved by diverting buffer flow through a 200 µl loop that contained the desired ACh solution by the activation of a four-way rotary Teflon injection valve (Alltech Associates, Deerfield, IL). Baseline efflux was calculated using the whole curve including points before the first stimulation, after the final stimulation and also between agonist stimulations.
Data Calculation and Analyses
All curve fits were conducted using the non-linear least squares algorithm in Sigma Plot 2001. [3H]Epibatidine saturation curves were analyzed using a two-site model: B = [Bm1*Epi/(Kd1 + Epi)] + [Bm2*Epi/(Kd2 + Epi)], where B is the specific [3H]epibatidine bound at each concentration of [3H]epibatidine (Epi), while Bm1 and Bm2 are the maximal high and low affinity sites with apparent binding affinities of Kd1 and Kd2, respectively. Inhibition of [3H]epibatidine binding by cytisine or d-tubocurarine was analyzed using a two-site model: B = [B1 / (1 + I / K1)] + [B2 / (1 + I/K2)], where B is the [3H]epibatidine bound at any concentration of inhibitor, I; B1 and B2 are the density of binding sites sensitive to inhibition with apparent inhibition constants, K1 and K2, respectively. Inhibition of [3H]epibatidine binding by αBgt was analyzed using the following model: B= B1/(1+I/Ki) + B2 where B1 is binding inhibited by αBgt with an apparent inhibition constant of Ki and B2 is the binding insensitive to inhibition by αBgt.
Cytisine-sensitive and cytisine-resistant components of the higher-affinity [3H]epibatidine binding sites were calculated from the difference in binding between samples containing no cytisine and those containing 100 nM cytisine using the two-site inhibition equation with the Ki values for the two cytisine sites (0.5 nM and 30 nM) fixed and the apparent IC50 values at these sites calculated using the Cheng-Prussoff equation (IC50 = Ki*(1+L/Kd), where L is the concentration of [3H]epibatidine and Kd is the average high affinity binding constant calculated from saturation binding experiments for the concentration of [3H]epibatidine used in a specific experiment (Marks et a., 1998; Perry et al., 2002). Lower-affinity, αBgt-sensitive sites were calculated as the difference in binding between samples containing no αBgt and those containing 100 nM αBgt. αBgt-resistant sites were calculated as the difference in binding between samples containing 100 nM αBgt and those containing 100 nM αBgt plus 300 µM d-tubocurarine.
ACh-stimulated 86Rb+ efflux was calculated as the increase in radioactivity above the basal efflux rate and normalized to basal efflux. Basal 86Rb+ efflux was calculated by fitting the counts before, between and after the stimulations to a two component exponential decay: EBt = EB0A*e-kA*t + EB0B*e-kB*t, where EBt is the basal 86Rb+ in each fraction at time t, EB0A and EB0B are the counts for the two components of efflux at time 0, and kA and kB are first order decay constants for the two components, respectively. ACh-stimulated efflux was calculated by subtracting the basal efflux calculated from the two-component exponential decay curve from the actual data. This difference was then divided by the basal efflux to yield a normalized response. ACh-stimulated 86Rb+ efflux was estimated by summing the points exceeding basal efflux during the time of agonist exposure. This method of calculation allowed comparison of response among regions and genotypes.
SPSSPC was used for statistical comparisons. In order to compare the effects of α4 genotype (+/+, +/− and −/−) on binding and efflux parameters in whole brain, a one-way analysis of variance (ANOVA) was used. To compare the effect of α4 gene deletion on binding and efflux in the fourteen brain regions, a two-way ANOVA (genotype by brain region) was used. Subsequently, data were analyzed using one-way ANOVA followed by Duncan’s post hoc test to evaluate the effect of genotype (gene deletion) on the parameters within each brain region.
RESULTS
Pharmacologically Identifiable [3H]Epibatidine Binding in Whole Brain Following Functional Deletion of the α4 nAChR Subunit Gene
The concentration dependence of [3H]epibatidine binding in whole brain is shown in Figure 1A. When a wide concentration range (0.005 to 40 nM) of ligand is used, [3H]epibatidine binding is distinctly biphasic as demonstrated by the Scatchard plots shown in Figure 1B. Apparent Kd values of 0.014 ± 0.001 nM and 7.2 ± 2.2 nM for the higher and lower-affinity sites, respectively, were estimated by nonlinear curve fitting and did not differ among the genotypes. Maximal binding for the higher and lower-affinity sites for wild-type mice were comparable (52.6±1.7 fmol/mg protein and 50.0±4.9 fmol/mg protein, respectively). A gene-dose dependent reduction following deletion of the α4 subunit was noted for [3H]epibatidine binding sites with either higher affinity (α4+/+= 52.6±1.7 fmol/mg protein; α4+/−=28.9±1.0 fmol/mg protein and α4−/− = 3.5±1.2 fmol/mg protein) or lower affinity (α4+/+ = 50.±4.9 fmol/mg protein; α4+/− = 29.7±2.1 fmol/mg protein and α4−/− = 19.6±2.3 fmol/mg protein). Deletion of α4 gene resulted in a greater reduction for the higher affinity sites (93%reduction) than for the lower affinity sites (61% reduction).
Figure 1. Effect of α4 nAChR gene deletion on [3H]epibatidine binding sites.
[3H]Epibatidine binding to whole brain particulate fractions of α4+/+ (black circles), α4+/− (gray circles) and α4−/− (white circles) mice. Each point is the mean±SEM of 4 separate experiments. Panel A shows saturation curves for [3H]epibatidine binding. Panel B shows Scatchard plots of the results in Panel A illustrating the biphasic binding pattern. Panel C shows inhibition of [3H]epibatidine (0.4 nM) binding by cytisine. Panel D shows inhibition of [3H]epibatidine (8 nM) binding by d-tubocurarine. All lines have been obtained by least squares curve fits of the results to the two site models described in the Methods.
Inhibition of [3H]epibatidine (0.4 nM) binding by cytisine is shown in FIGURE 1C. The inhibition profiles deviate from those expected for a single site and can be resolved into two components differentially sensitive to inhibition by cytisine. The IC50 values calculated for the higher affinity component (average IC50 = 2.7 nM) and the lower affinity component (average IC50 = 210 nM) did not differ among the genotypes. Targeting of the α4 subunit gene resulted in a gene dose-dependent reduction in the sites with higher affinity for cytisine (α4+/+ = 41.8 ± 4.1 fmol/mg protein; α4+/− = 18.6±3.3 fmol/mg protein; and α4−/− = 1.3±1.3 fmol/mg protein) and also the sites with lower affinity for cytisine (α+/+ = 10.1±4.3 fmol/mg protein; α4+/− = 8.0±3.4 14 fmol/mg protein; and α4−/− = 4.3±1.3 fmol/mg protein). The cytisine-sensitive sites were more affected by functional α4 gene deletion (98% reduction, sites remaining in α4−/− mice are not significantly different from zero) than were the cytisine-resistant sites (57% reduction).
Differential inhibition by d-tubocurarine estimates the number of lower-affinity sites (Marks et al., 1999, 2006) and is illustrated in Panel 1D. Inhibition of [3H]epibatidine (8 nM) by d-tubocurarine is biphasic. IC50 values for the sites with higher (average IC50 = 82 µM) and lower (average IC50 = 13 mM) affinity for d-tubocurarine did not differ among genotypes. α4 gene targeting reduced both the components with higher (α4+/+ = 28.1±4.4 fmol/mg protein; α4+/− = 17.7±2.1 fmol/mg protein; and α4−/− = 6.5±0.4 fmol/mg protein) and lower (α+/+ = 52.0±8.5 fmol/mg protein; α4+/− = 28.4±2.5 fmol/mg protein; and α−/− = 10.0±1.2 fmol/mg protein) affinity for d-tubocurarine. Binding to the lower-affinity [3H]epibatidine binding site when using 8 nM [3H]epibatidine is not maximal. However, binding to the higher affinity [3H]epibatidine sites is maximal and the density of these sites is comparable to the density measured with 0.4 nM [3H]epibatidine.
Effects of Deletion of α4 Gene on Nicotinic Binding in Fourteen Brain Regions
The experiments with whole brain homogenates described above demonstrate that the α4 subunit is essential for the expression of distinct subsets of both the higher and lower-affinity [3H]epibatidine binding sites. Several of the pharmacologically identifiable subtypes in whole brain are expressed at low levels making evaluation of the effects of α4 gene targeting on these [3H]epibatidine binding subsets technically difficult. Therefore, the effects of α4 gene targeting on both higher and lower affinity [3H]epibatidine binding was determined in fourteen brain regions (Table 1). Graphical illustration of the effects of α4 gene targeting on these sites is provided for thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus in Figure 2.
Table 1.
