Abstract
The mitotic arrest-deficient protein Mad1 forms a complex with Mad2, which is required for imposing mitotic arrest on cells in which the spindle assembly is perturbed. By mass spectrometry of affinity-purified Mad2-associated factors, we identified the translocated promoter region (Tpr), a component of the nuclear pore complex (NPC), as a novel Mad2-interacting protein. Tpr directly binds to Mad1 and Mad2. Depletion of Tpr in HeLa cells disrupts the NPC localization of Mad1 and Mad2 during interphase and decreases the levels of Mad1-bound Mad2. Furthermore, depletion of Tpr decreases the levels of Mad1 at kinetochores during prometaphase, correlating with the inability of Mad1 to activate Mad2, which is required for inhibiting APCCdc20. These findings reveal an important role for Tpr in which Mad1–Mad2 proteins are regulated during the cell cycle and mitotic spindle checkpoint signaling.
Keywords: Mad1, Mad2, mitotic spindle checkpoint, Tpr
The mitotic spindle checkpoint is a regulatory mechanism required for proper segregation of chromosomes during cell division (Musacchio and Salmon 2007). It acts by delaying sister chromatid separation until all the chromosomes have aligned at the metaphase plate. Components of the mitotic spindle checkpoint were first isolated in Saccharomyces cerevisiae and include Mad1, Mad2, Mad3/BubR1, Bub1, Bub3, and Mps1, all of which are conserved in higher eukaryotes. Mad1 is a positive regulator of the mitotic spindle checkpoint and is thought to recruit Mad2 to unattached kinetochores and facilitate Mad2’s checkpoint function (Li and Benezra 1996; Dobles et al. 2000). Mad2 arrests cells in prometaphase by inhibiting the activity of the anaphase-promoting complex (APC) through forming an inactive complex with Cdc20 and APC (Li et al. 1997; Fang et al. 1998). Here, we identified the translocated promoter region (Tpr), a component of the nuclear pore complex (NPC), as a novel Mad1- and Mad2-interacting protein and provide evidence that Tpr is important for the Mad1–Mad2-mediated mitotic spindle checkpoint in mammalian cells.
Results and Discussion
A C-terminal deletion mutant of Mad2, which does not bind to either Mad1 or Cdc20, fails to induce mitotic arrest following spindle disruption (Fang et al. 1998; Chen et al. 1999). To identify factor(s) that interacts with Mad2, we engineered HEK293 cell lines to stably express TAP-tagged wild-type (TAP-Mad2wt) or mutant Mad2, in which 20 residues in the C-terminal and 10 residues in the N-terminal regions (TAP-Mad2ΔC20/ΔN10) were deleted (Supplemental Material). We chose TAP-Mad2ΔC20/ΔN10 because of the low expression levels of TAP-Mad2ΔC20 (data not shown). Endogenous Mad1 and Cdc20 proteins were detected by mass spectrometric analysis only in TAP-Mad2wt complexes, indicating that the TAP-Mad2 proteins are functional. Strikingly, only one protein band with a relative molecular mass ∼270 kDa was detected visibly in the TAP-Mad2wt, but not the TAP-Mad2ΔC20/ΔN10 column (Supplemental Fig. 1A). The ∼270-kDa band was subjected to mass spectrometry; 56 peptide sequences were obtained, all of which were derived from the Tpr protein, a component of the NPC. Endogenous Tpr was readily immunoprecipitated from HEK293 cells transfected only with Myc-Mad2wt, but not with Myc-Mad2ΔC20 (Supplemental Fig. 1B). None of the peptides derived from other NPC components or nuclear transport machinery were detected by mass spectrometric analysis (data not shown). Furthermore, Myc-Mad2wt did not bind to other NPC components, as determined by immunoblot analysis using the mAB414 antibody that reacts with several of the NPC proteins, including Nup358, Nup214, Nup153, and Nup63 (data not shown).
