Abstract
Heterochromatin protein 1 (HP1) proteins are conserved in eukaryotes, with most species containing several isoforms. Based on the properties of Drosophila HP1a, it was proposed that HP1s bind H3K9me2,3 and recruit factors involved in heterochromatin assembly and silencing. Yet, it is unclear whether this general picture applies to all HP1 isoforms and functional contexts. Here, we report that Drosophila HP1c regulates gene expression, as (1) it localizes to active chromatin domains, where it extensively colocalizes with the poised form of RNApolymerase II (RNApol II), Pol IIoser5, and H3K4me3, suggesting a contribution to transcriptional regulation; (2) its targeting to a reporter gene does not induce silencing but, on the contrary, increases its expression, and (3) it interacts with the zinc-finger proteins WOC (without children) and Relative-of-WOC (ROW), which are putative transcription factors. Here, we also show that, although HP1c efficiently binds H3K9me2,3 in vitro, its binding to chromatin strictly depends on both WOC and ROW. Moreover, expression profiling indicates that HP1c, WOC, and ROW regulate a common gene expression program that, in part, is executed in the context of the nervous system. From this study, which unveils the essential contribution of DNA-binding proteins to HP1c functionality and recruitment, HP1 proteins emerge as an increasingly diverse family of chromatin regulators.
Keywords: HP1, WOC, ROW/CG8092, chromatin, Drosophila
The contribution of chromatin to the regulation of genomic functions is well established. Most frequently, regulation by chromatin involves the establishment of specific patterns of post-translational histone modifications, which result in recruitment of regulatory nonhistone proteins (Ruthenburg et al. 2007). Heterochromatin protein 1 (HP1) constitutes one of the best-studied examples (Hiragami and Festenstein 2005; Hediger and Gasser 2006; Lomberk et al. 2006), where a regulatory nonhistone protein is recruited to chromatin through the recognition of a specific histone modification, di- or trimethylation of Lys 9 on the histone H3 tail (H3K9me2,3). This interaction, which involves the N-terminal chromodomain of HP1, is known to play a fundamental role in the formation and maintenance of heterochromatic domains.
Except in budding yeast, HP1 is widely conserved in eukaryotes, with most species containing several isoforms (Hiragami and Festenstein 2005; Lomberk et al. 2006). HP1 proteins are characterized by a common structural organization consisting of two conserved domains, the N-terminal chromodomain and the C-terminal chromo-shadow domain, which are spaced by a variable nonconserved hinge domain. The existence of multiple isoforms suggests functional specialization, with different isoforms playing different functions. For instance, in Drosophila, three of the five HP1 isoforms (HP1a, HP1b, and HP1c) are ubiquitously expressed, while the other two (HP1d/Rhino and HP1e) are predominantly expressed in the germline (Vermaak et al. 2005). Moreover, ubiquitously expressed HP1 isoforms show differential chromosomal distributions, as HP1a is mainly associated to heterochromatin, while HP1c is excluded from centromeric heterochromatin and HP1b is found both at euchromatic and heterochromatic domains (Smothers and Henikoff 2001). A similar situation is observed in mammals, where the patterns of localization of the three HP1 isoforms (HP1α, HP1β, and HP1γ) overlap only partially and show differential dynamics during differentiation and cell cycle progression (Minc et al. 1999; Hayakawa et al. 2003; Dialynas et al. 2007). On the other hand, in the nematode Caenorhabditis elegans, two HP1 isoforms exist (HPL1 and HPL2), showing preferential euchromatic association, partially nonoverlapping expression and localization patterns, and distinct mutant phenotypes (Coustham et al. 2006; Schott et al. 2006). Interestingly, the only HP1 protein (TLF2/LHP1) of Arabidopsis thaliana appears to localize exclusively to euchromatin, where it colocalizes with H3K27me3 (Nakahigashi et al. 2005).
The molecular mechanisms that determine the differential distribution of the various HP1 isoforms, and their differential functional properties, remain largely unknown. Most of our knowledge about the mechanisms of action of HP1 proteins derives from studies addressing the functional properties of Drosophila HP1a or mammalian HP1α. From these studies, a general picture emerges by which, through the chromodomain, HP1 proteins bind chromatin regions enriched in H3K9me2,3 while, through the chromo-shadow domain, recruiting different factors resulting in various functional outcomes; namely, heterochromatin assembly and gene silencing (Hiragami and Festenstein 2005; Hediger and Gasser 2006; Lomberk et al. 2006). It is uncertain whether this general picture applies to all HP1 proteins and possible scenarios. Actually, Drosophila HP1a is known to play a more complex role(s) in the regulation of gene expression, as it is required for the expression of several heterochromatic genes (Hearn et al. 1991; Clegg et al. 1998; Lu et al. 2000), and certain euchromatic genes (Cryderman et al. 2005; De Lucia et al. 2005). Moreover, Drosophila HP1a is recruited to developmentally regulated genes and heat-shock-induced puffs in an RNA-dependent manner (Piacentini et al. 2003), and in an erythroid cell line, murine HP1γ was found associated to actively transcribed genes (Vakoc et al. 2005).
In this study, we report on the functional characterization of HP1c, a Drosophila HP1 protein of largely unknown properties. Our results show that HP1c extensively colocalizes with poised RNA polymerase II (RNApol II) and H3K4me3, a modification that correlates with active chromatin domains (Bernstein et al. 2005; Barski et al. 2007; Heintzman et al. 2007). Moreover, targeting HP1c to a reporter construct does not induce silencing but, on the contrary, results in increased expression of the reporter gene. Here, we also report on the interaction of HP1c with the zinc-finger proteins WOC (without children) (Wismar et al. 2000; Warren et al. 2001), and Relative-of-WOC (ROW), which are putative transcription factors. HP1c efficiently binds H3K9me2,3 in vitro, but its binding to chromatin strictly depends on both WOC and ROW. Moreover, expression profiling indicates that HP1c, WOC, and ROW extensively cooperate to regulate gene expression, especially in the context of the nervous system. These results unveil the essential contribution of sequence-specific DNA-binding proteins to functionality of HP1c and its recruitment to chromatin.
Results
HP1c localizes to active chromatin domains
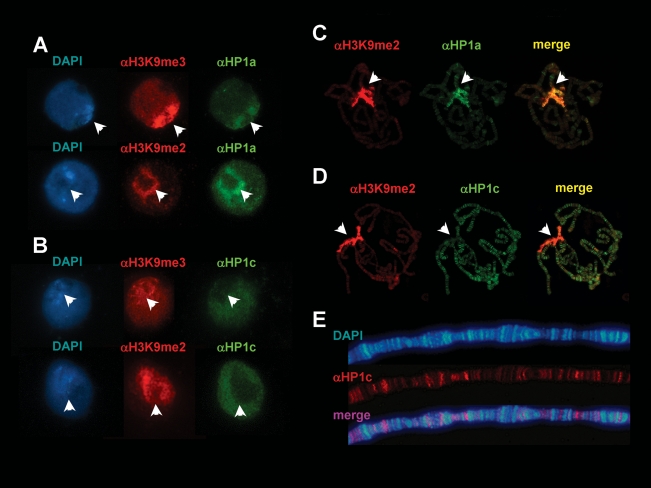
HP1c localization was determined both in cultured S2 cells and polytene chromosomes. In interphase S2 cells, heterochromatin regions, which are enriched in H3K9me2,3, show strong colocalization with HP1a (Fig. 1A) but very poor staining with αHP1c antibodies (Fig. 1B), indicating that HP1c preferentially localizes to euchromatin being mostly excluded from heterochromatin. Similar results were reported earlier by others (Smothers and Henikoff 2001). Similarly, in polytene chromosomes, HP1a and HP1c also show drastically different patterns of localization, as HP1a localizes to the heterochromatic chromocentre (Fig. 1C) while HP1c is exclusively found at the euchromatic chromosome arms (Fig. 1D). In addition to the chromocentre, HP1a is known to localize to a number of euchromatic sites (Fanti et al. 2003), where it shows no significant colocalization with HP1c (data not shown). These results indicate that, despite their strong similarity, HP1a and HP1c show mutually exclusive patterns of chromosomal distribution.
Figure 1.
HP1c localizes to multiple euchromatic sites. The patterns of immunolocalization of HP1a (A,C) and HP1c (B,D), in S2 cells (A,B) and polytene chromosomes (C,D), are shown (in green). Arrows indicate the position of the heterochromatic chromocentre determined by staining with αH3K9me3 and/or αH3K9me2 (in red). (E) The pattern of immunolocalization of HP1c (in red) is presented at a higher magnification to show its localization to interbands. DNA was stained with DAPI (in blue).
