Abstract
Bistability in developmental pathways refers to the generation of binary outputs from graded or noisy inputs. Signaling thresholds are critical for bistability. Specification of the left/right (LR) axis in vertebrate embryos involves bistable expression of transforming growth factor β (TGFβ) member NODAL in the left lateral plate mesoderm (LPM) controlled by feed-forward and feedback loops. Here we provide evidence that bone morphogenetic protein (BMP)/SMAD1 signaling sets a repressive threshold in the LPM essential for the integrity of LR signaling. Conditional deletion of Smad1 in the LPM led to precocious and bilateral pathway activation. NODAL expression from both the left and right sides of the node contributed to bilateral activation, indicating sensitivity of mutant LPM to noisy input from the LR system. In vitro, BMP signaling inhibited NODAL pathway activation and formation of its downstream SMAD2/4–FOXH1 transcriptional complex. Activity was restored by overexpression of SMAD4 and in embryos, elevated SMAD4 in the right LPM robustly activated LR gene expression, an effect reversed by superactivated BMP signaling. We conclude that BMP/SMAD1 signaling sets a bilateral, repressive threshold for NODAL-dependent Nodal activation in LPM, limiting availability of SMAD4. This repressive threshold is essential for bistable output of the LR system.
Keywords: SMAD1, NODAL, lateral plate mesoderm, left/right asymmetry, BMP, bistability
The left/right (LR) axial pathway establishes asymmetry in the patterning and morphogenesis of multiple internal organs, including the heart. In mammals, this pathway involves the breaking of molecular symmetry in or around the node, transfer of asymmetry information to the lateral plate mesoderm (LPM), propagation of molecular asymmetries throughout the LPM, and interpretation of these signals for proper organ morphogenesis (Nonaka et al. 2002). Recent experiments have demonstrated that the breaking of bilateral symmetry in the mouse node occurs via a process termed “nodal flow,” in which motile cilia in the node generate leftward movement of molecular determinants via lipoprotein vesicles (Hirokawa et al. 2006; Shiratori and Hamada 2006). NODAL, a transforming growth factor β (TGFβ) superfamily member, is a key molecule in the LR cascade. NODAL acts first in mesoderm specification and anterior–posterior axis formation, signaling through a membrane complex containing type I and II TGFβ serine/threonine kinase receptors, as well as GPI-anchored members of the EGF-CFC family. This complex phosphorylates intracellular SMAD2 and SMAD3, which associate with the common TGFβ/NODAL/BMP (bone morphogenetic protein) pathway SMAD, SMAD4, and forkhead transcription factor FOXH1, to regulate downstream target genes (Shen 2007). In the LR pathway, Nodal is first expressed in crown cells of the node at late primitive streak stages and this is required to initiate Nodal expression in the left LPM adjacent to the node, where it then mediates propagation of a left-sided cascade of gene expression cranially and caudally through a positive regulatory loop (Saijoh et al. 2000, 2003; Meno et al. 2001; Brennan et al. 2002; Norris et al. 2002). Current evidence suggests that NODAL expressed from the node is directly responsible for activating Nodal in the LPM (Tanaka et al. 2007). Nodal also activates Lefty1 and Lefty2, which encode feedback inhibitors of NODAL signaling, and Pitx2, encoding a homeodomain transcription factor that is thus far the only known effector for LR organogenesis (Brennan et al. 2002; Saijoh et al. 2003). The positive and negative NODAL feedback loops have the characteristics of a reaction diffusion and self-enhancing lateral inhibition system (Nakamura et al. 2006), and involve multiple extracellular and intracellular modulators, including miRNAs, that enable exquisite control over spatial and temporal signaling dynamics (Collignon 2007; Shen 2007). The midline plays an important role in LR asymmetry. Located at the level of the floor plate of the neural tube, the so-called “midline barrier” insulates left and right sides as molecular asymmetries develop, and its activity critically requires expression of the NODAL inhibitor LEFTY1 on its left side. In humans, mutations in LR pathway genes or those involved in development of crucial LR structures such as the axial midline, node, and nodal cilia, are found in approximately one in 10,000 live births (Levin and Palmer 2007).
BMPs have been implicated in LR patterning, but data on their precise roles have been highly contradictory, with results varying both within and between developmental models (Rodriguez Esteban et al. 1999; Yokouchi et al. 1999; Monsoro-Burq and Le Douarin 2001; Fujiwara et al. 2002; Piedra and Ros 2002; Schlange et al. 2002; Kishigami et al. 2004; Zhu and Scott 2004; Chocron et al. 2007; Mine et al. 2008). This is in part because BMPs act at distinct times and in distinct spatial domains to influence laterality (Kishigami et al. 2004). Genetic data have provided evidence that BMPs need to be inhibited for correct left-sided Nodal expression (Chang et al. 2000, 2002; Constam and Robertson 2000; Kishigami et al. 2004; Mine et al. 2008). However, the spatial and temporal requirement for BMP signaling in these models could not be precisely established as both the node and the LPM were affected, casting doubt on the integrity of nodal flow and/or the midline barrier.
In the present study we provide genetic evidence from analysis of Smad1-null and conditionally deleted embryos that BMP signaling plays a repressive role in the LR pathway in LPM. The LR system is an excellent example of a signaling pathway that displays bistability— the generation of a robust all-or-none (binary) output from graded or noisy inputs (Ferrell 2002). Signaling thresholds are essential for the integrity of bistable pathways. Our studies define a role for BMP/SMAD1 in setting a threshold for NODAL signaling in part by limiting availability of the common TGFβ/NODAL/BMP pathway effector, SMAD4. This threshold allows a robust bistable output from the LR system, while also conferring protection against intrinsic biological noise.
Results
Smad1-null embryos display laterality phenotypes
To gain insights into the role of BMP signaling in the embryonic LR axis in the mouse, we analyzed Smad1−/− embryos at 9.5 d post-coitum (dpc) for LR defects. Homozygous embryos typically displayed improper cardiac looping, abnormalities in embryo turning, and discordance between these events (n = 43). Examination using scanning electron microscopy (SEM) revealed hearts with rightward (30%), leftward (47%), and forward loops (23%) (Fig. 1A–D). Even in embryos with rightward loops, pronounced abnormalities were observed in the architecture of the loop, and some embryos were partially bifid (Fig. 1C), likely arising secondarily to defects in mesoderm or endoderm formation (Tremblay et al. 2001).
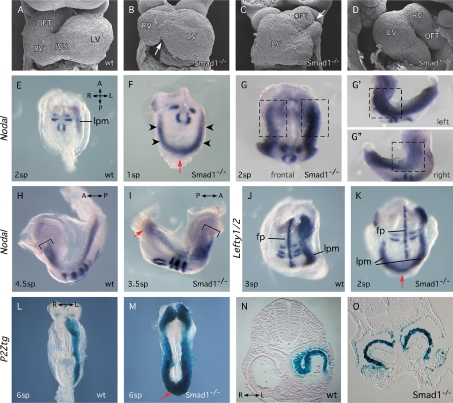
Figure 1.
Smad1-null embryos display LR defects. (A–D) SEM micrographs of hearts showing frontal (A–C) and lateral (D) views of wild-type (wt) (A) and Smad1−/− (B–D) embryos at 8.5–9.0 dpc. Arrows highlight abnormal interventricular sulcus (B) and bifid OFT (C). Hearts in C and D shows reversed and forward loops, respectively. (E–K) Whole-mount in situ hybridization showing expression of Nodal or Lefty1/2 in Smad1−/− mutant and wild-type embryos. Uncx4.1 signal indicates number of formed somite pairs (sp). Arrowheads (F) indicate robust precocious bilateral expression of Nodal at 1 sp. Red arrows (F,I,K) highlight expression of Nodal or Lefty2 across the caudal midline. Boxes in (G,G′,G″) highlight differences in Nodal expression levels in the anterior of Smad1−/− embryos. Brackets (H,I) indicate equivalent cranial extents of Nodal expression in wild type and Smad1−/− mutants. (L–O) LacZ staining in P2Ztg and P2Ztg;Smad1−/− embryos at 8.5 dpc highlighting molecular left isomerism in Smad1−/− mutants. (fp) Floorplate; (IVS) interventricular sulcus; (LV) left ventricle; (lpm) lateral plate mesoderm; (OFT) outflow tract; (RV) right ventricle.