Effects of α4 gene deletion on four pharmacologically identifiable [3H]epibatidine binding sites. Higher affinity [3H]epibatidine binding was measured using 0.4 nM ligand and cytisine-sensitive and cytisine-resistant sites were estimated as described in the Methods. Lower affinity [3H]epibatidine binding was measured using 15 nM ligand and αBgt-sensitive and αBgt-resistant sites were estimated as described in the Methods. All values represent mean ± SEM for 6–10 individual experiments and are expressed as fmol bound/mg protein. Values marked with an asterisk (*) differ significantly from those of wild-type mice.
| Higher Affinity [3H]epibatidine Sites | Lower Affinity [3H]epibatidine Sites | ||||
|---|---|---|---|---|---|
| Cytisine-Sensitive | Cytisine-Resistant | αBgt-Sensitive | αBgt-Resistant | ||
| Olfactory Bulbs | |||||
| α4+/+ | 9.11±3.05 | 21.75±7.36 | 4.77±3.93 | 17.03±4.78 | |
| α4+/− | 6.26±3.75 | 18.72±1.82 | 3.27±5.00 | 9.25±2.83 | |
| α4−/− | −0.58±2.04* | 17.01±1.49 | 2.93±2.81 | 9.28±4.41 | |
| Olfactory Tubercles | |||||
| α4+/+ | 39.70±4.90 | 13.33±2.74 | 16.70±4.27 | 20.03±3.37 | |
| α4+/− | 22.63±4.78* | 11.75±2.10 | 10.85±3.74 | 18.01±3.06 | |
| α4−/− | 2.25±1.23* | 6.28±0.67 | 12.63±3.43 | 11.36±3.96 | |
| Cerebral Cortex | |||||
| α4+/+ | 59.60±3.69 | 6.32±2.53 | 5.75±1.04 | 19.34±2.31 | |
| α4+/− | 28.61±2.33* | 5.33±1.04 | 8.38±1.48 | 7.91±1.74* | |
| α4−/− | 0.94±0.64* | 2.37±0.88 | 6.44±1.32 | 4.31±1.41* | |
| Hippocampus | |||||
| α4+/+ | 38.21±2.12 | 7.86±1.24 | 16.08±1.64 | 21.46±2.31 | |
| α4+/− | 20.06±1.91* | 3.53±1.60* | 17.65±1.49 | 11.46±1.87* | |
| α4−/− | −0.11±1.45* | 0.83±0.87* | 16.30±1.56 | 6.78±1.49* | |
| Striatum | |||||
| α4+/+ | 64.39±7.14 | 17.06±2.13 | 11.12±1.34 | 19.76±3.61 | |
| α4+/− | 41.46±3.45* | 13.38±1.23* | 9.50±2.25 | 14.71±3.99 | |
| α4−/− | 0.51±0.91* | 7.20±1.21* | 5.91±2.35 | 10.66±3.46 | |
| Hypothalamus | |||||
| α4+/+ | 37.73±3.89 | 13.55±1.96 | 19.71±1.86 | 17.70±4.33 | |
| α4+/− | 15.50±3.67* | 9.80±2.00 | 12.76±3.71 | 10.86±3.26 | |
| α4−/− | 1.41±1.53* | 3.50±2.16* | 15.13±3.76 | 10.73±2.22 | |
| Thalamus | |||||
| α4+/+ | 127.22±6.39 | 31.04±5.48 | 11.21±3.33 | 47.33±2.56 | |
| α4+/− | 60.97±2.99* | 18.97±2.78* | 12.03±2.51 | 24.60±1.59* | |
| α4−/− | −0.30±0.59* | 9.07±1.46* | 9.05±1.32 | 5.56±1.23* | |
| Habenula | |||||
| α4+/+ | 104.2±19.6 | 180.8±17.1 | 12.3±23.7 | 86.2±21.6 | |
| α4+/− | 26.5±5.3* | 159.5±23.1 | 6.4±16.7 | 67.3±8.9 | |
| α4−/− | −32.8±10.7* | 174.0±11.2 | 15.5±4.6 | 27.8±7.9* | |
| Interpeduncular Nucleus | |||||
| α4+/+ | 114.8±19.9 | 248.3±336.4 | 7.5±19.4 | 101.0±21.8 | |
| α4+/− | 77.2±16.0* | 273.3±32.6 | 5.6±18.6 | 82.1±21.2 | |
| α4−/− | 6.4±10.5* | 258.3±37.6 | 31.7±13.7 | 43.4±15.3 | |
| Midbrain | |||||
| α4+/+ | 95.37±5.98 | 21.03±4.27 | 15.86±3.60 | 34.28±3.66 | |
| α4+/− | 40.33±3.08* | 13.53±1.86 | 16.30±2.46 | 22.30±2.29* | |
| α4−/− | 1.11±1.07* | 4.64±1.28* | 11.32±1.06 | 12.78±2.43* | |
| Superior Colliculus | |||||
| α4+/+ | 109.27±16.15 | 68.8±10.34 | 27.44±8.78 | 64.51±9.80 | |
| α4+/− | 47.14±9.32* | 55.95±5.35 | 26.71±3.14 | 47.27±8.97 | |
| α4−/− | −0.64±3.11* | 41.85±5.91 | 22.18±5.61 | 28.92±7.06* | |
| Inferior Colliculus | |||||
| α4+/+ | 61.78±5.75 | 40.50±5.29 | 46.60±7.24 | 43.68±4.91 | |
| α4+/− | 19.00±2.54* | 30.44±3.47 | 45.39±5.74 | 21.60±7.50* | |
| α4−/− | 4.06±1.38* | 23.74±3.38* | 48.07±4.02 | 19.26±4.91* | |
| Hindbrain | |||||
| α4+/+ | 49.70±4.70 | 16.88±2.10 | 12.97±2.83 | 23.38±1.85 | |
| α4+/− | 23.32±2.61* | 10.60±2.06 | 9.73±1.65 | 13.23±2.51* | |
| α4−/− | 0.84±0.82* | 8.11±1.49* | 11.47±1.16 | 8.10±2.26* | |
| Cerebellum | |||||
| α4+/+ | 7.75±2.53 | 8.63±5.57 | 5.40±1.87 | 6.35±1.18 | |
| α4+/− | 2.75±1.85 | 6.15±3.23 | 4.31±2.05 | 4.38±1.14 | |
| α4−/− | −0.41±0.90* | 3.35±1.40 | 2.59±3.03 | 4.87±1.82 | |
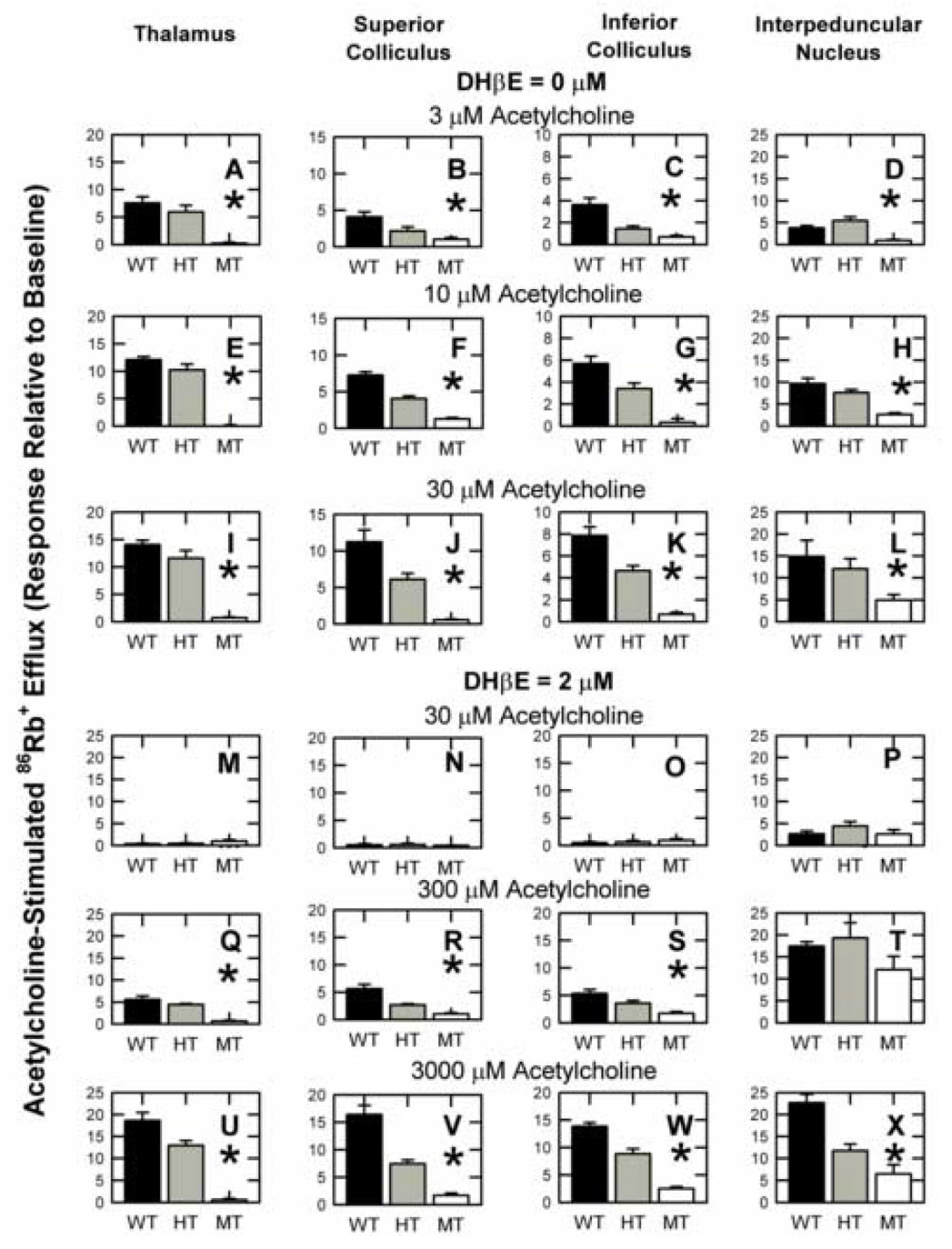
Figure 2. Effect of α4 nAChR gene deletion on pharmacologically identifiable [3H]epibatidine binding sites in four brain regions.
[3H]Epibatidine binding to particulate fractions prepared from thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus was measured using either 0.4 nM [3H]epibatidine to estimated the cytisine-sensitive, cytisine-resistant higher affinity sites or 15 nM [3H]epibatidine to measure the α-bungarotoxin-sensitive and α-bungarotoxin-resistant lower affinity sites as described in the methods. Each bar displays the mean ± SEM of 6–10 individual experiments for α4+/+ (black bars), α4+/− (gray bars) and α4−/− (white bars) mice. Those brain regions, in which a significant effect of α4 genotype on the indicated binding sites was detected by one-way ANOVA, are indicated with an asterisk (*). Results for each of the binding sites for these four regions and ten additional brain regions are summarized in Table 1.
Subsets of Higher-Affinity Binding Sites Following Functional α4 nAChR Gene Deletion
Cytisine-Sensitive, higher-affinity [3H]Epibatidine Binding Sites
The effect of α4 gene targeting on the cytisine-sensitive, higher affinity [3H]epibatidine binding sites in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus is illustrated in Figure 2, Panels A, B, C and D, respectively and is summarized for these and ten additional brain regions in Table 1.
