SUMMARY
Alzheimer’s disease (AD) is a genetically heterogeneous disorder characterized by early hippocampal atrophy and cerebral Aβ peptide deposition. Using TissueInfo to screen for genes preferentially expressed in the hippocampus and located in AD linkage regions, we identified a novel gene on 10q24.33 that we call CALHM1. We show that CALHM1 encodes a multipass transmembrane glycoprotein that controls cytosolic Ca2+ concentrations and Aβ levels. CALHM1 homomultimerizes, shares strong sequence similarities with the selectivity filter of the NMDA receptor, and generates a large Ca2+ conductance across the plasma membrane. Importantly, we determined that the CALHM1 P86L polymorphism (rs2986017) is significantly associated with AD in independent case-control studies of 3,404 participants (allele-specific OR=1.44, P=2×10−10). We further found that the P86L polymorphism increases Aβ levels by interfering with CALHM1-mediated Ca2+ permeability. We propose that CALHM1 encodes an essential component of a novel cerebral Ca2+ channel that controls Aβ levels and susceptibility to AD.
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by a massive loss of neurons in several brain regions and by the presence of cerebral senile plaques comprised of aggregated amyloid-β (Aβ) peptides (Mattson, 2004; Selkoe, 2001). The first atrophy observed in the AD brain occurs in the medial temporal lobe, which includes the hippocampus, and is the result of a massive synaptic degeneration and neuronal death (Braak and Braak, 1991; de Leon et al., 2007). Two major Aβ species are found, Aβ40 and Aβ42; both are produced from the sequential endoproteolysis of the amyloid precursor protein (APP) by BACE1/β-secretase and by presenilin (PS)/γ-secretase complexes. APP can also undergo a non-amyloidogenic proteolysis by α-secretase, which cleaves APP within the Aβ sequence and thereby precludes Aβ generation (Marambaud and Robakis, 2005; Wilquet and De Strooper, 2004).
The etiology of the disease is complex because of its strong genetic heterogeneity. Rare autosomal dominant mutations in the genes encoding APP, PS1, and PS2 cause early-onset AD, whereas complex interactions among different genetic variants and environmental factors are believed to modulate the risk for the vast majority of late-onset AD (LOAD) cases (Kennedy et al., 2003; Lambert and Amouyel, 2007; Pastor and Goate, 2004). To date, the only susceptibility gene unambiguously demonstrated worldwide is the ε4 allele of APOE on chromosome 19 (Strittmatter et al., 1993). However, epidemiological studies indicate that the presence of the APOE ε4 allele cannot explain the overall heritability of AD, implying that a significant proportion of LOAD cases is attributable to additional genetic risk factors (Lambert and Amouyel, 2007; Pastor and Goate, 2004). Supporting this observation, concordant evidence of linkage to LOAD has been observed in different chromosomal regions, including on chromosome 10 where a strong and consensual susceptibility locus is present (Bertram et al., 2000; Blacker et al., 2003; Ertekin-Taner et al., 2000; Farrer et al., 2003; Kehoe et al., 1999; Myers et al., 2000). However, despite intensive research efforts to characterize the genetic factor(s) located within the chromosome 10 region, no gene has been conclusively linked to LOAD risk (Bertram et al., 2006; Grupe et al., 2006; Kuwano et al., 2006; Minster et al., 2006).
A number of neurodegenerative disorders are caused by mutations in genes expressed principally in the central nervous system. This is the case for the brain proteins tau and α-synuclein, which are linked to autosomal dominant forms of frontotemporal dementia and Parkinson’s disease, respectively. Here we postulated that susceptibility to LOAD could come from genes predominantly expressed in affected brain regions, such as the hippocampus. We used TissueInfo (Skrabanek and Campagne, 2001) and the Alzgene database (Bertram et al., 2007) to screen for genes predominantly expressed in the hippocampus and located in linkage regions for LOAD, and identified CALHM1, a gene of unknown function, located on chromosome 10 at 1.6 Mb of the LOAD marker D10S1671 (Bertram et al., 2000). We found that CALHM1 homomultimerizes, controls cytosolic Ca2+ concentrations, and shares similarities with the predicted selectivity filter of the N-methyl-D-aspartate receptor (NMDAR). Voltage-clamp analyses further revealed that CALHM1 generates Ca2+ selective cation currents at the plasma membrane. Importantly, we determined that the frequency of the rare allele of the non-synonymous single nucleotide polymorphism (SNP) rs2986017 in CALHM1, which results in a proline to leucine substitution at codon 86 (P86L), is significantly increased in AD cases in five independent cohorts. Further investigation demonstrated the functional significance of the rs2986017 SNP by showing that the P86L mutation promotes Aβ accumulation via a loss of CALHM1 control on Ca2+ permeability and cytosolic Ca2+ levels. Here we propose that CALHM1 is a component of a novel cerebral Ca2+ channel family involved in Aβ metabolism and that CALHM1 variants may influence the risk for LOAD.
RESULTS
Gene discovery
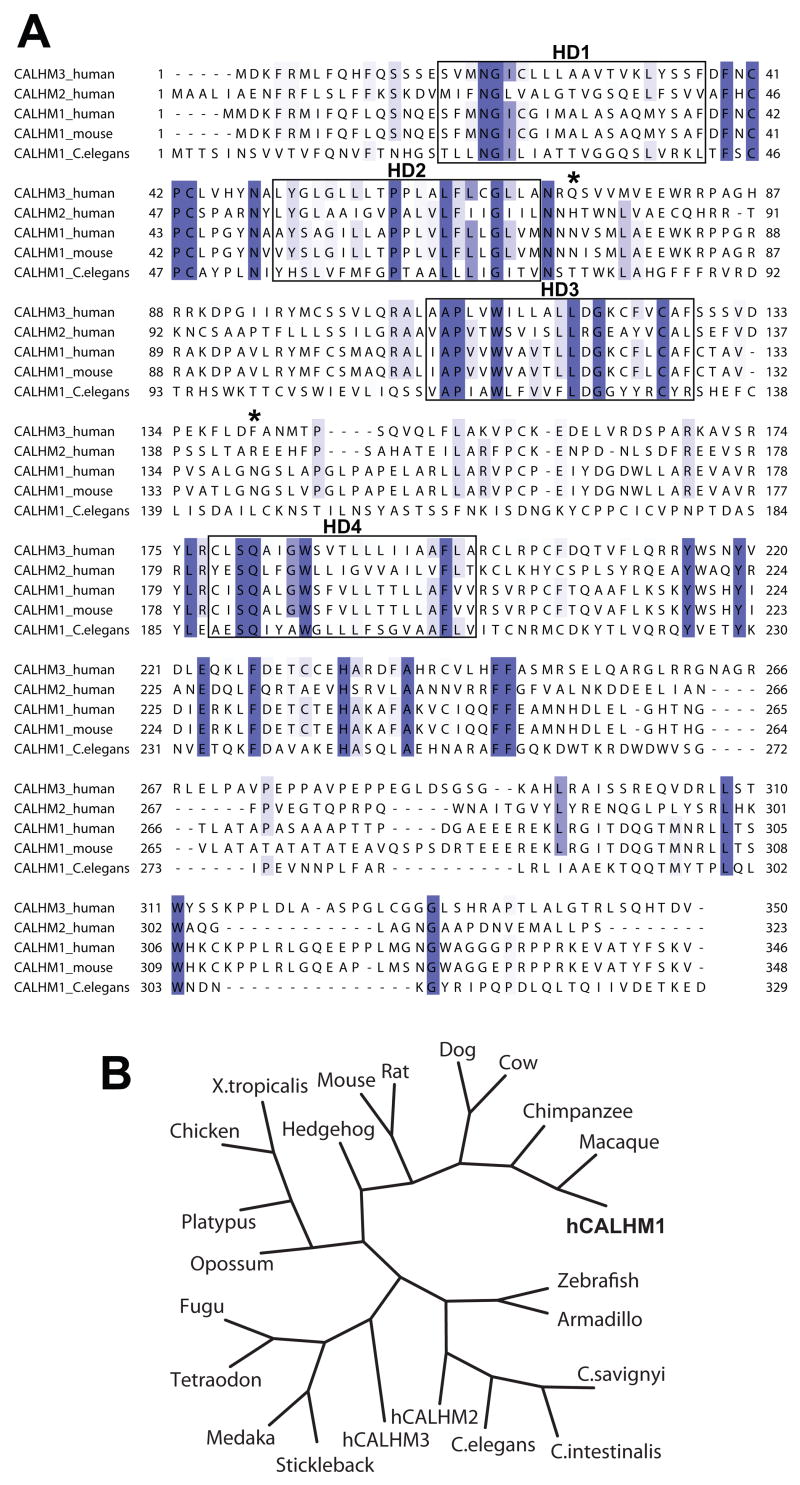
We screened the human genome with TissueInfo to annotate human transcripts with tissue expression levels derived from the expressed sequence tag database (dbEST) (Campagne and Skrabanek, 2006; Skrabanek and Campagne, 2001). Out of 33,249 human transcripts, the TissueInfo screen identified 30 transcripts, corresponding to 12 genes, with expression restricted to the hippocampus (Table 1). These transcripts matched either one or two ESTs sequenced from the hippocampus. Among these genes, one of unknown function, previously annotated as FAM26C, matched two hippocampal ESTs and mapped to the AD locus on 10q24.33 (Table 1). This gene, hereafter referred to as CALHM1 (calcium homeostasis modulator 1), encodes an open reading frame (ORF) of 346 amino acids and is predicted to contain four hydrophobic domains (HDs; TMHMM prediction) and two N-glycosylation motifs (NetNGlyc 1.0 prediction) (Fig. 1A). No significant amino acid sequence homology to other functionally characterized proteins was found. Sequence database searches identified five human homologs of CALHM1 (collectively identified as the FAM26 gene family). Two homologs of human CALHM1 with broader tissue expression profiles (see Supplemental Data), are clustered next to CALHM1 in 10q24.33 and are designated CALHM2 (26% protein sequence identity, previously annotated as FAM26B) and CALHM3 (39% identity, FAM26A) (Fig. 1A). CALHM1 is conserved across at least 20 species, including mouse and C. elegans (Figs. 1A and 1B).
