Summary
Recruitment of signaling molecules to the cytoplasmic domains of the CD3 subunits of the T cell receptor (TCR) is crucial for early T cell activation. These transient associations either do or do not require tyrosine phosphorylation of CD3 immune tyrosine activation motifs (ITAMs). Here we show that the non-ITAM-requiring adaptor protein Nck forms a complex with an atypical PxxDY motif of the CD3ε tail, which encompasses Tyr166 within the ITAM and a TCR endocytosis signal. As suggested by the structure of the complex, we find that Nck binding inhibits phosphorylation of the CD3ε ITAM by Fyn and Lck kinases in vitro. Moreover, the CD3ε/Nck interaction downregulates TCR surface expression upon physiological stimulation in mouse primary lymph-node cells. This indicates that Nck performs an important regulatory function in T lymphocytes by inhibiting ITAM phosphorylation and/or removing cell surface TCR via CD3ε interaction.
Keywords: CD3ε, Nck, nuclear magnetic resonance, phosphorylation, receptor internalization
Introduction
Engagement of a T cell receptor (TCR) by a peptide bound to a major histocompatibility complex (MHC) molecule on antigen-presenting cells (APCs) is an essential trigger for antigen-specific T cell activation. In αβ T cells, each TCR is a multi-subunit complex composed of one αβ clone-specific heterodimer in non-covalent association with three sets of invariant CD3 dimers (CD3εγ and CD3εδ heterodimers, and a CD3ζζ homodimer). While the αβ TCR heterodimer is responsible for specific immune recognition, its two subunits contain short cytoplasmic tails with no obvious capacity to mediate signal transduction1-5. In contrast, the CD3 dimers are known signal transducers, transferring information on the status of TCR ligation by peptide/MHC (pMHC) to intracellular signaling molecules6. Details about the nature of TCR-pMHC recognition, as well as the structures of extracellular domains of TCR components, have been provided by both X-ray crystallography and NMR spectroscopy1,5,7-11.
The interactions between the CD3 cytoplasmic tails and cytoplasmic signaling molecules are primarily activation-dependent. Associations involving an immunoreceptor tyrosine-based activation motif, ITAM (YxxL/I(x6−8)YxxL/I), present one in CD3ε and CD3γ and three in CD3ζ, have been most extensively studied12,13. Upon TCR-pMHC ligation, ITAMs are phosphorylated by Src family protein tyrosine kinases, such as Lck and/or Fyn, thereby creating binding sites for the protein tyrosine kinase ZAP70 or other SH2-domain containing signaling molecules14-16. The structures of Src kinases have been solved in several forms17, as has the structure of the ITAM-dependent signaling molecule ZAP7018,19, all of which have contributed to the understanding of ITAM-dependent TCR signaling.
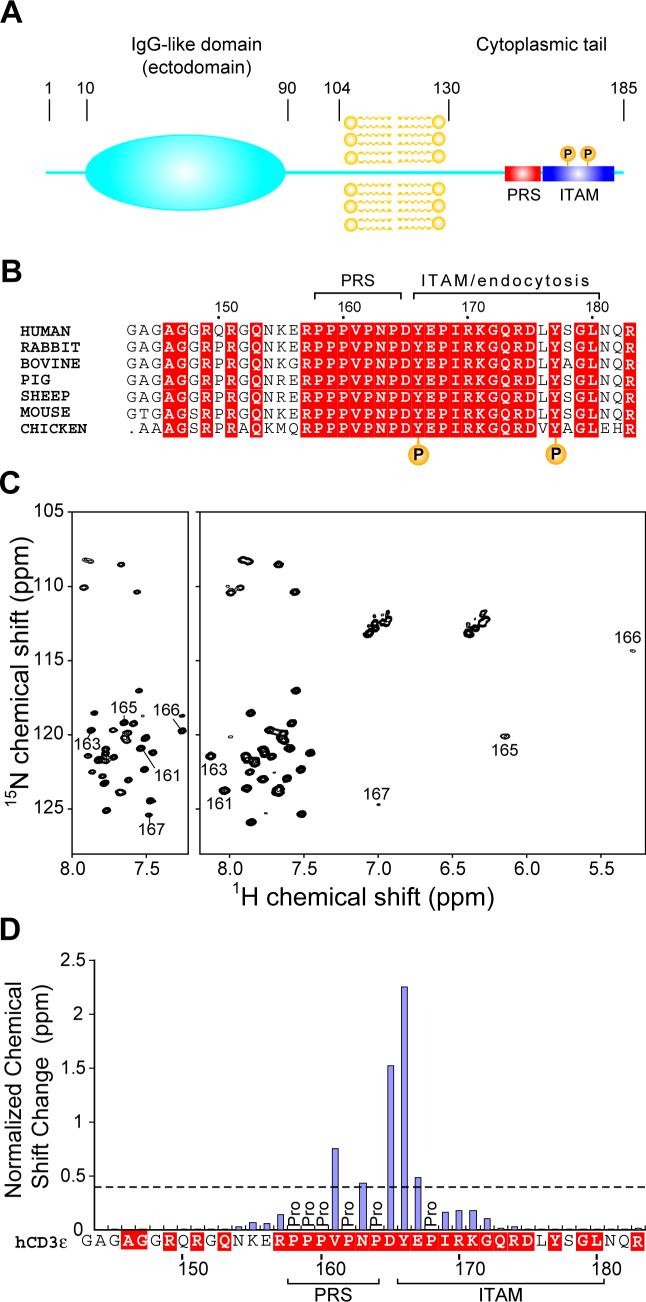
Evolutionally conserved segments other than ITAMs are also present within the cytoplasmic tails of CD3 subunits, however, implying important additional functionality. To date, little biological or structural information is available concerning such non-ITAM-requiring interactions. Moreover, the relationship between ITAM and non-ITAM-requiring interactions in T cell signaling remains unclear. In this regard, recruitment of an adaptor protein, Nck, to the cytoplasmic domain of CD3ε is known to involve non-ITAM sequences20,21. This interaction depends upon the proline-rich sequence (PRS) in CD3ε, which is N-terminal to the CD3ε ITAM, and the first SH3 domain of Nck (Figure 1A and B). Nck recruitment to CD3ε occurs prior to phosphorylation of the CD3ε ITAM and, as such, is proposed to be the earliest event after TCR-pMHC ligation20. However, the importance of CD3ε/Nck interaction involving the CD3ε PRS is still a matter of debate20,22. It is worth noting that another interesting feature of the Nck recognition is that it encompasses Tyr166, a phosphorylation site in the CD3ε ITAM, and tyrosine phosphorylation of the residue abolishes Nck-SH3 binding to CD3ε23.
Figure. 1.
Domain architecture and mapping of the Nck2 SH3.1 binding site on the cytoplasmic tail of CD3ε (A) Domain boundaries of human CD3ε. CD3ε is a single transmembrane protein with an IgG-like ectodomain (shown as a blue ellipse). The 55-residue cytoplasmic domain of human CD3ε contains a PRS (red box) and an ITAM (blue box). The two tyrosine-phosphorylation sites of the ITAM are indicated with “P”. (B) Sequence alignment of the cytoplasmic segments of CD3ε. The conserved residues are shown in white letters on red background. The locations of the PRS (residues 158−164), the ITAM (residues 166−180) and two phosphorylation sites in the ITAM are also indicated. ITAM is overlapping with the T cell endocytosis signal in CD3ε as indicated by 24. (C) 1H-15N HSQC spectra of 15N-labeled cytoplasmic segment of CD3ε without (left) and with (right) 4-fold excess of Nck2 SH3.1. The residues with significant chemical shift changes ((ΔδH2 + (ΔδN / 5)2)1/2 > 0.4 ppm) are labeled. (D) Plot of the normalized chemical shift changes of the CD3ε tail upon binding to Nck2 SH3.1. The sequence of human CD3ε is shown in the same representation as in (B).
