Abstract
Previously we created a paclitaxel-sensitive strain of Saccharomyces cerevisiae by mutating five amino acid residues in β-tubulin in a strain that has a decreased level of the ABC multidrug transporters. We have used site-directed mutagenesis to examine the relative importance of the five residues in determining sensitivity of this strain to paclitaxel. We found that the change at position 19 from K (brain β-tubulin) to A (yeast β-tubulin) and at position 227 from H (brain β-tubulin) to N (yeast β-tubulin) had no effect on the activity of paclitaxel. On the other hand, the changes V23T, D26G and F270Y, drastically reduced sensitivity of AD1-8-tax to paclitaxel. Molecular modeling and computational studies were used to explain the results.
Keywords: Tubulin, Paclitaxel, Saccharomyces cerevisiae, Mutagenesis
1. Introduction
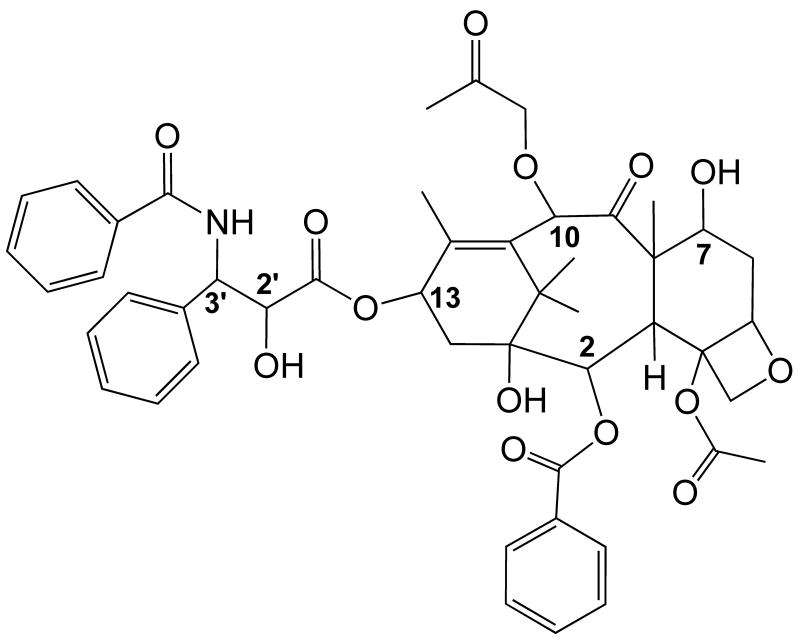
Paclitaxel (Taxol) (Fig. 1) is an anti-mitotic drug that has been shown to be effective against a variety of tumors, including breast, ovarian, lung and head and neck cancers [1,2]. It binds to the β-subunit of tubulin in microtubules, stabilizes the microtubules against depolymerization and decreases the dynamic nature of these polymers, leading to mitotic arrest and apoptosis [3]. Although paclitaxel is a successful anti-tumor agent, it does suffer from delivery problems due to poor aqueous solubility and to problems associated with the development of resistance. Because of these problems, there is interest in developing analogues that can overcome resistance due to the overexpression of multidrug transporters and analogues with more favorable aqueous solubility. The rational design of new analogues requires that the binding interactions between the drug molecule and tubulin be known in detail.
Fig. 1.
Structure of paclitaxel.
An electron microscopic crystal structure of tubulin at a 3.5 Å resolution has been obtained from a polymer induced to form by zinc ions and stabilized by paclitaxel [4,5]. Although this polymer consists of flat sheets of protofilaments rather than true microtubules, information obtained from this structure has provided valuable insights into the nature of the amino acid residues in β-tubulin that are in close proximity to the taxane molecule. This information, together with modeling studies and data from experiments using various physical techniques, have led to models of the conformation of bound paclitaxel as well as to speculation pertaining to the interaction of specific amino acids in β-tubulin with paclitaxel [6-9].
