Abstract
Protection afforded by HIV Tat-based vaccines has differed in Indian rhesus and Mauritian cynomolgus macaques. We evaluated native Tat and Ad-HIVtat priming/Tat-boosting regimens in both species. Both vaccines were immunogenic. Only the Ad-tat regimen modestly reduced acute viremia in rhesus macaques after SHIV89.6P challenge. Confounding variables uncovered in Mauritian macaques included significant associations of susceptibility to infection with MHC class IB and class II H2 and H5 haplotypes, and resistance to infection with class IB haplotypes H3 and H6. Although protection here was limited, Tat-based vaccines incorporating other HIV components have shown greater efficacy. Combination strategies should be further explored.
1. Introduction
The HIV pandemic is a major and urgent public health concern. At least 40 million people worldwide are infected with the virus. Thus, development of an effective vaccine continues to be a critical need. Among target HIV antigens for vaccine development is Tat, the potent transcriptional transactivator of HIV gene expression. Tat is produced early after infection [1, 2] and is indispensable for viral replication, transmission, and AIDS pathogenesis [3-6]. Release of Tat from infected cells and its uptake by infected and uninfected cells is critical to the biology of the virus [5, 7-10]. In infected cells, Tat promotes viral replication or transactivates the replication of tat-defective or latent proviruses [11]. In uninfected cells, Tat can modulate cellular gene expression [3, 12, 13], up-regulate HIV co-receptors [14, 15], and induce or inhibit apoptosis [16, 17]. Early inhibition of Tat function should contribute to control of viral replication and slowing of AIDS progression. In fact, Tat-specific CTLs are associated with control of virus replication early in infection [18] and both anti-Tat antibody and Tat-specific CTLs have been correlated with reduced viremia and slow progression to AIDS [19-21].
Conflicting results have been reported following vaccination with Tat-based vaccines. Immunizations of rhesus macaques with Tat protein, vectored tat, Tat toxoid or Tat peptides have elicited no protection [22, 23] or partial protection [24, 25] against SIVmac239, SHIV33, or SHIV89.6P challenges, while immunizations of cynomolgus macaques with native Tat protein or DNA encoding tat have shown strong, long-term protective efficacy against SHIV89.6P [26-29].These contrasting results might reflect species differences with regard to immunogenicity or host resistance factors, or differences in vaccine characteristics, vaccination routes, delivery systems, timing of immunizations or challenge protocols. Here we addressed these issues, eliminating the latter variables by conducting two identical immunization and challenge protocols in Indian rhesus and Mauritian cynomolgus macaques. The first approach replicated previous studies in cynomolgus macaques in which multiple immunizations with native HIV Tat protein were shown to elicit long-term protection against SHIV89.6P in Mauritian cynomolgus macaques [26, 29]. The second approach was based on a replication-competent Ad-recombinant vaccine strategy [30]. These replicating vaccines have been shown to elicit better cellular immune responses and prime higher titered antibodies, including functional antibodies, compared to replication-defective Ad-recombinants encoding the same HIV genes [31, 32]. When combined with envelope subunit boosting, the vaccine strategy has shown potent protection against virulent SIVmac251 challenge [33] and durable protective efficacy with no intervening boost [34].
Studies using both vaccine regimens were conducted in the two non-human primate models, and immunogenicity and protective outcomes following challenge with SHIV89.6P were compared. As the entire repertoire of MHC alleles can now be predicted for essentially all Mauritian cynomolgus macaques [35-37], we also determined the MHC genotypes of the study animals. These investigations revealed a new association of Mauritian MHC haplotype and susceptibility/resistance to SHIV89.6P infection. The association of particular MHC alleles with resistance of rhesus macaques to SIV and SHIV infection is well established [38-43]. Our results here extend the phenomenon to cynomolgus macaques of Mauritian origin. The significantly higher peak Tat-specific T cell proliferative responses seen in vaccinated macaques with the resistant haplotypes prior to challenge suggest cellular immunity should be further explored as a possible mechanism for the observed resistance. Results of the haplotype analysis and vaccine evaluations, together with reports showing that vaccines targeting Tat in combination with other viral proteins elicit good protective efficacy in non-human primates [44-46], suggest that HIV Tat vaccines might be best exploited in combination with other viral antigens.
2. Materials and methods
2.1. Vaccines
Escherichia coli- expressed HIVIIIB Tat protein (Advanced Bioscience Laboratories, Inc., (ABL) Kensington, MD), greater than 95% pure and retaining full biological activity [7], was lyophilized and stored at -70°C prior to use. To retain activity, Tat for immunizations was freshly reconstituted at 4 μg/μl in degassed phosphate buffered saline (PBS; Invitrogen) containing 0.1% BSA (Sigma-Aldrich) and 0.1mM dithiothreitol (DTT), capped, covered with foil, and kept on ice. All plasticware was pre-rinsed with PBS-BSA buffer. For subcutaneous administrations, Tat was diluted in cold PBS, mixed with an equal volume of alum and inoculated (10 μg/500 μl final concentration). For intradermal administrations, Tat was diluted in PBS to a concentration of 6 μg/250μl. Tat-immune stimulating complexes (ISCOMS) [47] were prepared by adding 200 μl of the ISCOM matrix to 50 μg lyophilized Tat, mixing, and incubating with slow stirring at room temperature for 30 minutes. The mixture was cooled on ice, diluted with PBS to 600 μl and administered (200 μl/dose) intramuscularly as soon after preparation as possible.
Replication-competent Ad5hr-HIVtat has been described [48]. Control immunogens included an empty Ad5hr E3-deleted vector, alum, and ISCOM matrix.
2.2. Animals, immunization and sample collection
Indian rhesus (Macaca mulatta) and Mauritian cynomolgus (Macaca fascicularis) macaques were maintained according to guidelines and protocols of the Animal Care and Use Committee, Washington National Primate Research Center, University of Washington (Seattle, WA). Identical immunization regimens were followed for both cynomolgus and rhesus macaques (Table 1). The schedule of Tat protein immunizations was published previously [26]. The experimental cynomolgus immunization groups contained 9 animals each, however, 2 macaques in the Ad5hr-HIVtat group died from anesthesia complications prior to challenge. Pre-challenge data are reported for all nine macaques and post-challenge data for the remaining seven. Control cynomolgus groups contained 3 macaques each. All cynomolgus macaques were males. The rhesus experimental immunization groups contained 8 macaques each; 3 macaques were in each control group. Five of eight macaques in the Tat protein and four of eight macaques in the Ad5hr-HIVtat groups were females. Control groups each contained two females and one male. All rhesus macaques were negative for Mamu A*01, but two were positive for Mamu B*17 (A02005 in the Ad5hr-HIVtat group and A02023 in the adjuvant control group).
Table 1.
Immunization and challenge protocol in cynomolgus and rhesus macaquesa.
| Week | Tat proteinb | Ad-Tatc | Ad vector control | Adjuvant control |
|---|---|---|---|---|
| 0 | Tat protein (SC/ID) | Ad-Tat (IN) | AdΔE3 (IN) | Alum (SC) |
| 2 | Tat protein (SC/ID) | Alum (SC) | ||
| 6 | Tat protein (SC/ID) | Alum (SC) | ||
| 11 | Tat protein (SC/ID) | Alum (SC) | ||
| 12 | Ad-Tat (IT) | AdΔE3 (IT) | ||
| 15 | Tat protein (SC/ID) | Alum (SC) | ||
| 21 | Tat protein (SC/ID) | Alum (SC) | ||
| 24 | Tat protein (SC) | Alum (SC) | ||
| 28 | Tat protein (SC/ID) | Alum (SC) | ||
| 32 | Tat protein (SC/ID) | Alum (SC) | ||
| 36 | Tat +ISCOM (IM) | Tat protein (SC) | Alum (SC) | ISCOM (IM) |
| 50 | SHIV89.6P (IV)d | SHIV89.6P (IV) | SHIV89.6P (IV) | SHIV89.6P (IV) |
Cynomolgus and rhesus macaques in each immunization group are listed in Fig. 2 and 3, respectively.