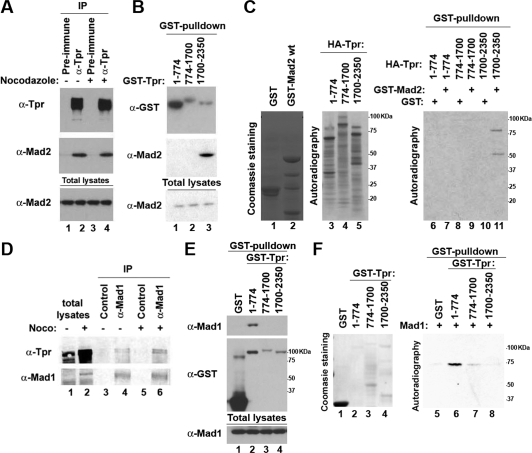
Endogenous Mad2 was present in the anti-Tpr immunoprecipitates from cell extracts of HeLa cells grown asynchronously or arrested in mitosis by treatment with the spindle damaging agent nocodazole (Fig. 1A), indicating that Tpr binds Mad2 in vivo. Next, to identify the region of Tpr that interacts with Mad2, HEK293 cells were transfected with a series of GST-tagged Tpr deletion mutants. As determined by GST pull-down analysis, endogenous Mad2 was readily precipitated from cells transfected with the C-terminal region of Tpr extending from residues 1700 to 2350, Tpr(1700–2350), while neither the large internal coiled-coil domain, Tpr(774–1700), nor the N-terminal coiled-coil domain, Tpr(1–774), did so (Fig. 1B). Next, to test whether Tpr binds to Mad2 directly, we expressed recombinant GST-Mad2wt or GST-Mad2ΔC20 in Escherichia coli and translated a series of Tpr deletion mutants in vitro in the presence of 35S-cysteine and 35S-methionine. In vitro translated Tpr proteins were individually mixed with recombinant GST or GST-Mad2 proteins and subjected to GST pull-down analysis. Only the in vitro translated Tpr(1700–2350) exhibited binding activity to recombinant GST-Mad2wt (Fig. 1C). Moreover, recombinant His-tagged Tpr(1700–2350) expressed in E. coli bound to recombinant GST-Mad2wt, but not to GST-Mad2ΔC20 (Supplemental Fig. 1C), indicating that Tpr binds to Mad2 directly through the C-terminal region of Tpr.
Figure 1.
Tpr binds to Mad1 and Mad2 directly. (A,D) Total cell lysates were prepared from HeLa cells grown asynchronously or arrested in mitosis by nocodazole followed by mitotic shake-off to allow collection, and then subjected to immunoprecipitation analysis. (B,E) The indicated proteins were expressed in HEK293 cells and subjected to GST pull-down analysis. (C, lanes 1,2) Coomassie staining of purified recombinant GST-tagged proteins. (Lane 3–5) Autoradiography of in vitro translated Tpr proteins. (Lanes 6–11) GST pull-down analysis was performed, and the bound proteins were visualized by autoradiography. (F, lanes 1–4) Coomassie staining of purified recombinant GST-tagged proteins. (Lanes 5–8) GST pull-down analysis was performed, and the bound proteins were visualized by autoradiography.
Since Mad1 forms a complex with Mad2, we tested whether Tpr also binds to Mad1. Endogenous Tpr was present in anti-Mad1 immunoprecipitates from cell extracts of HeLa cells (Fig. 1D), indicating that Tpr binds Mad1 in vivo. As determined by GST- pull-down analysis, endogenous Mad1 was only precipitated from cells transfected with GST-Tpr(1–774) (Fig. 1E). Furthermore, recombinant GST-Tpr(1–774) expressed in E. coli bound to in vitro translated Mad1 (Fig. 1F), indicating that Tpr binds to Mad1 directly, independent of the Mad2-binding region of Tpr.