Table 1.
TissueInfo expression screen1
| Chromosome | Band | Ensembl Transcript ID | Hit(s) | Hit(s) in Hippocampus2 | Tissue Summary | Gene Name/Other ID |
|---|---|---|---|---|---|---|
| 1 | p34.3 | ENST00000319637 | 2 | 2 | hippocampus | EPHA10 |
| 2 | p21 | ENST00000306078 | 2 | 1 | hippocampus | KCNG3 |
| 2 | q37.1 | ENST00000313064 | 2 | 1 | hippocampus | C2orf52 |
| 6 | q15 | ENST00000303726 | 3 | 1 | hippocampus | CNR1 |
| 6 | q25.3 | ENST00000308254 | 1 | 1 | hippocampus | Retired in Ensembl 46 |
| 6 | q27 | ENST00000322583 | 1 | 1 | hippocampus | NP_787118.2 |
| 9 | q21.33 | ENST00000298743 | 3 | 1 | hippocampus | GAS1 |
| 10 | q24.33 | ENST00000329905 | 3 | 2 | hippocampus | CALHM1 (FAM26C) |
| 11 | q24.1 | ENST00000354597 | 3 | 1 | hippocampus | OR8B3 |
| 17 | q25.3 | ENST00000326931 | 2 | 1 | hippocampus | Q8N8L1_HUMAN |
| 19 | p12 | ENST00000360885 | 1 | 1 | hippocampus | Retired in Ensembl 46 |
| X | q27.2 | ENST00000298296 | 1 | 1 | hippocampus | MAGEC3 |
One transcript is shown for each gene identified in the screen. Genomic location and number of hit(s) in dbEST are reported for each transcript.
Hit(s) in hippocampus indicates how many ESTs matching the transcript were sequenced from a cDNA library made from the hippocampus.
Figure 1. Alignment and phylogeny of CALHM1.
(A) Sequence alignment of human CALHM3, CALHM2, and CALHM1, and of murine and C. elegans CALHM1. Conserved sequences are highlighted in blue and sequence conservation is mapped in a color gradient, the darkest color representing sequences with absolute identity and lighter colors representing sequences with weaker conservation. Boxes denote hydrophobic domains 1–4 (HD1–4). Stars, predicted N-glycosylation sites on human CALHM1.
(B) Phylogenetic tree including human CALHM1 (hCALHM1).
CALHM1 characterization
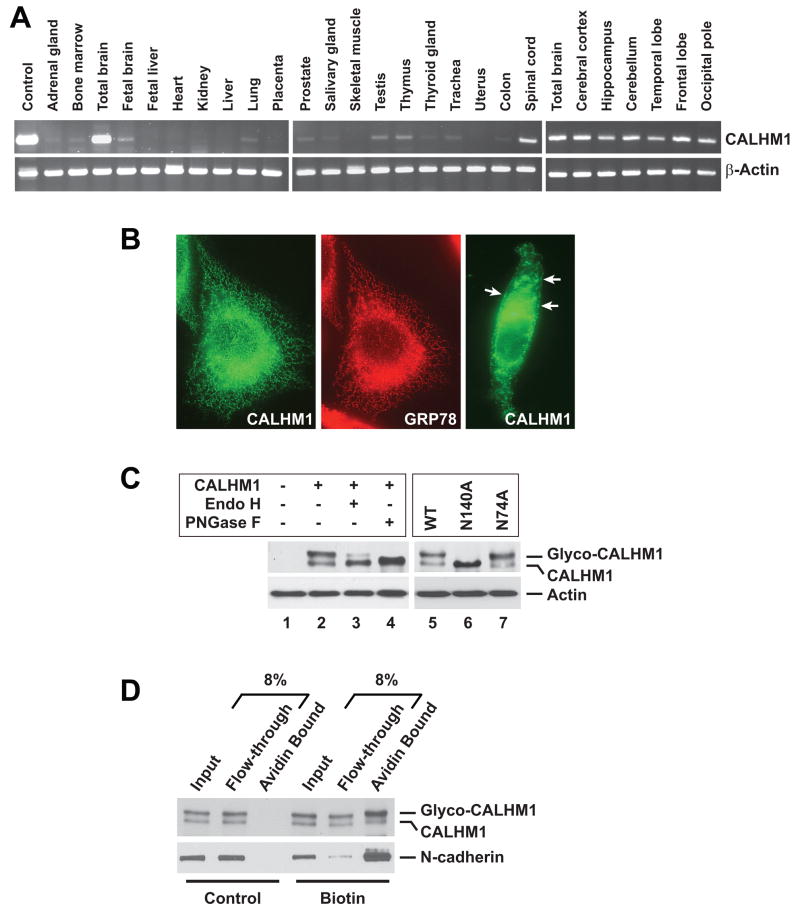
Using RT-PCR, we analyzed human CALHM1 expression in 20 tissues and six brain regions. The expression of CALHM1 was highest in the total adult brain and in all brain regions tested. CALHM1 expression was noticeably lower in all other tissues, including fetal brain (Fig. 2A). qRT-PCR revealed endogenous CALHM1 expression in retinoic acid-differentiated SH-SY5Y cells (Fig. S1A), suggesting that CALHM1 is a protein of neuronal origin. Immunofluorescence staining in transiently transfected cells revealed that CALHM1 strongly localized to the endoplasmic reticulum (ER) where it colocalized with the ER resident protein GRP78 (Fig. 2B, left and middle panels). However, some cells revealed immunoreactivity for CALHM1 at the cell surface, suggesting that a pool of CALHM1 was localized at or near the plasma membrane (Fig. 2B, right panel, arrows). Western blotting (WB) analyses revealed the presence of two immunoreactive bands in CALHM1-transfected cells (Fig. 2C, lanes 1 and 2). Because human CALHM1 is predicted to be N-glycosylated at asparagine residues N74 and N140 (see Fig. 1A, stars), we asked whether these bands might represent different N-glycosylated forms of the protein. Treatment of CALHM1-transfected cell lysates with N-glycosidase F, which cleaves all types of asparagine-bound N-glycans, completely eliminated the appearance of the higher molecular weight band and resulted in the accumulation of the lower band, which we conclude corresponds to the unmodified core protein (Fig. 2C, lanes 2 and 4). CALHM1 was partially resistant to endoglycosidase H-mediated deglycosylation (Fig. 2C, lanes 2 and 3), indicating that CALHM1 can reach the medial Golgi compartment and the cell surface, where proteins are terminally glycosylated and acquire resistance to endoglycosidase H. Plasma membrane expression of CALHM1 was also investigated by cell surface biotinylation. Figure 2D illustrates that a pool of CALHM1, enriched in glycosylated forms of the protein, was biotinylated and thus was present at the plasma membrane. We further determined that substitution of the N140 residue to alanine (N140A) completely prevented CALHM1 glycosylation, while N74A substitution had no effect (Fig. 2C, lanes 5–7). Thus, CALHM1 is a multipass transmembrane protein, N-glycosylated at the residue N140, predominantly expressed in the adult brain, and localized to the ER and plasma membranes. These data further indicate that the HD3-HD4 loop, which contains the N140 residue, is oriented toward the luminal side when CALHM1 is in the ER membrane and toward the extracellular space when CALHM1 reaches the plasma membrane.