HIVIIIB Tat protein: 10 μg given subcutaneously (SC) in alum + 6 μg given intradermally (ID) without adjuvant. Last immunization was 16 μg given with ISCOM intramuscularly (IM).
Ad-recombinant dose: 5X108 pfu each in PBS administered IN: intranasally; IT: intratracheally.
Intravenous (IV) challenge with SHIV89.6Pcyn243,15 MID50, for cynomolgus; SHIV89.6P, 30 MID50, for rhesus.
Peripheral blood mononuclear cells (PBMCs), collected before, during, and after immunization, were purified using lymphocyte separation medium (ICN Pharmaceuticals, Inc.) for rhesus and Ficoll-PaqueTM PLUS (Amersham Biosciences) for cynomolgus samples and used fresh for immunological assays. Plasma and sera were aliquoted and stored at -70°C until use.
2.3. Challenge virus
All macaques were challenged intravenously (IV) at week 50 with SHIV89.6P. Rhesus macaques received 30 MID50 of a SHIV89.6P stock [49] kindly provided by Drs. Norman Letvin and Keith Reimann, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA. Cynomolgus monkeys received 15 MID50 of a SHIV89.6P stock derived from a cynomolgus macaque inoculated with the original SHIV89.6P rhesus stock and termed SHIV89.6Pcy243 [26].
2.4. ELISPOT Assay
PBMCs secreting gamma interferon (IFN-γ) in response to overnight stimulation with a single pool of Tat 15-mers (1 μg/ml each) were enumerated using ELISPOT kits (U-Cytech, Utrecht, The Netherlands) as described [50]. Assays were performed in triplicate; background spots in wells containing only medium (RPMI 1640 containing 5% fetal calf serum, 1 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin) were subtracted. A positive response is defined as at least 10 spot forming cells (SFC) per million PBMC after subtraction of the mean SFC of control macaques plus two standard deviations at each time point evaluated.
2.5. T-Cell Proliferation Assay
Oxidized Tat was used for proliferative assays. Lyophilized Tat was reconstituted with degassed buffer as described above, exposed to light and air for 2 hours at room temperature, capped and exposed to light overnight, then aliquoted and stored at -70°C until use. Freshly isolated PBMC (105 cells/well) were cultured for five days in triplicate in 200 μl of RPMI 1640 medium containing 10% fetal calf serum (FCS), 1mM L-glutamine and 100 U/ml penicillin, 100μg/ml streptomycin (R-10) with 1μg of oxidized Tat /well at 37°C. On the fifth day, cells were pulsed overnight with 3H-thymidine (1μCi/well), harvested and counted as described [50]. Stimulation indices (SI) were calculated by dividing mean counts per minute (cpm) with Tat by mean cpm with R-10 plus degassed buffer. A positive response is defined as an SI of 2 or more after subtracting the mean SI of control animals + 2 standard deviations at each time point tested.
2.6. Antibody Assay
Serum binding antibodies to HIV Tat were determined by enzyme–linked immunosorbent assay (ELISA) [51]. Antibody titer was defined as the reciprocal of the serum dilution at which the absorbance of the test serum was twice that of a serum from a naïve macaque diluted 1:50.
2.7. Viral RNA and proviral DNA detection
Viral RNA in plasma was determined by nucleic acid sequence-based amplification (NASBA) as described [52]. Sensitivity of the NASBA assay is less than 2000 viral copies/input volume. A real time assay with a sensitivity of <50 copies/input volume [34] was used when plasma samples exhibited viral loads below the NASBA sensitivity level. For proviral DNA analysis, cellular DNA was purified using QIAamp DNA mini kits (QIAGEN Inc., Valencia, CA USA). SIV gag DNA was detected by nested PCR and confirmed by Southern blotting and hybridization to a 32P-labeled SIV gag probe as described [51]. The first PCR reaction consisted of 500 ng of purified DNA, 25 μl of 2X ready mix Go TaqR Green Master Mix (Promega, Madison, WI), 10 pmoles of each outer SIV gag primer, and distilled water to a final 50μl reaction volume. The second PCR reaction used a 10 μl aliquot of the first PCR product as DNA template and the inner primer pair. Thirty amplification cycles (1 minute denaturation at 94°C, 1 minute of primer annealing at 58°C, and 1.25 minute of extension at 72°C) were performed for each reaction followed by a final primer extension of 7 minutes. Positive controls (plasmid pCMV SIV-gag DX and a proviral-positive DNA sample) and negative controls (distilled water and DNA extracted from pre-challenge PBMC samples) were run concurrently with test samples.
2.8. MHC Microsatellite Haplotype Analysis
Microsatellite PCR assays were performed with genomic DNAs and a panel of 16 markers spanning the 5-Mb MHC region essentially as described previously [37, 53]. MHC haplotype predictions were generated based on the microsatellite profiles, inferring alleles for the class I and class II regions based on previously established haplotype-allele associations [36, 37].
2.9. Mafa-B*510101 sequence-specific PCR
PBMC RNA was isolated with a MagNA Pure LC RNA Isolation kit (Roche Applied Science, Indianapolis, IN). Complementary DNA (cDNA) was synthesized using a Superscript™ III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Real-time PCR was performed on a LightCyler 480 (Roche Applied Science, Indianapolis, IN) with cDNA templates and SYBR Green PCR Master Mix (Applied Biosytems, Foster City, CA) in a 10ul final volume. Amplification of a 140 bp cDNA-PCR product from the Mafa-B*510101 allele was achieved with the primer pair Mafa-B*510101-SPPF, 5’-CAAGGACGCCGCACAGT, and Mafa-B*510101-SPPR, 5’- GATACCCGCGGAGGAGGT. The thermal cycling conditions used were: activation at 95°C for 10 min, amplification between 60°C for 30 sec and 95°C for 30 sec x 40 cycles, and a final denaturation between 60°C and 95°C (30 acquisitions/sec) to generate melting profiles (PCR product Tm = 87.1 °C).
2.10. Statistical Analysis
Differences in peak and chronic phase viremia between immunization groups, species, and CD4 counts among the macaques pre- and post-challenge, were evaluated using the exact two tailed Wilcoxon rank sum test. CD4 decline between groups of cynomolgus macaques was compared using the two tailed Student’s t test. The Cochran-Armitage trend test was used for the comparisons of antibody titers and analysis of MHC haplotype distribution among macaques grouped by chronic viremia outcomes. Analysis of peak viremia levels, MHC haplotypes, and proliferative responses used the Kruskal-Wallis test and the Wilcoxon rank sum test.
3. Results
3.1. Pre-challenge immune responses to Tat
Strong humoral immunity was elicited in both species by both vaccine regimens. In cynomolgus monkeys (Fig. 1A), anti-Tat titers first appeared in the Tat protein group at week 4. By week 8, the titers were significantly higher than those in controls, and the difference persisted until the time of challenge (p = 0.0045). In contrast, Tat–specific antibody in the Ad5hr-HIVtat group first appeared after the second Ad5hr-HIVtat immunization and became significantly increased above control levels after the second Tat protein boost at week 36 (p = 0.018). This difference was maintained until week 48. The Tat protein group had significantly higher Tat-specific binding titers than the Ad5hr-HIVtat group from weeks 14 through 34 (p<0.0004). This higher titer was also observed at week 48, two weeks prior to challenge (p = 0.0056; Fig. 1A).