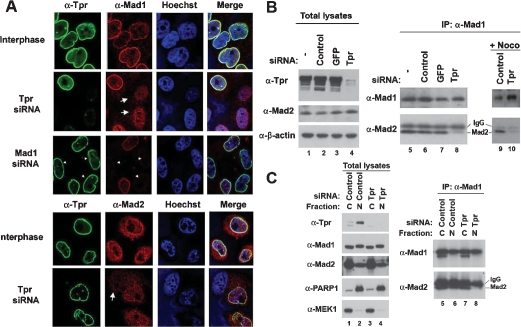
Tpr is localized in the nuclear basket of the NPC during interphase (Cordes et al. 1997). To directly explore the functional significance of the interactions between Tpr and Mad2, we conducted double-labeling and immunofluorescence microscopy studies in HeLa cells. As Mad1 and Mad2 proteins also localize to the NPC during interphase in yeast and mammalian cells (Campbell et al. 2001; Iouk et al. 2002; Scott et al. 2005), Mad1 colocalized remarkably with Tpr to the nuclear envelope (NE) during interphase and throughout the cell cycle (Fig. 2A; Supplemental Fig. 2). Likewise, Mad2 colocalized with Tpr to the NE, although it was also detected in the nucleus and the cytoplasm (Fig. 2A). Importantly, depletion of Tpr by siRNA transfection completely abolished the NPC localization of Mad1 and Mad2 proteins, whereas depletion of Mad1 did not affect the NPC localization of Tpr (Fig. 2A), suggesting that Tpr anchors Mad1 and Mad2 to the NPC during interphase. As Tpr binds to Mad2 directly, depletion of Mad1 did not change either the NPC localization of Mad2 or the ability of Tpr to bind Mad2 (Supplemental Fig. 3A,B). Notably, the NPC localization of Mad1 in HeLa cells transfected with siRNAs against other NPC components, including Nup50 and Nup98, did not change (Supplemental Fig. 4A). Moreover, depletion of Tpr did not cause a detectable change in NPC structures (Supplemental Fig. 4B), suggesting a specific role of Tpr to anchor Mad1 and Mad2 to the NPC. Furthermore, as determined by immunoprecipitation analysis using anti-Mad1 antibodies, depletion of Tpr decreased the protein levels of Mad2 bound to Mad1, particularly in the nuclear fraction (Fig. 2B,C), suggesting a potential role of Tpr scaffolding Mad1 and Mad2 at the nuclear side of NPC. Depletion of Tpr did not affect protein levels of Mad2 in the nucleus (Fig. 2C, lane 4), which excludes the possibility that loss of nuclear localization of Mad2 in interphase may lead to reduction in its association with Mad1. Moreover, it has been previously reported that depletion of Tpr from the NE does not lead to loss of other NPC basket proteins, and nuclear import mediated by a basic amino acid signal (NLS) is unaffected (Frosst et al. 2002).
Figure 2.
Tpr determines the NPC localization of Mad1 and Mad2. (A) Immunofluorescence analysis. HeLa cells transfected with siRNA were stained with indicated antibodies. The arrows indicate the interphase cells showing mislocalized Mad1 and Mad2 by depleting Tpr. The arrowheads indicate the Mad1-depleted interphase cells. (B,C) After siRNA transfection to HeLa cells, equal amount of total lysates were subjected to immunoprecipitation analysis using anti-Mad1 antibody, and the bound proteins were visualized with immunoblot analysis. (B) For lanes 9 and 10, HeLa cells transfected with siRNA were arrested in mitosis by nocodazole before immunoprecipitation analysis. (C) HeLa cells were separated to the nucleus (N) and the cytoplasm (C) prior to immunoprecipitation analysis. (Lanes 1–4) PARP1 and MEK1 were used as markers for the nuclear and the cytoplasmic fractions, respectively.
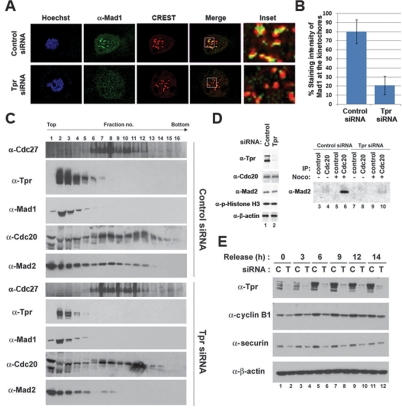
Mad1 localizes to kinetochores and subsequently recruits Mad2 to kinetochores during prometaphase (Li and Benezra 1996; Luo et al. 2002). We observed that Tpr colocalized with kinetochore proteins, including Bub3 and Mps1, during prometaphase (Supplemental Fig. 4C). As Tpr colocalizes with Mad1 throughout the cell cycle, we tested whether Tpr is also essential for the kinetochore localization of Mad1. Indeed, in the absence of Tpr, Mad1 protein at kinetochores during prometaphase markedly decreased by approximately fourfold compared with control (Fig. 3A,B), whereas other centromere and kinetochore markers, including CREST and CENP-E, did not (Supplemental Fig. 4D). As recruiting Mad2 to kinetochores by Mad1 permits Mad2 to form an inactive complex with APCCdc20 (Luo et al. 2002, 2004; De Antoni et al. 2005), we tested whether Tpr affects Mad2-APCCdc20 association by sucrose density gradient sedimentation analysis. In HeLa cells transfected with control siRNA and subsequently arrested in mitosis by nocodazole treatment, Mad2 comigrated with APCCdc20 in fractions 6–13, whereas depletion of Tpr led to the loss of Mad2 from APCCdc20 (Fig. 3B). Similarly, depletion of Mad1 caused the loss of Mad2 from APCCdc20 (Supplemental Fig. 5). Furthermore, depletion of Tpr decreased Cdc20-bound Mad2 levels as compared with controls (Fig. 3C). Checkpoint-defective cells containing catalytically active APCCdc20 are expected to prematurely degrade cyclin B1 and securin (Irniger et al. 1995; King et al. 1995; Sudakin et al. 1995). Indeed, when cells were synchronized at the G1/S boundary and released, Tpr-depleted cells failed to accumulate high expression levels of cyclin B1 and securin seen in control cells (Fig. 3D), whereas depletion of Tpr did not cause a measurable defect in cell cycle progression (data not shown). Taken together, these results suggest that Tpr is important for Mad1 and Mad2 to activate the mitotic spindle checkpoint.
