Abstract
Isochromosome 17q, or i(17q), is one of the most frequent nonrandom changes occurring in human neoplasia. Most of the i(17q) breakpoints cluster within a ∼240-kb interval located in the Smith-Magenis syndrome common deletion region in 17p11.2. The breakpoint cluster region is characterized by a complex architecture with large (∼38–49 kb), inverted and directly oriented, low-copy repeats (LCRs), known as REPA and REPB that apparently lead to genomic instability and facilitate somatic genetic rearrangements. Through the analysis of bacterial artificial chromosome (BAC) clones, pulsed-field gel electrophoresis (PFGE), and public array comparative genomic hybridization (array CGH) data, we show that the REPA/B structure is also susceptible to frequent meiotic rearrangements. It is a highly dynamic genomic region undergoing deletions, inversions, and duplications likely produced by non-allelic homologous recombination (NAHR) mediated by the highly identical SNORD3@, also known as U3, gene cluster present therein. We detected at least seven different REPA/B structures in samples from 29 individuals of which six represented potentially novel structures. Two polymorphic copy-number variation (CNV) variants, detected in 20% of samples, could be structurally described along with the likely underlying molecular mechanism for formation. Our data show the high susceptibility to rearrangements at the i(17q) breakpoint cluster region in the general population and exemplifies how large genomic regions laden with LCRs still represent a technical challenge for both determining specific structure and assaying population variation. The variant REPA/B structures identified may have different susceptibilities for inducing i(17q), thus potentially representing important risk alleles for tumor progression.
Structural aberrations involving chromosome 17 are frequently observed in human carcinomas. Isochromosome 17q, or i(17q), is one of the most frequent nonrandom genomic alterations ocurring in primitive neuroectodermal tumor/medulloblastoma (50%; Biegel 1997), chronic myeloid leukemia (CML), acute myeloid leukemia (AML), and myelodysplastic syndrome (MDS) (Babicka et al. 2006). According to the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman), i(17q) can be found in ∼2.5% of hematologic malignancies, particularly in CML, in which it can be found in ∼12% of cases. In neuroblastomas and hematologic malignancies, amplification of 17q is a significant predictive factor for adverse outcome (Bown et al. 1999). In a prospective clinical trial of patients with AML aged 60 yr and above, abnormal 17p and 17q were among the most frequent findings (82%–100%) with complex karyotypes (van der Holt et al. 2007).
Remarkably, i(17q) breakpoints frequently cluster in the genome at the short arm of chromosome 17, specifically at the 17p11 region (Fioretos et al. 1999; Scheurlen et al. 1999; Babicka et al. 2006; Mendrzyk et al. 2006). Babicka et al. (2006) showed that the 17p11 is the only region that has been frequently affected in all three hematologic malignancies (AML, CML, MDS), indicating such abnormality as having a pathogenic role during progression and clonal evolution of those diseases.
Emerging evidence strongly associates the 17p11 breakpoint predisposition to the 17p11 genomic architectural features. The 17p11, particularly the 17p11.2 sub-band, is a highly genetically unstable region and presents a unique genomic architecture, marked by several dispersed as well as adjacent and both directly oriented and inverted low-copy repeats (LCRs). The Smith-Magenis syndrome (SMS; MIM 182290) is caused by a recurrent ∼4-Mb deletion generated by Non-Allelic Homologous Recombination (NAHR) that is mediated by two LCRs, the so-called proximal and distal SMS-REPs (Chen et al. 1997; Park et al. 2002). Alternatively, NAHR between the same LCRs can produce a reciprocal duplication, which is the molecular cause of the Potocki-Lupski syndrome (PTLS; MIM 610883) (Potocki et al. 2000; Shaw et al. 2002; Bi et al. 2003; Potocki et al. 2007). Both diseases have been reported also in patients carrying nonrecurrent deletions/duplications potentially stimulated by other LCRs in the region (Stankiewicz et al. 2003; Shaw et al. 2004; Potocki et al. 2007). The 17p11.2 region is also frequently subject to other structural variations, including a huge inversion involving the SMS-REP distal and SMS-REP middle (Database of Genomic Variants [DGV] http://projects.tcag.ca/variation/).