Figure 1. Tat-specific binding antibody induced in cynomolgus and rhesus macaques by vaccination prior to challenge.
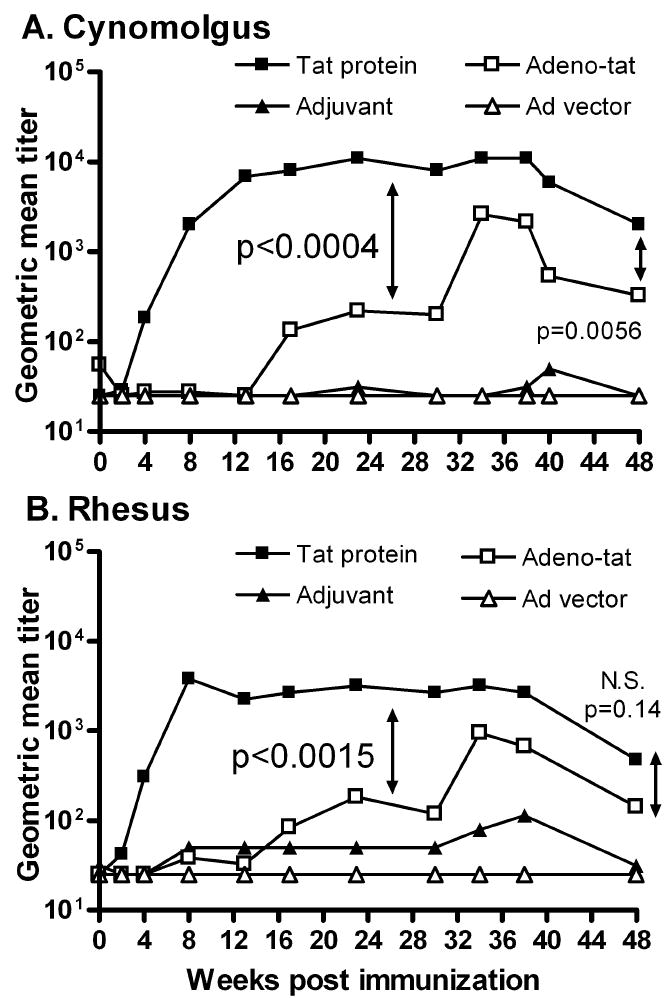
Geometric mean titers for each immunization group are shown for Mauritian cynomolgus (Panel A) and Indian rhesus (Panel B) macaques. P values shown in panels A and B represent the significant difference in antibody titer between Tat protein and Ad5hr-HIVtat groups for weeks 14-34 (panel A) and for weeks 4-30 (panel B). P values at week 48 represent differences prior to challenge at week 50.
A similar pattern was observed in the rhesus macaques (Fig. 1B). The Tat protein group showed significantly higher Tat antibody titers compared to the adjuvant controls beginning at week 4 until the time of challenge (weeks 4-38, p = 0.0061; week 48, p = 0.012) The Ad5hr-HIVtat group first exhibited elevated titers compared to controls at week 34, and the difference was maintained until week 48 (p = 0.024). Compared to the Ad5hr-HIVtat group, the Tat protein group had consistently elevated anti-Tat titers (weeks 4 – 30, p<0.0015). The higher titers were maintained, although the significant difference disappeared by week 48, p = 0.14; Fig. 1B).
Overall, the Tat protein vaccine elicited higher anti-Tat titers in cynomolgus compared to rhesus macaques, beginning at week 14 and over the immunization course (p values from 0.039 to 0.0023). The difference at week 48 prior to challenge was significant at the p = 0.018 level. The Ad5hr-HIVtat regimen also elicited slightly higher anti-Tat titers in cynomolgus compared to rhesus macaques, reaching a significant difference at week 48 prior to challenge (p = 0.037).
Cellular immunity was elicited by both vaccine regimens in both animal models, although less potent relative to the induced humoral immunity (Table 2). Numbers of Tat-specific IFN-γ secreting cells induced were low in both species, as was the frequency of positive responses. The Ad5hr-HIVtat regimen elicited two- to five-fold higher mean peak ELISPOT responses than the Tat protein regimen in the cynomolgus and rhesus models respectively, but similar percentages of responding macaques were seen in both species. Most macaques of both species also exhibited T cell proliferative responses induced by both vaccine regimens, although with a low frequency similar to the ELISPOT results (Table 2). Overall, neither non-human primate model displayed a consistently better cellular immune response to the vaccines.
Table 2.
Pre-challenge Tat–specific cellular immune responses.
| ELISPOT | Proliferation | ||||||
|---|---|---|---|---|---|---|---|
| Macaques | Vaccination group | Peaka (SFC) |
Respondersb (%) |
Frequencyc (Mean %) |
Peaka (SI) |
Respondersb (%) |
Frequencyc (%) |
| Cynomolgus | |||||||
| Tat protein | 26
(0 -77) |
67 | 7 | 5.2
(<2 – 15.9) |
78 | 12 | |
| Adeno-Tat | 51
(0 – 237) |
78 | 14 | 4.7
(<2 – 12.4) |
67 | 10 | |
| Rhesus | |||||||
| Tat protein | 22
(0 - 93) |
38 | 6 | 6.8
(<2 – 14.4) |
88 | 34 | |
| Adeno-Tat | 109
(0 - 260) |
88 | 22 | 4.7
(<2 – 13.2) |
88 | 13 | |
Mean (range) of peak positive responses, weeks 2-50.
Percent of macaques exhibiting a positive response, weeks 2-50.
Mean of frequency of positive responses for each macaque over the 8 to 10 time points evaluated.
3.2 Outcome of SHIV89.6P challenge
Following SHIV89.6Pcy243 challenge, all cynomolgus macaques became infected (Fig. 2A-D). The majority of animals exhibited high peak viremia followed by a decline in the chronic phase of infection. However, one adjuvant control (04020, Fig. 2B) and two macaques each in the Tat protein (04016, 04017, Fig. 2A) and the Ad5hr-HIVtat groups (04022, 04023, Fig. 2C) never exhibited detectable viremia, although PBMC from these five macaques were positive for SIV gag proviral DNA at one or more time points (Fig. 2A-C). The aviremia in adjuvant control 04020 and the rapid viremia control in Ad5hr control 04026 (Fig. 2D) made it impossible to attribute aviremia in the immunized macaques to the vaccine or a host control mechanism.
Figure 2. Level of plasma viremia following intravenous challenge of Mauritian cynomolgus macaques with SHIV89.6P.