Figure 3.
Tpr is important for activating Mad1 and Mad2. (A) Immunofluorescence analysis. HeLa cells transfected with siRNA were arrested in mitosis by nocodazole for 6 h and stained with indicated antibodies. (B) The staining intensity of Mad1 at kinetochores (n = 300) was quantified from control or Tpr-depleted cells (n = 50, each) selected at random in prometaphase stage. (C,D) HeLa cells transfected with siRNA were arrested in mitosis by nocodazole followed by mitotic shake-off to allow collection. (C) Whole-cell extracts were prepared and resolved on sucrose density gradient by centrifugation. Gradients were separated into 16 fractions and precipitated with TCA. The indicated protein was identified with immunoblot analysis. (D) Immunoprecipitation analysis. (Lanes 3–10) Equal amounts of HeLa cell lysates were subjected to immunoprecipitation with anti-Cdc20 antibody, and bound Mad2 was determined by anti-Mad2 antibody. (E) HeLa cells transfected with siRNA were synchronized at the G1/S boundary by thymidine double block. Cells were harvested at the indicated times after release from the G1/S boundary and subjected to immunoblot analysis.
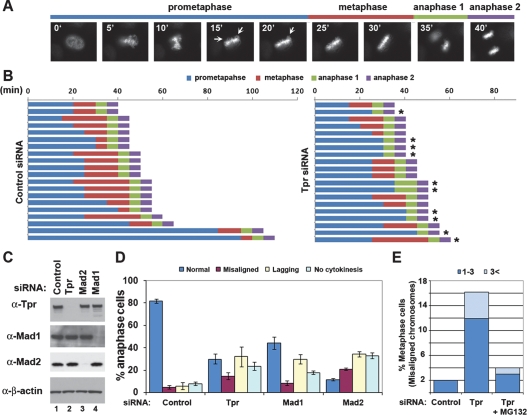
Next, to determined the effects of depleting Tpr on mitotic progression, siRNAs were cotransfected to HeLa cells together with a plasmid expressing GFP-tagged Histone H2B (GFP-H2B), and mitotic progression was monitored using time-lapse fluorescence microscopy. As previously shown in Mad1- or Mad2-depleted cells (Meraldi et al. 2004), 45% of Tpr-depleted cells proceeded into anaphase despite the presence of maloriented and unaligned chromosomes, whereas cells transfected with control siRNA did not proceed into anaphase until the completion of chromosome alignment on the spindle equator (Fig. 4A,B; Supplemental Fig. 6). About 70% of Tpr-depleted cells in mitosis contained errors in chromosome segregation, which were also observed in Mad1- or Mad2-depleted cells (Fig. 4D). Furthermore, inducing a transient mitotic arrest by treating Tpr-depleted cells with MG132, a proteasome inhibitor, markedly reduced the population of cells in metaphase containing misaligned chromosomes (Fig. 4E), suggesting that the mitotic defects in Tpr-depleted cells are not due to a direct role of Tpr in kinetochore function. However, it is important to note that Tpr-depleted cells did not result in significant acceleration of mitosis, whereas Mad2-depleted cells shortened the time of anaphase onset as previously described (Supplemental Fig. 6; Meraldi et al. 2004). Taken together, our findings suggest that Tpr is important for controlling the mitotic spindle checkpoint, but not mitotic timing.