Figure 2. Tissue expression, subcellular localization, and N-glycosylation of human CALHM1.
(A) Total RNA was used for RT-PCR analyses targeting CALHM1 and β-actin transcripts in multiple human tissues and brain regions.
(B) Immunofluorescence staining in CHO cells transfected with human Myc-tagged CALHM1 using anti-Myc (green) and anti-GRP78 (red) antibodies.
(C) Lysates from HT-22 cells transfected with wild type (WT) or mutated (N140A and N74A) Myc-CALHM1, were incubated in the absence (−) or presence (+) of endoglycosidase H (Endo H) or N-glycosidase F (PNGase F). Cell lysates were probed with anti-Myc (upper panels) and anti-actin (lower panels) antibodies.
(D) Cell surface-biotinylated proteins from Myc-CALHM1-transfected HT-22 cells were precipitated using immobilized avidin and probed with anti-Myc (upper panel) and anti-N-cadherin (lower panel, cell surface positive control) antibodies.
CALHM1 controls cytosolic Ca2+ levels
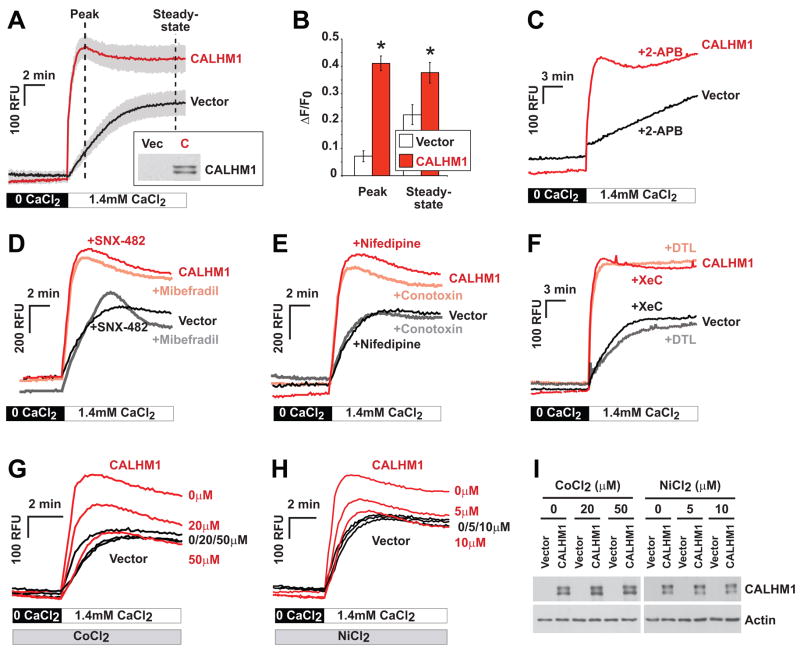
The predicted membrane topology of CALHM1 suggests the presence of one re-entrant hydrophobic loop that does not cross the membrane bilayer and three membrane-spanning segments (TMHMM prediction). In the absence of significant homology to other characterized proteins, we postulated from the predicted topology that CALHM1 could function as an ion channel component. This is in part based on a suggestive similarity with the topology of ionotropic glutamate receptors, which also contain three transmembrane segments and a re-entrant loop that forms the lining of the ion channel pore region (Wollmuth and Sobolevsky, 2004). Because some ionotropic glutamate receptors are Ca2+-permeable membrane proteins (Gouaux and Mackinnon, 2005), we asked whether CALHM1 could control cytoplasmic Ca2+ levels. Measurements of intracellular Ca2+ concentration ([Ca2+]i) were conducted under resting conditions in the presence of physiological concentrations of extracellular Ca2+. To reveal possible changes in the rate of Ca2+ entry in CALHM1-expressing cells, [Ca2+]i measurements were also performed under extracellular “Ca2+ add-back” conditions. These conditions are obtained after a transient external Ca2+ depletion that generates a driving force for Ca2+ entry. Using the Ca2+ fluorescent dye Fluo-4 in mouse hippocampal HT-22 cells, no robust changes in fluorescence measurements were found under resting conditions following CALHM1 expression (not shown). However, CALHM1 expression resulted in a strong and sustained increase in [Ca2+]i after extracellular Ca2+ add-back (Fig. 3A). CALHM1 expression significantly increased the initial rate of change in [Ca2+]i producing a peak of fluorescence at ~2 min after Ca2+ addition (Figs. 3A and 3B, Peak). CALHM1 expression also induced a significant elevation in the steady-state [Ca2+]i, compared to control conditions (Figs. 3A and 3B, Steady-state). To measure absolute [Ca2+]i, we also determined the effect of CALHM1 on [Ca2+]i by using the ratiometric Ca2+ indicator Fura-2. We confirmed that, under Ca2+ add-back conditions, CALHM1 expression induced a significant elevation of [Ca2+]i from 106 ± 4 nM (prior to Ca2+ add-back) to 264 ± 48 nM at the peak (following Ca2+ addition), whereas control cells showed no significant changes in [Ca2+]i (from 105 ± 5 nM to 110 ± 6 nM; Fig. S2). Because massive Ca2+ influx can be cytotoxic, we evaluated the viability of cells expressing CALHM1. Figure S3 illustrates that, in both normal and Ca2+ add-back conditions, no noticeable cell viability impairments or cytotoxicity were observed following CALHM1 expression.
Figure 3. CALHM1 controls Ca2+ influx by a mechanism that does not promote VGCC or SOCE channel activation.
(A) Cytoplasmic Ca2+ measurements using Fluo-4 loading and Ca2+ add-back assays in HT-22 cells transiently transfected with Myc-CALHM1 or control vector. Cells were first incubated in Ca2+-free buffer (0 CaCl2) and then challenged with physiological extracellular Ca2+ concentrations (1.4 mM CaCl2) to monitor the progressive restoration of basal [Ca2+]i. Traces illustrate the mean relative fluorescence units (RFU) +/− S.D. (shaded areas) of three independent experiments. Inset, WB of the corresponding cell lysates probed with anti-Myc antibody (Vec, vector; C, CALHM1).
(B) Peak and steady-state of [Ca2+]i measurements as in (A) expressed in ΔF/F0 (*, P<0.001; Student’s t test).
(C–H) Cytoplasmic Ca2+ measurements as in (A) in cells pretreated with 2-APB [50 μM, (C)], SNX-482 [0.5 μM, (D)], mibefradil [1 μM, (D)], nifedipine [10 μM, (E)], ω-conotoxin MVIIC [Conotoxin, 5 μM, (E)], dantrolene [DTL, 10 μM, (F)], xestospongin C [XeC, 2 μM, (F)], or with the indicated concentrations of CoCl2 (G) and NiCl2 (H). Traces in (C–H) illustrate representative measurements of 2–3 independent experiments.
(I) WB with anti-Myc (upper panels) and anti-actin (lower panels) antibodies of protein extracts obtained from cells treated as in (G) and (H).