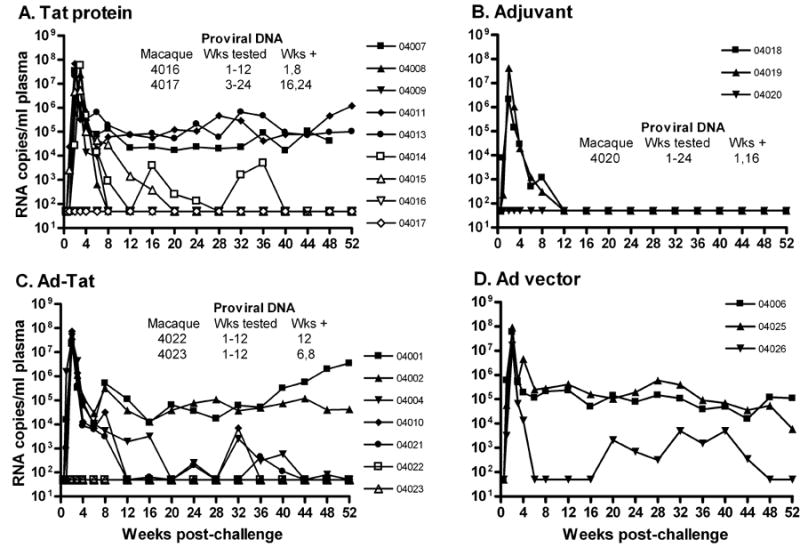
Panels A-D show viral loads for individual macaques in each immunization group. Proviral DNA (results shown in panels A,B,C) was evaluated on available PBMC samples collected post-challenge at weeks.1, 2, 3, 4, 6, 8, and monthly thereafter. PBMC obtained at 6 time points were assayed for each macaque where weeks tested are listed as 1-12. PBMC at 7 and 9 time points were assayed for each macaque where weeks tested are listed as 3-24 and 1-24, respectively.
Following SHIV89.6P challenge, all rhesus macaques became productively infected (Fig. 3A-D). In both the Tat protein group and adjuvant controls high peak viral burdens declined to variable set points. Two controls and one immunized macaque rapidly controlled viremia to undetectable levels (Fig. 3A, B), but overall no protection was observed. However, the Ad5hr-HIVtat regimen resulted in significant reduction in geometric mean peak viremia compared to controls (4 X 107 versus 3 X 108 SIV RNA copies/ml plasma; p = 0.024). This modest protective effect was not maintained in the chronic phase (Fig. 3C, D).
Figure 3. Level of plasma viremia following intravenous challenge of Indian rhesus macaques with SHIV89.6P.
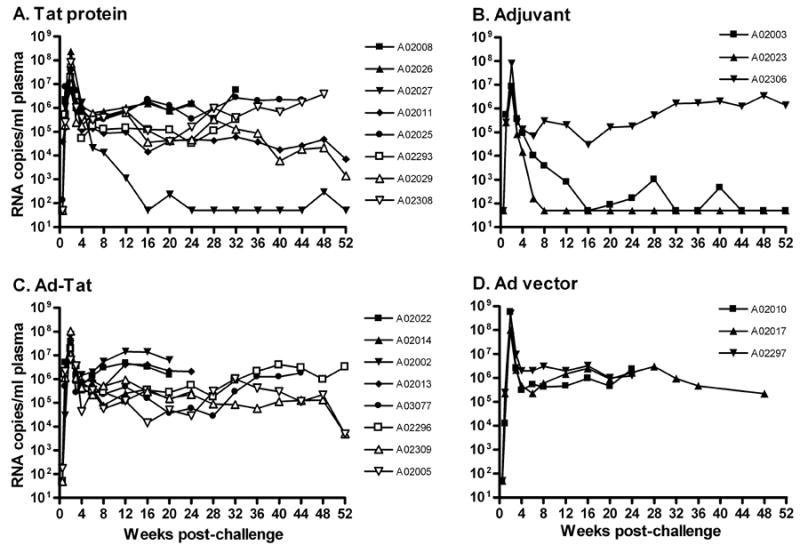
Panels A-D show viral loads for individual macaques in each immunization group.
The CD4+ T cell counts in the two species reflected the viral burdens (data not shown). No differences were observed between the counts of immunized macaques of either species and their respective control groups. Overall, the cynomolgus macaques maintained higher CD4 counts over weeks 3-18 post-challenge compared to the rhesus macaques, whether they were immunized with Tat protein (p = 0.0025) or the Ad5hr-HIVtat regimen (p = 0.0037).
3.3 Analysis of MHC haplotypes
The characterization of six common MHC haplotypes in feral Mauritian cynomolgus macaques [37] allowed determination of MHC genotypes for the cynomolgus macaques studied here. Microsatellite allele profiles were used to infer haplotypes spanning the 5–Mb MHC region and deduce complete genotypes for class I and class II alleles (Fig. 4). All MHC haplotypes observed were consistent with those previously reported for feral Mauritian-origin cynomolgous macaques except for two macaques (04022 and 04019) that shared a novel microsatellite profile for the MHC class I region of one of the paired haplotypes. Additional genotyping of several hundred feral Mauritian cynomolgus macaques has confirmed that this rare MHC haplotype, designated H7, is present in approximately 1% of the feral Mauritian population (RWW, JAK & DHO, unpublished results). In several cases, MHC alleles for two alternative haplotypes could not be distinguished with current microsatellite markers for ambiguous chromosomal regions flanking recombination breakpoints (hatched areas in Fig. 4). However, two of the four ambiguous class IB regions were tentatively resolved using a sequence-specific cDNA/PCR assay for Mafa-B*510101, an allele encoded on the H3 haplotype. cDNA from animal 04020 contained the Mafa-B*510101 allele but this allele was not detected in animal 04010, suggesting the presence and absence of the class IB H3 haplotype, respectively (Fig. 4).
Figure 4. Susceptibility and resistance to SHIV89.6P infection correlates with MHC class IB haplotypes of Mauritian cynomolgus macaques.
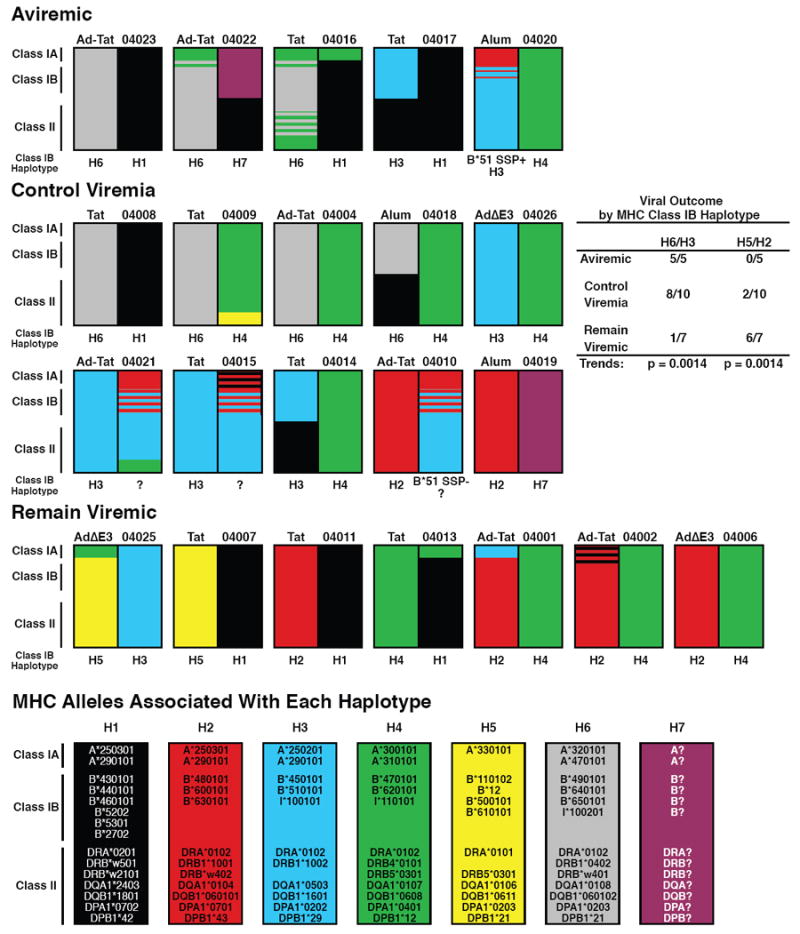
Viral outcomes were defined as aviremic: never exhibiting a plasma viral load greater than 50 SHIV89.6P RNA copies per ml over the entire 52 week observation period; control viremia: viremia level during the chronic phase of infection dropped to an undetectable level (<50 copies/ml) on two or more occasions; remain viremic: plasma viremia persisted above detectable levels over the entire observation period.