Gene-dose dependent reductions in the density of cytisine-sensitive [3H]epibatidine binding sites occurred in all four brain regions. The binding remaining in the α4−/− mice was not significantly different from zero. In addition, binding site density in the α4+/− mice was intermediate between that in wild-type and null mutant mice.
A gene-dose dependent decrease in binding site density was noted in all fourteen regions assayed (Table 1). Highly significant effects of differential α4 nAChR subunit expression on the cytisine-sensitive high affinity [3H]epibatidine binding sites were found for genotype [F(2, 270)=368.05, p <0.001] and brain region [F(13,270)=24.43, P<0.001). The significant two-way interaction indicated that gene targeting differentially affected these binding sites across brain regions. The number of cytisine-sensitive binding sites measured in α4−/− mice did not differ from zero in any region. Therefore, within the limits of the assay, cytisinesensitive, higher affinity [3H]epibatidine binding is entirely dependent on the expression of the α4 subunit. The density of cytisine-sensitive sites in α4+/− mice was intermediate between that of wild-type and null mutant mice. The average density of these sites in the fourteen brain regions of α4+/− mice was 49.2% of that of wild-type mice and binding site density did not significantly differ from 50% in any brain region.
Cytisine-Resistant, Higher Affinity [3H]Epibatidine Binding
The effect of α4 gene targeting on the cytisine-resistant component of higher affinity [3H]epibatidine binding in Th, superior colliculus, inferior colliculus and interpeduncular nucleus is illustrated in Panels E, F, G and H of Figure 2 and summarized in Table 1 for these four and ten additional brain regions.
The results shown for thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus in Figure 2, Panels E, F, G and H, respectively illustrate the range of effects elicited by α4 gene targeting on cytisine-resistant [3H]epibatidine binding. A significant 71% decrease in cytisine-resistant [3H]epibatidine binding was noted in thalamus following α4 gene deletion. The 39% reduction in superior colliculus did not quite attain statistical significance (F(2,18), P = 0.065), but the 41% decrease in inferior colliculus was statistically significant (F(2,27) = 4.15, P = 0.028). The α4 subunit does not appear to be required for the expression of any of these sites in interpeduncular nucleus, since binding in mutant mice was essentially the same as that of wild-type mice (4% higher). In those brain regions where an effect of α4 gene deletion was noted the effect was gene-dose dependent since binding in α4+/− mice was intermediate between that of the wild-type and null mutant mice.
Analysis of the effects of α4 gene deletion on the cytisine-resistant sites further illustrated that these sites are composed of diverse subtypes and the expression of which varies markedly among brain regions (F(13,270)=149.5, P < 0.001). A modest overall effect of gene targeting across the fourteen brain regions was noted (F(2,270) = 3.21, P = 0.042) reflecting the fact that deleting the α4 subunit did not reduce cytisine-resistant [3H]epibatidine binding in every brain region. Significant reductions were observed in 7 of 14 brain regions assayed (Table 1). In contrast to the elimination of the cytisine-sensitive [3H]epibatidine binding sites following deletion of α4, cytisine-resistant sites in α4−/− mice were significantly greater than zero in twelve of the fourteen regions assayed (residual binding in hippocampus and hypothalamus did not differ significantly from zero). The density of the cytisine-resistant sites in α4+/− mice was intermediate between that of wild-type and null mutant mice. In the seven brain regions, for which a significant effect of α4 gene deletion was noted, average cytisine resistant site density was 47.3% that of wild-type mice.
Subsets of Lower-Affinity [3H]Epibatidine Binding Sites Following Functional α4 nAChR Gene Deletion
αBgt-Sensitive, Lower-Affinity [3H]Epibatidine Binding Sites
Graphical representation of the effects of α4 gene deletion on the density of αBgt-sensitive, lower-affinity [3H]epibatidine binding sites in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus is shown in Figure 2, Panels I, J, K and L, respectively. Effects of α4 gene deletion on these binding sites in these four and ten additional brain regions are summarized in Table 1.
The histograms in Figure 2, Panels I, J, K and L illustrate that α4 gene deletion did not significantly affect on αBgt-sensitive, lower-affinity [3H]epibatidine binding sites in thalamus, superior colliculus, inferior colliculus or interpeduncular nucleus.
Significant differences among brain regions in αBgt-sensitive, lower-affinity [3H]epibatidine site density were observed [F(13, 270)=8.26, P<0.001). α4 gene deletion had no significant effect on these sites [F(2,270)=0.22, P>0.05]. Subsequent analysis of individual brain regions confirmed that deletion of α4 had no significant effect on αBgt-sensitive [3H]epibatidine binding (Table 1).
αBgt-Resistant, Lower-Affinity [3H]Epibatidine Binding Sites
The effects of α4 gene deletion in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus are illustrated graphically in Panels M, N, O and P, respectively. The pattern observed following α4 gene deletion is similar in each region with a gene-dose dependent reduction in the density of αBgt-resistant, lower-affinity [3H]epibatidine sites, although the effect in interpeduncular nucleus is not statistically significant (P = 0.13). These figures also illustrate that these binding sites are heterogeneous, since α4 gene targeting did not completely eliminate expression (binding in each brain region of α4−/− mice was significantly greater than zero).
The density of the αBgt-resistant, lower-affinity [3H]epibatidine binding sites differed significantly among the fourteen brain regions [F(13, 270)=24.30, P<0.001) and was reduced by deletion of the α4 subunit. [F(2,270)=35.68, P<0.001] as summarized in Table 1. Deletion of α4 gene also tended to reduce the density of αBgt-resistant, lower-affinity [3H]epibatidine sites in most brain regions and these reductions were statistically significant in cerebral cortex, hippocampus, habenula, midbrain, and hindbrain in addition to thalamus, superior colliculus and inferior colliculus. However, αBgt-resistant sites remaining in every brain region of α4−/− mice were all significantly greater than zero. Binding site density in the α4+/− mice was generally intermediate between that of wild-type and null mutant mice with an average of 36.6% of the α4- dependent sites remaining in the α4+/− mice.
Comparison of the Effects of α4 Gene Targeting to Those of β2 and β4 Gene Targeting
The effects of β2 or β4 gene targeting on subsets of [3H]epibatidine binding sites has been determined previously (Marks et al., 1999, 2000, 2006 and unpublished results). Therefore, the effects of the β2 and β4 gene targeting can be compared to those of the α4 gene deletion presented here. These comparisons are made in Figure 3. The values plotted in this figure compare the quantitative effects of α 4 gene deletion to the quantitative effects of β2 or β4 gene deletion.
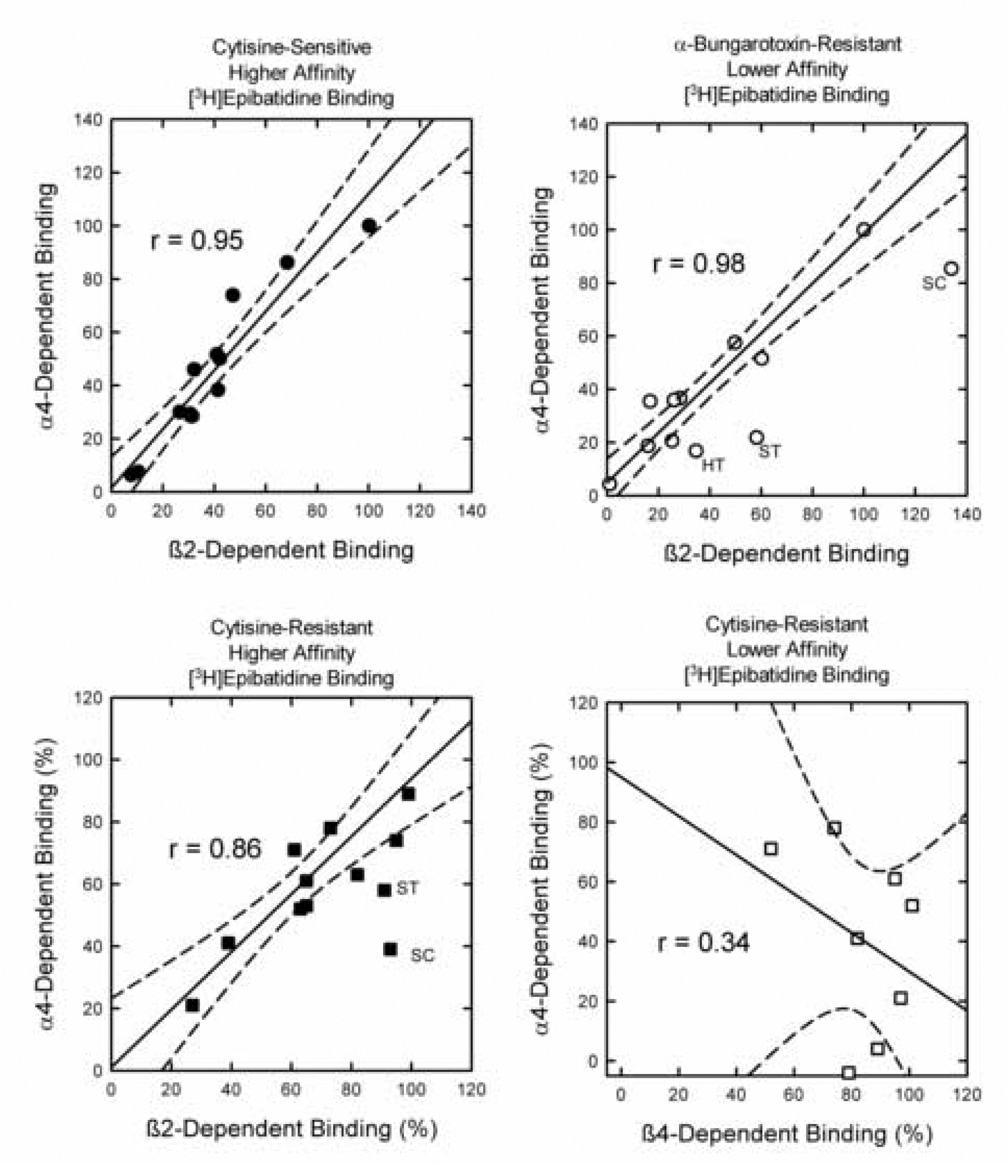
Figure 3. Comparison of the Effects of α4 Gene Deletion on [3H]Epibatidine Binding Sites to the Effects of β2 or β4 Gene Deletion.