One important mechanism of Ca2+ entry coupled to ER Ca2+ release is called store operated Ca2+ entry (SOCE). In excitable cells, such as neurons, voltage-gated Ca2+ channels (VGCCs) represent another critical mechanism of Ca2+ influx during membrane depolarization (Berridge et al., 2003). Inhibition of SOCE by the use of 2-APB did not prevent CALHM1 from affecting [Ca2+]i (Fig. 3C). Similarly, selective blockage of the different subtypes of VGCCs with SNX-482 (R-type VGCC inhibitor), mibefradil (T-type), nifedipine (L-type), or ω-conotoxin MVIIC (N-, P-, Q-types) did not block the rise of [Ca2+]i induced by CALHM1 expression (Figs. 3D and 3E). Because cytosolic Ca2+ can be released from intracellular stores via activation of the inositol 1,4,5-triphosphate receptors (InsP3Rs) or the ryanodine receptors (RyRs) at the ER membrane (Berridge et al., 2003), we next asked whether CALHM1 expression promotes InsP3R or RyR activation. The InsP3R inhibitor xestospongin C and the RyR inhibitor dantrolene were found to have no effect on the CALHM1-driven [Ca2+]i increase (Fig. 3F), indicating that ER Ca2+ release via InsP3Rs or RyRs did not account for the effect of CALHM1 on cytosolic Ca2+ levels. Because presenilins were recently proposed to form ER calcium leak channels (Tu et al., 2006), we also investigated whether CALHM1 requires the presence of PS1 or PS2 to control cytosolic Ca2+ levels. We found that CALHM1 expression caused similar increases in [Ca2+]i in WT fibroblasts and in fibroblasts deficient for both PS1 and PS2 (Fig. S4), showing that CALHM1 controls cytosolic Ca2+ levels independently of presenilins. We found, however, that the increase of [Ca2+]i observed following CALHM1 expression was blocked by cobalt (Co2+) and nickel (Ni2+), two nonspecific Ca2+ channel blockers. Indeed, Figures 3G and 3H show that 50 μM Co2+ or 10 μM Ni2+ were sufficient to completely inhibit the rise of intracellular Ca2+ induced by CALHM1 without causing changes in CALHM1 expression (Fig. 3I). Because Ni2+ does not penetrate the cells (Shibuya and Douglas, 1992), these results suggest that the two inorganic Ca2+ channel blockers acted at the plasma membrane to block Ca2+ entry. Collectively, these results strongly indicate that CALHM1 expression promotes Ca2+ influx via activation of a cell surface ion channel that is distinct from known VGCC or SOCE channels.
CALHM1 has ion channel properties
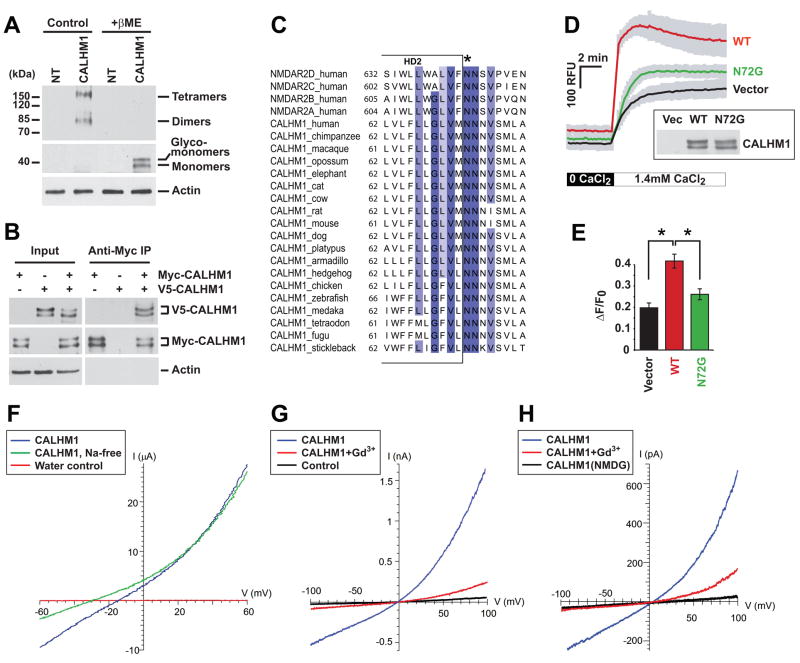
Because many channels multimerize to form an ion pore, and because monomeric CALHM1 cannot create a functional pore with three transmembrane segments, we asked whether CALHM1 could form multimers. WB analyses of CALHM1-transfected cells under non-reducing conditions revealed the presence of immunoreactive bands with molecular weights compatible with dimers and tetramers of CALHM1 (Fig. 4A). To test the possibility that CALHM1 self-associates, we co-expressed in cells two different tagged versions of the protein and used co-immunoprecipitation experiments to determine whether the two versions of CALHM1 form a complex. We found that immunoprecipitation of Myc-tagged CALHM1 co-precipitated V5-tagged CALHM1 (Fig. 4B), indicating that CALHM1 homomultimerized to form dimeric and possibly tetrameric structures.
Figure 4. Ion channel properties of CALHM1.
(A) Lysates from non-transfected (NT) and Myc-CALHM1-tranfected HEK293 cells were analyzed by WB in the absence (Control) or presence of β-mercaptoethanol (+βME) using anti-Myc (two upper panels) and anti-actin (lower panel) antibodies.
(B) Lysates from HEK293 cells transfected (+) or not (−) with V5-tagged CALHM1 (V5-CALHM1) or Myc-CALHM1, were immunoprecipitated with anti-Myc antibody. Total lysates (Input, left panels) and immunoprecipitates (Anti-Myc IP, right panels) were analyzed by WB using antibodies against V5 (upper panels), Myc (middle panels), and actin (lower panels).
(C) Partial sequence alignment of human NMDAR NR2 (NMDAR2) subunits A–D and CALHM1 from various species. Sequence conservation is highlighted in a blue gradient as described in Fig. 1A. Star denotes Q/R/N site.
(D) Cytoplasmic Ca2+ measurements in HT-22 cells transiently transfected with control vector and WT or N72G-mutated Myc-CALHM1. Cells were treated and results analyzed as in Fig. 3A (n = 3 independent experiments). Inset, WB of the corresponding cell lysates with anti-Myc antibody.
(E) Peak of [Ca2+]i measurements as in (D) expressed in ΔF/F0 (*, P<0.001; Student’s t test).
(F) Representative current traces during voltage ramps in Xenopus oocytes injected with CALHM1 cRNA (blue and green traces) or water (red trace) in normal LCa96 solution (blue and red traces) or in Na+-free LCa96 solution (replaced with equimolar N-methyl-D-glucamine (NMDG); green trace).
(G) Whole-cell currents in CALHM1-expressing (blue and red traces) or control (black trace) CHO cells in response to voltage ramps before (blue trace) and after (red trace) perfusion with 100 μM Gd3+. Bath contained 120 mM NaCl, pipette solution contained 122 mM CsCl (see Experimental Procedures). Cell capacitances of the CALHM1-expressing and control cells were 18.5 pF and 13.0 pF, respectively.
(H) Whole-cell currents in CALHM1-expressing CHO cells (uncorrected for leakage currents) in response to voltage ramps in bi-ionic Ca2+/Cs+ solutions (20 mM Ca-aspartate in bath, 120 mM Cs-aspartate in pipette; see Experimental Procedures) before (blue trace) or after (red trace) bath addition of 100 μM Gd3+(Cm = 24.1 pF). Reversal potential Vrev = +8.3 ± 2.9 mV (n = 7) after correction for liquid junction potential and leakage current, indicating PCa : PCs = 5. No currents were observed in CALHM1-expressing cells with NMDG-aspartate in bath and pipette solutions (black trace; Cm = 20.5 pF).
Ionotropic glutamate receptors are ion conducting membrane proteins with specific ion selectivity properties (Gouaux and Mackinnon, 2005). Recent advances in the structural analysis of ion channels have determined that the ion selectivity of some ion channels is controlled by a short amino acid sequence called the selectivity filter, which forms a narrow constriction in the pore across the membrane bilayer (Gouaux and Mackinnon, 2005). The predicted selectivity filter of ionotropic glutamate receptors is located in a re-entrant loop called M2 and is critical for Ca2+ permeability (Dingledine et al., 1999; Wollmuth and Sobolevsky, 2004). By manual inspection, we screened ionotropic glutamate receptor subunit sequences for similarities with CALHM1 and found a short sequence in C-terminus of CALHM1 HD2 that aligns with the predicted ion selectivity filter of NMDAR NR2 subunits (Fig. 4C). Previous studies have determined that the asparagine (N) residue in the so-called Q/R/N site of NMDAR NR2 subunits is critical for ion selectivity and permeation (see Fig. 4C, star) (Wollmuth and Sobolevsky, 2004). By sequence comparison, we identified a highly conserved N72 residue in human CALHM1 that aligns with the Q/R/N site at the C-terminus end of the second hydrophobic domain of both CALHM1 and NMDAR (Fig. 4C, star). Importantly, we found that mutagenesis of the N72 residue to glycine (N72G) resulted in a significant inhibition of the effect of CALHM1 on [Ca2+]i (Fig. 4D and 4E).