An intriguing MHC genotype/phenotype correlation emerged when the animals were grouped according to virological outcome after SHIV89.6P challenge rather than their immunized or control status. Three clear categories were observed: A) those that never exhibited detectable viremia, B) those that exhibited acute viremic but controlled chronic viremia to below 50 SHIV89.6P RNA copies/ml, and C) those that maintained high chronic viremia. All seven cynomolgus macaques possessing the H6 class 1B MHC haplotype remained aviremic or controlled chronic viremia (Fig. 4). Likewise, six of seven animals that possessed the H3 1B haplotype were aviremic or controlled viremia, while only 1 remained viremic. Thus, 100% of macaques that remained aviremic and 80% of those that controlled viremia possessed either an H3 or H6 IB haplotype, while only 14 % of macaques that remained viremic had either of these haplotypes (significant trend for resistance, p = 0.0014; Fig. 4). Conversely, macaques that possessed either the H2 or H5 class IB haplotype appeared more susceptible to SHIV89.6P infection: 0 of 5 were aviremic and 2 of 10 controlled viremia, while 6 of the 7 remaining viremic animals carried one of these two haplotypes (trend analysis: p = 0.0014; Fig. 4).
These correlations of MHC haplotype with susceptibility/resistance to SHIV89.6P challenge were supported by longitudinal analysis of viral loads (Fig. 5A). Macaques with the H2/H5 haplotype exhibited peak viral loads significantly elevated 2 to 3 logs compared to H1/H4 and H3/H6 macaques respectively (peak acute viremia weeks 1-4 for H2/H5 macaques versus all others: p = 0.0006). The peak acute viral load for H3/H6 macaques vs all others has a p value of 0.015. These differences persisted during chronic infection (weeks 8 – 52) where H2/H5 macaques maintained higher viral loads than the others (p = 0.013) and H3/H6 macaques exhibited lower viral loads than the others (p = 0.023).
Figure 5. Viral loads of Mauritian cynomolgus macaques with H2/H5 and H3/H6 MHC class IB haplotypes are significantly correlated with susceptibility and resistance to SHIV89.6P infection respectively.
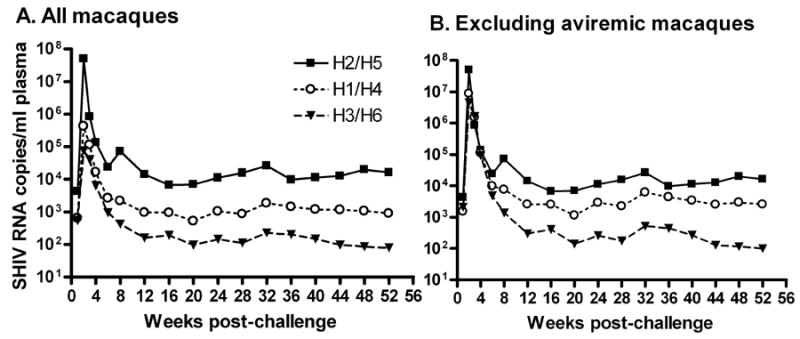
Geometric mean viral loads are plotted for macaques grouped according to the indicated haplotypes.
An effect of the MHC class IB haplotypes on pathogenesis was seen by a greater CD4 decline in animals that became productively infected after SHIV89.6P challenge. Cynomolgus macaques with H3 and H6 haplotypes had a smaller drop in CD4 counts over weeks 0 and 28 (621 ± 93) compared to non-H3 and H6 macaques (911 ± 69; p = 0.027). The CD4 decline of H2 and H5 macaques (904 ± 79) vs all others (655 ± 90) was not statistically significant (p = 0.067).
These correlations suggested that the H2/H5 and H3/H6 haplotypes had a strong effect on susceptibility/resistance to SHIV89.6P infection and may have obscured a protective effect of the vaccine regimens on challenge outcome. After challenge, 69% of the 16 immunized cynomolgus monkeys were aviremic or controlled viremia. Similarly, 67% of the 6 controls were aviremic or controlled viremia. A retrospective analysis of a larger cohort of vaccinated and control Mauritius cynomolgus macaques is underway to further examine the role of MHC class 1B haplotypes on SHIV89.6P infection and vaccine-induced protection (Ensoli et al, in preparation).
3.4 Analysis of cellular immunity by haplotype
The correlation of viral burden with MHC haplotype implicates cellular immunity in chronic viremia control. ELISPOT responses of macaques grouped by MHC haplotype revealed no differences pre- or post-challenge in Tat-specific IFN-γ secretion with respect to MHC haplotypes in the vaccinated macaques or post-challenge in the controls (data not shown). However, peak Tat-specific T cell proliferative responses pre-challenge were higher in vaccinated macaques with the H3 or H6 haplotype compared to all others (Table 3; p = 0.011). Post-challenge the H3 and H6 macaques continued to display higher peak SI (Table 3) although not significantly different from non-H3/H6 macaques, even when the aviremic macaques which lacked continual stimulation in vivo were excluded. Further, post-challenge the control macaques did not exhibit differences in SI with respect to MHC haplotype. These results suggest that vaccination against Tat of H3/H6 macaques rather than non-H3/H6 macaques might elicit Tat-specific T cell proliferative responses and better control of chronic viremia, a hypothesis currently being explored in a larger cohort of animals (Ensoli et al, in preparation).
Table 3.
Tat-specific T cell proliferation by MHC class IB haplotype in vaccinated macaques.
| Peak Stimulation indices
|
|||||
|---|---|---|---|---|---|
| Macaque | Pre-challenge | Post-challengea | Pre- challenge | Post-challengea | |
|
H3 Haplotype
|
|||||
| 4014 | 11.4 | 11.4 | |||
| 4015 | 15.9 | 19.3 | |||
| 4017* | 5.3 | 12.9 | |||
| 4021 | 7.7 | 6.7 | H3 mean | 10.2 ± 2.3 | 12.6 ± 2.6 |
|
| |||||
|
H6 Haplotype
|
|||||
| 4004 | 4.7 | 5.2 | |||
| 4008 | 0 | 3.0 | |||
| 4009 | 5.9 | 151.4 | |||
| 4016* | 3.1 | 0 | |||
| 4022* | 12.4 | 0 | |||
| 4023* | 6.5 | 0 | H6 mean | 5.4 ± 1.7 | 26.6 ± 25.0 |
|
| |||||
| H3/H6 mean | 7.3 ± 1.5 | 21.0 ± 14.6 | |||
|
|
|||||
|
Non-H3/H6 Haplotypes
|
|||||
| 4001 | 0 | 0 | |||
| 4002 | 5.5 | 7.7 | |||
| 4007 | 2.9 | 0 | |||
| 4010 | 0 | 0 | |||
| 4011 | 2.0 | 2.9 | |||
| 4013 | 0 | 0 | Non-H3/H6 mean | 1.7 ± 0.9 | 1.8 ± 1.3 |
|
| |||||
| H3/H6 vs Non-H3/H6b | p = 0.011 | ||||
Negative proliferative responses are recorded as 0. Aviremic macaques are marked by an asterisk.