We assayed the onset of expression of LR genes Nodal, Lefty1, Lefty2, and Pitx2. Simultaneous detection of Uncx4.1 expression allowed accurate determination of the number of somite pairs (sp). Asymmetric expression of Nodal in the left LPM is normally initiated in embryos at the 2-sp or 3-sp stage (Fig. 1E). In Smad1−/− mutants, Nodal expression was initiated prematurely at the 1-sp stage as well as bilaterally in three of four embryos examined (Fig. 1F, arrowheads). Caudal expression extended across the midline (arrow). In 1–2-sp mutants, expression in the left LPM was stronger and/or broader than on the right side, particularly in the anterior region (Fig. 1G–G″), or when compared with the left side of wild-type embryos (Fig. 1E,F). At 3–5 sp, the Nodal expression domain extended anteriorly to the same degree in both wild-type and mutant embryos (Fig. 1H,I). However, expression always extended further posteriorly in mutants compared with wild-type embryos, reaching the base of the allantois and crossing the midline (Fig. 1H,I; see below). Lefty2 expression in the LPM was similarly deregulated including expression across the midline caudally (Fig. 1J,K).
We crossed a Pitx2c isoform-specific lacZ transgene, 17-P1 (Shiratori et al. 2001), hereafter referred to as P2Ztg, into the Smad1 mutant background. This transgene accurately recapitulated endogenous LR expression of Pitx2c (M.B. Furtado, C. Biben, H. Hamada, and R.P. Harvey, in prep.). P2Ztg staining was bilateral in all Smad1-null embryos (Fig. 1L–O), extending throughout the heart (Fig. 1N,O), across the midline caudally at the base of the allantois (Fig. 1M), and into the allantois itself (Fig. 4L,M, below; data not shown). In Smad1−/− embryos at 9.5 dpc, P2Ztg expression encompassed the entirety of the atria/atrioventricular canal (AVC) and most of the myocardium of the primitive ventricle (Fig. 2C,F; arrowheads). We conclude that loss of Smad1 leads to a fully penetrant left isomerism.
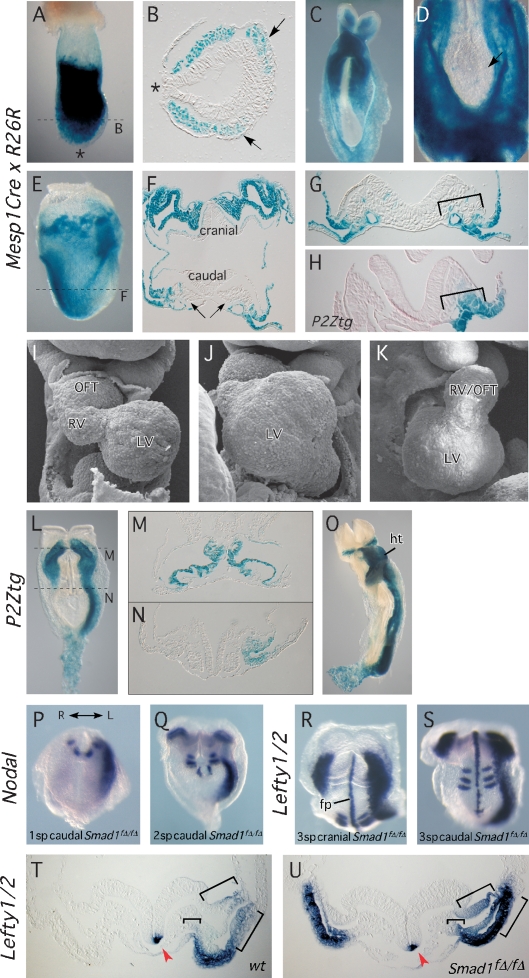
Figure 4.
Mesp1Cre conditional deletion of Smad1. (A–F) LacZ reporter studies revealing the expression pattern of the Mesp1Cre driver. (A,B) Whole-mount Mesp1Cre/+;R26R embryo at 7.5 dpc and section showing LacZ staining in anterior mesoderm although not the node or primitive streak (asterisks). Arrows indicate cardiac mesoderm. (C,D) Whole-mount views of Mesp1Cre/+;R26R 8.5 dpc embryos (embryo in D is overstained) showing strongest LacZ staining in the anterior LPM and lack of staining in node, notochord, and paraxial mesoderm. (E,F) All cranial mesoderm is LacZ-positive in 8.0 dpc embryos, whereas caudally, extraembryonic mesoderm is LacZ-positive while embryonic mesoderm shows highly mosaic expression (arrows). (G,H) Comparison of Mesp1Cre-driven R26R recombination (G) and the NODAL-responsive area revealed by P2Ztg expression (H) in comparable caudal sections at 8.0 dpc. (I–K) SEM views of 9.5 dpc Mesp1Cre/+;Smad1f/f (Smad1fΔ/fΔ) embryos, showing defects in cardiac looping and chamber morphogenesis. Compare with control heart in Figure 1A. (L–V) Bilateral activation of LR pathway genes in the anterior LPM in Smad1fΔ/fΔ embryos. (L–N) Ectopic lacZ staining from P2Ztg in the right anterior LPM and allantois of Smad1fΔ/fΔ embryos at 8.0 dpc. (O) Ventral whole-mount view of P2Ztg expression in Smad1fΔ/fΔ embryos at 8.5 dpc. (P,Q) Nodal expression initiates earlier (1 sp) in the left LPM of Smad1fΔ/fΔ mutants (P), before being established bilaterally in the anterior LPM at 2 sp (Q). (R,S) Cranial and caudal views, respectively, of a 3 sp embryo showing Lefty1/2 expression. Note normal expression of Lefty1 in floorplate, and of Lefty2 bilaterally in the anterior LPM, strongest on the left. The wild-type control is shown in Figure 1J. (T,U) Transverse cranial sections of wild-type and Smad1fΔ/fΔ 8.0 dpc embryos showing Lefty1/2 expression. Brackets indicate domains in which Lefty2 expression is expanded in the cranial left LPM in mutants. Arrowhead indicates Lefty1 expression on the left side of the floorplate. (fp) Floorplate; (ht) heart; (L) left; (LV) left ventricle; (OFT) outflow tract; (R) right; (RV) right ventricle.
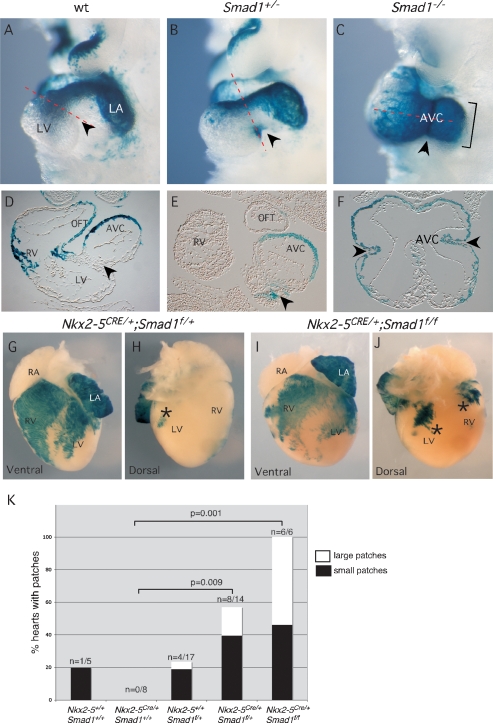
Figure 2.
Laterality pathway is sensitive to Smad1 gene dosage. (A–F) Whole-mount and section views showing LacZ staining from the P2Ztg allele in wild-type (wt) and mutant embryos at 9.5 dpc. Arrowheads indicates dorsocaudal aspect of the AVC. (G–J) Ventral (G,I) and dorsal (H,J) views of newborn Nkx2-5Cre/+;Smad1flox/+ and Nkx2-5Cre/+;Smad1flox/flox hearts after staining for LacZ expressed from P2Ztg. Asterisks indicate ectopic LacZ patches. (K) Table showing quantification of number of embryos with patches for each genotype (significance determined by Fisher’s exact test). Note trend, albeit nonsignificant with numbers of hearts analyzed, toward more patches and larger patches in homeozygotes compared with heterozygotes. (AVC) Atrioventricular canal; (LA) left atrium; (LV) left ventricle; (OFT) outflow tract; (RA) right atrium; (RV) right ventricle.