Two-electrode voltage clamping of Xenopus oocytes was employed to determine the effects of CALHM1 expression on plasma membrane conductance. Oocytes were injected with either water or CALHM1 cRNA and conductance was recorded 24–72 hours later in a normal Na+-containing bath. In water-injected oocytes, the resting membrane potential Vm was −38 ± 1 mV (n = 74), and the membrane conductance, measured as the slope conductance around the reversal potential Vrev, was 1.5 ± 0.2 μS (n = 7). In contrast, the Vm in CALHM1-expressing oocytes was depolarized to −16 ± 0.3 mV (n = 96; P < 0. 0001) and membrane conductance was enhanced to 422 ± 78 μS (n = 12) (P < 0.005). The current-voltage (I/V) relation was outwardly rectifying (slope conductances of 372 ± 110 and 670 ± 131 μS at -55 mV and +55 mV, respectively) (Fig. 4F). Depolarization of the resting Vm suggested that the CALHM1-enhanced conductance was contributed by a Na+ permeability. Isosmotic replacement of bath Na+ with NMDG hyperpolarized Vm by 7 ± 0.8 mV (Fig. 4F; n = 12; P < 0.0001). These results demonstrate that expression of CALHM1 conferred a novel constitutive Na+ conductance in Xenopus oocyte plasma membrane.
Expression of CALHM1 in CHO cells also generated an outwardly rectifying current in whole cell recordings with Cs+ in the pipette (cytoplasmic) and Na+ in the bath (Fig. 4G). The current reversed ~0 mV, indicating that the relative permeabilities of Cs+ and Na+ were similar (PNa: PCs = 0.8). The current was not observed in either untransfected or EGFP-transfected cells (Fig. 4G, control), and it was eliminated when the monovalent cations in the bath and pipette solutions were replaced with NMDG (Fig. 4H), indicating that the current was carried by Cs+ and Na+. The CALHM1-induced slope conductance measured around the Vrev was 360 ± 60 pS/pF (n = 42) compared with 74 ± 17 pS/pF (n = 11) in control cells. Gd3+ (100 μM) nearly completely inhibited the CALHM1-induced current (Fig. 4G). With bath Na+ replaced by NMDG and 20 mM Ca2+, an outwardly rectifying, Gd3+-sensitive current was observed in the CALHM1-expressing cells that reversed at +8.3 ± 2.9 mV (n = 7), indicating PCa : PCs = 5 (Fig. 4H). Thus, expression of CALHM1 conferred a novel constitutive Ca2+ selective cation current in CHO cell plasma membrane.
In summary our studies show that a region of CALHM1 shares sequence similarities with the selectivity filter of NMDAR and that the N72 residue is a key determinant in the control of cytosolic Ca2+ levels by CALHM1. Furthermore, electrophysiological analyses in CALHM1-expressing Xenopus oocytes and CHO cells demonstrated that CALHM1 induced a novel plasma membrane Ca2+ selective cation current. Together with the observation that the effect of CALHM1 had properties that did not overlap those of known Ca2+ channels, these results suggest that CALHM1 may be a novel pore-forming ion channel.
CALHM1 controls APP processing
Because cytosolic Ca2+ is critical for the regulation of APP processing (LaFerla, 2002), we asked whether CALHM1 expression influences Aβ levels. Figure 5A shows that, under resting conditions with physiological concentrations of extracellular Ca2+, CALHM1 expression in APP-transfected mouse neuroblastoma N2a cells had no noticeable effect on extracellular Aβ levels (panel a, lanes 1–4). Under Ca2+ add-back conditions, however, expression of CALHM1 strongly and significantly decreased total extracellular Aβ accumulation (Fig. 5A, panel b, lanes 1–4), including Aβ1-40 and Aβ1-42 (Fig. 5B). Importantly, the decrease in Aβ levels triggered by CALHM1 expression was accompanied by an elevation of sAPPα levels, while cellular full-length APP remained unchanged (Fig. 5A, panels c and d, lanes 1–4). The effect of endogenous CALHM1 on APP processing was studied in differentiated neuroblastoma cells. We found that neuronal differentiation with retinoic acid, a condition which we showed induces endogenous CALHM1 expression in SH-SY5Y cells (Fig. S1A), resulted in a robust decrease of extracellular Aβ levels (Figs. S1B and S1C). Although the inhibitory effect of neuronal differentiation on Aβ accumulation was observed both in the presence and absence of Ca2+ add-back conditions, it was significantly potentiated upon Ca2+ add-back conditions after 6 days of differentiation (Fig. S1C), suggesting that Aβ levels were regulated by Ca2+ influx in these conditions. Strikingly, inhibition of endogenous CALHM1 expression by RNA interference, led to a significant increase of Aβ levels in differentiated cells following Ca2+ add-back (Figs. 5C–5E). These data demonstrate that endogenous CALHM1 contributes to the inhibitory effect of neuronal differentiation on Aβ accumulation. Thus, CALHM1 expression controls APP processing by interfering with extracellular Aβ accumulation and by promoting sAPPα accumulation. The effects of CALHM1 on the regulation of Aβ and sAPPα levels are in line with its effect on [Ca2+]i, indicating that CALHM1 controls APP proteolysis in a Ca2+-dependent manner.
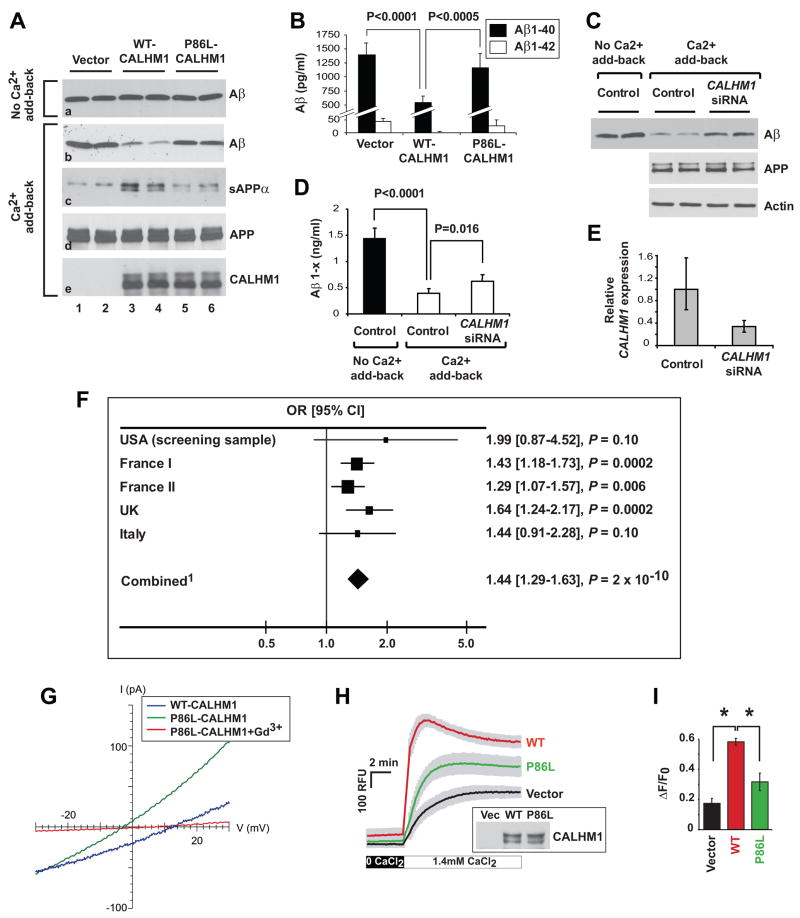
Figure 5. The CALHM1 P86L polymorphism influences Ca2+ homeostasis, APP processing, and AD risk.
(A and B) SwAPP695-N2a cells were transiently transfected with control vector or with WT or P86L-mutated Myc-CALHM1. Six and half hours post-transfection, medium was changed and cells were incubated for 60 min in the absence or presence of Ca2+ add-back conditions as described in Experimental Procedures. Total secreted Aβ and sAPPα, and cellular APP and Myc-CALHM1 were analyzed by WB (A). Secreted Aβ1-40 and Aβ1-42 were analyzed by ELISA in the presence of Ca2+ add-back conditions (n = 12; Student’s t test) (B).
(C–E) APP695-SH-SY5Y cells differentiated for 15 days with retinoic acid were treated for 3 days with Accell siRNAs directed against human CALHM1. Medium was then changed and cells were incubated for 90 min in the absence or presence of Ca2+ add-back conditions. Total secreted Aβ and cellular APP and actin were analyzed by WB (C). Total secreted Aβ1-x was quantified by ELISA (n = 3; Student’s t test) (D). CALHM1 mRNA levels were assayed by real-time qRT-PCR analysis. Histogram illustrates the mean relative CALHM1 expression ± S.D. (control, n = 4; CALHM1 siRNA, n = 3) (E).