Responses weeks 1-8 post-challenge.
H3/H6 macaques were combined for this analysis, as proliferative responses did not differ between groups.
Viral loads by haplotype groupings were examined after omitting aviremic macaques to eliminate reduced acute phase viral burdens mediated by unknown mechanisms. H2/H5 macaques continued to display elevated chronic viremia, 0.5 to 1.5 logs higher than H1/H4 and H3/H6 macaques respectively (Fig. 5B). During acute infection (weeks 1 – 4) the H2/H5 macaques still displayed higher peak viremia compared to all others (p = 0.0079). Viremia of H2/H5 macaques two-weeks post-challenge was higher than that of all others (p = 0.0010), while that of H3/H6 macaques was lower (p = 0.036). Three of the six H3/H6 macaques vaccinated with Tat exhibited delayed peak viremia, and two of these three exhibited an anamnestic T cell proliferative response (data not shown) implying a vaccine effect.
T cell proliferation is a surrogate for MHC class II-restricted CD4 T helper cell responses [54]. The majority of Mauritian macaques with H3 or H6 MHC class IB haplotypes also had H3 and H6 class II haplotypes (5 of 7 for both), and all macaques with H2 or H5 class IB haplotypes also had H2 and H5 class II haplotypes (Fig. 4). Ten of eleven macaques with H3 or H6 class II haplotypes were aviremic or controlled viremia, while 1 of 11 remained viremic, a non-significant trend for resistance (p = 0.15). But macaques with H2 and H5 class II haplotypes exhibited a significant trend for susceptibility (p = 0.0014): none were aviremic, 2 of 8 controlled viremia, and 6 of 8 remained viremic. Analysis of longitudinal viral loads by MHC class II haplotypes showed that macaques with the H2 or H5 class II haplotype had higher viremia levels (data not shown). Acute viremia in macaques with class II H2/H5 haplotypes vs non-H2 and H5 macaques was higher when all macaques were included (p = 0.0006) and when aviremic macaques were excluded (p = 0.0079), as was chronic viremia: (p = 0.025 with all macaques included; p = 0.015 with aviremic macaques excluded).
4. Discussion
In this study identical vaccine protocols in Mauritian cynomolgus and Indian rhesus macaques addressed previously reported disparate outcomes of Tat-based vaccine regimens in these animal models. Strong anti-Tat antibodies were elicited in both species, with the highest titers seen in the cynomolgus macaques immunized with Tat protein. In contrast, weak cellular immunity was elicited in both species by both vaccine regimens. As strong induction of IFN-γ secreting cells by the Ad5hr-HIVtat recombinant was previously seen in mice [48], fewer Tat T cell epitopes may be recognized in non-human primates. Epitope mapping could resolve this question. Tat-specific proliferation was also low compared to results of previous cynomolgus monkey studies [26], but no basis for this difference could be discerned.
Following the SHIV89.6P challenges only rhesus macaques vaccinated with the Ad5hr-HIVtat regimen showed a transient 1 log reduction in acute viremia. When corrected for multiple comparisons in a multi-arm vaccine study, this protection was no longer statistically significant [44]. In the cynomolgus model, however, MHC class IB haplotypes were seen to influence the course of SHIV89.6P infection. Animals carrying the H6 or H3 class IB haplotypes displayed chronic phase viral loads near the limit of detection after SHIV89.6P challenge, while animals with H2 or H5 class IB haplotypes, maintained chronic viremia ~20-fold higher than the cohort as a whole. Higher viremia was also seen in macaques with H2 and H5 class II haplotypes. As most of the cynomolgus macaques in this study were concordant for class IB and class II haplotypes (Fig. 4), it will be important to examine whether both contribute to SHIV89.6P control and the immunologic mechanisms responsible. Recently, class II alleles have been shown to influence SIV viremia levels in rhesus macaques [55]. Here, vaccinated macaques with H3 or H6 haplotypes exhibited higher peak proliferative responses to Tat prior to challenge, suggesting that cellular immunity may contribute to the resistant phenotype. The remarkably simple MHC genetics of the geographically isolated Mauritian cynomolgus population [37] can be exploited in prospective studies to further explore the relationship between MHC haplotypes, immune response, and susceptibility to infection with SHIV89.6P and other SHIV isolates or SIV strains.
The lack of protection in the cynomolgus macaques immunized with Tat protein contrasted with earlier results in which the identical vaccine regimen protected against the same SHIV89.6Pcy243 stock [26]. The reason for this difference is not known, but the 10 MID50 challenge dose instead of the 15 MID50 used here might have played a role. Challenge dose effects will be explored in depth in a large retrospective cohort study (Ensoli et al, in preparation).
The association of MHC class IB haplotypes with viremia control in Mauritian cynomolgus macaques is not surprising. In humans, HLA-B alleles exert a dominant influence on the outcome of HIV infection, with particular HLA-B allele expression associated with control of viremia, CD4 count, and rate of disease progression [56]. HLA-B*27 and B*57 are associated with delayed AIDS progression, while HLA-B*35 is associated with accelerated AIDS onset [57]. In rhesus macaques, Mamu-B*17 is associated with reduced plasma viremia and slowed disease progression following infection with SIVmac239 [40, 42], although by itself it does not guarantee better disease outcome [58]. Mamu-B*08 positive rhesus macaques display reduced chronic phase viremia following SIVmac239 infection, and the allele is overrepresented in elite controllers [38]. The particular Mafa-B sequences within the Mauritian H3 and H6 class IB haplotypes associated with resistance to SHIV infection and the identity of epitopes recognized remain to be identified. The basis for the association of the H2 and H5 class IB haplotypes with greater susceptibility to SHIV infection also needs elucidation. The HLA-B*35 allele has been reported to actively exert a negative effect [57]. Mafa-B sequences of the H2 and H5 class IB haplotype may behave similarly.
Susceptibility/resistance phenotypes of the Mauritian cynomolgus macaques may also be influenced by interactions of the highly polymorphic killer immunoglobulin-like receptors (KIR) present on natural killer (NK) cells and their equally polymorphic ligands, MHC class I molecules. NK cells provide a rapid initial defense against invading pathogens, and KIR by recognizing specific MHC class I molecules on target cells regulate their inhibition or activation. Specific interactions between distinct KIR3DL1 alleles and HLA-B loci have been shown to delay AIDS progression, contain HIV replication, and protect against opportunistic infections [59-62]. An absence of specific HLA ligands for inhibitory KIR has also been associated with the resistance of highly-exposed persistently seronegative individuals to HIV infection [63]. Similar interactions may be uncovered in the Mauritian cynomolgus macaques, a task that should be facilitated by the simple genetics of this population.
Our findings suggest an explanation for results reported earlier in which a majority of naïve Mauritian cynomolgus macaques naturally controlled SIV or SHIV replication [64]. The control was associated with early IFN-γ responses to Gag and Env peptides post-challenge. Here, only Tat responses were evaluated, so further studies are needed to examine other cellular responses by haplotype in depth. Confirmation of our findings will be important for future vaccine trials to allow selection of macaques that will exhibit susceptibility to SHIV infection, thus providing the sensitivity needed for low-dose challenge studies, while avoiding resistant animals that naturally control viral infection and confound vaccine experiments.