The effects of α4 deletion on subsets of [3H]epibatidine binding sites to effects of β2 or β4 gene deletion. The best fit regression is represented by the solid line and the 95% confidence limits by the broken lines. Panel A compares the effects of β2 deletion on cytisinesensitive higher affinity [3H]epibatidine binding to those of α4 deletion. Deletion of either subunit essentially eliminated these sites. Data points represent deleted binding site densities normalized to 100 for thalamus. Panel B compares the effects of β2 deletion on αBgt-resistant lower affinity [3H]epibatidine binding to those of α4 deletion. Data points represent deleted binding site densities normalized to values of 100 for thalamus. The regression line was calculated omitting the data for hypothalamus (HT),striatum (ST) and superior colliculus (SC). Panel C compares the percentage of the cytisine-resistant higher affinity [3H]epibatidine binding sites affected by deletion of either the β2 or α4. The regression line was calculated omitting the data for striatum and superior colliculus. Panel D compares the percentage of cytisine-resistant higher affinity [3H]epibatidine binding sites affected by deletion of either the β4 or α4. Only brain regions in which a significant effect of β4 gene deletion are included.
Deletion of either α4 or β2 virtually eliminates cytisine-sensitive, higher affinity [3H]epibatidine and the quantitative effects of the deletion of these two genes in the various brain regions is shown in Figure 3A. α4 and β2 gene deletion elicits very similar and highly correlated reductions in these binding sites as illustrated by the correlation coefficient of 0.95.
A significant subset of the αBgt-resistant, lower affinity [3H]epibatidine binding sites are eliminated by deletion of either α4 or β2. A scattergram comparing the effects of α 4 and β2 gene deletion on these sites across all brain regions yielded a correlation coefficient of 0.86. However, it was noted that the effect of β2 gene deletion on these lower affinity sites was significantly greater than that of α4 in three brain regions (hypothalamus, striatum and superior colliculus). In order to illustrate this point, the analysis of the effects of α4 and β2 gene deletion was conducted omitting the data from these regions and is represented by the regression line in Figure 3B (r = 0.98). Comparison of the data for hypothalamus, striatum and superior colliculus to the regression line demonstrates binding in these regions is significantly more affected by deletion of β2 than by deletion of α4, suggesting that a significant fraction of the β2*-dependent sites in these regions are not α4 β2*-dependent sites.
The cytisine-resistant, higher affinity [3H]epibatidine binding sites are heterogeneous, although deletion of either α4 or β2 affects them. Results in Figure 3C compare the percentage reduction of binding site density following deletion of either the α4 or β2 subunit. Regression analysis of all the data yielded a correlation coefficient of 0.68. However, as was the case with the αBgt-resistant, lower affinity [3H]epibatidine binding shown in Figure 3B, the significantly greater effects of β2 than α4 gene deletion were noted in striatum and superior colliculus. Therefore, as for Figure 3B, the correlation omitted data from these two regions (r = 0.86) and the regression line obtained from this analysis is shown in Figure 3C to illustrate that the effects of β2 deletion were significantly greater than those of α4 in striatum and superior colliculus.
Some of the cytisine-resistant, higher affinity [3H]epibatidine binding sites are reduced or eliminated by deletion of β4. The results in Figure 3D compare only the effects of α4 or β4 gene deletion in those regions with a significant effect of β4 gene deletion and presented as percent reduction from wild-type. The significant scatter and low correlation coefficient (r=0.34) indicate little overlap of the effects of α4 and β4 gene deletion.
ACh-Stimulated 86Rb+ Efflux in Whole Brain Following Functional Deletion of the α4 nAChR Subunit Gene
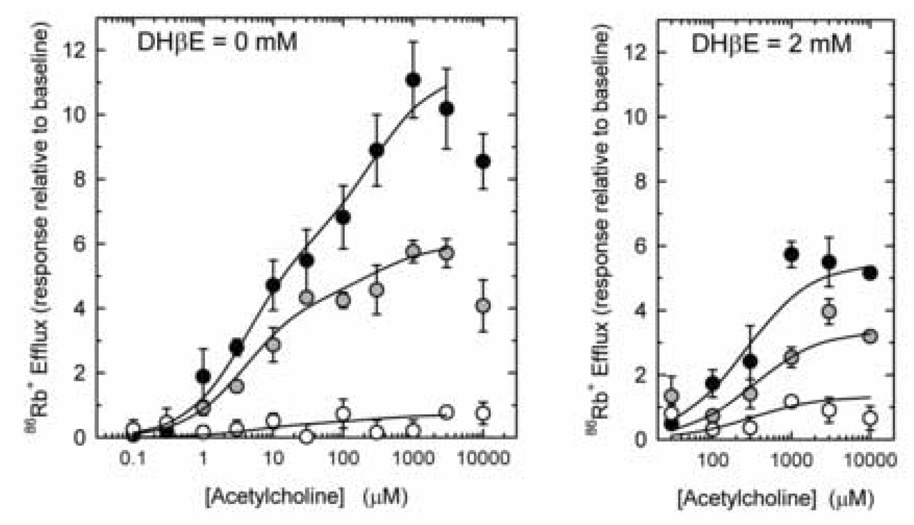
86Rb+ efflux has been used as a biochemical measurement of nAChR function in mouse brain synaptosomes (Marks et al., 1999; 2000, 2004). Nicotinic agonist stimulated 86Rb+ efflux from mouse brain synaptosomes is biphasic, a pattern that is similar to the biphasic ACh concentration-effect curves observed in heterologous expression systems (for example, Buisson and Bertrand, 2001; Nelson et al., 2003; Zhou et al., 2003; Zwart and Vijverberg, 1998). Nicotinic agonists vary in potency and efficacy in evoking 86Rb+ efflux (Marks et al., 1996). We used ACh in the studies described here because the biphasic nature of its concentration-effect curves is readily apparent and because its use allows better comparison to the results that have been reported on using heterologous expression systems. In order to obtain an initial estimate of the functional effects of α4 gene targeting, concentration effect curves for ACh stimulation of 86Rb+ efflux from whole brain synaptosomes were constructed for α4+/+, α4+/− and α4−/− mice.
Synaptosomes that had been loaded with 86Rb+ were exposed to various concentrations of ACh for 5 sec. The effect of α4 gene targeting on these responses is shown in Figure 4, Panel A. ACh-stimulated 86Rb+ efflux from wild-type synaptosomes is distinctly biphasic. Average EC50 values calculated from the two site fit of these curves are 3.3 µM and 380 µM for the components with higher and lower sensitivity to ACh, respectively. Targeting of the α4 subunit gene substantially reduced the maximal 86Rb+ efflux for both the higher (α4+/+ = 6.08±1.21; α4+/− = 4.11±0.71; and α4−/− = 0.42 ± 0.71) and lower (α4+/+ = 5.36±1.12; α4+/− = 1.79±0.67; and α4−/− = 0.05±0.65) sensitivity components of the ACh-stimulated responses.
Figure 4. Effect of α4 nAChR gene deletion on ACh-stimulated 86Rb+ efflux.
86Rb+ efflux stimulated by a 5-sec exposure to the indicated concentration of ACh was determined for whole brain synaptosomes prepared from α4+/+ (black circles), α4+/− (gray circles) and α4−/− (white circles) in the absence (Panel A) or presence (Panel B) of 2 µM DHβE. Each point is the mean±SEM of six separate experiments. The lines in panel A were calculated using a two site model and the lines in panel B were calculated using a one site model and represent the best fit determined by a non-linear least squares curve fit of the data.
The higher sensitivity component of ACh-stimulated 86Rb+ efflux is also more sensitive to inhibition by DHβE than is the component with lower ACh sensitivity (Marks et al., 1999). Consequently, agonist-stimulated 86Rb+ efflux measured in the presence of 2 µM DHβE and evoked by a short exposure to agonist primarily represents that component with lower sensitivity to activation by nicotinic agonists, including ACh. Therefore, in order to obtain an additional estimate of the effect of α4 gene targeting on the lower sensitivity component of ACh-stimulated 86Rb+ efflux, responses in the presence of 2 µM DHβE were determined and are shown in Figure 4B. The results of this analysis confirmed a gene-dose dependent reduction in the component with lower sensitivity for ACh activation (DHβE-resistant 86Rb+ efflux) (α4+/+ = 5.88±0.57; α4+/− = 3.69±0.58; and α4−/− = 0.37±0.47) by targeting of the α4 subunit gene. EC50 values were not significantly affected by α4 gene deletion (average EC50 = 370 µM)
Effects of α4 Gene Targeting on DH βE-Sensitive (Higher ACh Sensitivity Component) ACh-Stimulated 86Rb+ Efflux in Fourteen Brain Regions
In order to screen the effects of α4 gene targeting on the component of 86Rb+ efflux with higher sensitivity to ACh, crude synaptosomes loaded with 86Rb+ were stimulated by exposure to 3 µM, 10 µM and 30 µM ACh for 5 sec in the absence of DHβE. The results for all fourteen brain regions are summarized in Table 2 and graphical representations of the effect of α4 gene targeting in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus are shown in Figure 5.
Table 2.