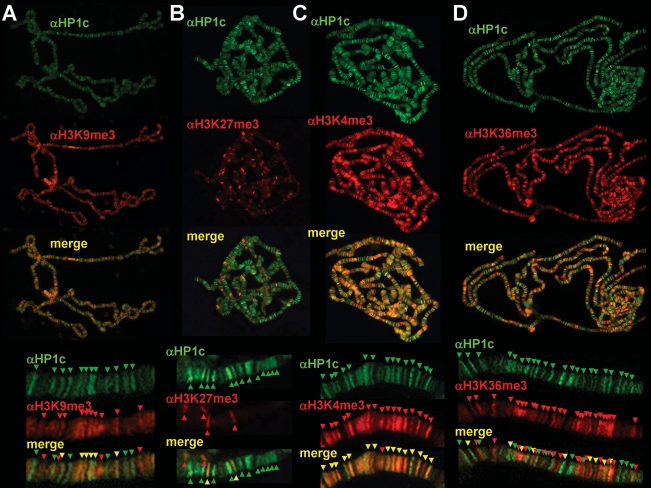
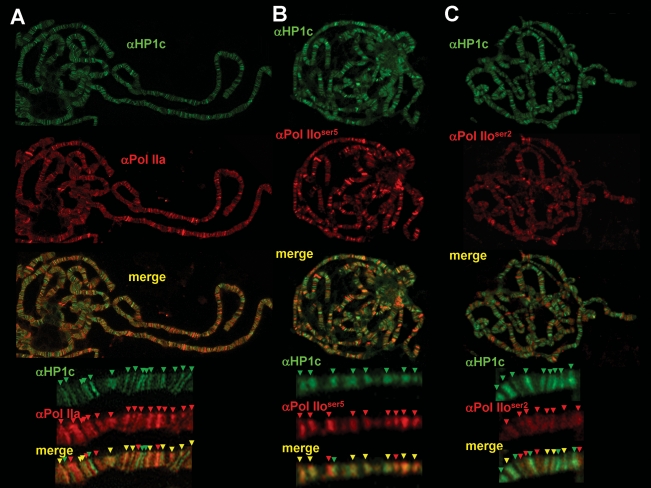
HP1c localization is restricted to interbands (Fig. 1E), which correspond to gene-rich regions that stain poorly with DAPI, suggesting a contribution to the regulation of gene expression. To analyze the potential contribution of HP1c to gene regulation, we determined the extent of colocalization of HP1c with different histone modifications that correlate with either gene silencing, H3K9me3 and H3K27me3, or activation, H3K4me3 and H3K36me3 (Fig. 2). HP1c shows partial colocalization with H3K9me3 (Fig. 2A). Approximately, 50% of HP1c bands colocalize with H3K9me3, and most frequently, intense HP1c bands show no significant αH3K9me3 reactivity. On the other hand, colocalization with H3K27me3 is only very scarce (Fig. 2B). In contrast, HP1c shows a strong colocalization with H3K4me3 (Fig. 2C), suggesting that HP1c might be involved in transcriptional regulation. Consistent with this hypothesis, HP1c strongly colocalizes with RNApol II (Fig. 3). Colocalization with the poised form of RNApol II phosphorylated at Ser 5, Pol IIoser5, is most extensive, as the majority of bands positive for αHP1c are also positive for αPol IIoser5 (Fig. 3B). On the other hand, colocalization with the elongating form of RNApol II phosphorylated at Ser 2, Pol IIoser2, is much weaker (Fig. 3C), suggesting that HP1c participates at the initial stages of transcription regulation rather than during elongation. Consistent with this hypothesis, HP1c shows only a weak colocalization with H3K36me3 (Fig. 2D), a modification that occurs during elongation and extends all through transcribed regions (Krogan et al. 2003; Bell et al. 2008).
Figure 2.
HP1c localizes to active chromatin domains. The extent of colocalization of HP1c (in green) and H3K9me3 (A), H3K27me3 (B), H3K4me3 (C), and H3K36me3 (D) (in red), is determined in polytene chromosomes. Enlarged images are shown at the bottom of each panel. Arrows indicate distinct αHP1c (green) and αH3K9me3, αH3K27me3, αH3K4me3, and αH3K36me3 (red) signals. Yellow arrows indicate colocalization.
Figure 3.
HP1c colocalizes with the poised form of RNApol II, Pol IIoser5. The extent of colocalization of HP1c (in green) and unphosphorylated RNApol II (A), Pol IIoser5 (B), and Pol IIoser2 (C) (in red), is determined in polytene chromosomes. Enlarged images are shown at the bottom of each panel. Arrows indicate distinct αHP1c (green) and αRNApol IIa, α Pol IIoser5, and α Pol IIoser2 (red) signals. Yellow arrows indicate colocalization.
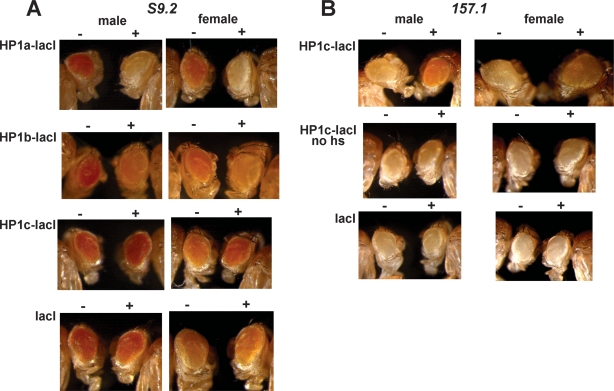
To further analyze the contribution of HP1c to gene expression, we determined the effects of targeting HP1c to a reporter construct (Fig. 4). In these experiments, we took advantage of transgenic flies carrying a reporter white transgene containing multiple binding sites for the lacI repressor at the regulatory region (Li et al. 2003). Lines S9.2 and 157.1 were used, which contain 46 and 256 lacI-repeats respectively. In these lines, expression of fused proteins carrying the lacI-DNA-binding domain results in their targeting to the reporter gene. Previous studies showed that, in general, targeting HP1 proteins to a reporter gene induces silencing (Ayyanathan et al. 2003; Li et al. 2003; Verschure et al. 2005; Smallwood et al. 2007). Actually, in line S9.2, expression of HP1a-lacI results in silencing of the reporter gene, which is stronger in males (Fig. 4A). Similar results were reported earlier by others (Li et al. 2003). Expression of HP1b-lacI also induces silencing of the reporter gene, but in contrast, expression of HP1c-lacI does not (Fig. 4A). On the contrary, a slightly increased expression of the reporter white gene is observed. These results were confirmed when HP1c-lacI was expressed in line 157.1, which is more suitable to detect activating effects, as background expression of the reporter gene is low. As shown in Figure 4B, white expression is increased in flies expressing HP1c-lacI. In these experiments, expression of the fused proteins was driven by the hsp70-promoter and, therefore, was induced by repeated heat-shock treatments. The observed activation of the reporter gene is specific of HP1c, as no effect is observed in the absence of heat shock (Fig. 4B) or in flies expressing the lacI-DNA-binding domain alone (Fig. 4A,B).
Figure 4.
Targeting HP1c to a reporter white transgene increases its expression. (A) The eye phenotype of S9.2 flies expressing the indicated fused proteins (+) is compared with that of siblings where no fused protein is expressed (−). Expression was induced by heat-shock treatment. (B) Similar experiments as in A, but performed in 157.1 flies. Results are presented when expression of HP1c-lacI was induced by heat shock (top) and when no heat-shock treatment was applied (middle).
HP1c interacts with the zinc-finger proteins WOC and ROW
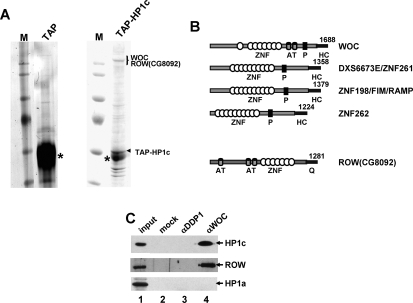
Altogether, the results reported above strongly suggest that Drosophila HP1c contributes to transcriptional regulation. Next, we asked about the molecular basis of this contribution. To address this question, multiprotein complexes containing HP1c were purified to identify interacting factors that could account for its functional properties. For this purpose, stable S2-cell lines expressing a TAP-HP1c fusion protein were obtained. Purified complexes contain, in addition to HP1c-TAP, a major band of high molecular weight (Fig. 5A, lane HP1c-TAP), which is absent in mock purifications performed from cells expressing the TAP domain alone (Fig. 5A, lane TAP). A few additional weak bands of lower molecular weight are also observed. Next, we proceeded to identification of the high-molecular-weight band mentioned above. LC/MS analysis provided two major hits—WOC (Mascot score of 1031, with 24 identified peptides corresponding to 11% sequence coverage) (Wismar et al. 2000; Warren et al. 2001), and CG8092, which we propose to name Relative-of-WOC (ROW) (Mascot score of 1239, with 66 identified peptides corresponding to 30% sequence coverage) (Supplemental Tables S1, S2). A few other polypeptides were also identified but with much lower scores. Domain structure and organization strongly suggest that both WOC and ROW are transcription factors (Fig. 5B), as they contain multiple zinc-finger and AT-hook domains for sequence-specific DNA binding and, in addition, ROW contains a C-terminal domain that, enriched in glutamine (Q) residues, resembles transactivation domains of some transcription factors (Triezenberg 1995). WOC is homologous to three human zinc-finger proteins: DXS6673E/ZNF261 (van der Maarel et al. 1996), ZNF198/FIM/RAMP (Popovici et al. 1998; Xiao et al. 1998), and ZNF262 (Sohal et al. 1999). Most remarkably, the C-terminal domain (∼300 amino acids) is highly conserved in the three human proteins as well as in WOC. On the other hand, ROW shows no significant homology with any known protein.