Using solution NMR techniques, we determined the structure of the complex between the CD3ε cytoplasmic tail and the first SH3 domain of Nck2 (referred to hereafter as Nck2 SH3.1). The structure defines an atypical SH3 domain interaction such that Nck2 SH3.1 recognizes the 158-PPPVPNPDY-166 sequence in the CD3ε molecule, including the aforementioned Tyr166 within the CD3ε ITAM motif. This is also the first structure describing a non-canonical PxxDY sequence interaction with a SH3 domain. The structural characterization of the complex revealed the molecular basis by which phosphorylation of Tyr166 abolishes the binding between Nck and CD3ε. More importantly, however, the structure indicated that the CD3ε/Nck2-SH3.1 interaction inhibits the phosphorylation of the CD3ε ITAM by the Src protein tyrosine kinases, Fyn or Lck. Indeed, this was confirmed by in vitro CD3ε phosphorylation analysis using Src kinases. In addition, we show that the CD3ε/Nck interaction downregulates surface expression of the TCR. This provides the first direct evidence that the CD3ε/Nck interaction is involved in the control of TCR surface expression, consistent with an earlier report that the CD3ε tail contains an endocytosis signal24 providing mechanistic insight. The results presented here clearly revealed that the CD3ε/Nck interaction has a potential to regulate T cell development and activation under physiological stimulation, such as contact with peptide-MHC complexes, by inhibiting of ITAM phosphorylation and/or reducing TCR cell surface expression. Thus, in contrast to a role of Nck in establishing an optimal TCR signaling output, as proposed by Gil et al20, our data suggest that the Nck/ CD3ε interaction is also capable of downregulating T-cell activation via a non-ITAM-requiring mechanism.
Results and discussion
Structure of CD3ε in complex with Nck
To elucidate the significance of the Nck2/CD3ε interaction we decided to investigate its effect on T-cell function. To identify the extent of the Nck2-binding segment of the CD3ε tail we compared the main-chain NH resonance positions of a 41 residue cytoplasmic segment of human CD3ε containing (residues 143−183) in the presence and absence of Nck2 SH3.1 (Figure 1C). As shown in the left spectrum, the cytoplasmic domain of CD3ε has only limited dispersion in the absence of Nck2 SH3.1, indicating an overall unfolded structure. Upon binding to Nck2 SH3.1, however, the resonances of residues 161−167 experienced significant chemical shift changes, reflecting that this segment has become folded in the protein complex (Figure 1C, right). Figure 1D displays the normalized chemical shift changes for amide resonances of the CD3ε peptide upon binding to Nck2 SH3.1. While the regions N-terminal to Pro158 or C-terminal to Glu167 are not involved in any extensive interaction, with no significant chemical shift changes (<0.2 ppm), the segment from Val161 to Glu167 exhibits changes of more than 0.4 ppm. Thus, these residues appear to be responsible for binding to Nck2 SH3.1. Calculated from the concentration dependence of peak positions, the dissociation constant (KD) for the interaction was determined to be 40 ± 9 μM (Supplemental Figure 1A). The binding affinity was consistent with the affinity obtained from tryptophane fluorometry (33 ± 10 μM, Supplemental Figure 2). The most significant feature of the Nck recognition sequence of CD3ε is that it encompasses Tyr166, a phosphorylation site in the CD3ε ITAM and the phosphorylation of the residue abolishes Nck-SH3 binding to CD3ε.
Our selection of the Nck-binding sequences is consistent with a recent report by Kresti et al. who used a peptide matrix analysis of a series of mutated CD3ε fragments to identify the binding segment23. They report that Nck1-SH3.1 binding shows a dependence on the 159-PPVPNPDY-166 segment of CD3ε. This is slightly smaller than the segment identified in the NMR spectra but contains the residues most strongly affected. They first reported that the binding sequence contains Tyr166 of the ITAM and its phosphorylation abolishes Nck binding. They remark that the sequence contains an atypical PxxDY motif sequence (where x is any amino acid, here 162-PNPDY-166) that was first identified as a SH3-binding sequence for the Eps8 proteins26.
To obtain a more mechanistic understanding of the CD3ε/Nck interaction, we decided to determine the solution structure of CD3ε in complex with Nck2 SH3.1. Unfortunately, the rather high KD values (40 ± 9 μM) of the CD3ε/Nck interaction, and dissociation kinetics in the intermediate exchange regime cause line broadening of the interface resonances, which hampers a straightforward structural characterization when using isolated Nck2 SH3.1 complexed with a CD3ε segment. Thus, we designed a single-chain (sc) construct of the CD3ε/Nck2-SH3.1 (scCD3ε/Nck2-SH3.1) complex, where the Nck binding region of CD3ε is connected to the N-terminus of Nck2 SH3.1 with a 13 amino-acid linker. The singlechain approach has previously been found invaluable for stabilizing weak interactions and allowing for the accurate determination of the structure of several protein complexes7,27. The 1H-15N HSQC spectrum of the scCD3ε/Nck2-SH3.1 construct was identical to that of isolated Nck2 SH3.1 in complex with CD3ε, except for narrower line widths of interface residues and the additional peaks that derived from the sc linker. This indicates that the native binding mode is preserved (data not shown).
To determine the structure of the sc construct, standard NMR procedures were pursued (Table 1). The structure calculations were carried out with 100 random initial structures and 20 structures with the lowest energy function were chosen for final analysis. Figure 2A shows a set of converged structures of scCD3ε/Nck2-SH3.1. The N-terminal segment (residues 153−158 of CD3ε) and the linker regions (13 amino-acid single-chain linker plus residue 1−3 of Nck2 SH3.1) differ significantly among the calculated structures consistent with being mobile. The remaining part of the structures are well converged with a root mean square difference (RMSD) of 0.29 ± 0.07 Å for backbone atoms and 0.86 ± 0.09 Å for all heavy atoms (Table 1). Satisfactory values in Ramachandran and G-factor analysis indicate that the quality of structure is sufficient for structural analysis including sidechain conformations (Table 1).
Table 1.
Statistics for the final 20 NMR structures of CD3ε peptide in complex with Nck2 SH3.1a
| Number of NOE distance constraints | |
| Total | 1486 |
| Intra-residue | 706 |
| Short and Medium (|i-j| = 1 ∼ 4) | 402 |
| Long (|i-j| > 4) | |
| Intramolecular | 302 |
| “Intermolecular” | 76 |
| Backbone angular restraints (ϕ and φ) | 92 |
| Ramachandran Plot (Residues 158−166 for CD3ε and 4−59 for Nck SH3.1)b | |
| Most favored region (%) | 78.6 |
| Additionally allowed region (%) | 21.4 |
| Generously allowed and disallowed region (%) | 0.0 |
| G-factors | |
| Mainchain dihedral angles | −0.81 |
| χ1 dihedral angles | −0.65 |
| χ1/χ2 dihedral angles | −1.29 |
| Average RMSD to mean structure (Residues 158−166 for CD3ε and 4−59 for Nck2 SH3.1, Å) | |
| Backbone (N, Cα, C) | 0.29 ± 0.07 |
| All heavy atoms | 0.86 ± 0.09 |
None of these 20 structures exhibited distance violations >0.5 Å or dihedral angle violations >5°. There are no bad contacts in the structures. The RMSD for covalent bonds and angles relative to the standard dictionary is 0.004 Å and 0.7 degrees, respectively, with all covalent bonds and angles within 6.0 × RMSD for the structures.