Effects of α4 gene deletion on ACh-stimulated 86Rb+ efflux. ACh-stimulated 86Rb+ efflux was measured using synaptosomes prepared from fourteen brain regions in either the absence of DHβE using 3 µM, 10 µM and 30 µM ACh or in the presence of 2 µM DHβE using 30 µM, 300 µM and 3000 µM ACh. Values are mean ± SEM of 5–7 individual experiments and are expressed as peak size relative to baseline. Values differing significantly (P < 0.05) from those of wild-type mice are marked with an asterisk (*). Values for α4 null mutant mice differing significantly from zero (P < 0.05) are marked with a dagger (†).
| ACh-Stimulated 86Rb+ Efflux DHβE = 0 µM | ACh-Stimulated 86Rb+ Efflux DHβE = 2 µM | |||||
|---|---|---|---|---|---|---|
| [ACh] | 3 µM | 10µM | 30µM | 30µM | 300µM | 3000 µM |
| Olfactory Bulbs | ||||||
| α4+/+ | 1.7±0.3 | 2.3±0.5 | 3.2±0.4 | 0.3±0.2 | 1.9±0.4 | 5.7±0.8 |
| α4+/− | 0.6±0.1* | 1.1±0.1* | 1.5±0.3* | 0.3±0.1 | 1.9±0.2 | 3.0±0.2* |
| α4−/− | 0.5±0.2* | 0.3±0.1*† | 0.2±0.2* | 0.7±0.2† | 0.9±0.1*† | 1.1±0.2*† |
| Olfactory Tubercles | ||||||
| α4+/+ | 1.6±0.2 | 3.1±0.4 | 5.0±0.8 | 0.3±0.1 | 2.3±0.2 | 7.6±0.7 |
| α4+/− | 1.1±0.2 | 2.1±0.3* | 2.7±0.3* | 0.4±0.2 | 1.8±0.2 | 5.0±0.5* |
| α4−/− | 0.6±0.2*† | 0.2±0.1* | 0.6±0.1*† | 0.1±0.2 | 0.9±0.2*† | 2.6±0.4*† |
| Cerebral Cortex | ||||||
| α4+/+ | 2.2±0.5 | 3.5±0.3 | 5.0±0.5 | 0.1±0.1 | 1.7±0.4 | 6.7±0.9 |
| α4+/− | 1.4±0.4 | 2.9±0.3 | 3.6±0.4* | 0.4±0.1 | 1.3±0.2 | 4.6±0.5* |
| α4−/− | 0.4±0.1*† | 0.2±0.2* | 0.3±0.1*† | 0.2±0.1 | 0.4±0.1*† | 0.8±0.2*† |
| Hippocampus | ||||||
| α4+/+ | 2.1±0.7 | 3.4±0.8 | 4.8±0.4 | 0.3±0.2 | 1.8±0.3 | 5.8±0.7 |
| α4+/− | 1.1±0.2 | 2.0±0.2* | 3.5±0.3* | 0.3±0.1 | 1.2±0.2 | 2.9±0.5* |
| α4−/− | 0.2±0.1* | −0.1±0.2* | 0.3±0.1* | 0.4±0.1† | 0.9±0.3† | 0.4±0.1*† |
| Striatum | ||||||
| α4+/+ | 2.3±0.4 | 4.9±0.6 | 5.6±0.6 | 0.7±0.2 | 2.1±0.4 | 7.1±0.9 |
| α4+/− | 1.8±0.1* | 2.4±0.3* | 4.2±0.4* | 0.3±0.1 | 1.7±0.3 | 4.8±0.8* |
| α4−/− | 0.3±0.1*† | 0.0±0.1* | 0.7±0.2*† | 0.2±0.2 | 0.8±0.3*† | 0.6±0.2* |
| Hypothalamus | ||||||
| α4+/+ | 2.0±0.6 | 3.6±0.5 | 5.3±0.8 | 0.3±0.1 | 2.5±0.5 | 5.4±0.7 |
| α4+/− | 0.9±0.2 | 1.9±0.2* | 2.6±0.2* | 0.5±0.2 | 1.5±0.2* | 3.8±0.5* |
| α4−/− | 0.1±0.3* | 0.7±0.5* | 0.4±0.2* | 0.2±0.2 | 0.7±0.2*† | 0.6±0.1*† |
| Thalamus | ||||||
| α4+/+ | 7.5±1.2 | 12.9±0.6 | 14.1±0.5 | 0.4±0.1 | 5.6±0.8 | 18.6±1.9 |
| α4+/− | 5.9±1.2 | 10.2±1.0 | 11.6±1.4 | 0.5±0.1 | 4.4±0.2 | 12.9±1.1* |
| α4−/− | 0.2±0.2* | 0.1±0.1* | 0.7±0.2*† | 1.0±0.3† | 0.7±0.2*† | 0.6±0.3* |
| Habenula | ||||||
| α4+/+ | 11.0±0.7 | 18.5±1.4 | 19.9±1.9 | 0.6±0.2 | 6.1±1.5 | 22.5±2.6 |
| α4+/− | 9.4±1.2 | 19.8±1.7 | 19.8±1.5 | 0.9±0.2 | 5.9±1.1 | 16.4±2.0* |
| α4−/− | 0.2±.0.1* | 0.7±0.1*† | 0.3±0.3* | 0.3±0.1† | 1.0±0.1*† | 0.7±0.1*† |
| Interpeduncular Nucleus | ||||||
| α4+/+ | 3.8±0.5 | 9.6±1.2 | 14.9±3.7 | 2.1±0.6 | 17.2±1.1 | 22.5±2.3 |
| α4+/− | 5.4±0.9 | 7.6±0.7 | 12.1±2.3 | 4.4±1.1 | 19.3±3.4 | 11.8±2.1* |
| α4−/− | 1.0±0.2*† | 2.6±0.4*† | 5.0±1.3*† | 2.6±1.0 | 12.1±3.1† | 6.5±2.1*† |
| Midbrain | ||||||
| α4+/+ | 1.9±0.3 | 3.9±0.4 | 6.2±0.7 | 0.5±0.1 | 3.4±0.6 | 10.0±1.0 |
| α4+/− | 1.8±0.2 | 3.0±0.4 | 3.4±0.6* | 0.3±0.2 | 1.8±0.2* | 6.5±0.5* |
| α4−/− | 0.6±0.3* | 0.6±0.2*† | 0.5±0.3* | 0.2±0.1 | 0.6±0.1*† | 0.1±0.3* |
| Superior Colliculus | ||||||
| α4+/+ | 4.2±0.8 | 7.4±0.5 | 12.2±1.6 | 0.4±0.3 | 5.7±1.0 | 16.5±2.0 |
| α4+/− | 2.2±0.5* | 4.1±0.4* | 6.1±0.8* | 0.6±0.2 | 2.7±0.2* | 7.5±0.7* |
| α4−/− | 1.0±0.2*† | 1.3±0.2*† | 0.6±0.1*† | 0.4±0.1† | 1.1±0.2*† | 1.7±0.4*† |
| Inferior Colliculus | ||||||
| α4+/+ | 3.6±0.6 | 5.7±0.7 | 7.9±0.8 | 0.5±0.1 | 5.3±0.7 | 13.8±0.7 |
| α4+/− | 1.4±0.2* | 3.4±0.5* | 4.7±0.5* | 0.6±0.3 | 3.6±0.5* | 8.8±0.9* |
| α4−/− | 0.7±0.2*† | 0.4±0.3* | 0.9±0.2*† | 0.9±0.3† | 1.7±0.3*† | 2.5±0.4*† |
| Hindbrain | ||||||
| α4+/+ | 1.9±0.5 | 2.9±0.4 | 5.0±0.8 | 0.2±0.2 | 3.4±0.4 | 7.5±1.2 |
| α4+/− | 1.2±0.4 | 1.8±0.5 | 3.6±0.5 | 0.1±0.1 | 3.0±0.4 | 5.2±0.5 |
| α4−/− | 0.6±0.2† | 0.8±0.2*† | 0.2±0.3* | 0.3±0.1† | 0.6±0.1*† | 1.2±0.4*† |
| Cerebellum | ||||||
| α4+/+ | 0.5±0.1 | 0.6±0.1 | 0.8±0.2 | 0.4±0.2 | 1.3±0.4 | 2.0±0.3 |
| α4+/− | 0.7±0.3 | 0.6±0.2 | 1.1±0.3 | 0.2±0.1 | 0.8±0.2 | 1.8±0.2 |
| α4−/− | 0.2±0.1 | 0.6±0.4 | 0.7±0.3† | 0.4±0.2 | 1.1±0.1† | 1.2±0.3† |
Figure 5. Effect of α4-nAChR gene deletion on DHβE-sensitive and DHβE-resistant ACh-stimulated 86Rb+ efflux.
Crude synaptosomes were prepared from thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus of α4+/+ (black bars), α4+/− (gray bars) and α4−/− (white bars) mice. Each bar represents the mean ±SEM of 5–7 individual experiments. Synaptosomes loaded with 86Rb+ were exposed for 5 sec to 3 µM, 10 µM and 30 µM ACh in the absence of DHβE or to 30 µM, 300 µM or 3000 µM ACh in the presence of 2 µM DHβE as indicated. Those brain regions, in which a significant effect of α4 gene deletion was detected by one-way ANOVA, are marked with an asterisk (*). Results for these four regions and ten addition regions are summarized in Table 2.
Graphical representation of the effects of α4 gene deletion for thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus are shown in Figure 5. 86Rb+ efflux increased with increasing ACh concentration and targeting of the α4 gene significantly reduced ACh-stimulated 86Rb+ efflux in each brain region following stimulation with all three ACh concentrations. However, the magnitude of the reductions following α 4 gene targeting varied among the brain regions. These differences are most apparent for responses elicited by 30 µM ACh. A 95% reduction in response was observed for thalamus (Figure 5I) and superior colliculus (Figure 5J), while approximately one-third of the ACh-stimulated response persisted in the interpeduncular nucleus of α4−/− mice (Figure 5L) and 11% remained in inferior colliculus (Figure 5K). The residual 86Rb+ efflux stimulated by 30 µM ACh in all four regions of α4−/− mice was significantly greater than zero indicating that some ACh-stimulated 86Rb+ efflux with higher ACh sensitivity did not require expression of the α4 subunit.