(F) Five independent case-control studies were analyzed to assess the association of rs2986017 with AD risk. The allelic OR (T vs. C) was estimated in each population and in the combined one. 1Test for heterogeneity: χ2 = 2.84, df = 4, P = 0.59; Test for overall effect: Z = 6.06, P = 2.10−9 (Mantel-Haentzel method, fixed OR = 1.42 [1.27–1.59]).
(G) Whole-cell currents in CHO cells expressing WT- (blue trace; Cm = 13.2 pF) or P86L-CALHM1 (green trace; Cm = 22.9 pF) in same bi-ionic conditions as in Fig. 4H. P86L-CALHM1-expressing cells remained sensitive to block by 100 μM Gd3+ (red trace), but the reversal potential was shifted to more hyperpolarized voltages (Vrev = −8.9 ± 3.6 mV ; n = 6), indicating a reduced Ca2+ permeability (PCa : PCs = 2) compared with that of WT-CALHM1.
(H) Cytoplasmic Ca2+ measurements in HT-22 cells transiently transfected with control vector and WT or P86L-mutated Myc-CALHM1. Cells were treated and results analyzed as in Fig. 3A (n = 3 independent experiments). Inset, WB of the corresponding cell lysates with anti-Myc antibody. (I) Peak of [Ca2+]i measurements as in (H) expressed in Δ F/F0 (*, P<0.001; Student’s t test).
The CALHM1 P86L polymorphism is associated with LOAD and affects plasma membrane Ca2+ permeability, cytosolic Ca2+ concentration, and Aβ levels
Because CALHM1 maps to a chromosomal region associated with susceptibility for LOAD, we tested whether CALHM1 SNPs could be associated with the risk of developing the disease. Two non-synonymous SNPs were reported in databases, rs2986017 (+394 C/T; P86L) and rs17853566 (+927 C/A; H264N). We sequenced the entire CALHM1 ORF using genomic DNA from 69 individuals, including 46 autopsy-confirmed AD cases and 23 age-matched normal controls. The rs17853566 SNP was not observed in this group. However, we confirmed the presence of the rs2986017 SNP with a potential over-representation of the T allele in AD subjects (AD = 36%, controls = 22%; Table 2 and Fig. 5F; USA screening sample). We next assessed the impact of rs2986017 on the risk of developing AD in four other independent case-control populations (2,043 AD cases and 1,361 controls combined, Table 2). The T allele distribution was increased in AD cases as compared to controls in all the studies, with odds ratios (ORs) ranging from 1.29 to 1.99 (OR = 1.44, P = 2 × 10−10 in the combined population; Fig. 5F). This association was highly homogeneous among the different case-control studies (test for heterogeneity, P = 0.59, I2 = 0%). We also observed that the T allele frequency in autopsy-confirmed AD cases was similar to that observed in probable AD case populations (Table 2). In the combined population, the CT or TT genotypes were both associated with an increased risk of developing AD (respectively, ORCT vs. CC ranging from 1.18 to 1.64, OR = 1.37, P = 3 × 10−5 in the combined population and ORTT vs. CC ranging from 1.44 to 4.02, OR = 2.03, P = 2 × 10−7 in the combined population; Table 2). All these observations were independent of the APOE status (Table 2 and P for interaction = 0.26).
Table 2.
Allele and genotype distributions of the CALHM1 P86L polymorphism (rs2986017) in AD case and control populations
| Allele distribution (%) | Genotype distribution (%) | |||||
|---|---|---|---|---|---|---|
| n | C | T | CC | CT | TT | |
| USA screening sample1,2 | ||||||
| Controls | 23 | 36 (0.78) | 10 (0.22) | 14 (0.61) | 8 (0.35) | 1 (0.04) |
| Autopsied AD cases | 46 | 59 (0.64) | 33 (0.36) | 20 (0.44) | 19 (0.40) | 7 (0.16) |
| France I3,4 | ||||||
| Controls | 565 | 907 (0.80) | 223 (0.20) | 370 (0.65) | 167 (0.30) | 28 (0.05) |
| AD cases | 710 | 1051 (0.74) | 369 (0.26) | 410 (0.58) | 231 (0.32) | 69 (0.10) |
| France II5,6 | ||||||
| Controls | 483 | 716 (0.74) | 250 (0.26) | 271 (0.56) | 174 (0.36) | 38 (0.08) |
| AD cases | 645 | 888 (0.69) | 402 (0.31) | 303 (0.47) | 282 (0.44) | 60 (0.09) |
| UK7,8 | ||||||
| Controls | 205 | 320 (0.78) | 90 (0.22) | 127 (0.62) | 66 (0.32) | 12 (0.06) |
| AD cases | 365 | 504 (0.69) | 226 (0.31) | 193 (0.53) | 118 (0.32) | 54 (0.15) |
| Autopsied AD cases | 127 | 169 (0.66) | 85 (0.34) | 57 (0.45) | 55 (0.43) | 15 (0.12) |
| Italy9,10 | ||||||
| Controls | 85 | 131 (0.77) | 39 (0.23) | 52 (0.61) | 27 (0.32) | 6 (0.07) |
| AD cases | 150 | 210 (0.70) | 90 (0.30) | 74 (0.49) | 62 (0.41) | 14 (0.09) |
| Combined studies11,12 | ||||||
| Controls | 1,361 | 2,110 (0.77) | 612 (0.23) | 834 (0.61) | 442 (0.32) | 85 (0.06) |
| AD cases | 2,043 | 2,881 (0.71) | 1,205 (0.29) | 1,057 (0.52) | 767 (0.37) | 219 (0.11) |
P = 0.10;
P = ns;
P = 0.0002;
P = 0.001;
P = 0.006;
P = 0.01;
P = 0.0002;
P = 0.00002;
P = 0.10;
P = ns;
P = 2 × 10−10;
P = 7 × 10−9
OR (CT vs. CC) = 1.37, 95% CI [1.18–1.59], P = 3 × 10−5
OR (CT vs. CC) = 1.27, 95% CI [1.08–1.50], P = 0.004 adjusted for age, gender, APOE status, and center
OR (TT vs. CC) = 2.03, 95% CI [1.56–2.65], P = 2 × 10−7
OR (TT vs. CC) = 1.77, 95% CI [1.33–2.36], P = 9 × 10−5 adjusted for age, gender, APOE status, and center
ns, non-significant
It is important to note that the rs2986017 distribution was in Hardy-Weinberg equilibrium in the different control populations but not in the combined one (χ2 = 6.35, df = 1, P = 0.01; Table 2). Since we mainly used direct sequencing for genotyping, the potential for technical biases is limited. It is therefore possible that the slight deviation from the expected genotype distribution might be linked to a loss of heterozygosity by copy number variations (CNVs) in the CALHM1 gene. We found no evidence of common CNV encompassing the rs2986017 locus (see Supplemental Data). However, we cannot exclude that the deletion of a short-sized segment around rs2986017 disrupts the Hardy-Weinberg equilibrium for this marker.
Further evidence of the influence of the CALHM1 gene on the risk of developing AD comes from the observation that, in the France I population, patients bearing the TT genotype had an earlier age at onset compared with the CT and CC carriers (66.8 ± 8.5 vs. 68.7 ± 7.7 years; P = 0.05). We observed the same trend in the autopsied UK brain cohort (60.5 ± 6.4 vs. 65.2 ± 10.3 years, P = 0.12) and in the Italian population (70.6 ± 9.7 vs. 74.3 ± 8.5 years; P = 0.10), but not in the France II population (64.4 ± 8.8 vs. 64.6 ± 9.8 years; P = ns). When the AD case populations were combined, the TT genotype was still associated with an earlier age at onset compared with the CT and CC carriers (65.7 ± 8.8 vs. 67.1 ± 9.3 years; P = 0.03 adjusted for gender, APOE status, and center).