Figure 4.
Tpr-depleted cells proceed to anaphase despite the presence of unaligned chromosomes. (A) Live cell time-lapse imaging. HeLa cells were transfected with a plasmid expressing GFP-H2B. Using nuclear envelope breakdown (NEBD) as T0, the time of mitotic progression was measured from live cell movies. (B) Mitotic progression of 20 cells selected at random. Asterisk (*) indicates the cells containing errors in DNA segregation. (C) Immunoblot analysis. (D) Anaphase cells (n > 100) were scored as normal (blue), having misaligned chromosomes (purple), containing lagging chromosomes (yellow), and having failed to promote cytokinesis (light blue). The average from three independent experiments is shown with standard deviation. (E) HeLa cells were transfected with the indicated siRNA. Forty-eight hours after transfection, cells were incubated with MG132 (2 μM) for 3 h. Mitotic spindle and chromosomes were stained with anti-β-tubulin antibody and Hoechst 33258, respectively. The percentages of total metaphase cells (n > 100) displaying one to three (blue) or more than three (light blue) misaligned chromosomes (chromosomes located outside rectangular area possessing the central 30% of mitotic spindle), defined according to Meraldi and Sorger (2005), are shown.
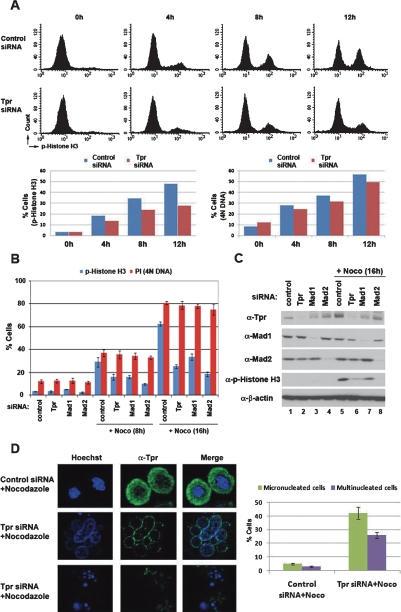
A high frequency of multinucleated and micronucleated DNA formations, apoptosis and a marked decrease in the mitotic index upon spindle damage were observed with Mad1- or Mad2-knockout cells (Dobles et al. 2000; Iwanaga et al. 2007). Treating control and Tpr-depleted HeLa cells with nocodazole for 12 h showed no significant difference in the population of cells possessing 4N DNA content (Fig. 5A, right bar graph). In contrast, when these cells were stained with phospho-histone H3, a mitosis-specific antigen, ∼48% of control cells were positive for phospho-histone H3, whereas only ∼27% of Tpr-depleted cells were (Fig. 5A, top panels and left bar graph; Supplemental Fig. 7A), indicating that approximately a half of Tpr-siRNA-transfected cells possessing 4N DNA content exited from mitotic arrest. Furthermore, when HeLa cells depleted of Tpr, Mad1, or Mad2 were challenged with nocodazole for 16 h, only 20%–30% cells were positive for phospho-histone H3, whereas ∼60% control cells were (Fig. 5B, blue bars). There was no significant difference in the population of cells possessing 4N DNA content (Fig. 5B, red bars), indicating that depletion of Tpr abrogates the mitotic checkpoint to a comparable extent to depletion of Mad1 or Mad2. Moreover, treating Tpr-depleted cells with nocodazole for 24 h markedly increased the population of cells containing a DNA content of less than 2N, indicating a higher incidence of apoptosis, compared with control cells (Supplemental Fig. 7B). In addition, when we determined cell and DNA morphology, only ∼30% of Tpr-depleted cells became rounded and contained condensed mitotic DNA, compared with ∼80% of control cells (Supplemental Fig. 7C). Moreover, ∼25% of Tpr-depleted cells were large and contained multiple nuclei in the presence of nocodazole (Supplemental Fig. 7C) and caused a variety of nuclear abnormalities, including micronuclei and multinucleated cells (Fig. 5D). In contrast, HeLa cells transfected with siRNAs against other NPC components efficiently rounded and became arrested in mitosis upon nocodazole treatment (Supplemental Fig. 8), suggesting specificity of Tpr for mitotic spindle checkpoint signaling. These phenotypes were reproduced by using a different siRNA against Tpr (see the Materials and Methods).