P2Ztg regulation is sensitive to Smad1 dosage
Pitx2 expression is sensitive to Nodal gene dosage (Meno et al. 1998, 2001; Lowe et al. 2001), so we assessed whether P2Ztg was misregulated in Smad1 heterozygotes. While P2Ztg expression was correctly restricted in the OFT, left and right ventricles, and left atrium (LA) in heterozygotes at 9.5 dpc (Fig. 2A,B), a patch of positive cells was observed in three of four embryos at the dorsocaudal surface of the AVC, likely corresponding to tissue derived from the right side of the LPM (arrowheads in Fig. 2A,B,D,E). This was never seen in wild-type embryos (n > 50).
We next used a conditional (floxed) allele of Smad1 (Smad1f/f) (Tremblay et al. 2001) to specifically delete the gene in cardiac cells expressing the homeodomain transcription factor Nkx2-5 (Stanley et al. 2002). Nkx2-5Cre/+;Smad1f/f mice were born in the correct Mendelian ratio (n = 6/32; 19% cf 12.5% expected), were viable at birth (n = 6) and 3 wk (n = 2), and had normal cardiac situs, suggesting that Nkx2-5-driven Cre does not delete Smad1 rapidly or broadly enough to disturb LR patterning in the heart. A compounding factor may be that Nkx2-5 is not expressed in the sinuatrial region of the forming heart tube in which Nodal and Pitx2 are most highly expressed (Mommersteeg et al. 2007). We nonetheless assessed P2Ztg expression in Nkx2-5Cre/+;Smad1f/f and control hearts at mid–late gestation and neonates to map potential focal misregulation of the NODAL pathway, as seen in Smad1+/− embryos. P2Ztg expression was roughly similar in mutants and wild-type controls, and there was no indication of left isomerism from LacZ staining in the atria (Fig. 2G,I; data not shown). However, the boundaries of LacZ expression domains in left and right ventricles were more variable (Fig. 2G,I), and multiple ectopic patches of LacZ staining were seen in the dorsal myocardial walls of the ventricles in both conditional heterozygotes (n = 8/14) and homozygotes (n = 6/6) (Fig. 2G–K) (P < 0.01 for each when compared with n = 0/8 patches in Nkx2-5Cre/+;Smad1+/+ control). This dorsal territory has its origins in the right LPM at earlier stages of development. There was a trend toward more patches and patches of a larger size in homozygotes. We also saw one or more patches in 5 of 17 Smad1f/+ embryos (30%), likely because the floxed Smad1 allele is hypomorphic, and we found a single small patch in one of five wild-type hearts examined (Fig. 2K). Collectively, our data confirm that the correct level of SMAD1 is critical for the integrity of the LR system and that a slight reduction or even stochastic fluctuation (as likely occurred in the wild-type embryo) can lead to the system becoming metastable.
Node and midline markers are not affected in Smad1-null embryos
Left isomerism can be generated by dysfunction in node morphogenesis (Hirokawa et al. 2006; Shiratori et al. 2006) and/or a disrupted midline barrier function (Meno et al. 2001). To assess node integrity in Smad1 mutants we examined expression of genes reflective of LR asymmetric patterning and/or function. In 0–1-sp Smad1 mutants (n = 5), Nodal expression around the node was normal (Fig. 3A,B), in three of these coincident with the emergence of precocious bilateral expression in the LPM (arrows). The left-sided bias in the Nodal pattern normally apparent at 5–7 sp was also evident in Smad1 mutants (Fig. 3C,D). Dante was expressed normally at the periphery of the node at 1–2 sp (n = 10) (Fig. 3E,F) and at 4–7 sp with a LR bias reciprocal to that of Nodal (Fig. 3G,H; Pearce et al. 1999). LPlunc1 was expressed normally in crown cells of the node with the normal strong leftward bias at 7 sp (n = 4) (Fig. 3I,J; Hou et al. 2004). Collectively, these results suggest that the node is properly formed and patterned in the absence of Smad1.
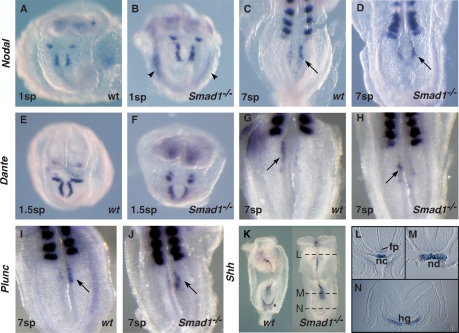
Figure 3.
Intact node and midline in Smad1-null embryos. (A–J) Ventral views of the node region (arrowed) of whole-mount wild-type (wt) and Smad1−/− embryos hybridized for Nodal, Dante, and Plunc expression. Included Uncx4.1 probe revealed number of formed somite pairs (sp). Note precocious Nodal expression in the LPM in a 1sp Smad1−/− mutant (arrowheads). (K–N) Whole-mount (K) and section views (L–N) showing expression of Shh in wild-type and Smad1−/− embryos. (fp) Floorplate; (hg) hindgut; (nc) notochord; (nd) node.
Signals emanating from the notochord are essential for patterning the ventral neural tube and formation of a midline barrier (Hamada et al. 2002). Mouse embryos lacking Sonic hedgehog (Shh) show disruption of the notochord and prechordal plate, and complete loss of Lefty1 expression in the floorplate (Tsukui et al. 1999). Normal Shh expression in Smad1 mutants (n = 6) revealed that the notochord was intact (Fig. 3K–N) and competent to induce Shh expression in anterior floorplate (Fig. 3L,M). There was weak down-regulation around the hindgut pocket in whole-mount embryos (Fig. 3K, asterisk) although expression within the hindgut floor was apparently normal on sections (Fig. 3N). Lefty1 was also correctly expressed with a leftward bias in the floorplate in null embryos (Figs. 1J,K, 4T,U). These data suggest that the midline is formed normally in Smad1 homozygotes and is likely competent to function as a LR midline barrier.
Smad1 is required in the LPM to suppress Nodal
While the node and midline appeared intact in Smad1-null embryos, our observations do not formally exclude a role for Smad1 in these structures. To test whether Smad1 is required specifically in the LPM to regulate Nodal, we conditionally deleted Smad1 using Cre recombinase driven by Mesp1 cis-regulatory elements (Mesp1Cre) (Saga et al. 1999). Previous analysis of the Mesp1Cre line demonstrated recombination favoring anterior mesoderm. To determine whether deletion occurred in the node and axial or paraxial mesoderm, we crossed the Mesp1Cre line to the R26R lacZ Cre-dependent reporter line and mapped cell lineages responsive to Cre. At 7.5 dpc, prior to the onset of LR signaling, LacZ staining was detected in the bilateral mesodermal wings several cell diameters from the primitive streak (Fig. 4A,B). No activity was observed in the node or axial mesoderm (asterisks). At 8.0–8.5 dpc, expression in cranial lateral and extraembryonic mesoderm was extensive (Fig. 4C–F) but only rare, isolated axial or paraxial mesodermal cells were LacZ-positive (Fig. 4D). Caudal LPM displayed chimeric LacZ expression (Fig. 4E–G) and comparison of sections from Mesp1Cre;R26R embryos with analogous sections of embryos from the P2Ztg line stained for LacZ demonstrated that the NODAL responsive domain in caudal LPM was significantly more extensive than the territory defined by Mesp1Cre activity (Fig. 4G,H, brackets).
We assessed Mesp1Cre–deleted Smad1 embryos at 9.0 dpc using SEM. Although the majority exhibited rightward looping (n = 19/25), three showed reversal and three forward looping, as observed in germline Smad1 nulls (Fig. 4I–K). Regardless of looping direction, cardiac morphology was abnormal in most conditional mutants (Prall et al. 2007).
We introduced the P2Ztg reporter into conditionally deleted Smad1f/f embryos and examined LacZ staining from 8.0–9.5 dpc (n = 9). At the linear heart tube stage, LacZ was correctly restricted to the left LPM in the posterior half of embryos (Fig. 4L–O), although it was bilaterally expressed in the anterior LPM. While inefficient deletion of Smad1 in the posterior LPM was anticipated from the Mesp1Cre;R26R analysis above, it was nonetheless striking that no right-sided P2Ztg expression was seen below the level of the sinus venosus.
Conditional loss of Smad1 led to bilateral Nodal expression in the LPM, although, as for P2Ztg, only in the anterior domain (Fig. 4P,Q). Surprisingly, we noted premature Nodal activation in the caudal left LPM in one of four embryos at 1 sp (Fig. 4P), whereas expression never occurred in wild-type controls at this stage (n = 9) (Fig. 2A). Furthermore, as for germline-deleted Smad1 embryos, there was ectopic expression of P2Ztg across the caudal midline and throughout the allantois from 8 dpc (n = 4/6) (Fig. 4L,O). These findings show that even mosaic deletion of Smad1 in the caudal left LPM has specific effects on the dynamics of the laterality pathway.