Tubulin from the budding yeast Saccharomyces cerevisiae does not bind paclitaxel [10,11]. Upon comparing the amino acid sequences of mammalian brain and S. cerevisiae β-tubulin we observed five differences at sites in β-tubulin purported to be important in paclitaxel binding. Changing these residues in yeast β-tubulin to those that occur in mammalian brain β-tubulin resulted in a mutated yeast tubulin that bound paclitaxel [12]. However, the strain carrying the mutated β-tubulin is not sensitive to paclitaxel, most likely because of the presence of multiple ABC drug transporters in the yeast plasma membrane. Subsequently, we introduced the mutated β-tubulin gene into a yeast strain that has diminished multi-drug transporter activity and created a strain, AD1-8-tax, which is sensitive to paclitaxel [13]. In order to determine the relative importance of the five amino acid residues to paclitaxel binding, we have now created new strains that contain different combinations of the five mutations, A19K, T23V, G26D, N227H, and Y270F. We tested the sensitivity of these strains to paclitaxel and found that the N227H and A19K mutations were not necessary to maintain sensitivity to paclitaxel. On the other hand, the T23V, G26D and Y270F mutations were essential for maximum activity. Molecular modeling and computational studies afforded explanations for these findings.
2. Materials and methods
2.1. Construction of mutant strains
Mutations were introduced into strain AD 12345678 (AD1-8) [14] having the genotype MATα, PDR1-3, ura3, his1, Δyor1::hisG, Δsnq2::hisG, Δpdr5::hisG, Δpdr10::hisG, Δpdr11::hisG, Δycf1::hisG, Δpdr3::hisG, Δpdr15::hisG. Two plasmids, pMG1, which contains the S. cerevisiae TUB2 gene with a His6 tag at the C-terminus [15], and pMG1-tax, which contains the tub2-His6-A19K-T23V-G26D-N227H-Y270F gene [12], were used to make the other strains used in this work. Appropriate restriction enzymes were used to cut the plasmids, the appropriate fragments were purified and ligated to a PCR product with the desired mutations producing new plasmids. These plasmids were amplified in E. coli XL1-Blue. A SacI/SphI fragment of each plasmid containing the tub2 gene of interest as well as the URA3 gene, was used to transform AD1-8 by homologous recombination using the lithium acetate method [16] as described previously [15]. Transformants were selected in SD medium (2% glucose, 0.67% yeast nitrogen base without amino acids, 0.001% histidine). Mutations were verified by DNA sequencing. Yeast strains were grown on YPD medium (2% glucose, 1% yeast extract, 2% peptone) at 30 °C.
2.2. Proliferation assays
The proliferation assays were performed in liquid medium in 96-well plates. Each well contained 100 μL of YPD supplemented with 100 U of penicillin, 100 μg of streptomycin, and paclitaxel at concentrations ranging from 0 to 25 μM. In other experiments benomyl was present at concentrations 0 to 80 μM. The wells were inoculated with 2000 cells in a 20 μL volume. The plates were placed in a humidified chamber on a shaker and incubated at 30 °C for 24 h to 27 h. The plates were then agitated on a vortex plate shaker to ensure yeast cells were completely suspended in solution and the optical density read at 620 nm in a 96-well plate reader (Bio-tek Instruments, Inc., Winooski, VT). Experiments were done in triplicate.
2.3 Molecular modeling
To help explain the different effects of the β-tubulin mutations on paclitaxel activity we modeled the T-taxol conformation [6] into the binding sites of the mutated proteins. We assumed that the basic conformation for paclitaxel would be largely conserved relative to the posited T-taxol structure. Thus, we constructed our models by projecting T-taxol onto the taxol-stabilized bovine tubulin crystal structure [4], and obtained preliminary models of the relevant complexes after applying the pertinent modifications to the original crystal structure. To allow for relaxation effects specific to each complex, the ligand plus all residues within 8.0 Å of the ligand were permitted to relax for 100 molecular mechanics steps, with the remainder of the tubulin structure held fixed. The position of paclitaxel was further optimized by allowing it to relax to molecular mechanics convergence with the entire receptor held fixed.
All of the above simulations were performed via SYBYL (SYBYL 7.3, the Tripos Associates, St. Louis MO, 2007) with the Tripos force field [17] and Gasteiger-Marsili electrostatics [18] within a non-bonding threshold of 8.0 Å. All other energetic and convergence parameters were left at default values. Free energies of paclitaxel binding to tubulin for each of the mutants and wild type strains were computed using the PMF scoring function [19].