In 2004, Barbouti and colleagues structurally characterized an ∼240-kb region in 17p11.2 spanning the i(17q) breakpoint cluster region in patients with hematological malignancies CML, AML, and MDS. This region is located within the 4-Mb SMS common deletion interval, specifically between the middle and proximal SMS-REPs. It is characterized by a complex architecture with large (∼38–49 kb) LCRs, known as REPA and REPB. In the reference genome, two copies of REPA are present in an inverted orientation (REPA1 and A2); they share 99.8% identity with each other. REPB is present in three copies, REPB1, B2, and B3, also sharing 99.8% of sequence identity. B1 and B3 copies are in direct orientation, whereas the B2 copy is in an inverted orientation with respect to REPB1, therefore potentially forming a cruciform structure. Five genes from the SNORD3@ cluster (also known as U3a1, U3a2, U3b1, U3b2, U3b3 or, according to the snoRNABase [http://www-snorna.biotoul.fr/] U3-2, U32b, U3-4, U3, U3-3, respectively) are located in the center of the palindromic structure and at the head portion of each REPA and B repeat. The SNORD3@ cluster includes evolutionarily conserved motifs (the snoRNABase) and belongs to a RNU3 gene family of small nuclear RNAs required for the processing of the pre-18S rRNA (Gao et al. 1997). Based on its complex genomic features, it has been suggested that the REPA/B structure can cause genomic instability and elicit susceptibility to genetic rearrangements (Barbouti et al. 2004; Mendrzyk et al. 2006). The formation of i(17q) appears to result from interchromatid mispairing of the direct repeats, producing a dicentric chromosome and an acentric chromosome which is lost during cell division.
Our objective was to systematically investigate the structure of the REPA/B region and determine if it varies within the population. According to the DGV, the REPA/B region is susceptible to frequent meiotic rearrangements, as 36% of all HapMap samples showed copy number polymorphism therein. We performed a detailed statistical analysis of Redon et al. (2006) comparative genomic hybridization (CGH) data on the REPA/B region across the HapMap samples. The results revealed the high frequency of variation existent at the REPA/B loci worldwide. Such numbers, however, do not elucidate how the structure can be genomically organized. Thus, we selected bacterial artificial chromosome (BAC) clones spanning the REPA/B region from the RPCI-11 library. The digestion pattern of each clone was obtained by pulsed-field gel electrophoresis (PFGE) and the REPA/B content was assessed by target-specific probe hybridizations. Empirically determined physical maps were compared to the Human Genome database. Surprisingly, we assembled two BAC-based REPA/B structures, each one with a different total size and REPA/B content. Novel different REPA/B structures could also be assessed after applying the PFGE assay on the analysis of a group of SMS patients, hemizygous for the region, and another group of random samples. Our results suggest that the variation can be far more complex than previously thought, possibly reflecting a wide spectrum of genomic rearrangements therein (inversions, duplications, deletions). Finally, we propose that the mechanism underlying the different REPA/B structures and rearrangements is likely NAHR between the highly similar SNORD3@ gene cluster.
Results
Statistical analysis of the Database of Genomic Variants: Evidence for copy number polymorphism at the REPA/B region across the HapMap samples
Redon et al. (2006) analyzed the entire human genome for copy-number variation (CNV) using two different array-based copy number analysis (ABCNA) platforms: Affymetrix GeneChip Human Mapping 500K (500K EA) and Whole Genome TilePath (WGTP) array Comparative Genomic Hybridization (aCGH) with large-insert BAC clones. The CNV in the REPA/B region was detected only by the WGTP platform whereas the 500K EA was not able to detect any variation. REPA/B was classified as a complex locus, whose underlying molecular basis could not be reliably inferred (Redon et al. 2006).