Lefty1 was expressed normally on the left side of the floor plate in conditional mutants (Fig. 4R–U, arrowheads). Similar to the pattern of Nodal, Lefty2 was expressed bilaterally in the cranial LPM only (Fig. 4R,S). As seen in germline-deleted mutants (Fig. 1K), cranial Lefty2 expression in conditional mutants was expanded on the left compared with the right side, and compared with the left side of normal embryos (Fig. 4T,U, brackets), showing that even left-sided NODAL signaling is enhanced in the absence of Smad1.
During establishment of laterality, BMP signaling in the LPM is symmetrical
Active phosphorylated (p) SMAD2 can be transiently detected in the left LPM at the time of Nodal and Lefty2 expression (Meno et al. 2001). We examined whether BMP signaling was similarly asymmetric immediately after Nodal expression becomes detectable in the LPM using antibody specific for activated phosphorylated BMP-SMADs 1, 5, and 8. However, we found uniform bilateral pSMAD1/5 staining (Smad8 is not expressed at this stage) in the LPM at 3–4 sp in wild-type embryos (n = 15), confirming published data (Mine et al. 2008), as well as in conditionally deleted (n = 8) and germline-deleted (n = 5) Smad1 mutants irrespective of the severity of morphological phenotype (Supplemental Fig. 1A–F). The robust bilateral pSMAD staining in Smad1 mutants is likely due to pSMAD5 expression and it is noteworthy that Smad5 mutants also show bilateral activation of the LR pathway, albeit in the presence of a defective midline (Chang et al. 2000). Genes encoding BMP2, BMP4, and BMP7 were also expressed symmetrically at 8–8.5 dpc in wild-type and mutant embryos (Supplemental Fig. 1G,H), as in the chick (Yokouchi et al. 1999). FGF-8 and other TGFβ family members including ACTIVIN and GDF1 are capable of inducing Nodal and Pitx2 when overexpressed in the right LPM (Wall et al. 2000). GDF-1 is an essential cofactor for NODAL in laterality signals (Rankin et al. 2000; Tanaka et al. 2007). Gdf1 and Fgf8 were also symmetrically expressed in both wild-type and mutant embryos at 8.5 dpc (Supplemental Fig. 1I,J; data not shown), suggesting that these genes do not fall directly under LR pathway control in the mouse (Tanaka et al. 2007).
NODAL contributes to bilateral expression of Pitx2 in Smad1 mutants
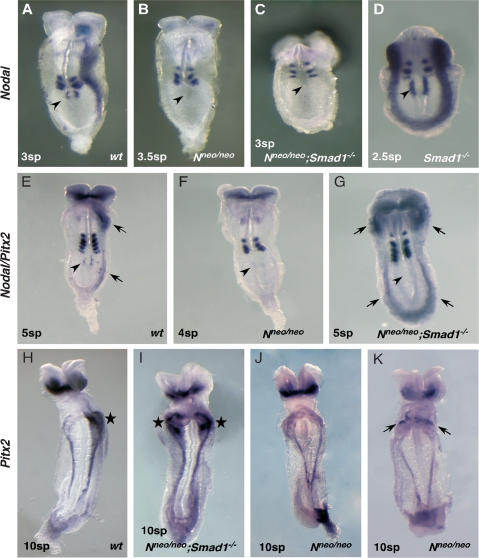
Nodal flow generates an inherently noisy input into the LR pathway, and NODAL signaling is activated in the right LPM, albeit abortively (Nakamura et al. 2006). We hypothesized that pSMAD1 functions to antagonize the LR pathway in both the left and right LPM, setting a threshold for activation of NODAL signaling and containing intrinsic biological noise. To test whether Nodal expression from the node is involved in the molecular left isomerism seen in Smad1 mutants, we generated Smad1-null embryos that were also homozygous for the Nodal-neo targeted allele (Saijoh et al. 2003). The Nodal-neo allele carries an insertion of a neomyocin resistance gene that disrupts Nodal expression in the node. Saijoh et al. (2003) showed that 64% of Nodalneo/neo embryos (7.0–8.2 dpc; n = 7/11) lack detectable Nodal in the node, with the remaining embryos showing only very weak expression insufficient to activate Nodal, Pitx2, Lefty2 or a sensitive Lefty2-lacZ transgene in the left LPM. We performed our initial analysis at 2–3 sp when Nodal is normally expressed in both the node and LPM (Fig. 5A). Confirming previous findings, we found that Nodal expression was undetectable in both the node and LPM in the majority (n = 6/7) of Nodalneo/neo embryos (Fig. 5B), with one showing very weak Nodal expression around the node insufficient to activate Nodal in the LPM (data not shown). In Smad1−/− controls Nodal was strongly bilateral in both the node and LPM (Fig. 5D). Findings in later stage Nodalneo/neo embryos were similar. At 4–5 sp, Nodal expression was undetectable in the node and Pitx2 remained off in the LPM in the majority of embryos (n = 9/12) (Fig. 5F), although in a few (n = 3/12) there was weak, bilateral expression in the LPM restricted to the region of the cardiac sinus venosus (data not shown). This weak and partially penetrant Pitx2 expression in the LPM is likely due to the earlier weak expression of NODAL around the node in some embryos (Saijoh et al. 2003; see above) that can eventually activate the LR pathway at a low level with disturbed left-sided dominance (Nakamura et al. 2006). At 8–12 sp, Nodalneo/neo embryos similarly showed no Pitx2 expression in the LPM in most cases (n = 7/10; Fig. 5J), with weak bilateral or right-sided expression in three out of 10 (Fig. 5K).
Figure 5.
LR pathway activation in Nodalneo/neo/Smad1−/− embryos. (A–D) At 2–4 sp, Nodal expression is robust in the node (arrowheads) and left LPM in wild-type embryos (A) but is bilateral in the LPM in Smad−/− mutants (D). No Nodal expression is seen in most Nodalneo/neo (Nneo/neo) or Nneo/neo;Smad1−/− embryos (see the text). (E–G) Whole-mount 4–5 sp embryos hybridized with a mixed Nodal/Pitx2 probe. Nodal is barely detectable in the node at this stage (arrowheads), although Pitx2 is expressed strongly in the anterior LPM and weakly in the caudal LPM (arrows). (F) Neither is it expressed in most Nneo/neo embryos. (G) In Nneo/neo; Smad1−/− embryos, Nodal expression is undetectable in the node but Pitx2 is bilaterally expressed cranially and caudally. (H–K) By 8.5 dpc (10 sp), Pitx2 expression in wild-type embryos is restricted to the left sinus venosus region of the heart (star) (H), and this pattern becomes bilateral in Nodalneo/neo;Smad1−/− double mutants (I). (J) Most Nneo/neo embryos show Pitx2 expression in extraembryonic tissues only. (K) A minority of Nneo/neo; Smad1−/− embryos show weak right-sided or bilateral expression (arrows) in sinus venosa.
Doubly homozygous Nodalneo/neo;Smad1−/− embryos at 2–3 sp showed absent Nodal expression in both the node and LPM in most cases (n = 3/4) (Fig. 5C), a situation very similar to that seen in Nodalneo/neo embryos. One embryo had very weak Nodal expression in the left LPM although not in the node (data not shown). However, in contrast to Nodalneo/neo embryos, at 4–7 sp Nodalneo/neo;Smad1−/− embryos showed strong bilateral Pitx2 expression in the LPM in all cases (n = 5/5) (Fig. 5E,G), while Nodal expression in the node remained undetectable (arrowheads). The same situation was seen in 8–12 sp Nodalneo/neo;Smad1−/− mutants, in which Pitx2 expression was strong and bilateral in the LPM in all cases (n = 9/9; Fig. 5H,I). We conclude that loss or severe down-regulation of Nodal expression in the node, imposed by the hypomorphic Nodalneo/neo allele, leads to a profound delay (1–2 sp or 1.5–3.0 h) in the precocious bilateral activation of the LR pathway in the LPM in Nodalneo/neo;Smad1−/− mutants. Thus, Nodal expression on both left and rights sides of the node makes a definitive contribution to the timing of bilateral activation of the LR pathway in Smad1 mutants. The right LPM of Smad1 mutants is hypersensitive to NODAL expression from the right side of the node.