The effects of α4 gene targeting on the DHβE-sensitive (higher ACh sensitivity) responses in the fourteen brain regions assayed are summarized in Table 2. The magnitude of ACh-stimulated 86Rb+ efflux measured at each ACh concentration varied among the brain regions (3 µM-F(13, 216) = 40.36; 10 µM-F(13, 216) =111.63; and 30 µM-( F(13,216) = 47.61; P<0.001 for each concentration) and deletion of the α4 subunit significantly reduced the response at each ACh concentration (3 µM-F(2,216) = 95.08; 10 µM-F(2,216) = 256.63; and 30 µM-F(2,216) = 163.63; P<0.001 for each concentration). Targeting of the α4 gene significantly reduced ACh-stimulated responses in every brain region at each ACh concentration with the exception of cerebellum, in which the low responses elicited by ACh were unaffected by deletion of α4. Targeting of the α4 gene virtually eliminated the86Rb+ efflux stimulated by 30 µM ACh in olfactory bulbs, hippocampus, hypothalamus, habenula, midbrain, and hindbrain, but in olfactory tubercles, cerebral cortex, striatum, thalamus, interpeduncular nucleus, superior colliculus, inferior colliculus and cerebellum residual 86Rb+ efflux was significantly greater than zero.
Effects of Functional α4 Gene Deletion on DHβE-Resistant (Lower ACh Sensitivity) ACh-Stimulated 86Rb+ Efflux in Fourteen Brain Regions
In order to screen the effects of α4 gene targeting on the component of 86Rb+ efflux with lower affinity for ACh, crude synaptosomes loaded with 86Rb+ were stimulated by exposure to 30 µM, 300 µM and 3000 µM ACh for 5 sec in the presence of 2 µM DHβE. This concentration of DHβE inhibits the higher sensitivity ACh-stimulated 86Rb+ efflux (Marks et al., 1999). The results for all fourteen brain regions are summarized in Table 2 and graphical representations of the effect of α4 gene targeting in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus are shown in Figure 5.
86Rb+ efflux increased with increasing ACh concentration and deletion of the α4 gene significantly reduced ACh-stimulated 86Rb+ efflux in each brain region following stimulation with 300 µM and 3000 µM, but not the 30 µM, ACh concentrations. Note that for each of the four brain regions, the 86Rb+ efflux measured in thalamus, superior colliculus, inferior colliculus and interpeduncular nucleus with 30 µM ACh for each α4 genotype in the presence of DHβE (Panels 5M, 5N, 5O and 5P, respectively) are similar to the 86Rb+ efflux measured following stimulation with 30 µM ACh in the absence of DHβE for these four regions of the α4−/− mice (Panels 5I, 5J, 5K and 5L). This similarity suggests that the 86Rb+ efflux persisting in the α α4−/− mice at 30 µM ACh is relatively insensitive to inhibition by DHβE. When synaptosomes prepared from these four brain regions were stimulated with 300 µM or 3000 µM ACh, significant effects of α4 gene deletion were noted. However, the magnitude of the reductions following α4 gene deletion varied among the brain regions. Targeting of the α4 gene significantly reduced 86Rb+ efflux stimulated by 300 µM or 3000 µM ACh in thalamus, superior colliculus and inferior colliculus, but the effect of gene deletion in interpeduncular nucleus was only significant when 3000 µM was used. The increase in 86Rb+ efflux observed in superior colliculus, inferior colliculus and interpeduncular nucleus with increasing ACh concentration indicates that the nonα4-nAChR in these brain regions have relatively low sensitivity to activation by ACh, while the persistent low activity in thalamus suggests that these residual nonα4-nAChR have relatively high sensitivity to ACh. The fraction of DHβE-resistant 86Rb+ efflux stimulated by 3000 µM ACh in α4−/− mice represents 3% of that in α4+/+ mice in thalamus, 10% in superior colliculus, 18% in inferior colliculus and 29% in interpeduncular nucleus (Panels 5U, 5V, 5W and 5X, respectively).
The results for DHβE-resistant ACh-stimulated responses in all fourteen brain regions are summarized in Table 2. No effect of α4 gene deletion was noted in any brain region following stimulation with 30 µM ACh. The activity measured for the three α4 genotypes under these conditions was very similar to that of the α4−/− stimulated with 30 µM ACh in the absence of DHβE. Deletion of α4 significantly reduced DHβE-resistant 86Rb+ efflux stimulated by 3000 µM ACh in every brain region but cerebellum, while the reduction following deletion of α4 was also found for 300 µM ACh in olfactory tubercles, cerebral cortex, striatum, hypothalamus, thalamus, habenula, midbrain, superior colliculus, inferior colliculus and hindbrain.
Comparison of Effects of α4 and β2 Gene Deletion on DHβE-Sensitive (Higher ACh Sensitivity) and DHβE-Resistant (Lower ACh Sensitivity) 86Rb+ Efflux
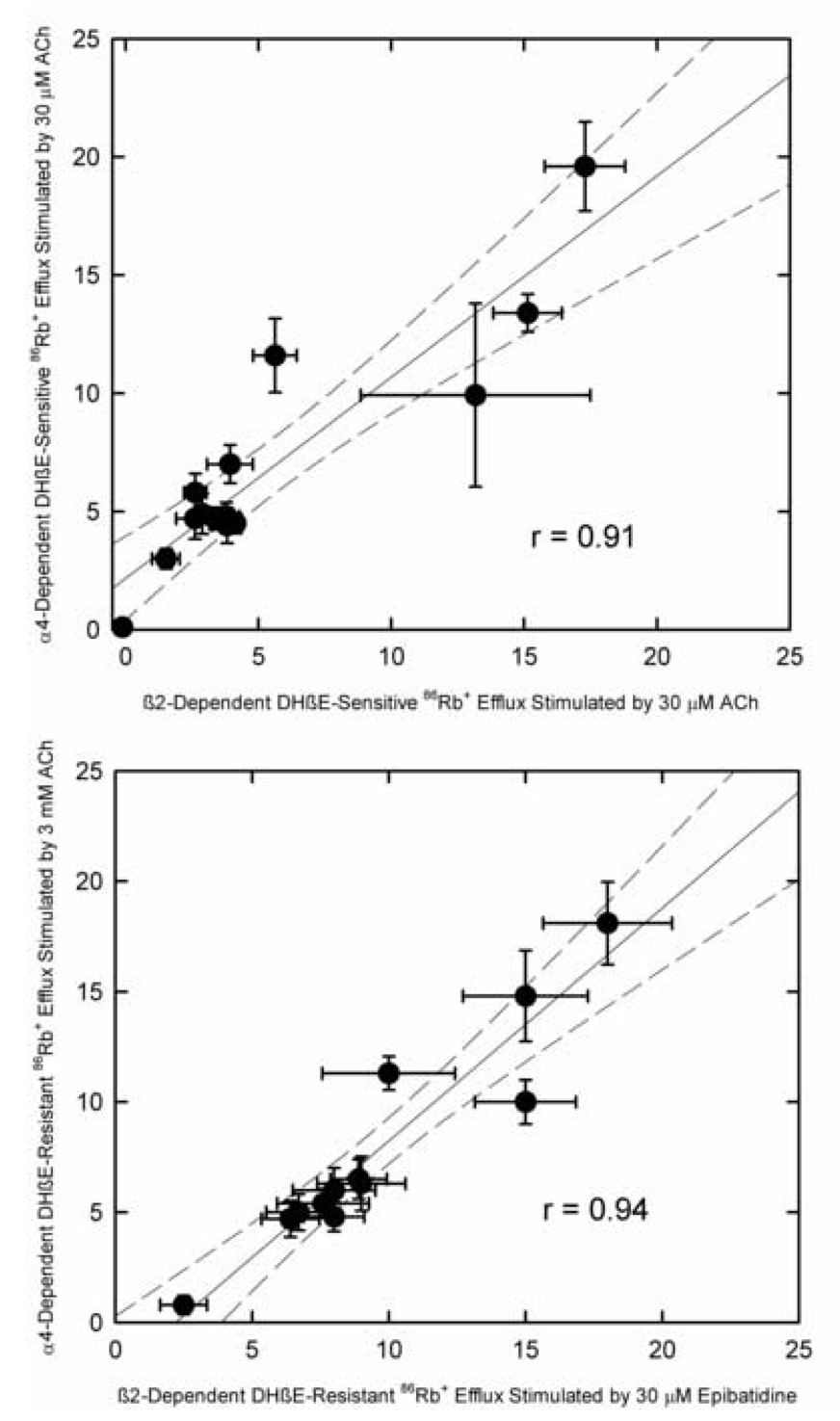
Previous results have demonstrated that deletion of the β2 subunit substantially reduces agonist-stimulated 86Rb+ efflux (Marks et al., 1999,2000 and unpublished observations). Thus, the effects of β2 deletion can be compared to those of α4 deletion for both DHβE-sensitive (higher ACh sensitivity) and DHβE-resistant (lower ACh sensitivity) 86Rb+ efflux as shown in Figure 6. These results compare the portion of the agonist-stimulated 86Rb+ efflux eliminated by α4 gene deletion to that eliminated by β2 gene deletion in the various brain regions (that is the difference in response between wild-type and null mutant mice). β2 and α4 gene deletion have very similar effects on both DHβE-sensitive (r=0.91) and DHβE-resistant (r=0.94) 86Rb+ efflux.
Figure 6. Comparison of the Effects of α4 or β2 Gene Deletion of 86Rb+ Efflux.
Effects of α4 gene deletion on DHβE-sensitive (Panel A) and DHβE-resistant (Panel B) 86Rb+ efflux are compared to the effects of β2 gene deletion. Each point represents the mean ±SEM for individual brain regions of the agonist-stimulated response sensitive to deletion of α4 or β2, that is the difference between responses measured for wild type and null mutant mice. Solid lines indicate the best-fit regression lines and dotted lines the 95% confidence limits for those lines.