While little protection was elicited here by the Ad5hr-HIVtat or Tat protein regimens, protection was observed in previous Tat vaccine studies [26, 28] in which a lower challenge dose (10 MID50) was used. Tat combined with other HIV antigens might better confer protection at higher challenge doses. Immunizations with Tat plus Rev and Tat plus other non-structural HIV gene products have shown protection against SIV [45, 46]. A potential synergy between Tat and Env leading to enhanced protective efficacy in rhesus macaques against SHIV89.6P was recently reported [44]. Prospective studies using Tat-based vaccine strategies are being conducted in non-human primates typed to control for host susceptibility/resistance factors. Further, human phase II trials of the HIV Tat vaccine and a phase I trial combining HIV Tat and HIV Env are about to begin. The outcome of these studies will determine the value of Tat as an HIV vaccine candidate.
Acknowledgments
We thank Drs. Norman Letvin and Keith Reimann, Beth Israel Deaconess Medical Center, Harvard Medical School, for the rhesus SHIV89.6P challenge stock, Kristine Aldrich and Ersell Richardson for excellent technical assistance, and Dr. Nancy Miller, DAIDS, NIAID, for helpful discussion. The following reagent was provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: complete peptide set for HIV consensus B Tat. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute; by the NIH, NIAID, Simian Vaccine Evaluation contract N01-AI-15431 to the University of Washington; by NIH, NIAID contract N01-AI-40088 to the University of Wisconsin, and NIH grant 1R21AI068488-01A2 to DHO, University of Wisconsin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arya SK, Guo C, Josephs SF, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III) Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 2.Fisher AG, Feinberg MB, Josephs SF, Harper ME, Marselle LM, Reyes G, et al. The transactivator gene of HTLV- III is essential for virus replication. Nature. 1986;320:367–71. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- 3.Chang HK, Gendelman R, Lisziewicz J, Gallo RC, Ensoli B. Block of HIV-infection by a combination of antisense tat RNA and TAR decoys: a strategy for control of HIV-1. Gene Ther. 1994;1:208–16. [PubMed] [Google Scholar]
- 4.Dayton AI, Sodroski JG, Rosen CA, Goh WC, Haseltine WA. The trans-activator gene of Human T-Cell Lymphotropic virus Type III is required for replication. Cell. 1986;44:941–47. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- 5.Ensoli B, Cafaro A. HIV-1 Tat vaccines. Virus Res. 2002;82:91–101. doi: 10.1016/s0168-1702(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein G. HIV-1 Tat protein as a potential AIDS vaccine. Nature Med. 1996;1:960–64. doi: 10.1038/nm0996-960. [DOI] [PubMed] [Google Scholar]
- 7.Chang HC, Samaniego F, Nair BC, Buonaguro L, Ensoli B. HIV-1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix- associated heparan sulfate proteoglycans through its basic region. AIDS. 1997;11:1421–31. doi: 10.1097/00002030-199712000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Ensoli B, Barillari G, Salahuddin SZ, Gallo RC, Wong-Staal F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi’s sarcoma lesions of AIDS patients. Nature. 1990;345:84–6. doi: 10.1038/345084a0. [DOI] [PubMed] [Google Scholar]
- 9.Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, et al. Release, uptake and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67:277–87. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, et al. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi’s sarcoma. Nature. 1994;371:674–80. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 11.Frankel AD, Pabo CO. Cellular uptake of the Tat protein from Human immunodeficiency virus. Cell. 1988;55:1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 12.Conant K, Ma M, Nath A, Major E. Extracellular Human immunodeficiency virus type 1 Tat protein is associated with increaseing both NF-kB binding and protein kinase C activity in primary human astrocytes. J Virol. 1996;70:1384–9. doi: 10.1128/jvi.70.3.1384-1389.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li CJ, Ueda Y, Shi B, Borodyansky L, Huang L, Li YZ, et al. Tat protein induces self-perpetuating permissivity for productiveHIV-1 infection. Proc Natl Acad Sci USA. 1997;94:8116–20. doi: 10.1073/pnas.94.15.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Bosch I, Hofmann W, Sodroski J, Pardee AB. Tat protein induces human immunodeficiency type 1(HIV-1) coreceptors and promotes infection with both macrophage-tropic and T-lymphotropic HIV-1 strains. J Virol. 1998;72:8952–60. doi: 10.1128/jvi.72.11.8952-8960.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Secchiero P, Zella D, Capitani S, Gallo RC, Zauli G. Extracellular HIV-1 Tat protein upregulates the expression of surface CXC-chemokine receptor 4 in resting CD4 T cells. J Immunol. 1999;162:2427–31. [PubMed] [Google Scholar]
- 16.Li CJ, Frieman DJ, Wang C, Metelev V, Pardee AB. Induction of apoptosis in uninfected lymphocytes by HIV-1 tat protein. Science. 1995;268:429–31. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 17.Zauli G, Gibellini D, Milani D, Mazzoni M, Borgatti P, La Placa M, et al. Human immunodeficiency type 1 Tat protein protects lymphoid,epithelial and neuronal cell lines from death by apoptosis. Cancer Res. 1993;53:4481–5. [PubMed] [Google Scholar]
- 18.Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips MN, Habeeb K, et al. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV -1 infected individuals. Proc Natl Acad Sci USA. 2001;98:1781–6. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Re MC, Vignoli M, Furlini G, Gibellini D, Colangeli V, Vitone F, et al. Antibodies against full-length Tat protein and some low-molecular weight Tat–peptide correlate with low or undetectable viral load in HIV-1 seropositive patients. J Clin Virol. 2001;21:81–9. doi: 10.1016/s1386-6532(00)00189-x. [DOI] [PubMed] [Google Scholar]
- 20.van Baalen CA, Pontesilli O, Huisman RC, Geretti AM, Klein MR, de Wolf F, et al. Human immunodeficiency virus type 1 Rev-and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J Gen Virol. 1997;78:1913–8. doi: 10.1099/0022-1317-78-8-1913. [DOI] [PubMed] [Google Scholar]
- 21.Zagury JF, Sill A, Blattner W, Lachgar A, Le Buanec H, Richardson M, et al. Antibodies to the HIV-1 Tat protein correlated with nonprogression to AIDS: a rationale for the use of Tat toxoid as an HIV-1 vaccine. J Hum Virol. 1998;1:282–92. [PubMed] [Google Scholar]
- 22.Allen TM, Mortara L, Mothé BR, Liebl M, Jing P, Calore B, et al. Tat-vaccinated macaques do not control simian immunodeficiency virus SIVmac239 replication. J Virol. 2002;76:4108–12. doi: 10.1128/JVI.76.8.4108-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvera P, Richardson MW, Greenhouse J, Yalley-Ogunro J, Shaw N, Mirchandani J, et al. Outcome of Simian-human immunodeficiency virus strain 89.6P challenge following vaccination of rhesus macaques with human immunodeficiency virus Tat protein. J Virol. 2002;76:3800–9. doi: 10.1128/JVI.76.8.3800-3809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein G, Manson K, Tribbick G, Smith R. Minimization of chronic plasma viremia in rhesus smacaques immunized with synthetic HIV-1 Tat peptides and infected with a chimeric simian/human immunodeficiency virus (SHIV33) Vaccine. 2000;18:2789–95. doi: 10.1016/s0264-410x(00)00085-2. [DOI] [PubMed] [Google Scholar]
- 25.Pauza CD, Trivedi P, Wallace M, Ruckwardt TJ, Le Buanec H, Lu W, et al. Vaccination with Tat toxoid attenuates disease in simian/HIV challenged macaques. Proc Natl Acad Sci USA. 2000;97:3515–9. doi: 10.1073/pnas.070049797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cafaro A, Caputo A, Fracasso C, Maggiorella MT, Goletti D, Baroncelli S, et al. Control of SHIV-89.6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nature Med. 1999;5:643–50. doi: 10.1038/9488. [DOI] [PubMed] [Google Scholar]
- 27.Cafaro A, Maggiorella MT, Baroncelli S, Fracasso C, Pace M, Borsetti A, et al. SHIV89.6P pathogenecity in cynomolgus monkeys and control of viral replication and disease onset by human immunodeficiency virus type 1 Tat vaccine. J Med Primatol. 2000;29:193–208. doi: 10.1034/j.1600-0684.2000.290313.x. [DOI] [PubMed] [Google Scholar]