In order to gain insight into the relevance of the rs2986017 SNP to the disease, we first investigated the effect of the corresponding P86L substitution on Ca2+ permeability and [Ca2+]i. Similar to WT-CALHM1, P86L-CALHM1 expressed in CHO cells generated a Gd3+-sensitive outwardly-rectifying cation current that reversed near 0 mV with Cs+ and Na+ in the pipette and bath solutions, respectively (not shown). However, with bath Na+ replaced by NMDG and 20 mM Ca2+, the current reversed at −8.9 ± 3.6 mV (Fig. 5G; n = 6), ~17 mV hyperpolarized compared with the currents recorded in WT-CALHM1-expressing cells, resulting in a reduced Ca2+ selectivity, PCa: PCs = 2 (Fig. 5G). Thus, the P86L polymorphism significantly reduced CALHM1-induced Ca2+ permeability. In addition, we observed that the P86L mutation caused a significant inhibition of the effect of CALHM1 on [Ca2+]i (Figs. 5H and 5I), reducing its values at the peak following Ca2+ add-back, from 264 ± 48 nM to 192 ± 34 nM (Fig. S2). We then asked whether the P86L polymorphism affects the control of APP processing by CALHM1. Whereas expression of WT-CALHM1 was found to stimulate sAPPα accumulation and to repress total Aβsecretion, the ability of the P86L-mutated CALHM1 to control APP processing was greatly impaired (Fig. 5A, panels b and c), while WT- and P86L-CALHM1 were expressed at comparable levels (Fig. 5A, panel e). Consequently, compared to WT-CALHM1, P86L-CALHM1 reduced sAPPα accumulation (Fig. 5A, panel c, lanes 3–6), and led to a significant elevation of total secreted Aβ levels (Fig. 5A, panel b, lanes 3–6), including Aβ1-40 and Aβ1-42 (Fig. 5B), indicating that the P86L mutation significantly impaired the effect of CALHM1 on the extracellular accumulation of sAPPα and Aβ. Collectively, these data show that the P86L polymorphism causes a partial loss of CALHM1 function by interfering with its control of Ca2+ permeability, cytosolic Ca2+ concentration, APP metabolism, and Aβ levels.
DISCUSSION
Using a bioinformatics strategy to screen for genes predominantly expressed in the hippocampus and located in linkage regions for LOAD, we identified CALHM1 on chromosome 10 (Table 1). CALHM1 was found to encode an integral membrane glycoprotein with key characteristics of a Ca2+ channel. CALHM1 controls cytosolic Ca2+ levels, homomultimerizes, and shares important sequence similarities with the predicted selectivity filter of NMDAR (Figs. 3 and 4). Significantly, we have also demonstrated that CALHM1 contains a functionally important N residue at position 72 that aligns with the Q/R/N site of the NMDAR selectivity filter (Fig. 4). Thus, NMDAR and CALHM1 share important structural similarities at the sequence level in a region that was previously described as a critical determinant of Ca2+ selectivity and permeability in glutamate receptor ion channels (Wollmuth and Sobolevsky, 2004). The potential role of CALHM1 in ion permeability was further investigated by voltage-clamping using two different cell models. This approach demonstrated that expression of CALHM1 generates a novel constitutive Ca2+ selective cation current at the plasma membrane. Additional studies that will examine the topology of CALHM1 and more precisely the organization of the region containing the critical N72 residue, will help us to clearly identify the role of CALHM1 in ion permeation.
In the present report, we have provided compelling evidence that the rs2986017 SNP in CALHM1, which results in the P86L substitution, is associated with both an increased risk for LOAD and a significant dysregulation of Ca2+ homeostasis and APP metabolism (Table 2 and Fig. 5). Specifically, we have shown that the P86L polymorphism impairs plasma membrane Ca2+ permeability, reduces cytosolic Ca2+ levels, affects sAPPα production, and concomitantly derepresses the effect of CALHM1 on Aβ accumulation. A large body of literature supports the notion that a deranged intracellular Ca2+ signaling occurs in AD and may be involved in the deregulation of APP processing and neurodegeneration (Khachaturian, 1989; LaFerla, 2002). APP metabolism involves a complex series of events and the direct influence of Ca2+ signaling on this process is still poorly understood (LaFerla, 2002). The present work provides strong support for the Ca2+ hypothesis of AD and is also an important step towards understanding the potential pathological cross talk between Ca2+ signaling disturbances and pathways of Aβ accumulation. Moreover, the identification of CALHM1 as a key modulator of Ca2+ homeostasis will allow us to further dissect the precise mechanism by which cytosolic Ca2+ modulates APP metabolism.
Screening the human genome for genes predominantly expressed in the hippocampus successfully prioritized CALHM1 among the many genes found in LOAD loci and thus demonstrates the utility of tissue expression profiling in the identification a novel candidate genes for LOAD. Candidate genes located in LOAD regions are often considered based on their potential implication in known AD biology (e.g., IDE). The strategy used in this study can therefore complement these approaches and suggest candidates, including those of unknown function, worthy of consideration. Supporting this notion, recent data have shown that tissue expression profiles can be used to effectively prioritize candidate genes in another neurodegenerative genetic disorder (F. Campagne, Program No. 446.12, San Diego, CA: Society for Neuroscience, 2007).
In summary, we propose that CALHM1 is a pore component of a novel cerebral ion channel family and that variants in the CALHM genes may constitute robust risk factors for LOAD. These results not only provide important new insights into the pathophysiology of Ca2+ homeostasis and APP metabolism in the central nervous system but also represent a strong genetic evidence of a channelopathy contribution to AD etiology. Finally, given its cell surface ion channel properties and its restricted expression, our work further establish CALHM1 as a potentially important molecular target for an anti-amyloid therapy in AD.
EXPERIMENTAL PROCEDURES
Bioinformatics analyses
Tissue expression profiles
We generated whole genome human tissue expression profiles by using TissueInfo (http://icb.med.cornell.edu/crt/tissueinfo/index.xml), information in Ensembl (human build NCBI35), and dbEST. TissueInfo profiling was done as previously described (Campagne and Skrabanek, 2006; Skrabanek and Campagne, 2001). Whole genome profiles were filtered with InsightfulMiner 7.0 (Insightful Corp.) to extract the subset of transcripts annotated by TissueInfo as ‘specific to hippocampus’.
LOAD locus screen
The 30 transcripts predicted to be specific to the hippocampus by TissueInfo were annotated with their genomic location using EnsMart/Biomart (Kasprzyk et al., 2004) and data from Ensembl. Chromosome numbers and FISH band locations were used to identify those transcripts that matched a locus of susceptibility to AD, as documented in AlzGene (Bertram et al., 2007).
CALHM1 subcloning and mutagenesis
Human CALHM1 cDNA was obtained from ATCC. The ORF was subcloned in frame with a carboxy-terminal Myc-His or V5 tag into the pcDNA3.1 vector for expression experiments. P86L, N72G, N74A, and N140A mutations were introduced using the QuikChange II site-directed mutagenesis kit (Stratagene) and confirmed by sequencing.
Ca2+ measurements and Ca2+ add-back assays
Free cytosolic Ca2+ was measured in transiently transfected HT-22 cells plated in 6 well plates using the fluorescent Ca2+ indicator Fluo-4. Five and half hours post-transfection, cells were loaded with Fluo-4 as per manufacturer’s recommendations (Fluo-4 NW Ca2+ Assay Kit, Molecular Probes). For Ca2+ add-back assays, cells were washed with Ca2+/Mg2+-free phosphate-buffered saline (PBS) and incubated for 10 min in the absence or presence of the indicated inhibitors in Ca2+/Mg2+-free Hanks’ balanced salt solution (HBSS), supplemented with 20 mM HEPES buffer, 0.5 mM MgCl2, and 0.4 mM MgSO4. Ca2+ was then added back to a final concentration of 1.4 mM. Fluorescence measurements were carried out at room temperature using a Tecan GENios Pro plate reader at 485 nm excitation and 535 nm emission. Cells were then washed with PBS and analyzed by WB.
WB and ELISA
For APP processing analysis, APP-transfected cells were challenged with Ca2+ add-back conditions, as described above. Conditioned medium and cells were harvested after the indicated times of incubation at 37°C in a humidified 5% CO2 incubator. Secreted Aβ WB was performed as previously described (Marambaud et al., 2005). WB of sAPPα and APP was performed using 6E10 (Signet) and LN27 (Zymed) antibodies, respectively. For ELISA, secreted Aβ1-40, Aβ1-42, and Aβ1-x levels were quantified as per manufacturer’s recommendations (IBL-America). ELISA plates were read on a Tecan GENios Pro reader at 450 nm.
CALHM1 sequencing
CALHM1 ORF was resequenced using genomic DNA preparations obtained from 23 control individuals (age at study = 71.9 ± 16.0 years, 43% male) and 46 autopsy-confirmed AD patients (age at study = 77.8 ± 8.1 years, 55% male). Subjects and genomic DNA preparations were described elsewhere (Conrad et al., 2002). ORFs were amplified by PCR using primers described in Supplemental Data and PCR products were sequenced by GeneWiz.
SNP analyses
Populations
See Supplemental Data.