Smad1 silences the ASE element of the Nodal gene in vitro
We explored how SMAD1 might negatively regulate Nodal in the LPM using an in vitro model in which the NODAL pathway is constitutively active. We first confirmed that BMP and NODAL/TGFβ cognate Flag-SMADs could be activated by phosphorylation in COS7 cells. Expression of constitutively active BMP ALK6 receptor (CA-ALK6) phosphorylated Flag-SMAD1 but not Flag-SMAD2, while constitutively active TGFβ ALK5 receptor (CA-ALK5) specifically phosphorylated Flag-SMAD2 (data not shown). Importantly, at the level of SMAD protein phosphorylation, TGFβ and BMP pathways could be simultaneously activated in this assay (data not shown). We subsequently used CA-ALK5 to mimic NODAL stimulation.
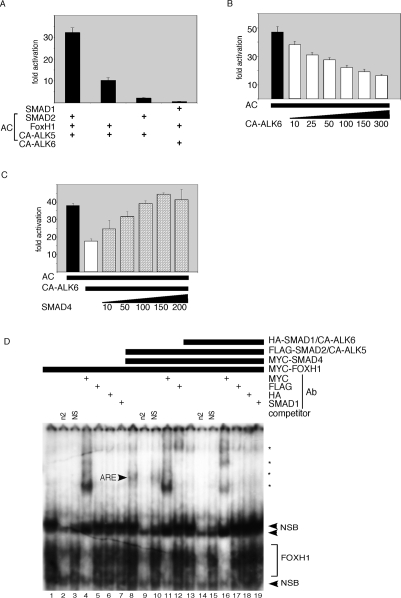
The asymmetric enhancer element (ASE) of Nodal drives asymmetric expression in the left LPM. The essential elements of the ASE are two closely spaced DNA-binding sites for the forkhead transcription factor FOXH1 (Saijoh et al. 2000). FOXH1 contains a SMAD interaction domain, and can activate the ASE element after complexing with active pSMAD2 and SMAD4. We used a reporter driven by seven tandem repeats of one of the FOXH1 sites present in the Nodal ASE element [(n2)7-luc], in transient transfection assays in 293T cells. The (n2)7-luc element recapitulates the specific left-sided LPM expression of Nodal in transgenic mice (Saijoh et al. 2000). Full activation of the (n2)7-luc reporter was dependent on the presence of FOXH1, SMAD2, and CA-ALK5 (hereafter referred to as activation cocktail [AC]) (Fig. 6A). As expected, constitutively active BMP receptor CA-ALK6 failed to activate the (n2)7-luc reporter. However, CA-ALK6 inhibited (n2)7-luc activity elicited by AC in a dose-dependent manner (Fig. 6B). Antagonism of the CA-ALK5/pSMAD2 pathway by pSMAD1 might be explained in part by competition between pSMAD2 and pSMAD1 for limiting amounts of their common transcriptional cofactor SMAD4. Indeed, ramped overexpression of SMAD4 completely reversed the inhibition imposed by the active BMP pathway in a dose-dependent manner (Fig. 6C).
Figure 6.
BMP/SMAD1 represses the NODAL pathway through competition for SMAD4. (A) The AC, composed of transfected SMAD2, CA-ALK5, and FOXHI vectors, activates the (n2)7-luciferase reporter 30-fold in 293T cells. Activation of the BMP pathway with transfected SMAD1 and CA-ALK6 shows no response. (B,C) CA-ALK6 vector represses AC-dependent activation of (n2)7-luciferase (B), which is rescued by Smad4 vector in a dose-dependent manner (C). Ten to 200/300 indicate amount of transfected DNA in nanograms. (D) EMSA analysis of DNA complexes bound to the n2 FOXH1-binding site oligonucleotide with and without activation by CA-ALK5 or CA-ALK6 (see the text). Proteins encoded by transfected vectors, competitor oligonucleotides, and antibodies used for supershift analysis are indicated above. Asterisks represent MYC–FOXH1/n2 DNA/anti-MYC antibody supershifted complexes. ARE indicates the complex composed of pSMAD2/3, FOXH1, and n2 DNA. (ARE) Activin responsive element; (n2) specific competitor; (NS) nonspecific competitor; (NSB) nonspecific bands.
We performed electrophoretic mobility shift assay (EMSA) to determine whether BMP/SMAD1 signaling could displace the AC from the n2 DNA element (Fig. 6D). FOXHI formed a complex on radiolabeled n2 oligonucleotide (Fig. 6D, lane 1) that could be competed off by excess cold n2 (Fig. 6D, lane 2), but not by nonspecific oligonucleotide (NS) (Fig. 6D, lane 3). Incubation with anti-MYC antibody led to multiple “supershifted” FOXH1 bands (Fig. 6D, lane 4; asterisks), likely different oligomeric configurations of FOXH1 and antibody on input DNA. The basal FOXH1 complex formed independently of NODAL/TGFβ pathway activation. However, the expression of AC components and SMAD4 (Fig. 6D, lane 8) led to formation of a slowly migrating complex referred to as the ARE (activin-responsive element complex). The ARE has been extensively characterized, and is composed of FOXH1, pSMAD2, and SMAD4 bound to the FOXH1 element (Inman and Hill 2002), confirmed here by the appearance of supershifted bands following incubation with anti-MYC (specific for FOXH1 and SMAD4) (Fig. 6D, lane 11) and anti-Flag (specific for SMAD2) (Fig. 6D, lane 12) antibodies. Cotransfection with CA-ALK6, leading to BMP pathway activation, resulted in abolition of the ARE, but not the basal FOXH1 complex (Fig. 6D, lane 13). Direct binding of SMAD1 to n2 was never detected (Fig. 6D, lanes 13,18,19).
Smad4 derepresses LR signaling in the right LPM
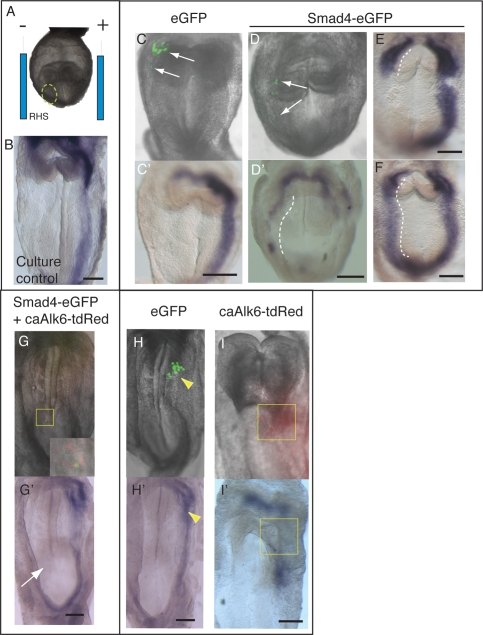
To further test the notion that BMP/pSMAD1 signaling limits availability of SMAD4, we microinjected DNA expression vectors into the right LPM of 4–6 sp embryos, before electroporation and in vitro culture (Fig. 7A). This stage avoided the confounding effects of culture on earlier (2–4 sp) embryos that led to randomization of the LR pathway, potentially due to disturbed nodal flow. After the culture period (7–9 h), Pitx2 expression was strictly left-sided in virtually all (n = 10/11) embryos (Fig. 7B). We first expressed CMV-Smad4-IRES-eGFP (Smad4-eGFP) or control CMV-IRES-eGFP (eGFP) vectors in the right LPM. Expression of enhanced green fluorescent protein (eGFP) from the internal ribosome entry site (IRES) allowed live embryo visualization of injection sites, and the localization of Smad4-expressing cells in some embryos at the end of in vitro culture (Fig. 7C,D).
Figure 7.
Increased SMAD4 activates the LR pathway in the right LPM in vivo. Whole-embryo electroporation. (A) Expression vectors were microinjected into the right anterior LPM (circled) of 4–6 sp embryos, contralateral to the normal left-sided expression domain of Pitx2 (B), then embryos were electroporated with the site of microinjection positioned next to the negative electrode. (C,C′) Control eGFP vector electroporation into the right LPM showing distribution of fluorescence (green) at the end of the culture period (arrows, C) but no change in Pitx2 expression (C′). (D,D′) Electroporation of Smad4-eGFP vector into the right LPM showing distribution of electroporated cells (arrows, D) and ectopic expression of Pitx2 in the right LPM (dashed line) as well as partial down-regulation of Pitx2 in the left LPM (D′). (E,F) Embryos showing different degrees of ectopic Pitx2 expression in the right LPM while displaying normal Pitx2 expression in the left LPM. (G,G′) Embryo electroporated with both Smad4-eGFP and caAlk6-tdRed vectors into the right LPM (see Supplemental Table 1) in which both green and red fluorescence were visible after the culture period (see inset showing enlargement of boxed region). All such embryos lacked Pitx2 expression in this region (arrow). (H–I′) Examples of embryos electroporated with eGFP or caAlk6-tdRed into the left LPM. Note the gap in left-sided Pitx2 expression in cells inheriting caAlk6-tdRed (boxed) but not eGFP vector (arrowheads). Bar, 50 μm.