Figure 5.
Tpr is important for mitotic checkpoint signaling. HeLa cells transfected with siRNA against Tpr or control nonsilencing siRNA. (A,B) FACS analysis. HeLa cells transfected with siRNA were treated with nocodazole for the indicated times and stained with anti-phospho-Histone H3 for the population of cells in mitosis and propidium iodide (PI) for the population of cells containing 4N DNA content. (C) Immunoblot analysis. (D) Confocal microscopy images. The cells were treated with nocodazole for 48 h and stained with anti-Tpr antibody and Hoeschst 33342. The graph summarizes the results of three independent experiments (n = 200–250); bars indicate standard deviations.
Recently, Nup358 (Ran-BP2) was found to localize to kinetochores during mitosis (Salina et al. 2003). Depletion of Nup358 by siRNA perturbs the localization of Mad1 and Mad2 from kinetochores. However, it is unlikely that Nup358 recruits Mad1 and Mad2 to the kinetochore from the NPC, as Nup358 resides on the cytoplasmic face of NPC during interphase, whereas Mad1 and Mad2 are located on the nucleoplasmic side of NPC, and no physical interaction has been found between Nup358 and Mad1–Mad2 proteins. In contrast, we demonstrate here that Tpr physically binds to Mad1 and Mad2 and that it is localized to the nuclear side of the NPC together with Mad1 and Mad2. Importantly, depletion of Tpr leads to mislocalization of Mad1 and Mad2 to the nucleus from the NPC (Fig. 2A), indicating that Tpr connects Mad1 and Mad2 to the NPC during interphase.
Although no physical interaction of Mad1p and Mad2p has been demonstrated with Mlp1p and Mlp2p, the yeast homologs of mammalian Tpr, it appears that Mad1p interacts with Mlp1p and Mlp2p, as yeast strains lacking both Mlp1p and Mlp2p fail to concentrate Mad1p at the nuclear periphery (Scott et al. 2005). Thus, together with our study here, the role of Tpr anchoring Mad1 to the NPC may be conserved throughout species. However, the localization of Mad1p to the NPC is largely maintained during mitosis, whereas most of Mad2p is released from the NPC and recruited to the kinetochores (Iouk et al. 2002; Gillett et al. 2004). In addition, Mad1p and Mad2p are found at kinetochores only in the presence of spindle damage and are required only when mitosis is defective in yeast, whereas Mad1 and Mad2 are essential during every division cycle in mammalian cells. Furthermore, Mlp1p and Mlp2p stay in the NE throughout cell cycle as the NE and NPC are not disassembled during mitosis in yeast, and the direct binding of Mad2p to Mlp1p and Mlp2p is unlikely, as association of the Mad1p–Mad2p complex with the NPC requires Mad1p. In contrast to yeast, we found that Tpr is dissociated from the NE during mitosis and colocalizes with Mad1 throughout all phases in the cell cycle (Supplemental Fig. 2). Furthermore, Tpr is capable of binding to Mad1 and Mad2 directly (Fig. 1; Supplemental Fig. 1), suggesting a distinct role of Tpr in regulating the Mad1–Mad2 complex in mammalian cells. In this sense, it is a critical question whether the factor that mediates the binding of Mad1 and Mad2 to the NPC is also important for their localization to kinetochores as well as for their function in the mitotic spindle checkpoint. We show here that Tpr-depleted HeLa cells fail to recruit Mad1 to the kinetochores and to impose the mitotic spindle checkpoint upon spindle damage, suggesting that Tpr is also important for the kinetochore localization of Mad1 and for imposing Mad1–Mad2-dependent mitotic spindle checkpoint signaling in mammalian cells.
Our results also suggest that Tpr may function as a platform for regulating the assembly of the Mad1–Mad2 complex at the NPC. Notably, Tpr directly binds to Mad1 and Mad2 through the N-terminal and C-terminal regions, respectively (Fig. 1). Conversely, depletion of Tpr dissociates Mad1 and Mad2 from the NPC (Fig. 2A) and decreases the levels of Mad1-bound Mad2, particularly in the nucleus (Fig. 2B,C), supporting the idea that Tpr may recruit Mad2 to the NPC, where Mad1 is located during interphase, and this may facilitate the formation of Mad1–Mad2 complex at the NPC. Notably, a mislocalization of Mad1–Mad2 proteins from NPC by depletion of Tpr does not activate the mitotic spindle checkpoint spontaneously, indicating that Tpr may also function in priming the Mad1–Mad2 complex during interphase for their subsequent function in the spindle checkpoint during mitosis.