- 28.Cafaro A, Titti F, Fracasso C, Maggiorella MT, Baroncelli S, Caputo A, et al. Vaccination with DNA containing tat coding sequences and unmethylated CpG motifs protects cynomolgus monkeys upon infection with SHIV89.6P. Vaccine. 2001;19:2862–77. doi: 10.1016/s0264-410x(01)00002-0. [DOI] [PubMed] [Google Scholar]
- 29.Maggiorella MT, Baroncelli S, Michelini Z, Fanales-Belasio E, Moretti S, Sernicola L, et al. Long-term protection against SHIV89.6P replication in HIV-Tat vaccinated cynomolgus monkeys. Vaccine. 2004;22:3258–69. doi: 10.1016/j.vaccine.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Malkevitch NV, Robert-Guroff M. A call for replicating vector prime/protein boost strategies in HIV vaccine design. Exp Rev Vaccines. 2004;Suppl. 3:89–101. doi: 10.1586/14760584.3.4.s105. [DOI] [PubMed] [Google Scholar]
- 31.Gomez-Roman VR, Florese RH, Peng B, Montefiori D, Kalyanaraman VS, Venzon D, et al. An adenovirus-based HIV subtype-B prime/boost vaccine regimen elicits antibodies mediating broad antibody-dependent cellular cytotoxicity against non-subtype B HIV strains. J Acquir Immune Defic Syndr. 2006;43:270–7. doi: 10.1097/01.qai.0000230318.40170.60. [DOI] [PubMed] [Google Scholar]
- 32.Peng B, Wang LR, Gómez-Román VR, Davis-Warren A, Montefiori DC, Kalyanaraman VS, et al. Replicating rather than nonreplicating adenovirus-human immunodeficiency virus recombinant vaccines are better at eliciting potent cellular immunity and priming high-titer antibodies. J Virol. 2005;79:10200–9. doi: 10.1128/JVI.79.16.10200-10209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gomez-Roman VR, Wang L, et al. Protection against mucosal simian-immunodeficiency virus SIVmac251 challenge by using replicating Adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol. 2004;78:2212–21. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malkevitch N, Patterson LJ, Aldrich MK, Wu Y, Venzon D, Florese RH, et al. Durable protection of rhesus macaques immunized with a replicating adenovirus-SIV multigene prime/protein boost vaccine against a second SIVmac251 rectal challenge: Role of SIV-specific CD8+ T cell responses. Virology. 2006;353:83–98. doi: 10.1016/j.virol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Krebs KC, Jin ZY, Rudersdorf R, Hughes AL, O’Connor DH. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol. 2005;175:5230–9. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- 36.O’Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, et al. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–62. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, et al. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cyomolgus macaques from Mauritius. J Virol. 2007;81:349–61. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–32. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mothé BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, et al. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:40–2736. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor DH, Mothe BR, Weinfurter JT, Fuenger S, Rehrauer WM, Jing P, et al. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol. 2003;77:9029–40. doi: 10.1128/JVI.77.16.9029-9040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, et al. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–7. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang ZQ, Fu TM, Casimiro DR, Davies ME, Liang X, Schleif WA, et al. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J Virol. 2002;76:12845–54. doi: 10.1128/JVI.76.24.12845-12854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, et al. A replication-competent Adenovirus-human immunodeficiency virus (Ad-HIV) tat and Ad–HIV env priming/Tat and envelope protein boosting regimen elicits enhanced protective efficacy against simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol. 2007;81:3414–27. doi: 10.1128/JVI.02453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hel Z, Tsai WP, Tryniszewska E, Nacsa J, Markham PD, Lewis MG, et al. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J Immunol. 2006;176:85–96. doi: 10.4049/jimmunol.176.1.85. [DOI] [PubMed] [Google Scholar]
- 46.Osterhaus ADME, van Baalen CA, Gruters RA, Schutten M, Siebelink CHJ, Hulskotte EGJ, et al. Vaccination with Rev and Tat against AIDS. Vaccine. 1999;17:2713–4. doi: 10.1016/s0264-410x(98)00498-8. [DOI] [PubMed] [Google Scholar]
- 47.Davis D, Morein B, Akerblom L, Lovgren-Bengtsson K, van Gills M, Bogers W, et al. A recombinant prime, peptide boost vaccination strategy can focus the immune response onto more than one epitope even though these may not be immunodominant in the complex immunogen. Vaccine. 1997;15:1661–9. doi: 10.1016/s0264-410x(97)00084-4. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Voltan R, Peng B, Davis-Warren A, Kalyanaraman VS, Alvord WG, et al. Enhanced cellular immunity to SIV Gag following co-administration of adenovirus encoding wild-type or mutant HIV Tat and SIV Gag. Virology. 2005;342:1–12. doi: 10.1016/j.virol.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 49.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, et al. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–8. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson LJ, Malkevitch N, Pinczewski J, Venzon D, Lou Y, Peng B, et al. Potent, persistent induction and modulation of cellular immune responses in rhesus macaques primed with Ad5hr-simian immunodeficiency virus (SIV)env/rev,gag, and or nef vaccines an d boosted with SIV gp120. J Virol. 2003;77:8607–20. doi: 10.1128/JVI.77.16.8607-8620.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buge SL, Richardson E, Alipanah S, Markham P, Cheng S, Kalyan N, et al. An Adenovirus-simian immunodeficiency virus env vaccine elicits humoral, cellular, and mucosal immune responses in rhesus macaques and decreases viral burden following vaginal challenge. J Virol. 1997;71:8531–41. doi: 10.1128/jvi.71.11.8531-8541.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham P, et al. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J Virol Methods. 2000;86:61–70. doi: 10.1016/s0166-0934(99)00184-6. [DOI] [PubMed] [Google Scholar]
- 53.Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, et al. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics. 2007;60:37–46. doi: 10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrop R, Ryan MG, Golding H, Redchenko I, Carroll MM. Monitoring of human immunological responses to vaccinia virus. Methods. Mol Biol. 2004;269:243–66. doi: 10.1385/1-59259-789-0:243. [DOI] [PubMed] [Google Scholar]
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, et al. The Major Histocompatibility Complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of SIV-infected rhesus macaque elite controllers. J Virol. 2007;82:859–70. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–74. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 57.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 58.Wojcechowskyj JA, Yant LJ, Wiseman RW, O’Connor SL, O’Connor DH. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J Virol. 2007;81:406–10. doi: 10.1128/JVI.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin MP, Gao X, Lee J, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nature Gen. 2002;31:429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 60.Qi Y, Martin MP, Gao X, Jacobson L, Goedert JJ, Buchbinder S, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Path. 2006;2:0741–5. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nature Gen. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, Nkenzasong JN, et al. Cutting Edge: Resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–92. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 64.Reimann KA, Parker RA, Seaman MS, Beaudry K, Beddall M, Peterson L, et al. Pathogenicity of Simian-human immunodeficiency virus SHIV89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J Virol. 2005;79:8878–85. doi: 10.1128/JVI.79.14.8878-8885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