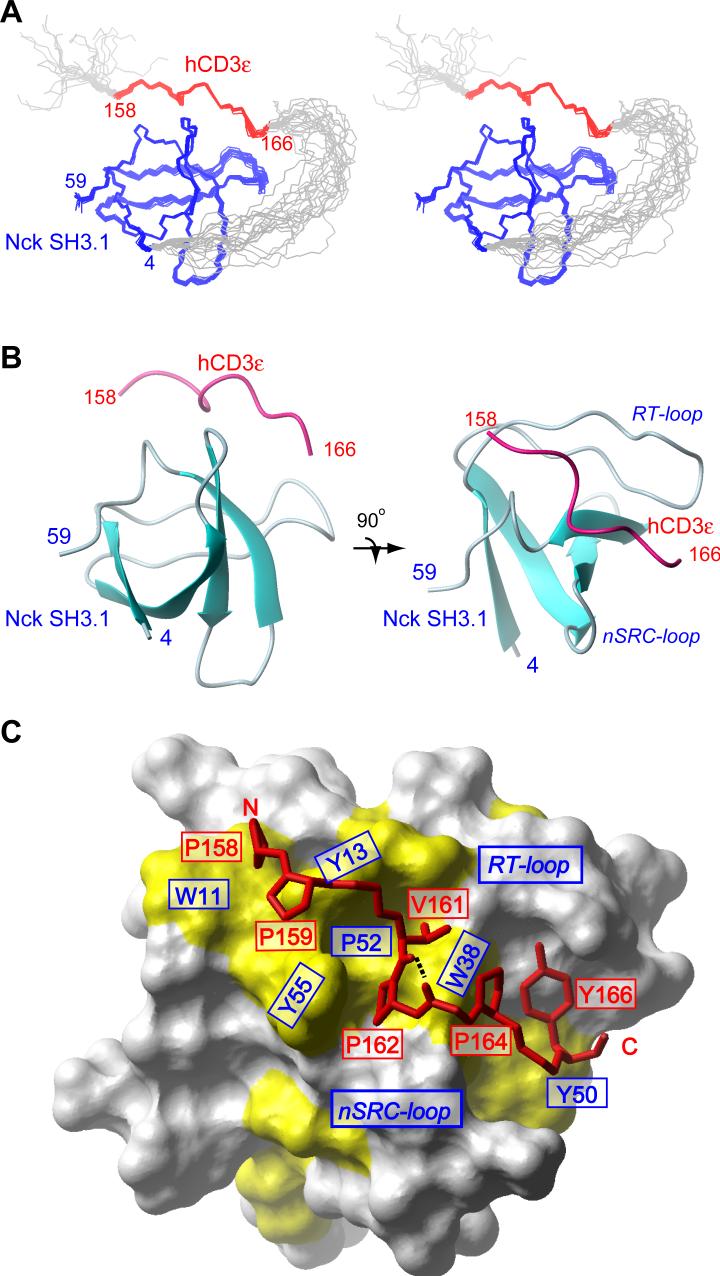
Figure 2.
Structure of the CD3ε/Nck2-SH3.1 complex. (A) Stereo view of an ensemble of 20 final NMR structures of the CD3ε/Nck2-SH3.1 complex. Blue, red, and grey lines show the backbone traces of Nck2 SH3.1 (residues 4−59), CD3ε(residues 158−166), and disordered regions (N-terminal and linker). (B) Ribbon representation of the CD3ε/Nck2-SH3.1 complex. The complex of Nck2 SH3.1 (blue) and CD3ε (red) are shown in two orientations. (C) Interface of the CD3ε/Nck2-SH3.1 interaction. The Nck2 SH3.1 surface is shown in white except for hydrophobic residues, which are colored in yellow. The backbone of CD3ε tail and the sidechains interacting with Nck Sh3.1 are depicted with red sticks. The hydrogen bond between the Val161 carbonyl oxygen and Asn163 amide proton is shown with a black dotted line.
The SH3 fold of Nck2 SH3.1 is retained after binding to CD3ε. The backbone RMSD between free28 and bound Nck2 SH3.1 is 1.4 Å. The CD3ε binding site is located between the RT and n-Src loops (Figure 2B). The 158-PPPVPNPDY-166 sequence of the CD3ε is well defined in the interaction with Nck2 SH3.1 showing a good convergence between the structures (Figure 2A and C).
Architecture of the CD3ε/Nck interface
There are two known isoforms for the human Nck protein, Nck1 and Nck2, and the homologous proteins are found in a variety of species. As shown in Supplemental Figure 3A, all Nck2 SH3.1 residues that are within 5 Å from bound CD3ε are conserved among Nck proteins, thus the structure of CD3ε/Nck2-SH3.1 complex appears to represents the CD3ε/Nck interactions in general. The sidechains of Pro158, Pro159, Val161, Pro162, Pro164, and Tyr166 of CD3ε interact with conserved hydrophobic regions on the Nck2 SH3.1 domain (Figure 2C, yellow surface).
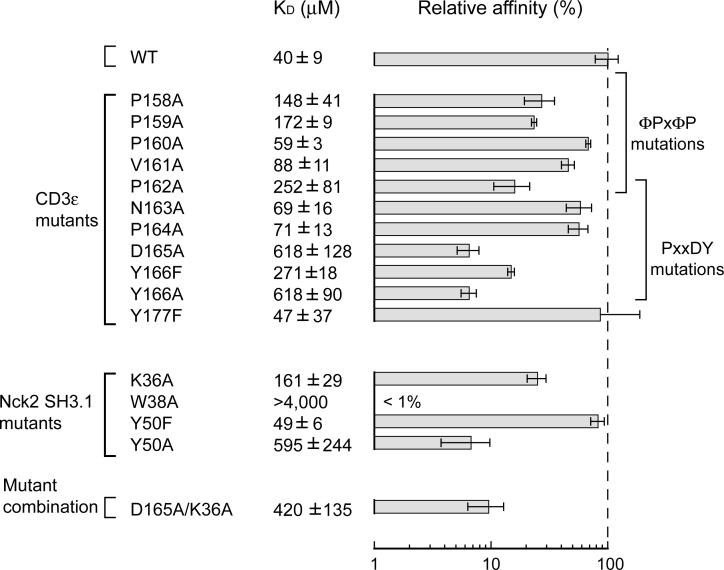
As discussed above, the Nck binding sequence of CD3ε contains an atypical PxxDY sequence (162-PNPDY-166). So far, no structural information has been available for any SH3-domain interaction involving this atypical binding motif, and our data present the first example of such a complex. The N-terminal half of the Nck binding sequence (158-PPPVP-162) can also be recognized as a partial sequence of typical Class II (ΦPxΦPxR/K) SH3-interacting motifs, which contains the ΦPxΦP motif but lacks the critical basic residues at positions +2 relative to the ΦPxΦP sequence (where x is any amino acid and Φ is a hydrophobic residue)29. Indeed, the ΦPxΦP sequence forms a typical polyproline II (PPII) helix upon binding to Nck2 SH3.1, in the same way as the Class II PRS-SH3 domain interactions. As expected from the structure, mutations of Pro158, Pro159, Val161, or Pro162 result in a larger reduction of the binding affinity than the exposed Pro160 (Figure 3). It is also important to note that the Nck recognition sequence of CD3ε seems to include Pro158, which was overlooked by the peptide matrix analysis carried out by Kesti et al., simply because the residue was outside of their mutated region23. Any single mutations in the ΦPxΦP sequence cause only moderate reduction of the affinity compared to the fact that the corresponding mutations for a typical Class II PRS-SH3 domain interaction reduce the binding affinity to less than 10%30. The result suggests that the ΦPxΦP sequence is less important in the CD3Φ/Nck2-SH3.1 interaction. In fact, the peptide that only contains the PRS segment of CD3ε (158-PPPVPNP-164) failed to bind to Nck2 SH3.1 (data not shown). On the other hand, mutations of the conserved residues (Asp164 or Tyr166) in the PxxDY sequence have a much more significant effect on the CD3ε/Nck2-SH3.1 interaction (the binding affinities of both mutants are reduced to 6.5% compared to WT). Thus, the PxxDY interactions, especially Asp165 and Tyr166 interactions, appear to be essential for the CD3ε/Nck-SH3.1 interaction rather than the ΦPxΦP motif. This is important to recognize since mutations of the less significant N-terminal part of the Nck-binding motif have been used to make conclusions about Nck's role in T-cell activation22.