In summary, we report a novel role of Tpr in which Mad1–Mad2 proteins are regulated at the NPC, and we suggest that Tpr is involved in controlling the Mad1–Mad2-dependent mitotic spindle checkpoint. It is interesting to note that the Tpr-Met oncogene, a carcinogen-induced chromosomal rearrangement with a protein dimerization domain of Tpr fused to the receptor tyrosine kinase domain of Met, has been described (Peschard and Park 2007). It is attractive to suggest that chromosomal rearrangement of Tpr could lead to chromosomal instability in certain tumors.
Materials and methods
Expression and purification of recombinant proteins
Recombinant GST proteins were expressed in E. coli strain BL21(DE3)pLysS and purified with glutathione agarose beads (Amersham). HA-tagged Tpr deletion mutants and Mad1 were translated in vitro in the presence of 35S-cysteine and 35S-methionine label mix (Perkin-Elmer) using reticulocyte lysate system (Promega).
GST pull-down assay
Each GST-tagged protein was loaded onto glutathione agarose beads in NP-40 cell lysis buffer (50 mM Tris-HCl at pH 8.0, 120 mM NaCl, 1% NP-40) containing 1 mM DTT, Complete Mini (Roche), and Phosphatase inhibitor mix I and II (Sigma) for 30 min at 4°C. The beads were washed with NP-40 cell lysis buffer and incubated with in vitro translated proteins overnight at 4°C. The matrix was washed prior to addition of sample buffer.
Immunofluorescence
Cells grown on poly-L-lysine coated cover slips were fixed with 4% paraformaldehyde for 10 min. After permeabilization in 0.1% Triton X-100, cells were blocked in PBS containing 3% bovine serum albumin (BSA) and then incubated with antibodies to Tpr (Bethyl Laboratory), Mad1 (9B10, Santa Cruz Biotechnologies), Mad2 (17D10, Santa Cruz Biotechnologies), CREST (Immunovision), or CENP-E (Santa Cruz Biotechnologies) for 2 h followed by isotype-specific antibodies coupled to Alexa 488 and Texas Red (Molecular Probes) for 1 h in blocking buffer. Fixed slides were stained with Hoechst 33,258. Images were acquired using confocal microscopy and deconvoluted by LSM 5 image software.
Synchronization, mitotic index, and cell cycle analysis
Control nonsilencing and siRNA oligonucleotides against Tpr (SI00750232 and SI00052808), Mad1 (SI00052808), or Mad2 (8102653847) were purchased (Qiagen) and transfected twice in 24-h intervals using Lipofectamine Plus (Invitrogen) at a final concentration of 100 nM. Twenty-four hours after the second siRNA transfections, cells were placed in a media containing nocodazole (200 ng/mL) for the indicated time prior to analysis. To synchronize cells at the G1/S boundary, thymidine was added to 2 mM in the final concentration and incubated for 16 h. Cells were refed with fresh medium for 10 h. Then, thymidine was added and incubated for 14 h. Cells were washed and refed with fresh medium containing nocodazole (200 ng/mL) and collected at the indicated time points. The mitotic index was measured with the Mitotic Index HitKit reagents (Cellomics). For cell cycle analysis, cells were fixed in 70% ethanol and stained with propidium iodide (5 μg/mL). For flow cytometry analysis of Histone H3 phosphorylation, cells were fixed and prepared by following the manufacturer’s instructions (Upstate Biotechnologies; H2A.X Phosphorylation Assay Kit) with an exception of using FICT-conjugated anti-phospho-Histone H3 antibody (Cell Signal). The cell cycle profile and the levels of phospho-Histone H3 were analyzed with a FACSCaliber with Cellquest software (Becton Dickinson).