Figure 5.
HP1c interacts with WOC and ROW. (A, lane TAP-HP1c) Multiprotein complexes containing TAP-HP1c were purified and analyzed by SDS-PAGE. Lane TAP corresponds to a mock purification performed from cells expressing the TAP domain alone. Lanes M correspond to molecular-weight markers. The position of the high-molecular-weight band that, as shown by LC/MS analysis, corresponds to WOC and ROW is indicated. The position of the TAP-HP1c bait is also indicated. Bands labeled with an asterisk correspond to IgGs. (B) Structural organization of WOC, ROW, and the three human homologs of WOC (DXS6673E/ZNF261, ZNF198/FIM/RAMP, and ZNF262). The positions of the zinc-finger (ZNF) and AT-hook domains (AT) are indicated. P indicates a proline-rich repeat conserved in WOC and its human homologs. HC corresponds to the highly conserved C-terminal domain present in WOC and its human homologs. Q corresponds to the C-terminal glutamine (Q)-rich domain present in ROW. (C) HP1c and ROW coimmunoprecipitate with WOC. Nuclear extracts were subjected to immunoprecipitation with αWOC antibodies (lane 4) or with control αDDP1 antibodies (lane 3), and analyzed by Western-blot using αHP1c (top), αROW (middle), and αHP1a antibodies (bottom). Lane 2 corresponds to a mock immunoprecipitation where no antibodies were added. Lane 1 corresponds to 10% of the input material used for immunoprecipitation.
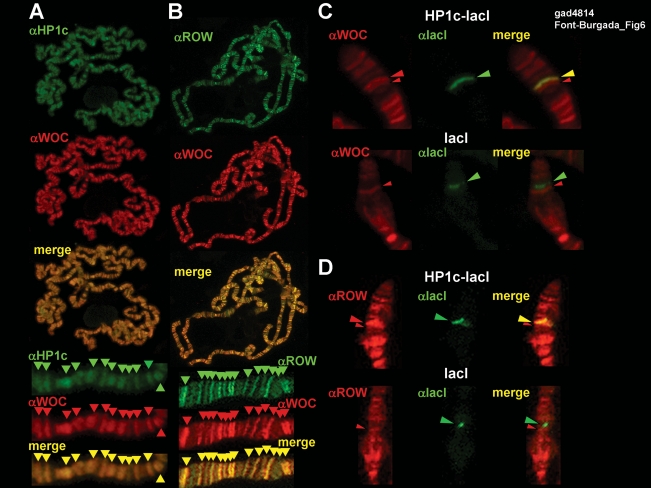
Coimmunoprecipitation experiments confirmed the interaction between HP1c, WOC, and ROW, as immunoprecipitation with αWOC antibodies results in quantitative coprecipitation of both HP1c and ROW (Fig. 5C). No coimmunoprecipitation of HP1c and ROW was observed when the extracts were treated with unrelated antibodies (αDDP1) or when no antibodies were added. Moreover, addition of αWOC antibodies does not result in coprecipitation of HP1a (Fig. 5C). Consistent with these results, both HP1c and ROW show the most extensive colocalization with WOC, with virtually all bands positive for αHP1c and αROW being also positive for αWOC and vice versa (Fig. 6A,B). In addition, targeting HP1c-lacI to the lacI-repeats of line 157.1 results in ectopic recruitment of both WOC and ROW, as additional αWOC and αROW signals are detected at the position corresponding to the lacI-repeats (Fig. 6C,D). No such recruitment is observed in flies expressing the lacI-DNA-binding domain alone (Fig. 6C,D).
Figure 6.
HP1c and ROW colocalize with WOC. (A,B) The extent of colocalization of WOC (in red) with HP1c (A) and ROW (B) (in green) is presented. Enlarged images are shown at the bottom. Arrows indicate distinct αHP1c (green), αROW (green), and αWOC (red) signals. Yellow arrows indicate colocalization. (C,D) Targeting of HP1c-lacI to the lacI insertion of 157.1 flies results in ectopic recruitment of WOC (C) and ROW (D). The patterns of immunolocalization of WOC and ROW (in red) are presented at the region of the lacI insertion in flies expressing HP1c-lacI (panel HP1c-lacI) and lacI alone (panel lacI). The patterns obtained with αlacI antibodies (in green) are also presented. Big arrows indicate the position of the αlacI signal, marking the position of the lacI insertion that colocalizes with additional αWOC and αROW signals when HP1c-lacI is expressed. Small arrows indicate the position of an endogenous αWOC/αROW signal, which does not colocalize with any αlacI signal.
WOC and ROW mediate chromosomal association of HP1c
The results discussed above indicate that HP1c interacts with the zinc-finger proteins WOC and ROW, which are likely to bind DNA sequences specifically. Therefore, it is possible that binding of HP1c to chromatin is mediated by WOC and ROW. To test this hypothesis, we analyzed chromosomal association of HP1c in woc and row mutants. For this purpose, we used wocRNAi and rowRNAi knockdown transgenic flies, which carry a UASGAL4 construct expressing a synthetic hairpin from the coding region of either woc or row that, upon crossing with flies expressing GAL4, generates siRNAs to silence expression of the corresponding gene. wocRNAi and rowRNAi show strong silencing of the corresponding gene when crossed to flies carrying a ubiquitous Actin5C-GAL4 driver, so that, at the third-instar larvae stage, corresponding mRNA levels are reduced to ∼40% of those observed in control larvae (Supplemental Fig. S1A,B). Under these conditions, both mutations show high lethality at the pupae stage, which is stronger in wocRNAi.
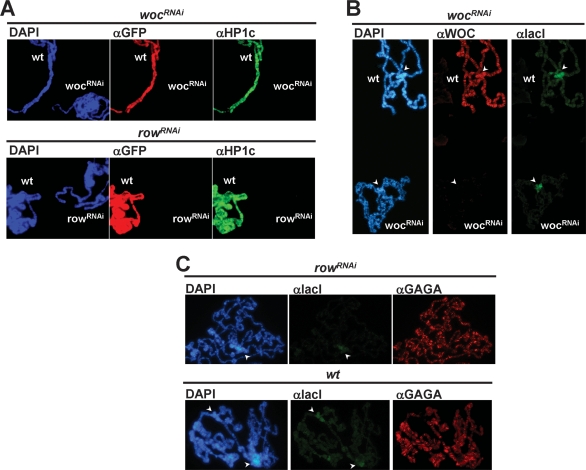
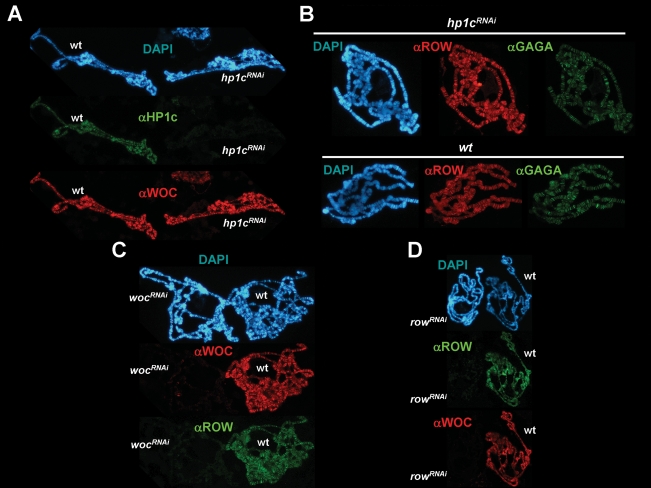
Chromosomal association of HP1c is strongly compromised in wocRNAi and rowRNAi mutant polytene chromosomes (Fig. 7A). In these experiments, depletion of WOC and ROW was induced in salivary glands by crossing wocRNAi and rowRNAi lines to a lio-GAL4 line, where the limbo promoter drives GAL4 expression. Mutant salivary glands were then mixed and squashed together with glands prepared from control wild-type larvae, which carry a UASGAL4-H2B∷GFP construct and, therefore, incorporate H2B-GFP in polytene chromosomes. In this way, mutant and control polytene chromosomes, which are identified by their staining with αGFP antibodies, are immunostained and observed under exactly the same experimental conditions. As shown in Figure 7A, wocRNAi and rowRNAi mutant chromosomes show highly reduced reactivity with αHP1c antibodies while control polytenes show strong αHP1c reactivity, strongly suggesting that both WOC and ROW are required for binding of HP1c to chromatin.