Figure 3.
Dissociation constants and relative affinities of CD3ε and Nck2 SH3.1 mutants. Dissociation constants (KD) of the indicated constructs to WT Nck2 SH3.1 or CD3ε segment (143−183) were determined by fitting the chemical shift changes due increasing concentration of binding partner (see Supplemental Figure 1 for detailed explanations). The bottom row shows the CD3ε/Nck2-SH3.1 mutant combination. Each value presents the average ± standard error of at least two measurements. Relative binding affinity (%) compared to the wt CD3ε/Nck2-SH3.1 interaction was calculated from the average dissociation constants. The dashed line indicates 100% relative affinity.
The PxxDY segment forms a kinked structure with a hydrogen bond between the Val161 carbonyl oxygen and the Asn163 amide proton (dotted line in Figure 2C). Trp38 of Nck2 SH3.1 is sandwiched by two proline residues (Pro162 and Pro164) in the PxxDY sequence of CD3ε, and W38A mutation completely abolished the interaction (Figure 3). P162A or P164A mutation also reduced the binding affinity to 16% or 57% relative to WT (Figure 3).
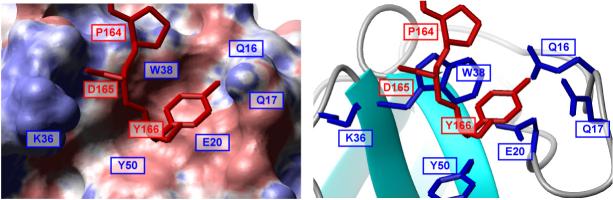
One of the conserved residues in the PxxDY sequence, Asp165, is in close proximity to a basic residue, Lys36, of Nck2 SH3.1 (Figure 4), and mutation of Lys36 to alanine decreased the binding constant of Nck2 SH3.1 to 25%. Thus, it seems that there is a sidechain-sidechain interaction between the acidic Asp165 and the basic Lys36. This interaction is also supported by double mutation analysis. While D165A mutation in CD3ε decreased the affinity to WT Nck2 SH3.1 to 6.5%, the same CD3ε mutation decreased the affinity to the K36A mutant of Nck2 SH3.1 only to 38% (Figure 3). The last residue in the PxxDY sequence, Tyr166, is accommodated by a groove formed by hydrophobic Trp38 and Tyr50, hydrophilic Gln16 and Gln17, and acidic Glu20 residues located between the RT and n-Src loop (Figure 2C and 4). In this pocket, Tyr166 participates in an aromatic-aromatic interaction with Trp38 and Tyr50, which are known to be energetically favorable31. Indeed, alanine but not phenylalanine mutation of Tyr50 largely decreased the binding affinity to CD3ε (Figure 3). The hydroxyl group of Tyr166 is pointing to a small pocket formed by Gln16, Gln17 and Glu20 in the RT loop of Nck2 SH3.1. Interestingly, a phenylalanine mutation of Tyr166 also reduced the binding affinity to 15% (Figure 3), suggesting that not only the aromatic-ring, but also the hydroxyl group is critical for the interaction.
Figure 4.
Detailed view of Asp165-Tyr166 binding site. The left panel is an electrostatic surface representation where positive and negative potentials (calculated with the program MOLMOL in blue and red, respectively). The right panel shows the sidechain orientation of Nck2 SH3.1 at the site. CD3ε is depicted with red stick in both panels.
The corresponding residues that interact with Asp165 and Tyr166 in the PxxDY motif of CD3ε (Lys36 for Asp165, and Gln16, Gln17, Glu20, and Tyr50 for Tyr166 in human Nck2) are identical in all Nck proteins, except for Tyr50, which is “silently” mutated to phenylalanine in some cases, and maintained by similar residues in Eps8 proteins, another family of proteins that binds to the PxxDY sequence (Figure S3, A and B). On the other hand, the corresponding site is not conserved in other SH3 domains (Figure S3C). Thus, the PxxDY interactions seem to be essential determining the specificity of the CD3ε/Nck-SH3.1 interaction. The importance of residues corresponding to Lys36 in Nck2 had been proposed for the PxxDY interaction23,32, however the CD3 residues critical for this interaction had not yet been defined.
Regulation of the CD3ε/Nck interaction by ITAM phosphorylation
As discussed above, a recent biochemical experiments revealed that tyrosine phosphorylation of the PxxDY motif abolishes Nck-SH3 binding to CD3ε23. However, no structural analysis had been pursued, and the significance of this feature has remained elusive. Our biochemical and cell biological experiments demonstrated that the interaction between a synthetic CD3ε peptide and the Nck2 SH3.1 domain (Supplemental Figure 4A), as well as the endogenous CD3ε/Nck interaction in a functional TCR complex on the Jurkat cell surface (Supplemental figure 4B) is regulated by the phosphorylation of the CD3ε ITAM. In the complex structure, the hydroxyl moiety of Tyr166 is proximal to the negatively charged Glu20 of Nck2 SH3.1 (Figure 4), thus the phosphorylation of Tyr166 causes repulsion between CD3ε and Nck. Furthermore, phosphorylated Tyr166 is sterically hindered, since the binding pocket that accommodates the hydroxyl group of Tyr166 is too small for an additional phosphate group. Phosphorylation of the two tyrosines in positions 166 and 177 of CD3ε ITAM is essential for the recruitment of ZAP70, an activation kinase in T cell signaling33. Thus, phosphorylation of Tyr166 serves as a molecular switch that determines the binding partner of CD3ε such as Nck and ZAP70 in a mutually exclusive manner.
Inhibition of ITAM phosphorylation by the CD3ε/Nck interaction
It is known that activation-dependent association between Nck and CD3ε occurs prior to ITAM phosphorylation20. Thus it is possible that the CD3ε/Nck interaction interferes with subsequent phosphorylation of the CD3εITAM by Src kinases. The hypothesis is supported by our structure of the CD3ε/Nck complex, where Tyr166 in the ITAM is largely buried in the binding pocket of the Nck2 SH3.1. To determine whether the CD3ε/Nck interaction inhibits the CD3ε ITAM phosphorylation by Src kinases, we carried out an in vitro kinase assay using a cytoplasmic segment of CD3ε harboring the entire Nck binding sequence plus the ITAM. The cytoplasmic CD3ε segment was mixed with Fyn or Lck kinase, in the presence and absence of Nck2 SH3.1 and the phosphorylation level of the segment was monitored by immunoblotting using 4G10 Ab. As shown in Figure 5A, tyrosine phosphorylation is clearly observed in the presence of Fyn (lane 2), while tyrosine phosphorylation is not observed in the absence of Fyn (lane 1), or in the presence of Src kinase inhibitor PP2 (lane 3). With increasing concentration of Nck2 SH3.1, the tyrosine phosphorylation of CD3ε was largely inhibited. The same tendency was observed for another Src kinase, Lck (data not shown). According to densitometry quantification, Nck2 SH3.1 can achieve 50% inhibition of CD3ε phosphorylation at concentrations of 20−30 μM, which is in good agreement with the affinity between CD3ε and Nck2 SH3.1 determined by NMR titration experiments (40 ± 9 μM, see Figure 3).