3. Results and discussion
3.1 Effect of mutations on doubling time and benomyl sensitivity
In order to determine the relative contributions of the five mutations to paclitaxel binding we have now constructed new strains with different combinations of the five mutations (K19A, V23T, D26G, H227N and F270Y). For simplicity, we refer to the five positions as either B for brain or Y for yeast. For example, strain BBBBB would have amino acids found in brain β-tubulin at the five positions, whereas strain YYYYY would have amino acids found in yeast β-tubulin at the five positions. To determine whether any of the combinations of mutations had major effects on cellular microtubules we determined the doubling times of the strains at 30 °C and 15 °C and the sensitivity to the microtubule destabilizing drug benomyl. A change in microtubule stability would be expected to alter the doubling time at 15 °C compared to the wild type since microtubules depolymerize at low temperatures. An increase in microtubule stability would result in a larger ID50 value for benomyl while a decrease in stability would lower the ID50. The doubling times ranged from 2.1 h to 2.4 h at 30 °C and 9.9 to 12.3 h at 15 °C. The ID50 values for benomyl ranged from 20 μM to 30 μM. Thus, based on these criteria, there was not a large difference in microtubule stability amongst the mutant and wild type strains.
3.2 Effect of mutations on cytotoxicity of paclitaxel
The sensitivity to paclitaxel was tested in a liquid assay (Table 1). The drug was as potent against the BBBYB and YBBBB strains as it was against strain BBBBB. The other strains were not affected by paclitaxel up to a paclitaxel concentration of 25 μM. Limited solubility of paclitaxel precluded the use of higher concentrations. These results indicate that paclitaxel cytotoxicity is diminished substantially by mutations at positions 23, 26 and 270. On the other hand, the K19A and H227N mutations had no apparent effect on paclitaxel activity.
Table 1.
Growth inhibition by paclitaxel
| Strain | Percenta | ID50, μM |
|---|---|---|
| BBBBB | 97 | 6.3 |
| YYYYY | 0 | ----- |
| YBBBB | 92 | 5 |
| BYBBB | 0 | ----- |
| BBYBB | 0 | ----- |
| BBBYB | 100 | 4.5 |
| BBBBY | 0 | ----- |
At 25 μM paclitaxel. Growth was for 23 to 27 h.
3.3 Molecular modeling
Several models have been proposed for the conformation of the tubulin bound form of paclitaxel, but evidence appears to support the so-called T-taxol conformation [6,20-23]. The original modeling of the T-taxol conformer in the binding site showed close interactions of the CH2 groups of K19 and D26 and the isopropyl side chain of V23 with the 3′-benzamido phenyl ring [6]. These results are consistent with photoaffinity labeling studies with a paclitaxel analogue bearing an azido group in the 3′-benzamido ring that labeled the β-tubulin peptide containing amino acids 1-31 [24]. F270 also plays a role in paclitaxel binding. The side chain of this amino acid is part of a hydrophobic basin that accommodates the baccatin III ring. The phenyl ring of F270 is in van der Waals contact with the methyl group of the C-4 acetate and the 3′-phenyl group (6). In the T-taxol conformation, H227 plays a prominent role with its imidazole ring positioned between the 2-benzoyl phenyl and the 3′-benzamido phenyl rings of paclitaxel (6).
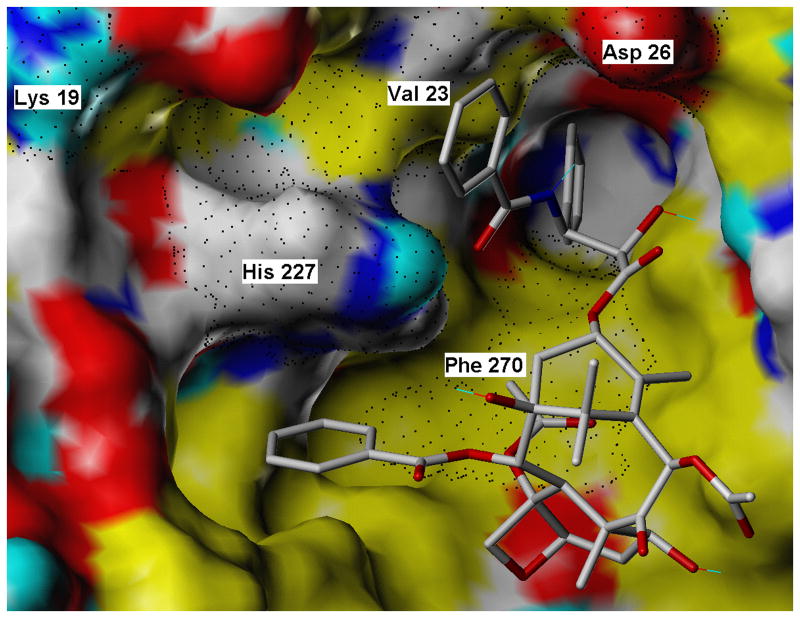