Pitx2 expression was used as readout for the LR pathway. With Smad4-eGFP vector, there was a striking increase in the frequency of ectopic Pitx2 expression (bilateral expression: n = 22/53; 42%) when compared with eGFP vector (bilateral expression: n = 1/11; 9%; P < 0.05, χ2 test) (Fig. 7C–F; Supplemental Table 1). Among embryos in which there was detectable GFP fluorescence at the end of the culture period, six of eight showed bilateral Pitx2 expression compared with one of seven for eGFP controls.
We next coelectroporated Smad4-eGFP with CMV-caAlk6-IRES-tdTomato (caAlk6-tdRed), a vector expressing CA-ALK6 as well as tdTomato red fluorescent protein (tdRED). Equivalent amounts of Smad4-eGFP and caAlk6-tdRed reduced the proportion of embryos expressing robust levels of Pitx2 in the right LPM, and increased the number showing weak expression, localized loss or a fragmented pattern (P < 0.001; χ2 test) (Supplemental Table 1). It is important to note that increased SMAD4 allows induction and propagation of the LR pathway in the right LPM in a non-cell-autonomous manner (via NODAL expression and diffusion), while CA–ALK6 can only inhibit this via a local (cell-autonomous) mechanism. In the five embryos in which both green and red fluorescent cells were codetected in the same region after culture, two showed very weak expression of Pitx2, while three showed none at all (Fig. 7G,G′). These findings strongly suggest that superactivation of the BMP pathway ameliorates the effects of overexpressed SMAD4.
A superactivated BMP pathway should also antagonize the normal NODAL pathway in the left LPM. Consistent with findings in frog and chick embryos (Esteban et al. 1999; Yokouchi et al. 1999; Zhu et al. 1999), electroporation of caAlk6-tdRed into the left LPM led to local inhibition of Pitx2 expression (weak, localized, or fragmented expression) in most embryos (n = 27/32; 84%), while eGFP vector had no effect (n = 12; P < 0.001) (Fig. 7H-I′; Supplemental Table 1).
Discussion
The first evidence for a repressive role for BMPs in the LR pathway came from overexpression studies in Xenopus (Ramsdell and Yost 1999). Inhibition of the BMP–ALK2–SMAD1/5/8 pathway using a truncated receptor led to bilateral Nodal expression and LR defects. Conversely, stimulation of the BMP pathway on the left repressed LR signaling. Mouse genetic studies have also suggested a repressive role for BMPs in LR signaling: embryos lacking SMAD5, SPC/PACE4, or ALK2 showed bilateral activation of Nodal and Pitx2 in the LPM (Chang et al. 2000; Constam and Robertson 2000). However, these models lacked the discriminatory power to define when and where BMPs act in the pathway, and defects in the midline barrier were evident in mutant embryos.
Numerous other studies have suggested that BMPs have complex and multiple roles in the laterality pathway. In the chick, BMP4 acts in an early, right-sided signaling cascade around Hensen’s node, which represses Shh expression on the right, thus creating a left-sided bias in the LR cascade (Monsoro-Burq and Le Douarin 2001). In zebrafish and Xenopus, BMP4 also acts on the left side of the forming heart to control looping (Chen et al. 1997; Breckenridge et al. 2001). The discovery of chick Caronte (Car), encoding a secreted NODAL and BMP antagonist expressed in the left LPM downstream from Shh, promoted a series of overexpression experiments that led to the conclusion that CARONTE acts as a left-sided BMP inhibitor, and that BMP inhibition was necessary for LR pathway activation (Esteban et al. 1999; Yokouchi et al. 1999; Zhu et al. 1999). However, overexpression of CARONTE and other BMP pathway agonists and antagonists in these studies likely influenced the activity of BMP4 on the right side of Hensen’s node (Piedra and Ros 2002; Schlange et al. 2002). Using similar approaches, others found that BMPs, in fact, define tissue competence for the LR pathway by controlling genes for NODAL receptor components CFC/CRYPTIC and ACTRIIa (Fujiwara et al. 2002; Piedra and Ros 2002; Schlange et al. 2002).
Our genetic analysis of LR pathway dynamics in Smad1-null and conditionally deleted embryos provides proof that BMP/SMAD1 signaling is indeed essential in the LPM for proper LR axis formation. Smad1 deletion led to precocious and bilateral activation of LR genes in the LPM, and to abnormal cardiac looping and embryo turning. A key feature of our analysis was the specific deletion of Smad1 in the LPM using conditional gene targeting. Furthermore, our studies of Smad1 mutants on the Nodalneo/neo background clearly implicate the action of NODAL itself from both left and right sides of the node in the bilateral induction of LR genes in the LPM in Smad1 mutants. We discovered that the repressive influence of BMP/SMAD1 on NODAL signaling is mediated in part by limiting availability of SMAD4. Cell culture assays showed that SMAD4 overexpression rescued repression of the NODAL pathway by the active BMP receptor, and, more definitively, enforced expression of SMAD4 in the right LPM in embryos induced the LR pathway ectopically. However, other BMP-dependent processes may more directly inhibit the NODAL pathway and contribute to the repressive threshold. This was suggested by EMSA experiments, which showed that activation of BMP signaling inhibited formation of the ARE transcriptional complex even in the presence of overexpressed SMAD4.
We propose that the repressive functions of the BMP/SMAD1 pathway set a critical threshold for NODAL-dependent Nodal activation in the LPM. During LR axis specification, cells of the left and right LPM must discriminate between subtly different levels of morphogens generated by nodal flow and convert this to a bistable output (Nakamura et al. 2006). Developing fields use different strategies, often involving positive and negative feedback loops, for converting graded or noisy inputs into bistable outputs; i.e., the generation of robust all-or-nothing responses (Ferrell 2002; Tian et al. 2007). These mechanisms depend heavily on appropriate signaling thresholds: If the threshold is too high the system will be unresponsive, if too low it will be noisy and bistability may be lost or even reversed (Ferrell 2002)—features characteristic of the LR pathway when disturbed.
Our threshold model requires that BMP/SMAD1/SMAD4 signaling represses the NODAL pathway bilaterally, not just in the right LPM. Several experimental observations support this view: (1) BMP pathway activity, as determined by the distribution of phosphorylated SMADs1/5, was bilaterally symmetrical at the time of NODAL pathway activation. Subsequent repression of the BMP pathway extracellularly appears to be an additional mechanism for reducing the signaling threshold and enhancing the robustness of the LR response, as originally proposed for CARONTE (Esteban et al. 1999; Yokouchi et al. 1999; Zhu et al. 1999). While no mouse gene for CARONTE has been identified, it has recently been shown that BMP-antagonists NOGGIN and CHORDIN become enriched in the left LPM, and embryo manipulations and genetic data show an interaction between the NOGGIN, BMP4, and LR pathways (Mine et al. 2008). An important point for this study, however, is that NOGGIN asymmetry is established downstream from the LR pathway and so cannot modify the threshold for initial NODAL-dependent activation of NODAL signaling in the LPM. (2) In the absence of Smad1, expression of Nodal, Lefty2, and Pitx2 occurred precociously in both the left and right LPM. The fact that right-sided expression developed concomitantly with expression on the left in the presence of an intact node and midline argues against the possibility that LR morphogens from the left eventually diffuse across to the right to activate the pathway, as is the situation in Lefty1 knockout embryos in which the midline barrier is defective (Meno et al. 1998). (3) Precocious expression in the LPM on both left and right sides was partially dependent on NODAL expression around the node, as demonstrated by the delay in bilateral LR activation in Smad1−/−;Nodalneo/neo embryos. The implied presence of NODAL protein in right as well as the left LPM is consistent with previous observations that even with normal nodal flow in wild-type embryos, the LR pathway is activated on the right, albeit weakly, before being repressed by a lateral inhibition mechanism (Hirokawa et al. 2006; Nakamura et al. 2006). (4) In null and Mesp1-Cre conditionally deleted embryos, bilateral LPM expression of Nodal, Lefty2, and Pitx2 was strongest and more extensive in the left LPM compared with the right, and compared with the left side of normal embryos, consistent with there being stronger than normal NODAL signaling on the left in Smad1 mutants.