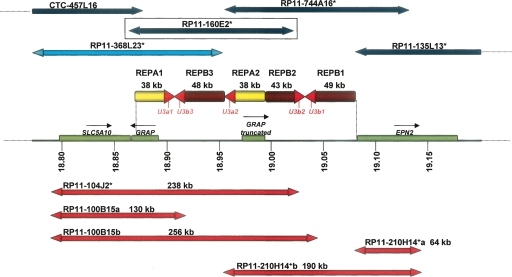
We downloaded the log2 probe ratio intensities available from the Copy Number Variation project (http://www.sanger.ac.uk/humgen/cnv/data/) for the 270 HapMap samples of each spanning REPA/B clone: RP11-368L23, RP11-104J2, RP11-160E2, RP11-744A16, and RP11-135L13. The estimated genomic position for each clone within the REPA/B region can be seen in Figure 1. Clone RP11-135L13 overlaps just the very end of the REPA/B region. Indeed, it shows very low variance (log2 intensity ratio < 0.01) across all the HapMap samples; hence, it was not informative for our analysis. The RP11-744A16 variance is lower than RP11-368L23, RP11-104J2, and RP11-160E2 variances, thus we considered it less informative as well. BAC clones, RP11-368L23, RP11-744A16, and RP11-104J2 span part of REPA/B plus either the upstream or downstream flanking regions with no copy change. It is uncertain how to interpret BAC array-based CGH results when the BAC clone partially overlaps the CNV region. Thus, in order to minimize false-negative assessments, we considered only the RP11-160E2 clone log2 ratio intensities in our analysis. Based on the reference genome, this clone includes 80% of the REPA/B region and it is constituted by REPA/B elements only (Fig. 1). Thus, all our statistical inferences on REPA/B within the worldwide population are based on the RP11-160E2 interrogating clone.
Figure 1.
The i(17)q cluster breakpoint genomic architecture at 17p11.2 is depicted; the positions are given relative to NCBI build 35. The horizontal line depicts the high-resolution physical map of the i(17q) breakpoint cluster region (REPA/B region) on chromosome 17 with coordinates (in Mb) listed below (Barbouti et al. 2004); genes are shown as green rectangles; yellow and brown arrows represent REPA and REPB, respectively; red arrowheads of REPAs and REPBs represent a nearly identical 4-kb sequence shared among both REPAs and REPBs that includes the SNORD3@ gene cluster (also known as U3). Above, horizontal dark blue arrows represent BAC clones positioned according to Barbouti et al. (2004); red arrows represent some of the BAC clones selected from RPCI-11 in this study. The BAC RP11-160E2, which was used in the statistical analysis, is boxed to emphasize its location. The double arrowheads represent the BAC ends. We sequenced the BAC forward and reverse ends and estimated their sizes and position along the genome. RP11-100B15 and RP11-210H14+ present two possible positions according to their end similarity to the reference genome, so we labeled these possibilities as a and b. Remarkably, clone RP11-210H14+ presented its forward and reverse ends matching to the same strand, suggesting an inversion of one of the REPB repeats. Clones marked with asterisks are present on the WGTP platform used in Redon et al. (2006).
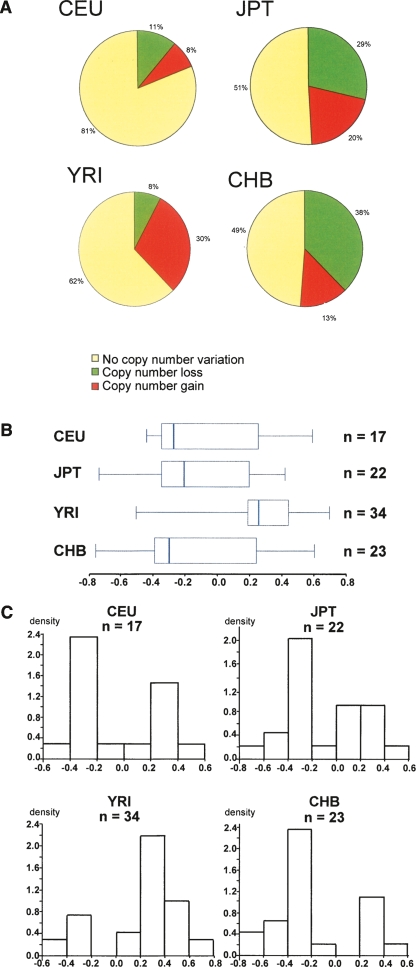
REPA/B CNV occurs in 96 of 269 (36%) HapMap samples, according to the DGV (variation 4025). Population-specific frequencies were 19% in CEU (US residents with Northern and Western European ancestry collected by the Center d'Etude du Polymorphisme Humain [CEPH]), 38% in Yoruba individuals from Nigeria (YRI), 49% in Japanese individuals from Tokyo (JPT), and 51% in Chinese individuals from Beijing , China (CHB) (Fig. 2A). It is important to note that, although CEU and YRI populations comprise parents and offspring, the results do not change if the offspring is removed. The CNV frequency distribution is statistically different among each sample pair (P < 0.008, χ2 test and Bonferroni adjusting for multiple test comparisons), except between JBT and CHB. CEU samples showed a similar frequency of losses and gains whereas JBT and CHB presented more losses and YRI more gains. The lower loss/gain frequencies observed in the CEU group may be explained by the fact that the reference genome used was from this group (Redon et al. 2006), thus, there may be a higher frequency of the same REPA/B structure (represented by the control individual used in the CGH experiment) within the CEU population. Alternatively, BAC-based CGH arrays are limited in their resolution capability and unable to detect subtle copy number changes of just one REP element. The BAC based-array has both reduced resolution and sensitivity that can contribute to increase false-negative variation rates when it comes to the analysis of such complex CNV loci.