Sucrose density gradient sedimentation analysis
HeLa cells were harvested in SB buffer (25 mM HEPES at pH 7.5, 1.5 mM MgCl2, 5 mM KCl) containing 1 mM DTT and the protease inhibitor cocktail Complete Mini (Roche). Then cells were subjected to three cycles of freeze–thawing, passage through a needle (26G3/8), and centrifugation at 5000 rpm for 5 min and 14,000 rpm for 1 h. Cell lysates were applied to a 15%–45% (w/w) sucrose density gradient in buffer A (25 mM Tris-HCl at pH 7.4 at 4°C, 25 mM NaCl, 5 mM MgCl2) and centrifuged at 4°C in a SW40 Ti Beckman rotor for 20 h at 30,000 rpm. Gradient analysis was performed as described (Lee and McCormick 2006).
Live cell time-lapse imaging
Cells were imaged every 5 min for 20 h using a Nikon TE2000E Inverted Microscope equipped with Perfect Focus with DIC, Phase, and Epi-fluorescence Opticsa, Photometrics Coolsnap HQ2 Camera, Applied Scientific Instrumentation MS-2000 Motorized Stage, Sutter Lambda LS Arc Lamp and Excitation Filter Wheel, Sutter Lambda 10-3 Emission Filter Wheel and In Vivo Scientific Incubator. All images were analyzed with NIS-Elements Advanced Research software.
Immunoprecipitation
Protein was prepared in NP-40 cell lysis buffer. For Figure 1, B and E, HEK293 cells were transfected with an equal amount of pDEST27 (Invitrogen) encoding indicated GST-tagged proteins. Forty-eight hours after transfections, cells were lysed in NP-40 cell lysis buffer, and extracts containing 500 μg of proteins were subjected to GST pull-down analysis. For Figure 1, A and D, and Figure 2B, HeLa cells were asynchronously grown or arrested in mitosis by nocodazole treatment for 24 h. For Figure 2C, HeLa cells were fractionated with N-Xtract (Sigma). For Figure 3C, HeLa cells were harvested by mitotic shake-off after nocodazole treatment, and Cdc20 proteins were immunoprecipitated with anti-Cdc20 antibody (H-175; Santa Cruz Biotechnologies). The following monoclonal and polyclonal primary antibodies were used: Mad2 (FL-205, SantaCruz Biotechnologies; PRB-452C, Covance), cyclin B1 (M-20, Santa Cruz Biotechnologies), securin (DSC-280) (Abcam), and β-actin (Sigma). The proteins were visualized using ECL reagents (Amersham).
Acknowledgments
We thank Drs. David Toczyski (University of California at San Francisco), David Morgan (University of California at San Francisco), Hideyuki Saya (Keio University), Mark Hannink (University of Missouri), and Hongtao Yu (University of Texas Southwestern), and Mrs. Mayumi Kitagawa (Daiichi Sankyo Co., Ltd) for providing many insightful comments. We thank Dr. Larry Gerace (Scripps) for Tpr cDNA, Dr. Kurk Thorn (University of California at San Francisco, Nickon Imaging Center) for assisting with time-lapse microscopy, and the University of California at San Francisco Cancer Center Cytometry Core facility for assisting with confocal microscopy analysis. The University of California at San Francisco Mass Spectrometry Facility (A.L. Burlingame, Director) was supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR015804, NIH NCRR RR001614, and NIH NCRR RR012961.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1677208.
References
- Campbell M.S., Chan G.K., Yen T.J. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Brady D.M., Smith D., Murray A.W., Hardwick K.G. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 1999;8:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach S., Rackwitz H.R., Franke W.W. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M., Benezra R., Sorger P. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P., Guan T., Subauste C., Hahn K., Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett E.S., Espelin C.W., Sorger P.K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T., Kerscher O., Scott R.J., Basrai M.A., Wozniak R.W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Piatti S., Michaelis C., Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y., Chi Y.H., Miyazato A., Sheleg S., Haller K., Peloponese J.M., Li Y., Ward J.M., Benezra R., Jeang K.T. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- King R.W., Peters J.M., Tugendreich S., Rolfe M., Hieter P., Kirschner M.W. A 20S complex containing CDC27 and CDC16 catalyses the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Lee S.H., McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea C., Mahaffey D., Rechsteiner M., Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl. Acad. Sci. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Sorger P.K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Peschard P., Park M. From Tpr-Met to Met, tumorigenesis and tubes. Oncogene. 2007;26:1276–1285. doi: 10.1038/sj.onc.1210201. [DOI] [PubMed] [Google Scholar]
- Salina D., Enarson P., Rattner J.B., Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., Wozniak R.W. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., Luca F.C., Ruderman J.V., Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]