In Smad1 heterozygotes, and embryos conditionally deleted for Smad1 in cardiac mesoderm, the BMP-dependent repression pathway became metastable, allowing stochastic activation of P2Ztg in myocardium of the heart derived originally from the right side of the embryo. We observed that such activation can also occur, albeit rarely, in wild-type embryos. Activation was abortive, occurring within only small patches. These data further support our conclusion that SMAD1 levels are precisely controlled for proper functioning of the LR pathway.
Our data lend strong support to the notion that BMP signaling via SMAD1 provides a competitive threshold for NODAL-dependent Nodal activation in the LPM. As noted above, BMPs may have multiple functions in laterality including establishment of node asymmetry, defining tissue competence for NODAL signaling, and guiding heart looping. In an evolutionary context, these multiple functions entwine LR signaling with other aspects of embryonic development, in which BMPs play multiple and overlapping roles. Thresholds are an aggregate of the efficiencies and dynamics of different biological and biophysical processes, and different organisms may have evolved innovative ways of tuning thresholds, as suggested for the LR pathway by the left-sided expression of Caronte in chick, and Noggin and Chordin in the mouse (Shen 2007; Mine et al. 2008), and the different handedness of expression of fgf8 and other LR genes in different models (Meyers and Martin 1999). The conserved regulation of dorsoventral axis formation in vertebrates by a BMP gradient is also subject to species-specific tuning (De Robertis 2008). The LR system is an exquisite example of how small differences in signaling experienced by two identical cell populations are converted into a robust bistable output. Transcription factor regulatory networks often use double-negative gates (repression of a repressor) to achieve spatiotemporal specificity and reliability of developmental processes (Ferrell 2002; Oliveri et al. 2008). This mechanism ensures that such processes are actively repressed in nonappropriate regions. A signal-dependent transcriptional process that works against a repressive threshold achieves the same goal. Mutual antagonism between the BMP and NODAL/TGFβ signaling pathways may be widespread in development and homeostasis, and deeper dissection of the intersection of these pathways will further illuminate how biological specificity is generated and how defects in thresholds might contribute to disease.
Materials and methods
Mouse lines
Smad1 and conditional Smad1 (Tremblay et al. 2001), Nodal-neo (Saijoh et al. 2003), Mesp1Cre (Saga et al. 1999), Nkx2-5IRESCre (Stanley et al. 2002), R26R (Soriano 1999), and P2Ztg (Shiratori et al. 2001) lines were as described. Genetic backgrounds were as follows: Smad1 lines: Swiss; Mesp1Cre, Nkx2-5IRESCre, and R26R lines: C57BL/6; P2Ztg: 129/SvJ.
In situ hybridization and immunohistochemistry
Whole-mount in situ hybridization was performed as described (Stennard et al. 2005) with minor modifications. Nodal, Lefty1/2, Dante, and Plunc antisense riboprobes were cloned by RT–PCR into pGEM-T Easy vector (Promega) from 8.5 dpc mouse embryo cDNAs. Details of other probes provided upon request. For immunohistochemistry, embryos were bleached in 3% H2O2/PBS–Triton X-100 (PBSTR), subjected to antigen retrieval (Vector labs), permeabilized with acetone and incubated overnight with α-phospho-SMAD1/5/8 antibody (Cell Signaling), followed by ABC signal amplification (Vector Laboratories), with detection using DAB tablets (Sigma).
SEM
Following incubation in Karnovsky’s fixative (2% paraformaldehyde, 2.5% gluteraldehyde, 0.1 M sodium phosphate at pH 7.2), samples were washed in distilled water and dehydrated in an alcohol series. Samples were critical point-dried, mounted, sputter-coated with gold, and viewed using a Cambridge S-360 microscope.
Cell transfections and Western blotting
COS-7 and 293T cells were transiently transfected using Lipofectamine Plus (Invitrogen). Cells were harvested after 48 h for luciferase determination (LARII-Promega). Normalization was to β-galactosidase activity from cotransfected LacZ plasmid (Galactostar-Tropix) (three independent experiments in triplicate). Western blots, performed using standard protocols, used PS-1 and PS-2 (Cell Signaling), monoclonal M2 α-Flag (Sigma) and HRP-conjugated α-mouse and α-rabbit (Amersham) antibodies. Detection was using ECL reagent (Amersham).
EMSA
Five micrograms to 10 μg of nuclear extract in binding buffer (25 mM HEPES at pH 7.4, 75 mM NaCl, 1 mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, 1 mM DTT, 10 μg/mL BSA, 0.2 mg/mL poly dI/dC) was incubated for 10 min at RT. Annealed oligonucleotides (5 pmol) were end-radiolabeled using [γ 32P] ATP and T4 polynucleotide kinase (New England Biolabs); 50,000 cpm were added to each sample and incubated for 20 min at room temperature. Ten microliters of mixture were run on 5% PAGE in 0.5× TBE, and the gel was dried and subjected to autoradiography. In supershift assays, 0.5–2 μg of antibody recognizing MYC, HA, Flag epitopes (Sigma) or Smad1 (Cell Signaling) were added to the reaction for 10 min before incubation with the radiolabeled oligonucleotide.
Embryo electroporation and culture
Embryos of the ARC/s strain were sorted according to somite number and initially kept in 100% rat serum in a 5% CO2 incubator at 37°C. Some embryos were randomly selected at this stage to be intact controls without microinjection or electroporation. In experimental embryos, 5–10 nL of vector (backbone vectors IRES2-EGFP [Clontech] or pcDNA3 [Invitrogen]) (4.24 μg/μL) was microinjected into the anterior right or left LPM. Embryos were positioned between electrodes of a BTX Electro Square Porator T820 electroporator that delivered 5 × 50 msec 12 V square-wave pulses, with a 1-sec interpulse gap. Efficiency of electroporation was 70%–86%. Whole embryos were cultured for 7–9 h in 75% rat serum, 25% DMEM at 37°C with rotation and continuous replenishment of the gas phase of 5% CO2, 20% O2, and 75% N2, then harvested for fluorescence imaging using a SPOT2 Slider digital camera and SPOT 32 software. Embryos were fixed in 4% PFA and processed for whole-mount in situ hybridization for Pitx2 expression.
Acknowledgments
We thank Hiroshi Hamada, Andrew McMahon, and Peter ten Dijke for reagents, and NHMRC, Australia, for funding (354400). M.J.S. and M.B.F. received support from NHLBI (F32-HL10389) and the National Heart Foundation, Australia (PB06S2915), respectively. P.P.L.T. is a Senior Principal Research Fellow of the NHMRC.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1682108.