Figure 5.
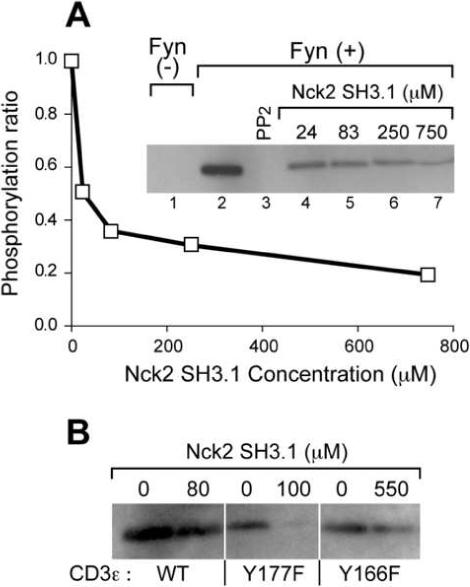
Nck SH3.1 down-regulates phosphorylation of CD3ε by Fyn (A) In vitro tyrosine phosphorylation of the CD3ε segment 143−183 by the Fyn kinase. The CD3ε segment was phosphorylated by recombinant Fyn kinase for 30 min at 37 °C in the presence of the indicated concentrations of Nck2 SH3.1 or 10 μM of the Src kinase inhibitor, PP2. The samples were subjected to immunoblotting with 4G10 Ab as shown in the inserted gel picture. The phosphorylation ratio of each Nck2 SH3.1 concentration relative to the condition in the absence of the protein was estimated by using Kodak 1D. These results are the representative of 2 individual experiments. (B) In vitro tyrosine phosphorylation of CD3ε mutants. The Tyr phosphorylation of CD3ε mutants by Fyn kinase in the presence and the absence of the indicated concentration of Nck2 SH3.1 were visualized by 4G10 Ab. The concentration of Nck2 SH3.1 was set two-fold higher than the equilibrium dissociation constants. The ratios of the band intensities with/without Nck SH3.1 are 40%, 10% and 80% for WT, Y177F, and Y166F, respectively. These results are the representative of 3 individual experiments.
Interestingly, densitometry analysis of the bands in Figure 5A showed that the phosphorylation level of CD3ε is decreased to ∼20% in the presence of higher concentration of Nck compared to the absence of the protein. This indicates that Nck binding not only inhibits the phosphorylation at Tyr166, but also that of Tyr177, which is 11 residues C-terminal to the Nck binding site. To confirm this observation, we performed the same experiment using CD3ε mutants that carry a single phosphorylation site in each construct (i.e. Y177F, which has only Tyr166, or Y166F that retains only Tyr177). Both mutants exhibited tyrosine phosphorylation in the absence of Nck2 SH3.1, confirming that Fyn can phosphorylate both Tyr166 and Tyr177 in CD3ε (Figure 5B). The phosphorylation of Tyr166 in the Y177F mutant was almost completely abolished at Nck2 Sh3.1 concentrations of roughly twice the KD (100 μM). For the Y166F mutant less tyrosine phosphorylation of Tyr177 was observed in the presence of 550 μM Nck2 SH3.1 (twice the KD of this mutant). The phosphorylation of the N-terminal tyrosine (Tyr166) was almost completely inhibited while the phosphorylation of the C-terminal tyrosine (Tyr177) was partially inhibited by Nck2 SH3.1 binding, possibly due to hindered steric accessibility for simultaneous Src kinase and Nck binding.
In a recent report, the mutually exclusive CD3ε binding of Nck and ZAP70 led to the conclusion that Nck binding has to be transient, followed by ITAM phosphorylation and ZAP70 recruitment23. Lacking the structural information we present here, the authors argued that the transient CD3ε/Nck interaction might facilitate recruitment of Src kinase and/or accelerate phosphorylation of the ITAM. However, based on the fact that the CD3ε/Nck interaction occurs before ITAM phosphorylation and the phosphorylations of both tyrosines in ITAM are required for ZAP70 recruitment, our structural and biochemical data indicate that the CD3ε/Nck interaction could also serve as a negative regulator in T cell activation by preventing phosphorylation of the CD3ε ITAM and subsequent ITAM-dependent recruitment of downstream signaling molecules. Therefore, we think that the role of CD3ε/Nck interaction in T cell activation has to be viewed more cautiously considering both positive and negative properties of Nck. Along these lines, it is also possible to imagine that Nck interaction regulates different ITAM phosphorylation levels in each od the CD3 components within the TCR complex12. A recent paper showed that the PRS in CD3ε is required for CD3Φ phosphorylation in the TCR complex of DP thymocytes, suggesting that the CD3ε PRS contributes to elevate the phosphorylation level of adjacent CD3Φ25. Considering their notion that Lck contains a putative binding site for Nck25, Nck might play a role in efficiently recruiting Lck to the unphosphorylated TCR complex, phosphorylating CD3Φ while maintaining CD3ε in an unphosphorylated state.
In the functional assays, we did not observe any significant difference in the kinetics of Ca2+ influx and T cell activation indicators, such as ERK phosphorylation, IFNγ expression, and CD69 expression, under conditions of forced Nck overexpression in Jurkat T cells (data not shown). In line with these observations, Szymczak et al. recently reported that the defects in T cell development and function of CD3ε-deficient ΔP mice could be rescued using a mutant CD3ε lacking the highly conserved proline residues (162-PPPVPNP-166 are mutated to 162-AAAVANA-166)20,22. However, one has to consider that the region involved in Nck binding shows the highest degree of conservation among CD3ε proteins. Thus, the apparent lack of a phenotype of the 162-AAAVANA-166 mutation might be due to the redundancy among the CD3 subunits (CD3εγδΦ) in the TCR complex that all support ZAP70 binding. These experiments make it difficult to reveal the role of individual CD3 subunits in T cell signaling and are not fully conclusive to claim the dispensability of the Nck binding site of CD3ε. In this regard, each of the CD3 subunits contains one or more ITAMs, whereas only CD3ε has an Nck binding motif. Thus, the functional modification provided by the CD3ε/Nck interaction might be masked by the other CD3 subunits. In addition, we think that the PRS-mutated CD3ε utilized by Szymczak et al.20,22 are not perfectly adequate considering our observation that the PRS in CD3ε is of minor importance for the CD3ε-Nck interaction. Indeed, we confirmed that the mutant CD3ε retains weak interaction (∼7 mM) to Nck SH3.1 (Supplemental Figure 5). This kind of weak interactions might not be identified by immunoprecipitation, however, could be physiologically significant34. Thus, the result obtained from the mutant CD3ε has to be interpreted cautiously.
Downregulation of TCR surface expression by the CD3ε/Nck interaction
In contrast to the functional redundancy among CD3 subunits in the T cell activation process, the proper trafficking of all individual subunits is required for normal surface expression of the TCR complex. While CD3δ and CD3γ can partially compensate for loss of expression of the other subunit, CD3ε is indispensable for TCR surface expression4. Thus, we reasoned that the significance of the CD3ε/Nck interaction in TCR expression might be observed more readily than its role in T cell activation. Interestingly, a endocytosis signal was found at residues 166−180 in CD3ε24, a segment overlapping with the Nck interaction site. Of note, Nck is known to be involved in actin cytoskeleton rearrangement, which plays a critical role in receptor-mediated endocytosis35. Thus, we hypothesized that the CD3ε/Nck interaction would affect TCR surface expression.