Figure 7.
Binding of HP1c to chromatin is mediated by WOC and ROW. (A) The pattern of immunolocalization of HP1c (in green) is presented in polytene chromosomes obtained from wocRNAi; lio-GAL4 (panel wocRNAi) and rowRNAi; lio-GAL4 mutant larvae (panel rowRNAi). Polytene chromosomes obtained from control UASGAL4-H2B∷GFP; lio-GAL4 larvae were mixed and squashed together with mutant polytene chromosomes. Control wild-type chromosomes (wt) are identified by their reactivity with αGFP antibodies (in red). (B) HP1c-lacI was overexpressed in wocRNAi; Act5C-GAL4 and in control gfpRNAi; Act5C-GAL4 flies. Binding of HP1c-lacI was determined in polytene chromosomes by immunostaining with αlacI antibodies (in green). Control wild-type chromosomes (wt) are identified by their reactivity with αWOC antibodies (in red). (C) Similar experiments as in B, but HP1c-lacI was overexpressed in rowRNAi; Act5C-GAL4 (panel rowRNAi) and in control gfpRNAi; Act5C-GAL4 flies (panel wt). In this case, mutant and control chromosomes were analyzed separately. As a control, the immunolocalization patterns obtained with αGAGA antibodies (in red) are shown. DNA was stained with DAPI (in blue). Arrows indicate the position of the chromocentre.
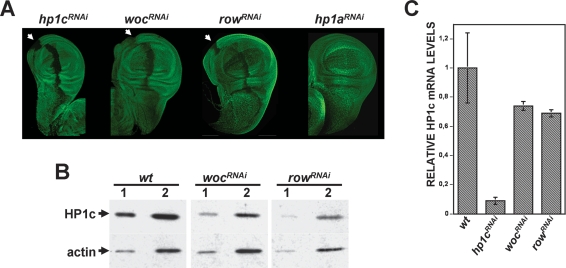
Lack of HP1c observed in wocRNAi and rowRNAi polytene chromosomes could also reflect a contribution to synthesis, and/or stability, of HP1c. To address this question, wocRNAi and rowRNAi lines were crossed to a ptc-GAL4 line, where GAL4 expression is driven by the patched promoter that, in the wing disc, is specifically active at the anterior/posterior (A/P)-boundary. Therefore, in these experiments, depletion of WOC and ROW was specifically induced at the A/P-boundary in the wing disc. Under these conditions, wocRNAi and rowRNAi mutant cells of the A/P-boundary show reduced HP1c levels (Fig. 8A). It must be noticed, however, that a much stronger effect is observed when similar experiments are performed in an hp1cRNAi mutant background (Fig. 8A), where depletion of HP1c is most efficient (Fig. 8C; Supplemental Fig. S1C). No effect is detected in a control hp1aRNAi line (Fig. 8A). Similar results were obtained when WOC and ROW were depleted ubiquitously, by crossing wocRNAi and rowRNAi to an Actin5C-GAL4 line, and the effects on HP1c levels determined by Western-blot (Fig. 8B). Under these circumstances, depletion of WOC and ROW results in a moderate decrease of HP1c. On the other hand, HP1c mRNA levels are only slightly reduced in wocRNAi and rowRNAi (Fig. 8C), strongly suggesting that reduced HP1c protein levels observed in the mutants reflects destabilization of the protein due, most likely, to its inability to bind chromatin. Similar effects were reported for HP1a and Polycomb (PC), when their binding to chromatin was impaired (Swaminathan et al. 2005; Tan et al. 2007).
Figure 8.
HP1c protein levels are decreased in wocRNAi and rowRNAi. (A) Immunostaining with αHP1c antibodies (in green) is presented in wing imaginal discs obtained from hp1cRNAi; ptc-GAL4 (panel hp1cRNAi), wocRNAi; ptc-GAL4 (panel wocRNAi), rowRNAi; ptc-GAL4 (panel rowRNAi), and hp1aRNAi; ptc-GAL4 larvae (panel hp1aRNAi). The arrows indicate the A/P-boundary where the ptc-promoter is specifically active. (B) HP1c levels are analyzed by Western-blot with αHP1c antibodies in extracts prepared from wocRNAi; Act5C-GAL4 (lanes wocRNAi), rowRNAi; Act5c-GAL4 (lanes rowRNAi), and control wild-type larvae (lanes wt). Two increasing amounts of extract were analyzed in each case (lanes 1,2). The signal obtained with αActin antibodies was used as loading control for normalization. (C) Quantitative RT–PCR analysis of the levels of HP1c mRNA observed in wocRNAi; Act5C-GAL4 (column wocRNAi), rowRNAi; Act5C-GAL4 (column rowRNAi), and control wild-type larvae (column wt). Results obtained with hp1cRNAi; Act5C-GAL4 larvae (column hp1cRNAi) are also included for comparison. Relative HP1c mRNA expression was determined in relation to GAP expression.
To further confirm the contribution of WOC and ROW to recruitment of HP1c, we performed overexpression experiments, where HP1c-lacI was expressed in wocRNAi and rowRNAi mutants, and its ability to bind polytene chromosomes compared with that observed when overexpression was carried out in control wild-type flies. When HP1c-lacI was expressed in wocRNAi, polytene chromosomes from mutant and control larvae were squashed together and immunostained with αlacI antibodies to determine binding of HP1c-lacI (Fig. 7B). In control polytenes, which are identified by their staining with αWOC antibodies, overexpressed HP1c-lacI binds to multiple euchromatic sites, as endogenous HP1c does. On the other hand, in wocRNAi chromosomes, binding of HP1c-lacI to euchromatin is impaired. Similar results were obtained when HP1c-lacI was expressed in rowRNAi (Fig. 7C). In this case, reactivity with αGAGA was used as a control for comparison since mutant and control wild-type chromosomes were observed separately. As in wocRNAi, no significant αlacI reactivity is observed at euchromatin in rowRNAi chromosomes. On the other hand, binding of WOC and ROW is not significantly affected in mutant hp1cRNAi polytene chromosomes (Fig. 9A,B), indicating that HP1c is not required for binding of WOC and ROW. Moreover, WOC and ROW are reciprocally required for binding to chromatin, as binding of ROW to chromatin is severely impaired in wocRNAi chromosomes (Fig. 9C) and, vice versa, binding of WOC is also strongly impaired in rowRNAi chromosomes (Fig. 9D). Altogether, these results strongly indicate that WOC and ROW form obligated hetero-oligomers that, in turn, bind HP1c and recruit it to specific genomic locations.
Figure 9.
WOC and ROW are reciprocally required for binding to chromatin. (A,B) The patterns of immunolocalization of WOC (A) and ROW (B) (in red) are presented in polytene chromosomes obtained from mutant hp1cRNAi; Act5C-GAL4 larvae. In A, polytene chromosomes obtained from control wild-type larvae were mixed and squashed together with mutant hp1cRNAi polytene chromosomes, which are identified by their lack of reactivity with αHP1c antibodies (in green). In B, mutant and control wild-type (wt) chromosomes were analyzed separately. As a control, the immunolocalization patterns obtained with αGAGA antibodies (in green) are shown. (C) The pattern of immunolocalization of ROW (in green) is presented in polytene chromosomes obtained from mutant wocRNAi; Act5C-GAL4 larvae. Polytene chromosomes obtained from control wild-type larvae were mixed and squashed together with mutant wocRNAi polytene chromosomes, which are identified by their lack of reactivity with αWOC antibodies (in red). (D) The pattern of immunolocalization of WOC (in red) is presented in polytene chromosomes obtained from mutant rowRNAi; Act5C-GAL4 larvae. Polytene chromosomes obtained from control wild-type larvae were mixed and squashed together with mutant rowRNAi polytene chromosomes, which are identified by their lack of reactivity with αROW antibodies (in green). DNA was stained with DAPI (in blue).
Interestingly, overexpression of HP1c-lacI results in its misincorporation to heterochromatin, as it is also detected at the heterochromatic chromocentre (Fig. 7B,C). Binding to heterochromatin, however, is not impaired in wocRNAi and rowRNAi chromosomes, suggesting that it occurs independently of WOC and ROW. It must also be noticed that mislocalization of HP1c-lacI to heterochromatin induces recruitment of WOC (Fig. 7B) that, otherwise, is excluded from heterochromatin (Fig. 6A).