Figure 2.
Comparative analysis of REPA/REPB variation among HapMap populations using BAC-array CGH data from the Copy Number Variation project (http://www.sanger.ac.uk/humgen/cnv/data/). (A) Frequency of CNV in all 269 HapMap samples. (B,C) Variation range of the copy number HapMap samples classified with losses and gains according to Redon et al. (2006) and DGV. (B) Box plot graphic of the log2 intensity ratios. (C) Histogram of the log2 intensity ratios versus density. The density was calculated as the relative frequency of the log2 value data that have been grouped into a specified numeric interval. As the interval is equally wide, the bar’s height represents the relative frequencies.
The samples classified with losses and gains according to Redon et al. (2006) were reanalyzed by us. The median of log2 ratio intensity values for each population sample group can be seen on the box plot represented in Figure 2B. All HapMap populations presented a high-variance range, suggesting a high variability of the REPA/B region. CEU, JBT, and CHB have their median skewed toward copy number losses. On the other hand, the YRI median is much higher than the other groups; nonparametric Mann-Whitney pairwise comparison indicates that the YRI sample median is statistically different from JBT and CHB median (P = 0.0016 and P = 0.000978, respectively, Bonferroni adjusted to P < 0.008), although not from CEU. About 50% of YRI log2 ratio intensity values are grouped between 0.2 and 0.4, as opposed to <0.2 values from CEU, CHB, and JPT, suggesting that the YRI samples show a bias toward REPA/B copy number gain (Fig. 2B,C).
Determining alternative genomic structures for the REPA/B region
Barbouti et al. (2004) determined a consensus REPA/B structure using overlapping BACs, P1-derived artificial chromosome (PACs), and fosmid clones from different individuals to generate a physical map for the reference haploid genome covering 240 kb of this genomic interval. The complex structure is formed by two REPA copies (∼38 kb each, 99.8% similarity) and three REPB copies (∼43–49 kb each, 99.8% similarity), constituting a long palindromic structure (Fig. 1). In the original paper two potential structures, differing by an inversion, were proposed.
CGH showed the high frequency of population variation at the REPA/B loci worldwide. Such results, however, do not reveal how the structure is varying on the genomic level. Thus, we sought to identify alternative REPA/B structures within the BAC library (RPCI-11), which was generated using a single individual (Osoegawa et al. 2001). The RPCI-11 BAC clones were selected from the Genome Browser Gateway at the University of California Santa Cruz (http://genome.ucsc.edu/cgi-bin/hgGateway, NCBI build 35), either fully or just end-sequenced, which overlapped the REPA/B region (chr17: 18869254–19081415, NCBI 35). We also included BAC clones once identified by hybridization screening of the RPCI-11 library in our previous work, which were mapped to the REPA/B region (Park et al. 2002). We used three restriction enzymes (NotI, PacI, and RsrII) and PFGE in order to compare the clone digestion patterns, followed by REPA- and B-independent probe hybridization. We resequenced both clone ends of all clones that did not reveal the expected digested pattern (based on the reference genome restriction map analysis).