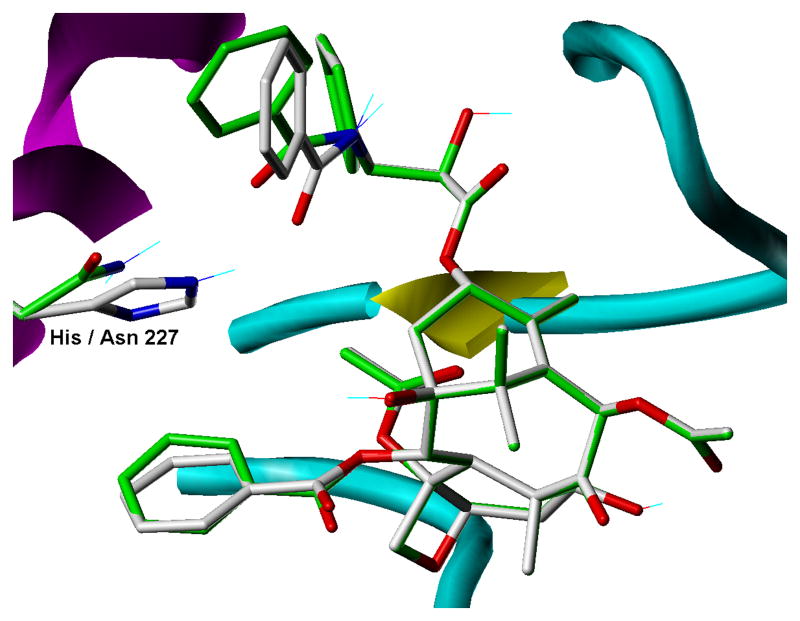
Our current study was undertaken to dissect out the relative contributions of the five residues (K19, V23, D26, H227 and F270) to the effectiveness of paclitaxel in inhibiting the proliferation of a yeast strain. The cytotoxicity results show that K19 plays little or no role in paclitaxel activity. Modeling of the paclitaxel site (Fig. 2) places the side chain of this residue far from paclitaxel (the closest K19 side chain atom is the β-carbon and it is 8.5 Å from the center of the 3′-benzamido ring) and it also does not appear to be a key component of any important intra-protein stabilizing features (salt bridges or H-bonds). The decrease in paclitaxel sensitivity that occurred in the V23T mutant is probably due to the loss of van der Waals interaction between the valine side chain and the benzamido phenyl ring. A D26G mutation would be expected to affect paclitaxel affinity because it would result in the loss of modest van der Waals interaction between the benzamido phenyl ring and the β-carbon of the D26 side chain. The importance of D26 to paclitaxel activity has also been demonstrated in a study that showed that a D26E mutation in a tumor cell line gave rise to paclitaxel resistance [25]. The F270Y mutation also led to a large decrease in the cytotoxicity of the taxane. F270 is in a hydrophobic pocket and is in close proximity to the 3′-phenyl ring (the p-carbon in the phenyl ring approaches within 3.5 Å of the nearest F270 ring carbon). The steric clash created by the added bulk of the OH group, together with its hydrophilic nature, undoubtedly explain the effect of the F270Y mutation. A F270V mutation has been associated with cell resistance to paclitaxel and with a decrease in the ability of paclitaxel to stimulate tubulin assembly [26]. The insensitivity of the strain with a H227N mutation to paclitaxel (Table 1) was surprising given the prominent role H227 plays in current models of the tubulin-paclitaxel complex. Obviously a substitution of aspargine for histidine at position 227 is tolerated. Molecular modeling and computations demonstrate that, although the van der Waals forces are weaker between the asparagine side chain and the 3′-benzamido ring of paclitaxel than between the histidine side chain and the benzamido ring, the asparagine side chain can form a H-bond with the benzamido carbonyl (distance = 2.22 Å) (Fig. 3). PMF binding free energies for paclitaxel complexed to wild type and each of the mutated tubulins are reported in Table 2. In general, the trend of the values is consistent with the cytotoxicity studies. BBBBB has a much higher negative free energy of binding than YYYYY. BBBYB and YBBBB have values in the same region as BBBBB. BBBBY shows a large drop in the negative value while BBYBB shows a smaller change. Only BYBBB does not seem to fit the trend. This could be a result of the fact that the PMF scoring function does not explicitly account for desolvation effects. The side chains of Val and Thr are similar in size and shape but in its predicted conformation (but in the absence of paclitaxel) Thr can form direct H-bonds to two water molecules that would have minimal commensurate interaction with Val. Thus, the paclitaxel binding event would incur a greater desolvation penalty in the case of BYBBB than for the BBBBB receptor.