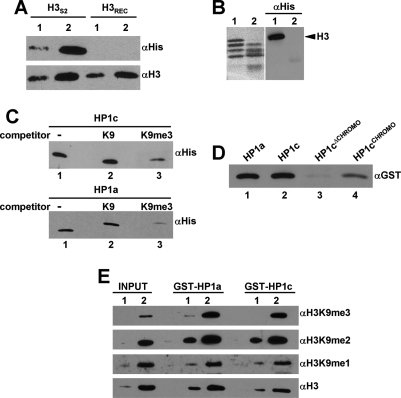
That binding of HP1c to chromatin is strictly dependent on the zinc-finger proteins WOC and ROW was unexpected, as HP1c contains an N-terminal chromodomain that, being homologous to that of HP1a, could mediate binding to H3K9me2,3 (Smothers and Henikoff 2001). In fact, all residues directly involved in this interaction are conserved in the chromodomain of HP1c. Actually, in vitro, HP1c specifically recognizes H3K9me2,3 through its chromodomain (Fig. 10). As judged by Far-Western analysis, HP1c binds endogenous, but not recombinant, histone H3 (Fig. 10A). This binding (1) depends on the N-terminal domain of histone H3, as it is abolished after mild digestion with trypsin to cleave the N-terminal tail (Fig. 10B); (2) involves recognition of H3K9me3, as it is competed by a H3K9me3-peptide but not by the equivalent unmethylated-peptide (Fig. 10C); and (3) is mediated by the chromodomain of HP1c, as no binding is observed with HP1cΔCHROMO, a truncated form missing the chromodomain, while the chromodomain by itself, HP1cCHROMO, shows significant binding (Fig. 10D). Moreover, HP1c is also capable of binding H3K9me2,3 in mononucleosomes (Fig. 10E). In these experiments, total mononucleosomes were subjected to pull-down assays with GST-HP1c and GST-HP1a fusion proteins. As shown in Figure 10E, mononucleosomes bound by GST-HP1c are strongly enriched in both H3K9me3 and H3K9me2 when compared with the input mononucleosomes. Enrichment is similar to that observed in mononucleosomes bound by GST-HP1a. On the other hand, mononucleosomes bound by both GST-HP1c and GST-HP1a show no significant enrichment in H3K9me1 as well as in a number of other histone modifications (i.e., H3K27me1,2,3, H4K20me1,2,3) (data not shown). These results indicate that, in vitro, HP1c and HP1a bind chromatin on the basis of a common molecular mechanism; namely, recognition of H3K9me2,3.
Figure 10.
HP1c binds H3K9me2,3 in vitro. (A) Binding of His-tagged HP1c to endogenous histone H3, obtained from S2 cells, (panel H3S2), as well as recombinant histone H3, expressed in bacteria, (panel H3REC), is analyzed by Far-Western using αHis antibodies (shown at top). Lanes 1 and 2 correspond to increasing amounts of H3. Western analysis with αH3 antibodies is presented at the bottom, as a loading control. (B) Nucleosomes prepared from S2 cells were treated with trypsin, to cleave the histone N-terminal tails (lanes 2) or not (lanes 1), and then binding of his-tagged HP1c was analyzed by Far-Western using αHis antibodies. Staining with Ponceau of the filter before hybridization with αHis antibodies is presented on the left. The position corresponding to histone H3 is indicated. (C) Binding of His-tagged HP1c (panel HP1c) or HP1a (panel HP1a) to endogenous histone H3 was analyzed by Far-Western using αHis antibodies. Binding was performed in the presence of 50 μM of a peptide from the H3 N-terminal tail trimethylated at K9 (lanes 3) or unmethylated (lanes 2). Lanes 1 correspond to the binding observed in the absence of any added competitor. (D) Binding to endogenous H3 of GST-tagged HP1c (lane 2) and truncated forms containing only the N-terminal chromodomain, HP1cCHROMO (amino acids 1–62) (lane 4), or missing it, HP1cΔCHROMO (amino acids 63–237) (lane 3), is analyzed by Far-Western using αGST antibodies. Lane 1 corresponds to binding of GST-tagged HP1a. (E) Pull-down assays of total mononucleosomes prepared from S2 cells were performed with GST-HP1c and GST-HP1a fused proteins, and bound mononucleosomes analyzed by Western using αH3K9me3, αH3K9me2, αH3K9me1, and αH3 antibodies. Lanes 1 and 2 correspond to increasing amounts of bound mononucleosomes. INPUT corresponds to 10% of mononucleosomes used for pull-down assays.
HP1c cooperates with WOC and ROW to regulate gene expression
The results reported above show that HP1c associates with the zinc-finger proteins WOC and ROW that, in addition, mediate its binding to chromatin. Next, we asked whether HP1c, WOC, and ROW are also functionally linked. Expression profiling was used to determine the actual contribution of HP1c, WOC, and ROW to gene expression. For this purpose, hp1cRNAi, wocRNAi, and rowRNAi lines were crossed to flies carrying an ubiquitous Act5C-GAL4 driver, and total RNA was prepared from blue staged male larvae. Under these experimental conditions, a number of nonspecific factors could also have an effect on gene expression, so that their contribution needed to be subtracted. These factors include two genomic insertions: the UASGAL4 construct expressing the synthetic hairpin against the corresponding gene and the Act5C-GAL4 insertion. Moreover, in the mutants, the RNAi pathway is hyperactivated and siRNAs are generated to a high concentration, which could also have an effect on gene expression. For each mutant condition, the original UASGAL4-hairpin line was used as control, to account for nonspecific effects due to the corresponding insertion. In addition, to account for nonspecific effects due to the Act5C-GAL4 insertion and hyperactivation of RNAi, two other controls were used: an Act5C-GAL4 line carrying an UASGAL4-hairpin construct against an unrelated gene (GFP), and the original line carrying the UASGAL4-hairpin insertion against GFP.
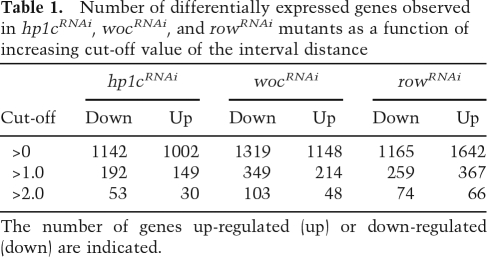
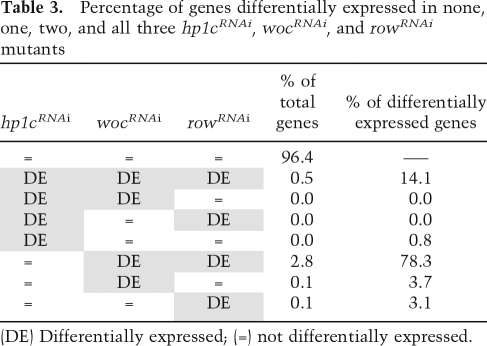
Differential expression was measured with an interval distance that provides a conservative approach to the analysis of complex samples that, as in the case described here, contain different sources of background noise (see the Supplemental Material for details). Briefly, for each gene, the interval of expression in the mutant (determined from the values observed in independent biological replicas) is compared with the interval of expression observed in the control samples (determined from the highest and lowest expression observed in all control samples). This defines, for each gene, a distance between intervals of expression in the mutant versus the controls, which is then divided by an estimate of the standard deviation. The distance is set to zero if the intervals overlap. Table 1 summarizes, for each mutant background, the number of differentially expressed genes observed at increasing cut-off values of the interval distance. At low cut-off values (>0), a similar number of differentially expressed genes are found in all three mutants. However, as the cut-off value is increased, and hence less false positives are expected, a higher number of genes is found in wocRNAi and rowRNAi than in hp1cRNAi, suggesting that WOC and ROW regulate more genes than HP1c.
Table 1.
Number of differentially expressed genes observed in hp1cRNAi, wocRNAi, and rowRNAi mutants as a function of increasing cut-off value of the interval distance
The number of genes up-regulated (up) or down-regulated (down) are indicated.