References
- Breckenridge R.A., Mohun T.J., Amaya E. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev. Biol. 2001;232:191–203. doi: 10.1006/dbio.2001.0164. [DOI] [PubMed] [Google Scholar]
- Brennan J., Norris D.P., Robertson E.J. Nodal activity in the node governs left–right asymmetry. Genes & Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Zwijsen A., Vogel H., Huylebroeck D., Matzuk M.M. Smad5 is essential for left–right asymmetry in mice. Dev. Biol. 2000;219:71–78. doi: 10.1006/dbio.1999.9594. [DOI] [PubMed] [Google Scholar]
- Chang W., Parra M., Ji C., Liu Y., Eickelberg O., McCarthy T.L., Centrella M. Transcriptional and post-transcriptional regulation of transforming growth factor β type II receptor expression in osteoblasts. Gene. 2002;299:65–77. doi: 10.1016/s0378-1119(02)01013-2. [DOI] [PubMed] [Google Scholar]
- Chen J.-N., van Eeden F.J.M., Warren K.S., Chin A., Nusslein-Volhard C., Haffter P., Fishman M.C. Left–right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Chocron S., Verhoeven M.C., Rentzsch F., Hammerschmidt M., Bakkers J. Zebrafish Bmp4 regulates left–right asymmetry at two distinct developmental time points. Dev. Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Collignon J. miRNA in embryonic development: The taming of nodal signaling. Dev. Cell. 2007;13:458–460. doi: 10.1016/j.devcel.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Constam D.B., Robertson E. SPC/PACE4 regulates a TGFb signaling network during axis formation. Genes & Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- De Robertis E.M. Evo-devo: Variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban C.R., Capdevila J., Economides A.N., Pascual J., Oritz A., Izpisua Belmonte J.C. The novel Cer-like protein Caronte mediates the establishment of embryonic left–right asymmetry. Nature. 1999;401:243–251. doi: 10.1038/45738. [DOI] [PubMed] [Google Scholar]
- Ferrell J.E.J. Self-perpetuating states in signal transduction: Positive feedback, double negative feedback and biostability. Curr. Opin. Cell Biol. 2002;6:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Dehart D.B., Sulik K.K., Hogan B.L. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left–right asymmetry in the mouse. Development. 2002;129:4685–4696. doi: 10.1242/dev.129.20.4685. [DOI] [PubMed] [Google Scholar]
- Hamada H., Meno C., Watanabe D., Saijoh Y. Establishment of vertebrate left–right asymmetry. Nat. Rev. Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Tanaka Y., Okada Y., Takeda S. Nodal flow and the generation of left–right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Hou J., Yashiro K., Okazaki Y., Saijoh Y., Hayashizaki Y., Hamada H. Identification of a novel left–right asymmetrically expressed gene in the mouse belonging to the BPI/PLUNC superfamily. Dev. Dyn. 2004;229:373–379. doi: 10.1002/dvdy.10450. [DOI] [PubMed] [Google Scholar]
- Inman G.J., Hill C.S. Stoichiometry of active smad-transcription factor complexes on DNA. J. Biol. Chem. 2002;277:51008–51016. doi: 10.1074/jbc.M208532200. [DOI] [PubMed] [Google Scholar]
- Kishigami S., Yoshikawa S., Castranio T., Okazaki K., Furuta Y., Mishina Y. BMP signaling through ACVRI is required for left–right patterning in the early mouse embryo. Dev. Biol. 2004;276:185–193. doi: 10.1016/j.ydbio.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Levin M., Palmer A.R. Left–right patterning from the inside out: Widespread evidence for intracellular control. Bioessays. 2007;29:271–287. doi: 10.1002/bies.20545. [DOI] [PubMed] [Google Scholar]
- Lowe L.A., Yamada S., Kuehn M.R. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- Meno C., Shimono A., Saijoh Y., Yashiro K., Mochida K., Ohishi S., Noji S., Kondoh H., Hamada H. lefty-1 is required for left–right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Meno C., Takeuchi J., Sakuma R., Koshiba-Takeuchi K., Ohishi S., Saijoh Y., Miyazaki J., ten Dijke P., Ogura T., Hamada H. Diffusion of nodal signaling activity in the absence of the feedback inhibitor Lefty2. Dev. Cell. 2001;1:127–138. doi: 10.1016/s1534-5807(01)00006-5. [DOI] [PubMed] [Google Scholar]
- Meyers E.N., Martin G.R. Differences in left–right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Mine N., Anderson R.M., Klingensmith J. BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left–right axis establishment. Development. 2008;135:2425–2434. doi: 10.1242/dev.018986. [DOI] [PubMed] [Google Scholar]
- Mommersteeg M.T.M., Brown N.A., Prall O.W.J., de Gier-de Vries C., Harvey R.P., Moorman A.F., Christoffels V.M. Pitx2 and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ. Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A., Le Douarin N.M. BMP4 plays a key role in left–right patterning in chick embryos by maintaining Sonic Hedgehog asymmetry. Mol. Cell. 2001;7:789–799. doi: 10.1016/s1097-2765(01)00223-4. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Mine N., Nakaguchi E., Mochizuki A., Yamamoto M., Yashiro K., Meno C., Hamada H. Generation of robust left–right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev. Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Nonaka S., Shiratori H., Saijoh Y., Hamada H. Determination of left–right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Brennan J., Bikoff E.K., Robertson E.J. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Oliveri P., Tu Q., Davidson E.H. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J.J., Penny G., Rossant J. A mouse cerberus/Dan-related gene family. Dev. Biol. 1999;209:98–110. doi: 10.1006/dbio.1999.9240. [DOI] [PubMed] [Google Scholar]
- Piedra M.E., Ros M.A. BMP signaling positively regulates Nodal expression during left right specification in the chick embryo. Development. 2002;129:3431–3440. doi: 10.1242/dev.129.14.3431. [DOI] [PubMed] [Google Scholar]
- Prall O.W.J., Menon M.K., Solloway M.J., Watanabe K., Zaffran S., Bajolle F., Biben C., McBride J.J., Robertson B.R., Chaulet H., et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls second heart field progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell A.F., Yost H.J. Cardiac looping and the vertebrate left–right axis: Antagonism of left-sided Vg1 activity by a right-sided ALK2-dependent BMP pathway. Development. 1999;126:5195–5205. doi: 10.1242/dev.126.23.5195. [DOI] [PubMed] [Google Scholar]
- Rankin C.T., Bunton T., Lawler A.M., Lee S.J. Regulation of left–right patterning in mice by growth/differentiation factor-1. Nat. Genet. 2000;24:262–265. doi: 10.1038/73472. [DOI] [PubMed] [Google Scholar]
- Rodriguez Esteban C., Capdevila J., Economides A.N., Pascual J., Ortiz A., Izpisua Belmonte J.C. The novel Cer-like protein Caronte mediates the establishment of embryonic left–right asymmetry. Nature. 1999;401:243–251. doi: 10.1038/45738. [DOI] [PubMed] [Google Scholar]
- Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J., Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Saijoh Y., Adachi H., Sakuma R., Yeo C.Y., Yashiro K., Watanabe M., Hashiguchi H., Mochida K., Ohishi S., Kawabata M., et al. Left–right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol. Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- Saijoh Y., Oki S., Ohishi S., Hamada H. Left–right patterning of the mouse lateral plate requires nodal produced in the node. Dev. Biol. 2003;256:160–172. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- Schlange T., Arnold H.H., Brand T. BMP2 is a positive regulator of Nodal signaling during left–right axis formation in the chicken embryo. Development. 2002;129:3421–3429. doi: 10.1242/dev.129.14.3421. [DOI] [PubMed] [Google Scholar]
- Shen M.M. Nodal signaling: Developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shiratori H., Hamada H. The left–right axis in the mouse: From origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Shiratori H., Sakuma R., Watanabe M., Hashiguchi H., Mochida K., Sakai Y., Nishino J., Saijoh Y., Whitman M., Hamada H. Two-step regulation of left–right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Shiratori H., Yashiro K., Shen M.M., Hamada H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development. 2006;133:3015–3025. doi: 10.1242/dev.02470. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stanley E.G., Biben C., Elefanty A., Barnett L., Koentgen F., Robb L., Harvey R.P. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- Stennard F.A., Costa M.W., Lai D., Biben C., Furtado M.B., Solloway M.J., McCulley D.J., Leimena C., Preis J.I., Dunwoodie S.L., et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Tanaka C., Sakuma R., Nakamura T., Hamada H., Saijoh Y. Long-range action of Nodal requires interaction with GDF-1. Genes & Dev. 2007;21:3272–3282. doi: 10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Harding A., Inder K., Plowman S., Parton R.G., Hancock J.F. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- Tremblay K.D., Dunn N.R., Robertson E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- Tsukui T., Capdevila J., Tamura K., Ruiz-Lozano P., Rodriguez-Esteban C., Yonei-Tamura S., Magallon J., Chandraratna R.A., Chien K., Blumberg B., et al. Multiple left–right asymmetry defects in Shh−/− mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc. Natl. Acad. Sci. 1999;96:11376–11381. doi: 10.1073/pnas.96.20.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall N.A., Craig E.J., Labosky P.A., Kessler D.S. Mesendoderm induction and reversal of left–right pattern by mouse Gdf1, a Vg1-related gene. Dev. Biol. 2000;227:495–509. doi: 10.1006/dbio.2000.9926. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y., Vogan K.J., Pearse R.V., Tabin C.J. Antagonistic signaling by Caronte, a novel Cerberus-related gene, establishes left–right asymmetric gene expression. Cell. 1999;98:573–583. doi: 10.1016/s0092-8674(00)80045-8. [DOI] [PubMed] [Google Scholar]
- Zhu A.J., Scott M.P. Incredible journey: How do developmental signals travel through tissue? Genes & Dev. 2004;18:2985–2997. doi: 10.1101/gad.1233104. [DOI] [PubMed] [Google Scholar]
- Zhu L., Marvin M.J., Gardiner A., Lassar A.B., Mercola M., Stern C.D., Levin M. Cerberus regulates left–right asymmetry of the embryonic head and heart. Curr. Biol. 1999;9:931–938. doi: 10.1016/s0960-9822(99)80419-9. [DOI] [PubMed] [Google Scholar]