To investigate this possibility, we utilized an in vitro antigen-specific primary T cell culture36 and compared the surface expression levels of the TCR under conditions of Nck overexpression vs. endogenous expression. Specifically, N15 TCR transgenic+/+ RAG2−/− (N15) mice lymph node (LN) T cells were transduced with Nck retrovirus (pMSCV) and reactivated with N15 TCR specific antigen (VSV8 peptide) loaded APCs. The expression of exogenous Nck was indicated by GFP expression derived from the IRES-containing pMSCV vector and the TCR surface expression level was measured by staining with the α-Vβ5 mAb MR9.4 specific for the N15 TCR β chain. Surprisingly, TCR surface expression was less in both resting and activated states for Nck overexpressing GFP+CD8+ T cells relative to the GFP+CD8+ T cells harboring the empty vector (Figure 6A, right panels). The reduced TCR staining was not due to either insufficient stimulation or staining, because GFP-CD8+ cells served as an internal control, with no difference in TCR surface expression among GFP-CD8+ cells. The downregulation of TCR expression was not observed for cells expressing Nck2 mutant with ≤10% CD3ε binding activity, W38A (Figure 6B). It is of interest that there is an apparent inverse correlation between TCR surface expression and affinities of Nck2 SH3.1 mutants. Not surprisingly, the same results were obtained by the stimulation with α-mCD3ε Ab 2C11 loaded on APCs (Supplemental Figure 6). Although not shown, similar results were observed for T cells from CD4 restricted TCR transgenic system (i.e. 5CC7 transgenic Rag2−/− mice36). Thus, the downregulation of TCR expression appears to depend on the CD3ε/Nck interaction.
Figure 6.
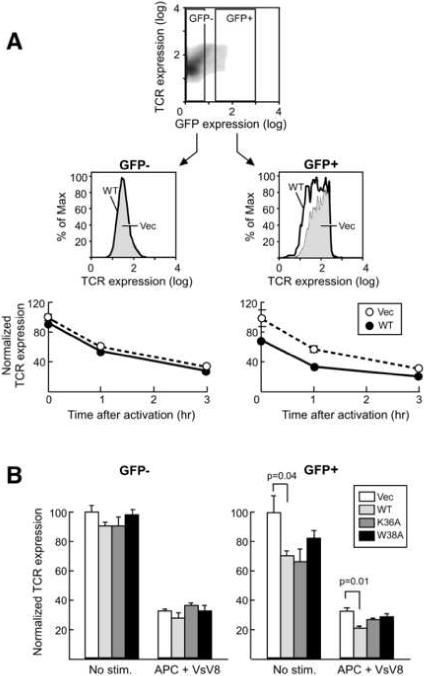
Nck overexpression downregulates TCR expression. (A) FACS analysis of TCR surface expression. The upper panel shows the density plot of GFP and TCR expression levels for CD8+ cells without stimulation. Nonretroviral gene-expressing cells (GFP-) and retroviral gene-expressing cells (GFP+) cells are gated as indicated in the panel. The middle panels show the overlaid TCR expression histograms for empty vector (thin boundary, shaded) or WT Nck (thick boundary, not shaded) transfected cells after 3hrs stimulation with VSV8-peptide (1 nM)-loaded APCs at 37 °C. Surface expression of TCR was monitored by PE-conjugated MR9.4 anti-TCR Vβ5 antibody. In the bottom panels, the mean fluorescence intensity of each sample relative to non-stimulated cells was calculated to determine the relative TCR expression. (B) TCR surface expression of LN CD8+ T cells transfected with either empty vector (Vec), wt Nck (WT), or mutants of Nck (K36A or W38A). Cells are un-stimulated or stimulated with VSV8-loaded APCs for 3 hrs at 37 °C. FACS analysis was performed in the same way as (A). The indicated values in (A) and (B) are averaged over three separate experiments with error bars representing ± SD of the mean. Statistical significance was evaluated by two-tailed Student's T-test.
TCR surface copy number under the Nck overexpression condition relative to the vector only was 70% in the resting state and further reduced to 60% at 1 hr post-activation, consistent with the enhancement of CD3ε/Nck interaction by TCR activation. The enhancement of TCR down regulation by Nck overexpression is reduced with longer activation (3hrs). It is of note that the enhanced TCR downregulation by Nck overexpression is not obvious with stronger activation using higher concentration of the VSV8 peptide (Figure 7). This indicates that stronger stimulation reduces the sensitivity of TCR internalization to a functional Nck/CD3ε interaction. This is consistent with reported data showing that the long treatment with much stronger stimulators, such as super antigen, is insensitive to Nck/CD3ε interaction22. Since Nck induced TCR downregulation is more effective for weaker stimulations, it seems that Nck provides a safety mechanism preventing erroneous T-cell activation by weak non-cognate stimuli. The TCR downregulation in the resting state was somewhat unexpected given that the CD3ε/Nck interaction is not observed constitutively at immunoprecipitation conditions (Supplemental Figure 4B). Perhaps binding is too weak and/or the accessibility is too limited to support co-immunoprecipitation under the condition we used. The transient exposure of endocytosis signal is also suggested for another TCR endocytosis mechanism in resting-state T cell, which involves the CD3γ di-leucine motif37. These data seem to be in conflict with results by Szymczak et al., showing that the TCR expression level in resting as well as super antigen-activated states was the same between the CD3ε-deficient ΔP mice rescued by the overexpression of wild type CD3ε and CD3ε mutant with PRS mutation22. However, one should keep in mind that comparing TCR surface expression levels under CD3 overexpression condition may not be physiologically relevant since it shows surface TCR concentrations that are different from endogenous expression38. Furthermore, and as already discussed above, the mutant utilized in these studies is not optimal for analyzing the effect of the CD3ε/Nck interaction, as it doesn't modify the key Nck-binding residues Asp165 and Tyr166. We also think that the specific peptide stimulation by a peptide-MHC complex is more physiologically relevant and suitable to analyze the effect of the CD3ε/Nck interaction compared to the strong and long activation procedure with superantigen used by Szymczak et al.22.
Figure 7.
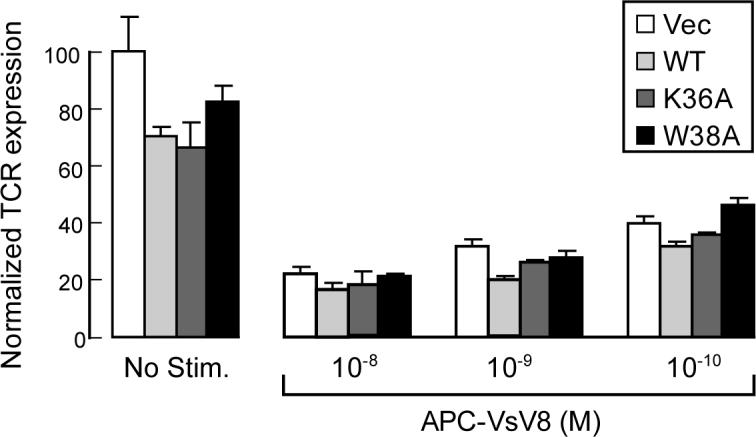
Downregulation of TCR expression by stimulation with different concentrations of the APC-VsV8 complex. TCR surface expression of LN CD8+ T cells transfected with either empty vector (Vec), wt Nck (WT), or mutants of Nck (K36A or W38A) were tested without stimulation and 3hrs after the activation with different concentration of VsV8-loaded APCs are shown. FACS analysis was done in the same way as in Figure 6. The indicated values are average of three separate experiments with error bars representing ± SD of the mean.
Since the molecular basis of TCR endocytosis remains to be evaluated37, the precise sequence of events by which the CD3ε/Nck interaction promotes TCR endocytosis is unclear. However, it is notable that Nck interacts with proteins involved in actin cytoskeletal rearrangement. These include the Wiskott Aldrich Syndrome Protein (WASP) and Pak1, which target SH3 domains distinct from SH3.1 (i.e. WASP and Pak1 binds to Nck SH3.3 and SH3.2 domains, respectively39). Since the actin cytoskeleton rearrangement appears to be required for TCR endocytosis40, our observations provide a potential mechanistic link between the CD3ε complex and the endocytosis machinery. The relationship between this CD3ε/Nck mechanism with that involving the CD3γ di-leucine motif37, remains to be investigated.