Fig. 2.
Binding mode for pactitaxel (CPK colored sticks) complexed with its brain β-tubulin receptor (solvent accessible surface). Surface coloring is as follows: red = O; blue = polar N; cyan = donatable H; white = polar (but not donatable) H, C; yellow = nonpolar H, C. Residues subjected to mutation in this study are labeled, and their surface extent is rendered with black dots.
Fig. 3.
Binding model comparison for paclitaxel binding to brain β-tubulin and the corresponding H227N mutant. For the wild type complex, carbon atoms in the ligand and in His227 are rendered in grey, while for the mutation the corresponding carbons are green. All other atoms are rendered according to standard CPK coloring. Other receptor features are indicated according to underlying secondary structure as follows: magenta helices, yellow sheets and cyan coils.
Table 2.
Free energies of bindinga
| Strain | Free energy, kcal/mol |
|---|---|
| BBBBB | -75.4 |
| YYYYY | -21.1 |
| YBBBB | -70.5 |
| BYBBB | -75.6 |
| BBYBB | -66.5 |
| BBBYB | -85.6 |
| BBBBY | -34.2 |
Calculated using the PMF method.
The creation of the various yeast strains carrying mutations at five amino acid sites known to be important in paclitaxel binding has provided us with a useful experimental tool with which we can assess the relative importance of these five residues in paclitaxel binding. However, we are aware that mutations can affect microtubule stability as well as paclitaxel binding and that changing microtubule stability can affect sensitivity of cell to anti-mitotic agents [26,27]. The lack of appreciable effects on doubling times and benomyl sensitivity argues against effects on microtubule stability. However, we plan to follow up these studies with studies related to the effects of the mutations on microtubule dynamics in vivo and in vitro.
Acknowledgments
This work was supported by NIH grant CA 105305.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mollinedo F, Gajate C. Microtubules, microtubule-interfering agents and apoptosis. Apoptosis. 2003;8:413–450. doi: 10.1023/a:1025513106330. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J, Giannakakou P. Targeting microtubules for cancer chemotherapy. Curr Med Chem – Anti- Cancer Agents. 2005;5:65–71. doi: 10.2174/1568011053352569. [DOI] [PubMed] [Google Scholar]
- 3.Jordan MA. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem - Anti-Cancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 4.Nogales E, Wolf SG, Downing KH. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 5.Löwe J, Li H, Downing KH, Nogales E. Refined structure of αβ-tubulin at 3.5 Å resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 6.Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. The binding conformation of Taxol in β-tubulin: a model based on electron crystallographic density. Proc Natl Acad Sci USA. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Poliks B, Cegelski L, Poliks M, Grycznski Z, Piszczek G, Jagtap PG, Studelska DR, Kingston DGI, Schaefer J, Bane S. Conformation of microtubule-bound paclitaxel determined by fluorescence spectroscopy and REDOR NMR. Biochemistry. 2000;39:281–291. doi: 10.1021/bi991936r. [DOI] [PubMed] [Google Scholar]
- 8.Geney R, Sun L, Pera P, Bernacki RJ, Xia S, Horwitz SB, Simmerling CL, Ojima I. Use of the tubulin bound paclitaxel conformation for structure-based rational drug design. Chem Biol. 2005;12:339–348. doi: 10.1016/j.chembiol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SA, Alcaraz AA, Snyder JP. T-taxol and the electron crystallographic density in β-tubulin. Org Letts. 2005;7:5549–5552. doi: 10.1021/ol051780p. [DOI] [PubMed] [Google Scholar]