Discussion
The results reported here indicate that the α4 subunit is required to form nAChRs that account for nearly 75% of total [3H]epibatidine binding in mouse brain and for expression of nAChRs that modulate both DHβE-sensitive (higher ACh sensitivity) and DHβE–resistant (lower ACh sensitivity) ACh-stimulated 86Rb+ efflux. Specifically, (1) Deleting α4 virtually eliminated cytisine-sensitive higher affinity [3H]epibatidine binding (approximately 45% of total brain [3H]epibatidine binding); (2) Deleting α4 reduced a fraction of cytisine-resistant [3H]epibatidine binding (approximately 2–3% of total brain [3H]epibatidine binding); (3) Deleting α4 eliminated a significant fraction of α-Bgt-resistant lower-affinity [3H]epibatidine binding (approximately 25% of total brain [3H]epibatidine binding) comparable to the effects of β2 deletion (Marks et al., 2006); (4) Deletion of α4 substantially reduces ACh-stimulated 86Rb+ efflux throughout the brain, but ACh-stimulated 86Rb+ efflux persists in several brain regions indicating that of nonα4β2*-nAChR mediate some ACh-stimulated 86Rb+ efflux; and (5) The extensive reduction of both nicotinic binding sites and functional responses by α4 deletion suggests that expression of previously unexpressed nAChR subtypes does not compensate for the deletion of α4.
[3H]Epibatidine Binding Sites
The elimination of cytisine-sensitive, high-affinity epibatidine binding sites by deletion of α4 (and β2, Marks et al., 2006) was expected given that immunochemical studies (Whiting and Lindstrom, 1989; Flores et al., 1992) and previous gene deletion studies (Picciotto et al., 1995; Marubio et al., 1999; Ross et al., 2000) established that sites with high agonist affinity are predominately, if not exclusively, α4β2*-nAChR. However, it had not previously been shown that α4 subunits contribute to the expression of lower affinity nAChRs in vivo. α4 gene deletion decreased lower affinity [3H]epibatidine binding in whole brain (≈60%) and in 8 of 14 regions (55% to 89%) (with a tendency to decrease expression in the remaining regions). Deleting the β2 gene produced similar effects (Marks et al., 2006 and data presented here). The prominent role of α4 and β2 subunits in the expression of both higher and lower affinity [3H]epibatidine binding sites is consistent with the observations that deletion of β2 (or α4) abolishes staining with a selective anti-α4 antibody, 125I-mAb299 (Whiteaker et al, 2006), and that [3H]epibatidine binding in cells transfected with the α4 and β2 nAChR subunits is biphasic with affinities for [3H]epibatidine very similar to those in mouse brain (Shafaee et al., 1999).
The nature of the lower affinity sites has not been definitively established, although there are several viable possibilities. One possibility is that the two sites with different affinity measure [3H]epibatidine binding to the desensitized state of α4β2-nAChRs with different α4/β2 stoichiometries that also exhibit different sensitivities for ACh activation (see below and Zwart and Vijverberg, 1998; Nelson et al., 2003; Zhou et al., 2003). A second possibility is that the lower affinity sites represent binding to ground state, rather than the desensitized state of the receptors. While possible, the fact that the incubations were conducted to equilibrium, which should have allowed ground state receptors to isomerize to the desensitized state (Lippiello et al., 1987; Marks et al., 1994), makes this unlikely. A third possibility is that the lower affinity sites represent binding to partially or improperly assembled receptors or to modified receptors or to receptors that are associated with other molecules. A fourth possibility is that the lower affinity receptors include an additional subunit. However, the relatively low expression of other nAChR subunits and the fact that biphasic [3H]epibatidine binding is observed in cells transfected with only α4 and β2 (Shafaee et al., 1999) suggests that incorporation of subunits that are neither α4 nor β2 cannot account for all of the lower affinity sites.
While the effects of α4 deletion on cytisine-sensitive higher affinity and αBgt-resistant lower-affinity [3H]epibatidine binding sites were significantly correlated across the 14 brain regions assayed (r=0.90), the relative ratio of these binding sites varied substantially among the brain regions (for example density of higher affinity sites was 7-fold greater than that of lower affinity sites in striatum, 3-fold greater in thalamus, but less than 2-fold greater in interpeduncular nucleus). This variability indicates that the α4-dependent higher and lower affinity sites are not merely different sites on the same receptor.
The gene-deletion studies also indicated that the α4 subunit contributed to some of the cytisine-resistant [3H]epibatidine binding sites. The studies described here do not yet establish possible subunit composition of these diverse sites. However, in some brain regions, such as striatum, olfactory tubercle and superior colliculus some of the cytisine-resistant, α4-containing higher affinity [3H]epibatidine binding sites correspond to sites that bind α-conotoxin MII (likely α4α6β2β3-nAChR, Champtiaux et al., 2003; Salminen et al., 2004; Gotti et al., 2004, 2005). It is certainly possible that assembly of α4 with other nAChR subunits contribute to these sites in other brain areas.
ACh-Stimulated 86Rb+ Efflux
The demonstration that α4 and β2 subunits are essential for expression of a significant fraction of both higher and lower affinity [3H]epibatidine binding sites does not establish the role of these subunits in nAChR mediated function. To address this issue, the functional consequences of α4 deletion were evaluated using ACh-mediated 86Rb+ efflux. Deletion of β2 reduces 86Rb+ efflux stimulated by lower and higher concentrations of ACh (corresponding to the DH βE-sensitive and DH βE-resistant responses, respectively) (Marks et al., 2000). Significant reductions in both responses were also observed following deletion of α4. The α4- dependent reduction in both DH βE-sensitive and DH βE-resistant 86Rb+ efflux is very similar to and highly correlated with the β2-dependent reduction throughout the brain (r=0.91 and r=0.94, respectively), supporting the assertion that most 86Rb+ efflux with both higher and lower sensitivity to activation by ACh is mediated by α4β2*-nAChR.
EC50 values for the two components of 86Rb+ efflux with different sensitivity to ACh are very similar to those measured in heterologous systems expressing α4β2-nAChR. These components have been postulated to be mediated by receptors differing in α4/β2 stoichiometry (higher sensitivity 2 α/3β, lower sensitivity 3α/2 β) (Zwart and Vijverberg, 1998; Nelson et al., 2003; Zhou et al., 2003). Inasmuch as deletion of α4 or β 2 (Marks et al., 2000) substantially reduced both the higher and lower sensitivity components of 86Rb+ efflux in mouse brain, it is distinctly possible that α4β2-nAChR with different stoichiometry similarly mediate biphasic ACh responses in native tissue.
Although α4β2*-nAChR is the predominant subtype that mediates both DH βE-sensitive and DH βE-resistant responses, residual activity persisted in 8 of 14 and 10 of 14 brain regions of α4−/− mice, respectively. This residual non α4 activity was generally small compared to that of wild-type mice. As was the case following β2 deletion, substantial activity with properties consistent with those of α3 β4-nAChR (Marks et al., 2002) was retained in interpeduncular nucleus and inferior colliculus. Residual DH βE-sensitive activity in olfactory tubercles and striatum of α4−/− mice was higher than that in β2−/− mice and may represent α6β2*-nAChR mediated responses that survive deletion of α4 but not β2 (Champtiaux et al., 2003; Salminen et al., 2004). The source of residual activity in other brain areas is presently unknown.
Acknowledgements
This study was supported by research grant DA03194 and animal resources grant DA15663 from the National Institute on Drug Abuse. The authors thank Julie J. Kuchinski, Jennifer A. Drapeau, Theresa DelVecchio and Esteban Loetz for animal care and genotyping.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkondon M, Albuquerque EX. Diversity of nicotinic acetylcholine receptors in rat hippocampal neurons. III. Agonist actions of the novel alkaloid epibatidine and analysis of type II current. J Pharmacol Exp Ther. 1995;274:771–782. [PubMed] [Google Scholar]
- Badio B, Daly JW. Epibatidine, a potent analgetic and nicotinic agonist. Mol Pharmacol. 1994;45:563–569. [PubMed] [Google Scholar]
- Bourin M, Ripoll N, Dailly E. Nicotinic receptors and Alzheimer's disease. Curr Med Res Opin. 2003;19:169–177. doi: 10.1185/030079903125001631. [DOI] [PubMed] [Google Scholar]
- Buisson B, Bertrand D. Chronic exposure to nicotine upregulates the human α4β2-nicotinic receptor function. J Neurosci. 2001;21:1819–1829. doi: 10.1523/JNEUROSCI.21-06-01819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champtiaux N, Gotti C, Cordero-Erausquin M, David DJ, Przbylski C, Lena C, Clementi F, Moretti M, Rossi FM, LeNovere N, McIntosh JM, Gardier AM, Changeux J-P. Subunit composition of functional nicotinic receptors in dopaminergic neurons expressed in dopaminergic neurons investigated with knock-out mice. J Neurosci. 2003;23:7820–7829. doi: 10.1523/JNEUROSCI.23-21-07820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani JA, DeBiasi M. Cellular mechanisms of nicotine addiction. Pharmacol Biochem Behav. 2001;70:439–446. doi: 10.1016/s0091-3057(01)00652-9. [DOI] [PubMed] [Google Scholar]
- Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of alpha 4 and beta 2 subunits and is up-regulated by chronic nicotine treatment. Mol Pharmacol. 1992;41:31–37. [PubMed] [Google Scholar]