Conclusion
In summary, we determined the structure of the CD3ε/Nck2-SH3.1 complex, the first described case for the recognition of an atypical PxxDY sequence by a SH3 domain. The structure revealed the mechanistic detail of a particularly interesting feature of the PxxDY interaction in the context of CD3ε/Nck binding such that the interaction is regulated by Tyr166 phosphorylation. More importantly, the structural and functional data suggest that the CD3ε/Nck interaction may have the potential to prevent ITAM phosphorylation that would arise from weak stimuli. This is supported by in vitro phosphorylation analysis carried out here. Furthermore, the CD3ε/Nck interaction mediates T cell receptor downregulation in resting and activated T cells. Since Nck is known as an adaptor linked to actin cytoskeleton rearrangements, the result provides an important coupling between CD3ε and TCR endocytosis. CD3ε/Nck binding is the first mechanistic interaction shown to regulate TCR expression levels in the resting T cell state. These findings raise the important possibility that the non-ITAM-requiring interaction serves as a negative regulator for T-cell activation by preventing the CD3ε ITAM phosphorylation and/or removing TCR from T cell surface. This observation is somewhat unexpected considering that former reports assign enhancing roles to the CD3ε/Nck interaction20,23. However, we cannot exclude the possibility of an indirect contribution of CD3ε/Nck interaction to T cell activation, such as recruiting activation molecules via the Nck second or third SH3 domain or its SH2 domain, all of which are distinct from the CD3ε interaction. The data presented here have established a clear role for Nck for TCR downregulation. Given the correlation between the cell fate in thymocytes selection and surface TCR expression level41,42, this may stimulate further research to reveal mechanistic details of TCR downregulation, the fate of the TCR after endocytosis, and its role in activation and development of T cells. Interestingly, a recent publication appeared independently of our completed study25 supporting the role of Nck in CD3ε endocytosis we establish here. Using “knock-in” mice carrying a deletion of the PRS in CD3ε, TCR surface expression level was found to be enhanced on DP thymocytes. Considering the clear role of Nck in TCR endocytosis, it is attractive to imagine that Nck serves as a physical link between CD3ε and the endocytosis machinery, playing an essential role in thymocyte development. The mechanism that recruits Nck to the CD3ε in an activation-dependent manner is not yet to befully understood. Further analyses addressing the structure of the CD3ε cytoplasmic tail in the TCR complex would be desirable for understanding the mechanistic detail of TCR signal transduction.
Materials and methods
All chemicals were purchased from Sigma unless otherwise noted. All stable-isotope-labeled materials were acquired from Cambridge Isotope laboratories. See supplemental material for the composition of buffers used in the experiments.
Expression and purification of Nck2 SH3.1
The gene for Nck2 SH3.1 consisting of 67 amino acid residues, was cloned into the pET30 vector (Novagen) and the protein expression and purification was performed as previously described28,43.
Preparation of the CD3ε peptides
The synthetic non-isotopically labeled CD3ε peptides were obtained from Tufts-New England Medical Center peptide synthesis facility (Boston, MA) (The sequences are shown in the supplemental material). The CD3ε segment (residue number 143−183) and the mutants of the segment were produced as GB1 fusion proteins in an E.coli strain, BL21 (DE3). The GB1 fusion protein has the PreScission™ protease recognition site, between GB1 and CD3ε segment, which produces the CD3ε segment with an additional LEHHHHHH purification tag at its C-terminus. The GB1 fusion protein was purified with Ni-NTA affinity chromatography and digested by PreScission protease. The digestion product was re-purified by a Ni-NTA column to obtain pure CD3ε segment.
Expression and purification of the scCD3ε/Nck-SH3.1 complex
The expression vector for the scCD3ε/Nck-SH3.1 complex was established by inserting a double strand DNA fragment (The sequence is shown in the supplemental material) in the NdeI site of the Nck2 SH3.1 expression vector. The resulting vector encodes a 93-residue protein with the Nck binding region of CD3ε (155-KERPPPVPNPDY-166) connected to the N-terminus of Nck SH3.1 through a 13 amino-acid linker (NSLKKGSLVKNLH). The sc protein was expressed and purified in the same way as Nck2 SH3.1.
NMR spectroscopy
All experiments were performed on a Bruker Avance 500 spectrometer equipped with a cryogenic probe. All spectra were collected using samples in 10 mM sodium phosphate buffer (pH 6.8), containing 100 mM NaCl and 10% D2O at 298 K. Spectra were processed using XWINNMR (Bruker) and analyzed with Sparky44 or CARA45. The resonance assignments for the scCD3ε/Nck2-SH3.1 complex were accomplished by HNcocaN and HNcocanH experiments46, HCcoNH experiment47, and standard triple resonance experiments48. The 15N NOESY experiments and 13C NOESY experiments specific for either aliphatic or aromatic regions were carried out with a 2 mM sample of the scCD3ε/Nck2-SH3.1 to obtain distance constraints. Dihedral angle restraints were obtained from TALOS analysis49.
Structure determination of CD3ε/Nck2-SH3.1 complex
The structure of CD3ε/Nck2-SH3.1 complex was determined with a simulated annealing protocol using the structural restraints derived from NMR spectroscopy. The final structures with statistical results shown in Table 1 were calculated using CYANA50. 100 sets of the final structure were calculated and 20 best structures with the lowest energy were used for further analysis. The structures were analyzed using PROCHECK-NMR51 and MOLMOL programs.
Src kinase assay
Recombinant human Fyn and Lck tyrosine kinases were purchased from Invitrogen. 5 μM of the CD3ε segment (residue 144−183) or mutants of the CD3ε segments were mixed with 1 μg/ml of Fyn or 5 μg/ml Lck in phosphorylation buffer and incubated in the presence or absence of Nck2 SH3.1 for 30 min at 37 °C. The reaction sample was subjected to SDS-PAGE and immunoblotting with 4G10 Ab. Kodak 1D was used for quantification in immunoblotting.
TCR internalization assay
N15 mice were generated as described previously42. C57BL/6 mice were purchased from Taconic Farms. All lines were maintained and bred under sterile barrier conditions in the animal facility of Dana Farber Cancer Institute (Boston, MA) as approved by the appropriate institutional review committee. The retrovirus infection and the first activation of N15 mice LN T cell suspension was done in the same way as described previously36. The activated LN cells (2 × 105 cells) were restimulated by 1 nM N15 TCR specific VSV8 peptide or 10 μg/ml 2C11 Ab loaded on APCs (2 × 105 cells) obtained from C57BL/6 mice for 1−3 hrs at 37 °C. The cells were surface stained by PE-conjugated MR9.4 Ab for monitoring surface expression level of TCR and CyChrome-conjugated α-CD8 antibody for gating CD8+ T cells, and subjected to flow cytometric analysis using a FACScan (BD bioscience) and FlowJo software (Three Star).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (grants AI37581 to GW and AI19807 to ER). Maintenance of the NMR instruments used here was supported by the NIH grants GM47467 and EB002026. K.T. is supported by Japan Society for Promotion of Science. We thank Drs. Linda K. Clayton, Kristine Brazin, Maki Touma, and Robert J. Mallis for fruitful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION CODES:
The coordinates of the scCD3ε/Nck-SH3.1 complex have been deposited at PDB (access code 2jxb).