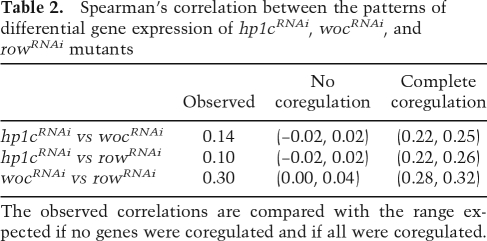
Given the interactions observed between HP1c, WOC, and ROW, we anticipate that these three factors regulate a common set of genes, so that the patterns of differentially expressed genes observed in the three mutants could correlate. To test this hypothesis, we measured the correlation between the patterns of differentially expressed genes in the mutants by computing Spearman’s rank correlation coefficient between the interval distances. As shown in Table 2, the patterns of differentially expressed genes show correlations that are much higher than those expected under the hypothesis that no genes are coregulated (P-value < 1.0e−8). Next, to determine to what extent the gene expression programs executed by HP1c, WOC, and ROW overlap, we assessed the range of Spearman’s correlation values that would be expected under complete coregulation. For this purpose, a simulation study was carried out under the assumption that all genes are coregulated and that the change in expression relative to the controls is the same for the three mutants. As shown in Table 2, the observed correlation between wocRNAi and rowRNAi falls within the expected interval for complete coregulation, supporting that WOC and ROW execute a common gene expression program. On the other hand, correlations observed between hp1cRNAi and either wocRNAi or rowRNAi are lower than expected for complete coregulation, an observation that could be anticipated from the higher number of differentially expressed genes observed in wocRNAi and rowRNAi than in hp1cRNAi (Table 1). More relevant, in this case, is the circumstance that virtually all differentially expressed genes in hp1cRNAi turn out to be differentially expressed in both wocRNAi and rowRNAi (Table 3). For this analysis, genes were classified into nine different categories according to whether they are not differentially expressed in any of the mutants or they change expression in one, two, or the three mutant conditions. As shown in Table 3, most of the genes are not differentially expressed in any of the three mutants. Consistent with the strong correlation observed between wocRNAi and rowRNAi, a high proportion (92.4%) of total differentially expressed genes change expression both in wocRNAi and rowRNAi. On the other hand, most of the genes that are differentially expressed in hp1cRNAi also change expression in both wocRNAi and rowRNAi, as only a low proportion are affected solely in hp1cRNAi and no genes are differentially expressed in hp1cRNAi and in only one of the other two mutants. Actually, 14.1% of total differentially expressed genes change expression in all three mutants, which accounts for 94.6% of the genes that are differentially expressed in hp1cRNAi. These results indicate that a vast majority of genes regulated by HP1c are also regulated by both WOC and ROW, which is fully consistent with the interactions described above. It must be noticed, however, that many differentially expressed genes (78.3%) change in wocRNAi and rowRNAi but not in hp1cRNAi.
Table 2.
Spearman’s correlation between the patterns of differential gene expression of hp1cRNAi, wocRNAi, and rowRNAi mutants
The observed correlations are compared with the range expected if no genes were coregulated and if all were coregulated.
Table 3.
Percentage of genes differentially expressed in none, one, two, and all three hp1cRNAi, wocRNAi, and rowRNAi mutants
(DE) Differentially expressed; (=) not differentially expressed.
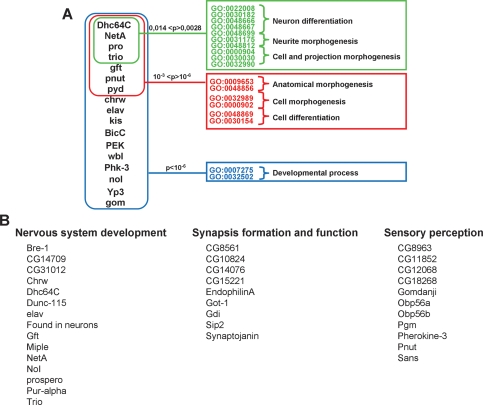
Next, we asked about the characteristics of the expression program coregulated by HP1c, WOC, and ROW. This analysis was restricted to the 158 genes that were found differentially expressed in the same direction in all three mutants, as they stand the highest probability of being truly coregulated by the three factors (Supplemental Table S3). Gene ontology (GO) analysis of this set of genes shows no statistically significant enrichment in any particular annotation after correction for multiple testing. To some extent, this observation was not unexpected considering the complexity of the samples used in our analysis, total RNA prepared from whole larvae. Clustering analysis, however, shows a number of associations of more than three genes sharing common GO annotations (Fig. 11A). Notably, a cluster of genes implicated in nervous system development and morphogenesis is observed. These include the transcription factor prospero (pros), the dynein motor Dhc64C, and the axon guidance factors, trio and NetrinA (NetA). Actually, a detailed examination of the 158 genes differentially expressed in the same direction in the three mutants, shows that, regardless of their actual GO annotations, 35 have attributed functions related to various aspects of development and functionality of the nervous system (Fig. 11B), including neurogenesis, synapsis formation and function, and sensory perception. Similar results are obtained when all genes differentially expressed in wocRNAi and rowRNAi mutants are analyzed. In this case, an additional cluster of genes involved in imaginal discs development is observed (data not shown). Altogether, these results suggest that, at least in part, HP1c, WOC, and ROW execute their genetic program in the context of the nervous system.
Figure 11.
HP1c, WOC and ROW coregulate a common gene expression program that, in part, is executed in the context of the nervous system. (A) Gene-cluster analysis of the 158 genes that are differentially expressed in the same direction in all three mutants. Associations of more than three genes sharing common GO annotations are presented. (B) Out of the 158 genes that are differentially expressed in the same direction in the three mutants, 35 have attributed functions related to development and functionality of the nervous system (neurogenesis, synapsis formation and function, and sensory perception).
Discussion
Here, we report on the interaction between Drosophila HP1c and the zinc-finger proteins WOC and ROW. Our results indicate that both WOC and ROW copurify with HP1c. Coimmunoprecipitation experiments corroborate the interaction of WOC with HP1c and ROW. Moreover, in polytene chromosomes, HP1c and ROW extensively colocalize with WOC, and targeting HP1c to an ectopic construct results in recruitment of both WOC and ROW. Altogether, these results indicate that HP1c, WOC, and ROW are components of a distinct multiprotein complex. HP1c–WOC interaction is likely to be direct, as WOC contains a canonical PxVxL motif (1536PHVLL1540), which is known to mediate binding to the chromo-shadow domain of HP1 proteins (Lechner et al. 2000, 2005; Smothers and Henikoff 2000). This motif is located within the highly conserved C-terminal HC domain of WOC, being also present in the three human homologs. ROW also contains several variant PxVxL motifs, suggesting that it might also bind directly to HP1c. In agreement with these observations, euchromatic localization of HP1c depends on the C-terminal chromo-shadow domain (Smothers and Henikoff 2001), strongly suggesting that it mediates interaction with WOC and ROW.
Binding of HP1c to chromatin relies on the zinc-finger proteins, WOC and ROW
Here, we also show that binding of HP1c to chromatin depends on WOC and ROW that, on the other hand, are reciprocally required for binding to chromatin. Domain structure and organization indicate that WOC and ROW are sequence-specific DNA-binding proteins. These results indicate that chromosomal association of HP1c is largely determined by the recognition of specific DNA sequences, which is in contrast to the situation observed in the case of Drosophila HP1a, or mammalian HP1α, where chromosomal association was found to depend on the recognition of a specific pattern of histone modifications; namely, H3K9 methylation (Hiragami and Festenstein 2005; Hediger and Gasser 2006). Several HP1 proteins, including HP1a and HP1α, were reported to interact with a number of transcription factors, replication proteins, and chromatin assembly complexes (Hiragami and Festenstein 2005; Hediger and Gasser 2006; Lomberk et al. 2006). Yet, it is unclear whether these interactions mediate recruitment of HP1 to specific sites, and/or in response to particular processes, or they actually take place after recruitment to regulate their functions. What is striking in the case of Drosophila HP1c is that its binding to chromatin is strictly dependent on DNA-binding proteins. Our results, however, also show that, through the chromodomain, HP1c efficiently binds H3K9me2,3 in vitro. Actually, overexpression of HP1c-lacI leads to its mislocalization to heterochromatin, likely reflecting binding to H3K9me2,3. In this context, it must be noticed that HP1c shows a partial colocalization with H3K9me3. However, binding of HP1c at these sites is in general weak, being also obliterated in the absence of WOC and ROW. HP1 proteins have been reported to interact with different histone methyltransferases (HMTs), being involved in their recruitment to specific sites (Hiragami and Festenstein 2005; Hediger and Gasser 2006; Lomberk et al. 2006). Therefore, it is possible that H3K9 methylation at these sites is actually the consequence of HP1c binding. Altogether, these observations indicate that recognition of H3K9me2,3 is not a major determinant of the association of HP1c with chromatin in vivo. Consistent with this interpretation, replacing the chromodomain of HP1c by that of HP1a does not alter its chromosomal distribution (Smothers and Henikoff 2001). The interaction with WOC and ROW does not appear to hinder the chromodomain from binding H3K9me2,3 since overexpression of HP1c-lacI brings both HP1c and WOC to heterochromatin. Several other possibilities can account for the inability of HP1c to bind H3K9me2,3 in vivo. The interaction of HP1c with WOC and ROW might be of higher affinity than the interaction with H3K9me2,3. In addition, post-translational modifications could regulate these interactions. It is also possible that HP1c is actively excluded from heterochromatin. Whether binding to H3K9me2,3 plays a role at any stage during development or cell cycle progression remains, however, to be determined.
HP1a and HP1b also localize to euchromatin, yet they show strong binding to heterochromatin. Therefore, WOC and ROW could also play a role in binding of HP1a and HP1b to euchromatin. In fact, euchromatic localization of HP1b is decreased in both wocRNAi and rowRNAi mutants (Supplemental Fig. S2). This effect, however, is much weaker than that observed in the case of HP1c. On the other hand, binding of HP1a to euchromatin is not grossly altered in wocRNAi and rowRNAi mutants (Supplemental Fig. S3), though its association to some specific loci, such as at the 31C region, appears to be affected (data not shown). On the other hand, binding of HP1a and HP1b to heterochromatin, which depends on H3K9me2,3, is not significantly affected in wocRNAi and rowRNAi mutants (Supplemental Figs. S2, S3). Similarly, mislocalization of overexpressed HP1c-lacI to heterochromatin, which likely reflects binding to H3K9me2,3, is not affected either in wocRNAi and rowRNAi mutants.