- 10.Barnes G, Louie KA, Botstein D. Yeast proteins associated with microtubules in vitro and in vivo. Mol Biol Cell. 1992;3:29–47. doi: 10.1091/mbc.3.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode CJ, Gupta ML, Jr, Reiff EA, Suprenant KA, Georg GI, Himes RH. Epothilone and paclitaxel: unexpected differences in promoting the assembly and stabilization of yeast microtubules. Biochemistry. 2002;41:3870–3874. doi: 10.1021/bi0121611. [DOI] [PubMed] [Google Scholar]
- 12.Gupta ML, Jr, Bode CJ, Georg GI, Himes RH. Understanding tubulin-Taxol interactions: Mutations that impart Taxol binding to yeast tubulin. Proc Natl Acad Sci USA. 2003;100:6394–6397. doi: 10.1073/pnas.1131967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foland TB, Dentler WL, Suprenant KA, Gupta ML, Jr, Himes RH. Paclitaxel-induced microtubule stabilization causes mitotic block and apoptotic-loke cell death in a paclitaxel-sensitive strain of Saccharomyces cerevisiae. Yeast. 2005;22:971–978. doi: 10.1002/yea.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 15.Gupta ML, Jr, Bode CJ, Dougherty CA, Marquez RT, Himes RH. Mutagenesis of β-tubulin cysteine residues in Saccharomyces cerevisiae: mutation of cysteine 354 results in cold-stable microtubules. Cell Motil Cytoskel. 2001;49:67–77. doi: 10.1002/cm.1021. [DOI] [PubMed] [Google Scholar]
- 16.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark M, Cramer RDI, Van Opdenbosch N. Validation of the general purpose Tripos 5.2 force field. J Comput Chem. 1989;10:982–1012. [Google Scholar]
- 18.Gasteiger J, Marsili M. Iterative partial equilization of orbital electronegativity: a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 19.Muegge I, Martin YC, Hajduk PJ, Fesik SW. Evaluation of PMF scoring in docking weak ligands to the FK506 binding protein. J Med Chem. 1999;42:2498–2503. doi: 10.1021/jm990073x. [DOI] [PubMed] [Google Scholar]
- 20.Ganesh T, Guza RC, Bane S, Ravindra R, Shanker N, Lakdawala AS, Snyder JP, Kingston DGI. The bioactive Taxol conformation on β-tubulin: Experimental evidence from highly active constrained analogs. Proc Natl Acad Sci USA. 2004;101:10006–10011. doi: 10.1073/pnas.0403459101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingston DGI, Bane S, Snyder JP. The taxol pharmacophore and the T-taxol bridging principle. Cell Cycle. 2005;4:279–289. [PubMed] [Google Scholar]
- 22.Alcaraz AA, Mehta AK, Johnson SA, Snyder JP. The T-taxol conformation. J Med Chem. 2006;49:2478–2488. doi: 10.1021/jm051119r. [DOI] [PubMed] [Google Scholar]
- 23.Paik Y, Yang C, Metaferia B, Tang S, Bane S, Ravindra R, Shanker N, Alcaraz AA, Johnson SA, Schaefer J, O'Connor RD, Cegelski L, Snyder JP, Kingston DGI. Rotational-echo double resonance NMR distance measurements for the tubulin-bound paclitaxel conformation. J Am Chem Soc. 2007;129:361–370. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao S, Krauss NE, Heerding JM, Swindell CS, Ringel I, Orr GA, Horwitz SB. 3′-(p-Azidobenzamido)taxol photolabels the N-terminal 31 amino acids of β-tubulin. J Biol Chem. 1994;269:3132–3134. [PubMed] [Google Scholar]
- 25.Hari M, Loganzo F, Annable T, Tan X, Musto S, Morilla DB, Nettles JH, Snyder JP, Greenberger LM. Paclitaxel-resistant cells have a mutation in the paclitaxel-binding region of β-tubulin (Asp26Glu) and less stable microtubules. Mol Cancer Ther. 2006;5:270–278. doi: 10.1158/1535-7163.MCT-05-0190. [DOI] [PubMed] [Google Scholar]
- 26.Giannakakou P, Sackett DL, Kang YK, Zhan Z, Buters JTM, Fojo T, Poruchynsky MS. Paclitaxel-resistant human ovarian cancer cells have mutant β-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem. 1997;272:17118–17125. doi: 10.1074/jbc.272.27.17118. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Garay ML, Chang L, Blade K, Menick DR, Cabral F. A β-tubulin leucine cluster involved in microtubule assembly and paclitaxel resistance. J Biol Chem. 1999;274:23875–23882. doi: 10.1074/jbc.274.34.23875. [DOI] [PubMed] [Google Scholar]