References
- 1.Wang J.-h., Reinherz EL. Structural basis of T cell recognition of peptides bound to MHC molecules. Molecular Immunology. 2002;38:1039–1049. doi: 10.1016/s0161-5890(02)00033-0. [DOI] [PubMed] [Google Scholar]
- 2.Qian D, Weiss A. T cell antigen receptor signal transduction. Curr Opin Cell Biol. 1997;9:205–12. doi: 10.1016/s0955-0674(97)80064-6. [DOI] [PubMed] [Google Scholar]
- 3.Werlen G, Palmer E. The T-cell receptor signalosome: a dynamic structure with expanding complexity. Current Opinion in Immunology. 2002;14:299–305. doi: 10.1016/s0952-7915(02)00339-4. [DOI] [PubMed] [Google Scholar]
- 4.Alarcon B, Gil D, Delgado P, Schamel WW. Initiation of TCR signaling: regulation within CD3 dimers. Immunol Rev. 2003;191:38–46. doi: 10.1034/j.1600-065x.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- 5.Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–9. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Hayes SM, Shores EW, Love PE. An architectural perspective on signaling by the pre-, alphabeta and gammadelta T cell receptors. Immunol Rev. 2003;191:28–37. doi: 10.1034/j.1600-065x.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 7.Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105:913–23. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 8.Sun ZY, et al. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci U S A. 2004;101:16867–72. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnett KL, Harrison SC, Wiley DC. Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment. Proc Natl Acad Sci U S A. 2004;101:16268–73. doi: 10.1073/pnas.0407359101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudolph MG, Luz JG, Wilson IA. STRUCTURAL AND THERMODYNAMIC CORRELATES OF T CELL SIGNALING. Annual Review of Biophysics and Biomolecular Structure. 2002;31:121–149. doi: 10.1146/annurev.biophys.31.082901.134423. [DOI] [PubMed] [Google Scholar]
- 11.Garcia KC, Teyton L, Wilson IA. STRUCTURAL BASIS OF T CELL RECOGNITION. Annual Review of Immunology. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 12.Pitcher LA, van Oers NS. T-cell receptor signal transmission: who gives an ITAM? Trends Immunol. 2003;24:554–60. doi: 10.1016/j.it.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Underhill DM, Goodridge HS. The many faces of ITAMs. Trends Immunol. 2007;28:66–73. doi: 10.1016/j.it.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Cantrell D. T cell antigen receptor signal transduction pathways. Annu Rev Immunol. 1996;14:259–74. doi: 10.1146/annurev.immunol.14.1.259. [DOI] [PubMed] [Google Scholar]
- 15.Alberola-Ila J, Takaki S, Kerner JD, Perlmutter RM. Differential signaling by lymphocyte antigen receptors. Annu Rev Immunol. 1997;15:125–54. doi: 10.1146/annurev.immunol.15.1.125. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Weiss A. T cell receptor signalling. J Cell Sci. 2001;114:243–4. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- 17.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–27. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 18.Hatada MH, et al. Molecular basis for interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature. 1995;377:32–8. doi: 10.1038/377032a0. [DOI] [PubMed] [Google Scholar]
- 19.Deindl S, et al. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–46. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 20.Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–12. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 21.Gil D, Schrum AG, Alarcon B, Palmer E. T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J Exp Med. 2005;201:517–22. doi: 10.1084/jem.20042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szymczak AL, et al. The CD3epsilon proline-rich sequence, and its interaction with Nck, is not required for T cell development and function. J Immunol. 2005;175:270–5. doi: 10.4049/jimmunol.175.1.270. [DOI] [PubMed] [Google Scholar]
- 23.Kesti T, et al. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3epsilon. J Immunol. 2007;179:878–85. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 24.Borroto A, et al. The CD3{epsilon} Subunit of the TCR Contains Endocytosis Signals. J Immunol. 1999;163:25–31. [PubMed] [Google Scholar]
- 25.Mingueneau M, et al. The proline-rich sequence of CD3epsilon controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat Immunol. 2008;9:522–32. doi: 10.1038/ni.1608. [DOI] [PubMed] [Google Scholar]
- 26.Mongiovi AM, et al. A novel peptide-SH3 interaction. Embo J. 1999;18:5300–9. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freund C, et al. Structural investigations of a GYF domain covalently linked to a proline-rich peptide. Journal of Biomolecular NMR. 2003;27:143–149. doi: 10.1023/a:1024983029700. [DOI] [PubMed] [Google Scholar]
- 28.Park S, Takeuchi K, Wagner G. Solution structure of the first SRC homology 3 domain of human Nck2. J Biomol NMR. 2006;34:203–8. doi: 10.1007/s10858-006-0019-5. [DOI] [PubMed] [Google Scholar]
- 29.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 30.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–7. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 31.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–8. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 32.Castagnoli L, et al. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567:74–9. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- 33.Gauen LK, et al. Interactions of p59fyn and ZAP-70 with T-cell receptor activation motifs: defining the nature of a signalling motif. Mol Cell Biol. 1994;14:3729–41. doi: 10.1128/mcb.14.6.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaynberg J, et al. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–23. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Qualmann B, Kessels MM. Endocytosis and the cytoskeleton. Int Rev Cytol. 2002;220:93–144. doi: 10.1016/s0074-7696(02)20004-2. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, Reinherz EL. CD2BP1 modulates CD2-dependent T cell activation via linkage to protein tyrosine phosphatase (PTP)-PEST. J Immunol. 2006;176:5898–907. doi: 10.4049/jimmunol.176.10.5898. [DOI] [PubMed] [Google Scholar]
- 37.Geisler C. TCR trafficking in resting and stimulated T cells. Crit Rev Immunol. 2004;24:67–86. doi: 10.1615/critrevimmunol.v24.i1.30. [DOI] [PubMed] [Google Scholar]
- 38.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–94. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 39.McCarty JH. The Nck SH2/SH3 adaptor protein: a regulator of multiple intracellular signal transduction events. Bioessays. 1998;20:913–21. doi: 10.1002/(SICI)1521-1878(199811)20:11<913::AID-BIES6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, et al. Antigen receptor-induced activation and cytoskeletal rearrangement are impaired in Wiskott-Aldrich syndrome protein-deficient lymphocytes. J Exp Med. 1999;190:1329–42. doi: 10.1084/jem.190.9.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebzda E, et al. Selection of the T cell repertoire. Annu Rev Immunol. 1999;17:829–74. doi: 10.1146/annurev.immunol.17.1.829. [DOI] [PubMed] [Google Scholar]
- 42.Ghendler Y, et al. Double-positive T cell receptor(high) thymocytes are resistant to peptide/major histocompatibility complex ligand-induced negative selection. Eur J Immunol. 1997;27:2279–89. doi: 10.1002/eji.1830270923. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi K, Ng E, Malia TJ, Wagner G. 1−13C amino acid selective labeling in a 2H15N background for NMR studies of large proteins. J Biomol NMR. 2007;38:89–98. doi: 10.1007/s10858-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. SPARKY 3 - NMR Assignment and Integration Software. University of California; San Francisco: 2006. [Google Scholar]
- 45.Keller R. The Computer Aided Resonance Assignment Tutorial. 2004 [Google Scholar]
- 46.Sun ZY, Frueh DP, Selenko P, Hoch JC, Wagner G. Fast assignment of 15N-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J Biomol NMR. 2005;33:43–50. doi: 10.1007/s10858-005-1284-4. [DOI] [PubMed] [Google Scholar]
- 47.Sun ZY, et al. High-resolution aliphatic side-chain assignments in 3D HCcoNH experiments with joint H-C evolution and non-uniform sampling. J Biomol NMR. 2005;32:55–60. doi: 10.1007/s10858-005-3339-y. [DOI] [PubMed] [Google Scholar]
- 48.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 49.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 50.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–98. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.