Altogether, these observations suggest that, in Drosophila, HP1 proteins could be recruited to chromatin by at least two independent mechanisms: (1) recognition of H3K9me2,3, which is instrumental in heterochromatin binding, and (2) interaction with sequence-specific DNA-binding proteins, which mediate euchromatic localization of HP1c and, perhaps, of HP1b and HP1a to some specific loci. Actually, the interaction of HP1 proteins with DNA-binding proteins might be more frequent than anticipated. In fact, in C. elegans, HPL-2 was found to interact with LIN-13 (Coustham et al. 2006), a sequence-specific DNA-binding protein containing multiple zinc-finger domains.
The contribution of HP1c to gene expression
Here, we report that HP1c localizes at multiple active chromatin domains and cooperates with WOC and ROW, which show features characteristic of transcription factors, to regulate gene expression. Moreover, targeting HP1c to a reporter construct promotes expression of the reporter gene. This effect is specific of HP1c, as targeting both HP1a and HP1b induce silencing. These results indicate that, rather than as a silencing factor, HP1c acts as a transcriptional regulator that is recruited to chromatin by sequence-specific DNA-binding proteins.
Other HP1 proteins have also been shown to contribute to the regulation of gene expression, yet their presence is generally associated to silencing. These include Drosophila HP1a, which is required for proper expression of most heterochromatic genes as well as a few euchromatic genes (Hearn et al. 1991; Clegg et al. 1998; Lu et al. 2000; Piacentini et al. 2003; Cryderman et al. 2005; De Lucia et al. 2005). Mammalian HP1γ has also been shown to localize at active genes in a murine erythroid cell line (Vakoc et al. 2005). In these cases, presence of HP1 appears to be implicated in stabilizing RNA transcripts, or in another RNA-processing event occurring during elongation. In the case of HP1c, however, colocalization with the poised RNApol II form, Pol IIoser5, is much stronger than with the elongating form, Pol IIoser2, suggesting that HP1c acts at the promoter level rather than during elongation. Consistent with this hypothesis, HP1c shows a much stronger colocalization with H3K4me3, a modification that occurs at promoters, than with H3K36me3, which occurs all through transcribed regions and incorporates during elongation. In full agreement with our results, WOC also shows extensive colocalization with Pol IIoser5, which is stronger than with Pol IIoser2 (Raffa et al. 2005). These results favor a contribution to the regulation of genes containing poised RNApol II. Actually, recent studies show that the presence of poised RNApol II at promoters is more frequent than anticipated, particularly on developmental control genes (Muse et al. 2007; Zeitlinger et al. 2007). Interestingly, a high proportion of genes coregulated by HP1c, WOC, and ROW act during development and morphogenesis.
The precise molecular mechanism(s) underlying the contribution of HP1c to transcription regulation remains to be determined. However, a contribution to RNApol II recruitment appears unlikely since, in the absence of WOC, RNApol II recruitment is not affected (Raffa et al. 2005). A contribution to the regulation of poised RNApol II is also uncertain, as no gross changes in the levels of Pol IIoser5 and Pol IIoser2 are observed in woc-null mutants by either immunostaining (Raffa et al. 2005) or Western analysis (data not shown). It is possible, however, that HP1c/WOC/ROW act only on a reduced subset of genes containing poised RNApol II. It must also be noticed that HP1c likely participates both in promoting and inhibiting transcription. In fact, among the 158 genes that are differentially expressed in the same direction in hp1cRNAi, wocRNAi, and rowRNAi mutants, the number of up-regulated and down-regulated genes is similar, 80 in front of 78. Moreover, consistent with a contribution to repression, out of the 35 genes that are coregulated by HP1c, WOC, and ROW in the context of the nervous system, a significantly higher number of genes are found up-regulated than down-regulated in the mutants, 22 versus 13. On the other hand, targeting HP1c to a reporter, though modestly, increases its expression. Altogether, these observations suggest that, depending on the actual functional/promoter context, HP1c can be engaged in either promoting or inhibiting transcription.
The patterns of differentially expressed genes observed in wocRNAi and rowRNAi mutants show a very strong correlation, indicating that WOC and ROW share a common gene expression program. In addition, our results show that genes regulated by HP1c are also regulated by both WOC and ROW. On the other hand, a high proportion (78.3%) of differentially expressed genes change expression both in wocRNAi and rowRNAi but not in hp1cRNAi, suggesting that WOC and ROW could also regulate gene expression independently of HP1c. However, the extensive colocalization observed between WOC, ROW, and HP1c argues against this possibility. On the other hand, our results show that HP1c protein levels are significantly decreased in wocRNAi and rowRNAi mutants. In this scenario, stronger synergistic effects would be expected in wocRNAi and rowRNAi mutants than in hp1cRNAi, which could result in more genes being differentially expressed. Consistent with this hypothesis, a major proportion of genes that are differentially expressed in all three mutants shows stronger changes in wocRNAi and rowRNAi than in hp1cRNAi. Actually, out of the 158 genes that are differentially expressed in the same direction in all three mutants, 77 had smaller changes in hp1cRNAi than in wocRNAi or rowRNAi, a statistically significant higher number than the 52.6 genes expected under the assumption that the magnitude of change is the same in all three mutants (P-value < 0.0001).
Clustering analysis suggests that the gene expression program coregulated by HP1c, WOC, and ROW is executed, at least in part, in the context of the nervous system. In agreement with this hypothesis, expression of woc and row is high in the nervous system during embryogenesis and larval development, and mutant larvae have reduced brains (Wismar et al. 2000; Brody et al. 2002). Furthermore, one of the human homologs of WOC, DXS6673E/ZNF261, is implicated in a form of X-linked mental retardation (van der Maarel et al. 1996).
HP1c and telomere function
Others reported that WOC regulates telomere function, as it is required to prevent chromosomal end-to-end fusions (Raffa et al. 2005). The physical and functional interactions between WOC, HP1c, and ROW suggest that HP1c and ROW might also regulate telomere function. Actually, at telomeres, colocalization of WOC with HP1c and ROW is also most extensive, with all detectable αWOC bands overlapping with αHP1c and αROW bands and vice versa (Supplemental Fig. S4). However, the incidence of telomere fusions in hp1cRNAi and rowRNAi mutants is low, being similar to that observed in control flies, carrying an UAS-hairpin construct against an unrelated gene, GFP (data not shown). This is likely the consequence of both the hypomorph character of the mutations and hyperactivation of the RNAi pathway, which is known to regulate telomere function (Ho et al. 2008). Consistent with this hypothesis, the frequency of telomere fusions is also low in wocRNAi. The contribution of ROW to telomere function was also analyzed in rowl(2)SH2172, which corresponds to a very strong mutation caused by a P-element insertion at the ATG-start codon (Supplemental Fig. S1B). The incidence of telomere fusions is significantly higher in homozygous rowl(2)SH2172 flies than in control wild-type flies (Supplemental Fig. S5), confirming its contribution to telomere function. The use of currently unavailable hp1c-null mutations is also likely to confirm the contribution of HP1c to telomere function.
HP1a is also known to regulate telomere function (Fanti et al. 1998). Several observations, however, indicate that the contribution of WOC/ROW/HP1c is not related to that of HP1a. On one hand, su(var)2-5 mutants show much stronger effects than either woc or row mutants (Fanti et al. 1998; Raffa et al. 2005). Furthermore, both telomere length and expression of the telomeric retrotransposons Het-A and TART are increased in su(var)2-5 mutants, but they are not affected in woc-null mutants (Perrini et al. 2004; Raffa et al. 2005). In addition, expression profiling data show that, in hp1cRNAi, wocRNAi, and rowRNAi mutants, expression of Het-A and TART is not significantly affected (data not shown). Similarly, other genes known to contribute to telomere function do not change expression in hp1cRNAi, wocRNAi, and rowRNAi mutants (data not shown). Altogether, these observations suggest that the contribution of WOC/ROW/HP1c to the regulation of telomere function is direct and independent of their contribution to the regulation of gene expression.
Materials and methods
For a detailed description of the Materials and Methods, see the Supplemental Material.
Fly stocks and antibodies
wocRNAi, rowRNAi, and hp1aRNAi were obtained from VDRC. rowl(2)SH2172 was obtained from Szeged Drosophila Stock Center. hp1cRNAi transgenic flies were generated according to standard procedures. For targeting experiments, HP1a, HP1b, and HP1c were fused at C terminus to the lacI-DNA-binding domain.