- Gotti C, Moretti M, Zanardi A, Gaimarri A, Champtiaux N, Changeux J-P, Whiteaker P, Marks MJ, Clementi F, Zoli M. Heterogeneity and selective targeting of neuronal nicotinic acetylcholine receptor (nAChR) subtypes expressed on retinal afferents of the superior colliculus and lateral geniculate nucleus: identification of a new native nAChR subtype alpha3beta2(alpha5 or beta3) enriched in retinocollicular afferents. Mol Pharmacol. 2005;68:1162–1171. doi: 10.1124/mol.105.015925. [DOI] [PubMed] [Google Scholar]
- Houghtling RA, Davila-Garcia MI, Kellar KJ. Characterization of (+/−)(−) [3H]epibatidine binding to nicotinic cholinergic receptors in rat and human brain. Mol Pharmacol. 1995;48:280–287. [PubMed] [Google Scholar]
- Kuryatov A, Olale F, Cooper J, Choi C, Lindstrom J. Human α6 AChR subtypes: subunit composition, assembly, and pharmacological responses. Neuropharmacology. 2000;39:2570–2590. doi: 10.1016/s0028-3908(00)00144-1. [DOI] [PubMed] [Google Scholar]
- Leonard S, Adler LE, Benhammou K, Berger R, Breese CR, Drebing C, Gault J, Lee MJ, Logel J, Olincy A, Ross RG, Stevens K, Sullivan B, Vianzon R, Virnich DE, Waldo M, Walton K, Freedman R. Smoking and mental illness. Pharmacol Biochem Behav. 2001;70:561–570. doi: 10.1016/s0091-3057(01)00677-3. [DOI] [PubMed] [Google Scholar]
- Lindstrom J. In: The structure of nAChRs, in Neuronal Nicotinic Receptors, Handbook of Experimental Pharmacology. Clementi F, Fornasari D, Gotti C, editors. Vol. 144. Berlin: Springer-Verlag; 2000. pp. 101–162. [Google Scholar]
- Lippiello PM, Sears SB, Fernandez KG. Kinetics and mechanism of L-[3H]nicotine binding to putative high affinity receptor sites in rat brain. Mol Pharmacol. 1987;31:392–400. [PubMed] [Google Scholar]
- Marks MJ, Grady SR, Yang J-M, Lippiello PM, Collins AC. Desensitization of nicotine-stimulated 86Rb+ efflux from mouse brain synaptosomes. J Neurochem. 1994;63:2125–2135. doi: 10.1046/j.1471-4159.1994.63062125.x. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Meinerz NM, Kachinski JJ, Drapeau JA, Drago J, Collins AC. Elimination of the alpha4 nicotinic receptor subunit reduces both high and low affinity agonist-stimulated 86Rb+ efflux in mouse brain synaptosomes. Program no. 248.4. Washington, DC: Society for Neuroscience. Online; 2003. 2003 Abstract Viewer/Itinerary Planner. [Google Scholar]
- Marks MJ, Smith KW, Collins AC. Differential agonist inhibition identifies multiple epibatidine binding sites in mouse brain. J Pharmacol Exp Ther. 1998;285:377–386. [PubMed] [Google Scholar]
- Marks MJ, Whiteaker P, Calcaterra J, Stitzel JA, Bullock AE, Grady SR, Picciotto MR, Changeux J-P, Collins AC. Two pharmacologically distinct components of nicotinic receptor-mediated rubidium efflux in mouse brain require the β2 subunit. J Pharmacol Exp Ther. 1999;289:1090–1103. [PubMed] [Google Scholar]
- Marks MJ, Stitzel JA, Grady SR, Picciotto MR, Changeux J-P, Collins AC. Nicotinic agonist stimulated 86Rb+ efflux and [3H]epibatidine binding of mice differing in β2 genotype. Neuropharmacology. 2000;39:2332–2645. doi: 10.1016/s0028-3908(00)00115-5. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Whiteaker P, Grady SR, Picciotto MR, McIntosh JM, Collins AC. Characterization of [125I]epibatidine binding and nicotinic agonist-mediated 86Rb+ efflux in interpeduncular nucleus and inferior colliculus of β2 null mutant mice. J Neurochem. 2002;81:1102–1115. doi: 10.1046/j.1471-4159.2002.00910.x. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Whiteaker P, Collins AC. Deletion of the α7, β2 or β4 Nicotinic Receptor Subunit Genes Identifies Highly Expressed Subtypes with Relatively Low affinity for [3H]epibatidine. Mol Pharmacol. 2006;70:947–959. doi: 10.1124/mol.106.025338. [DOI] [PubMed] [Google Scholar]
- Marritt AM, Cox BC, Yasuda RP, McIntosh JM, Xiao Y, Wolfe BB, Kellar KJ. Nicotinic cholinergic receptors in the rat retina: simple and mixed heteromeric subtypes. Mol Pharmacol. 2005;68:1656–1668. doi: 10.1124/mol.105.012369. [DOI] [PubMed] [Google Scholar]
- Marubio LM, del Mar Arroyo-Jimenez M, Cordero-Erausquin M, Lena C, Le Novere N, de Kerchove d’Exaerde A, Huchet M, Damaj MI, Changeu J-P. Reduced antinociception in mice lacking neuronal nicotinic receptor subunits. Nature (Lond) 1999;398:805–810. doi: 10.1038/19756. [DOI] [PubMed] [Google Scholar]
- Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Trans. 2003;31:869–874. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- Nelson ME, Kuryatov A, Choi CG, Zhou Y, Lindstrom J. Alternate stoichiometries of α4β2 nicotinic acetylcholine receptors. Mol Pharmacol. 2003;63:332–341. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Goldner FM, Saeki M, Lorenzo T, Goldberg T, DeBiasi M, Dani JA, Patrick JW, Beaudet AL. Mice deficient in the α7 neuronal nicotinic acetylcholine receptor lack α-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci. 1997;17:9165–9171. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MJ, Beck A, Luetje CW. Neuronal nicotinic receptor β2 and β4 subunits confer large differences in agonist binding affinity. Mol Pharmacol. 2001;54:1132–1139. [PubMed] [Google Scholar]
- Perry DC, Xiao Y, Nguyen HN, Musachio JL, Davila-Garcia MI, Kellar KJ. Measuring nicotinic receptors with characteristics of alpha4beta2, alpha3beta2 and alpha3beta4 subtypes in rat tissues by autoradiography. J Neurochem. 2002;82:468–481. doi: 10.1046/j.1471-4159.2002.00951.x. [DOI] [PubMed] [Google Scholar]
- Picciotto MR. Nicotine as a modulator of behavior: beyond the inverted U. Trends Pharmacol Sci. 2003;24:493–499. doi: 10.1016/S0165-6147(03)00230-X. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Zoli M, Lena C, Bessis A, Lallemand Y, LeNovere N, Vincent P, Pich E, Bruret P, Changeux J-P. Abnormal avoidance learning in mice lacking functional high-affinity nicotine receptor in the brain. Nature. 1995;374:65–67. doi: 10.1038/374065a0. [DOI] [PubMed] [Google Scholar]
- Quik M. Smoking, nicotine and Parkinson's disease. Trends Neurosci. 2004;27:561–568. doi: 10.1016/j.tins.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Ross SA, Wong JY, Clifford JJ, Kinsella A, Massalas JS, Horne MK, Scheffer IE, Kola I, Waddington JL, Berkovic SF, Drago J. Phenotypic characterization of an alpha4 neuronal nicotinic acetylcholine receptor subunit knock-out mouse. J Neurosci. 2000;20:6431–6441. doi: 10.1523/JNEUROSCI.20-17-06431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, Grady SR. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65:1526–1535. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Shafaee N, Houng M, Truong A, Viseshakul N, Figl A, Sandhu S, Forsayth JR, Dwoskin LP, Crooks PA, Cohen BA. Pharmacological similarities between native brain and heterologously expressed α4β2 nicotinic receptors. Br J Pharmacol. 1999;128:1291–1299. doi: 10.1038/sj.bjp.0702900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker P, Jimenez M, McIntosh JM, Collins AC, Marks MJ. Identification of a novel nicotinic binding site in mouse brain using [(125)I]epibatidine. Br J Pharmacol. 2000a;131:729–739. doi: 10.1038/sj.bjp.0703616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker P, Marks MJ, Grady SR, Lu Y, Picciotto MR, Changeux J-P, Collins AC. Pharmacological and null mutation approaches reveal nicotinic receptor diversity. Eur J Pharmacol. 2000b;393:123–135. doi: 10.1016/s0014-2999(00)00052-2. [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Peterson CG, Xu W, McIntosh JM, Paylor R, Beaudet AL, Collins AC, Marks MJ. Involvement of the alpha3 subunit in central nicotinic binding populations. J Neurosci. 2002;22:2522–2529. doi: 10.1523/JNEUROSCI.22-07-02522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker P, Cooper JF, Salminen O, Marks MJ, McClure-Begley TD, Brown RWB, Collins AC, Lindstrom JM. Immunolabeling demonstrates the interdependence of mouse brain α4 and β2 nicotinic acetylcholine receptor subunit expression. J Comp Neurol. 2006;499:1016–1038. doi: 10.1002/cne.21181. [DOI] [PubMed] [Google Scholar]
- Whiting P, Lindstrom J. Characterization of bovine and human neuronal nicotinic acetylcholine receptors using monoclonal antibodies. J Neurosci. 1988;8:3395–3404. doi: 10.1523/JNEUROSCI.08-09-03395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonnacott S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997;20:92–98. doi: 10.1016/s0166-2236(96)10073-4. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Kellar KJ. The comparative pharmacology and up-regulation of rat neuronal nicotinic receptor subtype binding sites stably expressed in transfected mammalian cells. J Pharmacol Exp Ther. 2004;310:98–107. doi: 10.1124/jpet.104.066787. [DOI] [PubMed] [Google Scholar]
- Xu W, Orr-Urtreger A, Nigro F, Gelber S, Sutcliffe CB, Armstrong D, Patrick JW, Role LW, Beaudet AL, De Biasi M. Multiorgan autonomic dysfunction in mice lacking the β2 and the β4 subunits of neuronal acetylcholine receptors. J Neurosci. 1999;19:9298–9305. doi: 10.1523/JNEUROSCI.19-21-09298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Nelson ME, Kuryatov A, Choi C, Cooper J, Lindstrom J. Human α4β2 acetylcholine receptors formed from linked subunits. J Neurosci. 2003;23:9004–9015. doi: 10.1523/JNEUROSCI.23-27-09004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoli M, Lena C, Picciotto MR, Changeux J-P. Identification of four classes of nicotinic receptors using beta2 mutant mice. J Neurosci. 1998;18:4461–4472. doi: 10.1523/JNEUROSCI.18-12-04461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart R, Vijverberg HP. Four pharmacologically distinct subtypes of α4β2 nicotinic acetylcholine receptor expressed in Xenopus laevis oocytes. Mol Pharmacol. 1998;54:1124–1131. [PubMed] [Google Scholar]