Genotyping
In the France I population, the P86L genotype was determined by genomic DNA amplification. The genotyping of 176 individuals was checked by direct sequencing. Only two discrepancies were observed between CC and CT genotypes. In the UK, France II, and Italy populations, the P86L genotype was entirely determined by direct sequencing. See Supplemental Data for experimental details.
Statistical analyses
Univariate analysis was performed using Pearson’s χ2 test. The review manager software release 5.0 (http://www.cc-ims.net/RevMan/) was used to test for heterogeneity between the different case-control studies and to estimate the overall effect (Mantel-Haentzel fixed odds ratio; Fig. 5F). For multivariate analysis, SAS software release 8.02 was used (SAS Institute, Cary, NC) and homogeneity between populations was tested using Breslow-day computation (Breslow et al., 1978). The association of the P86L polymorphism with the risk of developing AD was assessed by a multiple logistic regression model adjusted for age, gender, APOE status, and center. Interactions between age, gender, or APOE and the P86L polymorphism were tested by logistic regression. No significant statistical interactions were detected. Finally, the potential impact of the P86L polymorphism on age at onset was assessed using a general linear model adjusted for gender, APOE status, and center.
Electrophysiology
Xenopus oocyte plasma membrane conductance was recorded 24–72 hours after cRNA injection. Single oocytes were placed in a 1 ml chamber containing LCa96 solution. In some studies, Na+ was replaced with NMDG. Conventional two-electrode voltage clamp was performed. Pulse+PulseFit software (HEKA Elektronik) was used to ramp the applied transmembrane potential (Vm) at 10-s intervals from −80 to 80 mV at 16 mV/s, and acquire data. Vm was clamped at the resting membrane potential between voltage ramps. Transmembrane current (I) and Vm were digitized at 200 Hz and recorded directly to hard disk. To determine the reversal potential Vrev, a fifth-order polynomial was fitted to the raw I-Vm data acquired during each voltage ramp, using macros developed in Igor Pro software (WaveMetrics Inc.). Whole cell recordings of CHO cells were performed with 2–5 MΩ pipettes using an Axopatch 200-B amplifier (Axon Instr. Inc.). Current-voltage (I–V) relationships were acquired in response to voltage ramps (±100 mV, 2 s duration). The recording chamber was continuously perfused with bath solution (2 ml/min). See Supplemental Data for experimental details.
Data analyzed using macros developed in Igor Pro were corrected for leakage currents (determined from a linear fit of the currents recorded at −80–−100 mV in the presence of 100 μM GdCl3 extrapolated over the entire ramping voltage domain) and for measured junction potentials. I/V curves presented in Figures have not been corrected for leakage current. Calculated values are given as means ± SEM.
Accession numbers
Human CALHM1 (previously annotated FAM26C) has Ensembl release 43 accession code ENSG00000185933 (Uniprot Q8IU99). CALHM3 (ENSG00000183128). CALHM2 (ENSG00000138172). See Supplemental Data for additional accession numbers.
See also Supplemental Data for additional Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. G. Thinakaran (University of Chicago, Chicago, IL) for kindly providing us with SwAPP695-N2a cells; Dr. D. Schubert (Salk Institute, La Jolla, CA) for HT-22 cells; Dr. B. De Strooper (K.U. Leuven and VIB, Leuven, Belgium) for WT and PS-deficient fibroblasts; Dr. N.K. Robakis (Mount Sinai School of Medicine, New York, NY) for 33B10 antibody; Dr L. Buée (INSERM U815, Lille, France) for APP695-SH-SY5Y cells; Dr. D. Mak (University of Pennsylvania, Philadelphia, PA) for assistance with electrophysiology data analysis; Dr. King-Ho Cheung for assistance with the fura-2 imaging experiments; and Dr. A. Chan (North Shore-LIJ, Manhasset, NY) for assistance with microscopy studies. The authors are grateful to Dr. C. Clancy (Weill Medical College of Cornell University, New York, NY), and Drs. M. Symons and R. Ruggieri (North Shore-LIJ, Manhasset, NY) for helpful comments on the manuscript. This work was supported by the Alzheimer’s Association (P.M.) and the NIH grant R01 MH059937 (J.K.F.). F.C. acknowledges support from the resources of the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine and the David A. Cofrin Center for Biomedical Information at Weill Cornell.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- Bertram L, Hsiao M, Lange C, Blacker D, Tanzi RE. Single-nucleotide polymorphism rs498055 on chromosome 10q24 is not associated with Alzheimer disease in two independent family samples. Am J Hum Genet. 2006;79:180–183. doi: 10.1086/505032. author reply 183–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE, Halvorsen KT, Prentice RL, Sabai C. Estimation of multiple relative risk functions in matched case-control studies. Am J Epidemiol. 1978;108:299–307. doi: 10.1093/oxfordjournals.aje.a112623. [DOI] [PubMed] [Google Scholar]
- Campagne F, Skrabanek L. Mining expressed sequence tags identifies cancer markers of clinical interest. BMC Bioinformatics. 2006;7:481. doi: 10.1186/1471-2105-7-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Vianna C, Freeman M, Davies P. A polymorphic gene nested within an intron of the tau gene: implications for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2002;99:7751–7756. doi: 10.1073/pnas.112194599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon MJ, Mosconi L, Blennow K, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Tsui W, Saint Louis LA, Sobanska L, et al. Imaging and CSF studies in the preclinical diagnosis of Alzheimer’s disease. Ann N Y Acad Sci. 2007;1097:114–145. doi: 10.1196/annals.1379.012. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, Kauwe JS, Maxwell TJ, Cherny S, Doil L, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, et al. A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Kennedy JL, Farrer LA, Andreasen NC, Mayeux R, St George-Hyslop P. The genetics of adult-onset neuropsychiatric disease: complexities and conundra? Science. 2003;302:822–826. doi: 10.1126/science.1092132. [DOI] [PubMed] [Google Scholar]
- Khachaturian ZS. Calcium, membranes, aging, and Alzheimer’s disease. Introduction and overview. Ann N Y Acad Sci. 1989;568:1–4. doi: 10.1111/j.1749-6632.1989.tb12485.x. [DOI] [PubMed] [Google Scholar]
- Kuwano R, Miyashita A, Arai H, Asada T, Imagawa M, Shoji M, Higuchi S, Urakami K, Kakita A, Takahashi H, et al. Dynamin-binding protein gene on chromosome 10q is associated with late-onset Alzheimer’s disease. Hum Mol Genet. 2006;15:2170–2182. doi: 10.1093/hmg/ddl142. [DOI] [PubMed] [Google Scholar]
- LaFerla FM. Calcium dyshomeostasis and intracellular signalling in Alzheimer’s disease. Nat Rev Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Amouyel P. Genetic heterogeneity of Alzheimer’s disease: Complexity and advances. Psychoneuroendocrinology. 2007 doi: 10.1016/j.psyneuen.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Robakis NK. Genetic and molecular aspects of Alzheimer’s disease shed light on new mechanisms of transcriptional regulation. Genes Brain Behav. 2005;4:134–146. doi: 10.1111/j.1601-183X.2005.00086.x. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Zhao H, Davies P. Resveratrol promotes clearance of Alzheimer’s disease amyloid-beta peptides. J Biol Chem. 2005;280:37377–37382. doi: 10.1074/jbc.M508246200. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minster RL, DeKosky ST, Kamboh MI. Lack of association of two chromosome 10q24 SNPs with Alzheimer’s disease. Neurosci Lett. 2006;408:170–172. doi: 10.1016/j.neulet.2006.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, et al. Susceptibility locus for Alzheimer’s disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- Pastor P, Goate AM. Molecular genetics of Alzheimer’s disease. Curr Psychiatry Rep. 2004;6:125–133. doi: 10.1007/s11920-004-0052-6. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Shibuya I, Douglas WW. Calcium channels in rat melanotrophs are permeable to manganese, cobalt, cadmium, and lanthanum, but not to nickel: evidence provided by fluorescence changes in fura-2-loaded cells. Endocrinology. 1992;131:1936–1941. doi: 10.1210/endo.131.4.1327724. [DOI] [PubMed] [Google Scholar]
- Skrabanek L, Campagne F. TissueInfo: high-throughput identification of tissue expression profiles and specificity. Nucleic Acids Res. 2001;29:E102–102. doi: 10.1093/nar/29.21.e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer’s disease-linked mutations. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilquet V, De Strooper B. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Wollmuth LP, Sobolevsky AI. Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 2004;27:321–328. doi: 10.1016/j.tins.2004.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.