αHP1a, αHP1b, αHP1c, and αROW antibodies were rat polyclonal. Rabbit polyclonal αWOC antibodies are described by Raffa et al. (2005). Antibodies against histone modifications and RNApol II forms were commercial (Abcam, Upstate Biotechnologies).
Immunostaining experiments and analysis of telomere fusions.
Immunostaining experiments were performed in S2 cells and polytene chromosomes and imaginal wing discs from third instar larvae, according to standard procedures (see the Supplemental Material). Telomere fusions were analyzed in brain squashes from third instar larvae. Samples were visualized on a Nikon Eclipse E-800 inverted microscope with a Colorview 12 camera or in a confocal Leica TCS SP2-AOBS microscope.
Purification and characterization of multiprotein complexes containing HP1c
Purification of multiprotein complexes was performed from stable S2 lines expressing TAP-HP1c. LC/MS analysis was performed in the Proteomics Unit of the “Institut de Recerca de la Vall d’Hebrón” (Barcelona).
Coimmunoprecipitation experiments
Coimmunoprecipitation experiments were performed with crude S2 nuclear extracts by the addition of αWOC, αDDP1, or no antibodies, according to standard procedures.
Analysis of the interaction of HP1c with H3K9me2,3 in vitro
Analysis of the interaction of HP1c with H3K9me2,3 in vitro was performed with recombinant GST- and His-tagged proteins. Mononucleosomes and total endogenous histones were obtained from S2 cells. GST-pull-down assays and Far-Western analysis were performed according to standard procedures.
Expression profiling analysis
For expression profiling, Drosophila Genome 2.0 GeneChip (Affymetrix) were hybridized with cDNA prepared from total RNA obtained from male larvae. Duplicates were processed for each of the genotypes analyzed. For each mutant condition, differential expression was measured via the distance between the intervals containing the mutant and the control expression values. Lists of differentially expressed genes were obtained by setting several cut-offs for the absolute value of the interval distance. To measure the degree of coregulation between the mutants, we computed the pairwise Spearman’s rank correlation between interval distances. GO analysis was performed with GOtoolBox.
Acknowledgments
We are grateful to C.Antoniewski, J.Bernués, M. Batllé, M.L.Espinàs, M.Gatti, A. Jordan, K.Furukubo-Tokunaga, J.T. Lis, A.Veraksa, and L.Wallrath for materials, and to A. Casali, D. Huertas, and I. Garcia-Bassets for helpful advice and discussions. We are most thankful to M. Lloret-Llinares for preliminary studies on the effect of hp1cRNAi on gene expression. We are also thankful to E. Fuentes and L. Bardia for technical assistance. J.F. acknowledges receipt of a doctoral fellowship from MEC. This work was financed by grants from MEC (BMC2006-1627, CSD2006-00049) and the CIRIT (2001SGR00344).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.481408.
References
- Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell O., Conrad T., Kind J., Wirbelauer C., Akhtar A., Schübeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol. Cell. Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Brody T., Stivers C., Nagle J., Odenwald W.F. Identification of novel Drosophila neural precursor genes using a differential embryonic head cDNA screen. Mech. Dev. 2002;113:41–59. doi: 10.1016/s0925-4773(02)00010-2. [DOI] [PubMed] [Google Scholar]
- Clegg N.J., Honda B.M., Whitehead I.P., Grigliatti T.A., Wakimoto B., Brock H.W., Lloyd V.K., Sinclair D.A. Supressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of chromosome rearrangement. Genome. 1998;41:495–503. [PubMed] [Google Scholar]
- Coustham V., Bedet C., Monier K., Schott S., Karali M., Palladino F. The C. elegans HP1 homologue HPL-2 and the LIN-13 zinc finger protein form a complex implicated in vulval development. Dev. Biol. 2006;297:308–322. doi: 10.1016/j.ydbio.2006.04.474. [DOI] [PubMed] [Google Scholar]
- Cryderman D.E., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L. Role of Drosophila HP1 euchromatic gene expression. Dev. Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Ni J.Q., Vaillant C., Sun F.L. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005;33:2852–2858. doi: 10.1093/nar/gki584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G.K., Terjung S., Brown J.P., Aucott R.L., Baron-Luhr B., Singh P.B., Georgatos S.D. Plasticity of HP1 proteins in mammalian cells. J. Cell Sci. 2007;120:3415–3424. doi: 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo G., Berloco M., Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Fanti L., Berloco M., Piacentini L., Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica. 2003;177:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Haraguchi T., Masumoto H., Hiraoka Y. Cell cycle behavior of human HP1 subtypes: Distinct domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003;116:3327–3338. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- Hearn M.G., Hedrick A., Grigliatti T.A., Wakimoto B. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Gasser S.M. Heterochromatin protein 1: Don’t judge the book by its cover! Curr. Opin. Genet. Dev. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hiragami K., Festenstein R. Heterochromatin protein 1: A pervasive controlling influence. Cell. Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.Y., Murnane J.P., Yeung A.K., Ng H.K., Lo A.W. Telomeres acquire distinct heterochromatin characteristics during siRNA-induced RNA interference in mouse cells. Curr. Biol. 2008;18:183–187. doi: 10.1016/j.cub.2007.12.059. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C., et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M.S., Begg G.E., Speicher D.W., Rauscher F.J. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: Direct chromoshadow domain–KAP-1 corepressor interactions is essential. Mol. Cell. Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M.S., Schultz D.C., Negorev D., Maul G.G., Rauscher F.J. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Li Y., Danzer J.R., Alvarez P., Belmont A.S., Wallrath L.L. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–1824. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- Lomberk G., Wallrath L.L., Urrutia R. The heterochromatin protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.Y., Emtage P.C., Duyf B.J., Hilliker A.J., Eissenberg J.C. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E., Allory Y., Wormann H.J., Courvalin J.C., Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K., Jasencakova Z., Schubert I., Goto K. The Arabidopsis heterochromatin protein 1 homolog (TERMINAL FLOWER 2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 2005;46:1747–1756. doi: 10.1093/pcp/pci195. [DOI] [PubMed] [Google Scholar]
- Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., Berloco M., Turano C., Ferraro A., Pimpinelli S. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici C., Adelaide J., Ollendorff V., Chaffanet M., Guasch G., Jacrot M., Leroux D., Birnbaum D., Pebusque M.J. Fibroblast growth factor receptor 1 is fused to FIM in stem-cell myeloproliferative disorder with t(8;13) Proc. Natl. Acad. Sci. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G.D., Cenci G., Siriaco G., Goldberg M.L., Gatti M. The putative Drosophila transcription factor Woc is required to prevent telomeric fusions. Mol. Cell. 2005;20:821–823. doi: 10.1016/j.molcel.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A.J., Li H., Patel D.J., Allis D.C. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott S., Coustham V., Simonet T., Bedet C., Palladino F. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 2006;298:176–187. doi: 10.1016/j.ydbio.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Smallwood A., Estève P.O., Pradhan S., Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes & Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers J.F., Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Smothers J.F., Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal J., Chase A., Goldman J.M., Cross N.C. Assignment of ZNF262 to human chromosome band lp34-p32 by in situ hybridization. Cytogenet. Cell Genet. 1999;85:306–307. doi: 10.1159/000015282. [DOI] [PubMed] [Google Scholar]
- Swaminathan J., Baxter E.M., Corces V.G. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes & Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Yang X., Zhuang L., Jiang X., Chen W., Lee P.L., Karuturi R.K., Tan P.B., Liu E.T., Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg S.J. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- van der Maarel S.M., Scholten I.H.J.M., Hubert J., Philippe C., Suijkerbuijk R.F., Gilgenkrantz S., Kere J., Cremers F.P.M., Ropers H.-H. Cloning and characterization of DXS6673E, a candidate gene for X-linked mental retardation in Xq13.3. Hum. Mol. Genet. 1996;5:887–897. doi: 10.1093/hmg/5.7.887. [DOI] [PubMed] [Google Scholar]
- Vermaak D., Henikoff S., Malik H.S. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:e9. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure P.J., van der Kraan I., de Leeuw W., van der Vlag J., Carpenter A.E., Belmont A.S., van Driel R. In Vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell. Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J.T., Wismar J., Subrahmanyam B., Gilbert L.I. Woc(without children) gene control of ecdysone biosynthesis in Drosophila melanogaster. Mol. Cell. Endocrinol. 2001;181:1–14. doi: 10.1016/s0303-7207(01)00404-x. [DOI] [PubMed] [Google Scholar]
- Wismar J., Habtemichael N., Warren J.T., Dai J.-D., Gilbert L.I., Gateff E. The mutation without children (rgl) causes ecdysteroid deficiency in third-instar larvae of Drosophila melanogaster. Dev. Biol. 2000;226:1–17. doi: 10.1006/dbio.2000.9811. [DOI] [PubMed] [Google Scholar]
- Xiao S., Nalabolu S.R., Aster J.C., Ma J., Abruzzo L., Jaffe E.S., Stone R., Weissman S.M., Hudson T.J., Fletcher J.A. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat. Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]