In total, we collected and analyzed 19 BAC clones from the RPCI-11 library. Using Sequencher software and the Genome Browser Gateway at the University of California Santa Cruz, along with a combination of PFGE, restriction mapping, and DNA sequence analysis, we were able to assign most of the clones to their expected genomic positions. However, three out of 19 clones, RP11-104J2, RP11-100B15, and RP11-210H14 (Fig. 3), did not match the genomic structure previously proposed by Barbouti et al. (2004). The NotI digestion pattern showed unexpected bands for all three clones (Fig. 3A): RP11-104J2 clone yielded a band ≤169 kb rather than the expected 238-kb band; RP11-100B15 yielded a band between 169 and 182 kb rather than either the expected 130 kb or 256 kb. Finally, RP11-210H14 generated one band around 169 kb rather than producing either 64-kb or 190-kb bands. Blast analysis of the end sequences of this clone to the reference genome showed both ends matching the same 5′–3′ orientation. This observation suggests that one of the ends is actually in an inverted orientation with respect to the reference. RP11-210H14 was the key clone to determining how the entire REPA/B structure is organized.
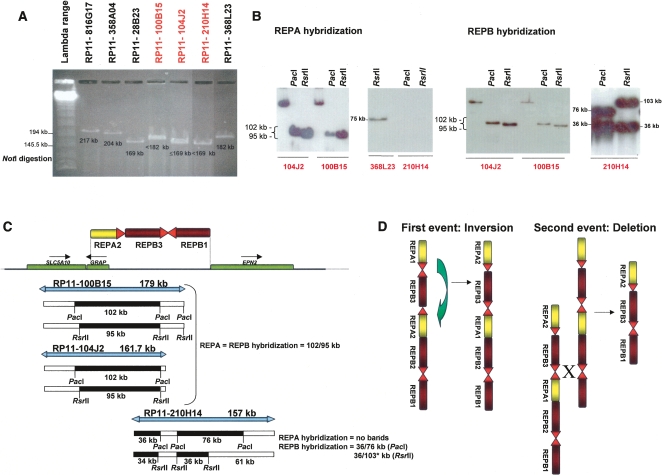
Figure 3.
RPCI-11 library clones presenting a new REPA/B structure. (A) NotI digestion profile in 1% agarose gel of some of the RPCI-11 library clones spanning the REPA/REPB region. NotI enzyme has just one restriction endonuclease recognition site per clone, thus we were able to estimate their overall size (shown below each band). Clones with black type were used as reference clones for band sizing whereas clones in red presented an unexpected digested band, suggesting a different genomic content compared to the reference genome. (B) PacI and RsrII digestion followed by specific REPA and REPB hybridization of the clones RP11-104J2, RP11-100B15, and RP11-210H14. These results confirmed the presence of REPA and REPB at the selected clones but in an unexpected copy number. RP11-104J2 and RP11-100B15 have just one copy of REPA and one copy of REPB; RP11-210H14 has no copies of REPA but two copies of REPB. (C) (Top) alternative genomic structure of REPA/B based on forward and reverse clone end sequences, restriction enzyme digestion profiles, and REPA and REPB hybridization. (Bottom) PacI and RsrII restriction maps for each clone according to that new alternative structure. *103 kb size (34 kb + 61 kb) is expected as the bacterial sequence part of the cloning BAC has no RsrII restriction site. The expected digestion patterns for all clones are consistent with those observed in B, so we can conclude that the individual used to construct the RP11 BAC library carries two different REPA/REPB copy numbers. (D) Molecular mechanisms and proposed model for the generation of the new structure based on Barbouti et al. (2004) REPA/REPB structure background. First event should be a large inversion involving REPAs; the presence of two REPA copies with 99.8% identity in inverted orientation would favor the occurrence of NAHR and produces inversion of the whole segment within. The second event is a deletion of one REPA and one REPB. This can occur by interchromosomal, intrachromosomal, or intrachromatid mispairing between the highly similar SNORD3@ gene clusters at the REPA/REPB and REPB/REPB heads, leading to a deletion of two REPs elements by the NAHR mechanism.
The PacI and RsrII restriction maps confirmed our results: the clones RP11-104J2, RP11-100B15, and RP11-210H14 do not present the expected reference genome digestion pattern (data not shown). We transferred the BAC digestions to a nylon membrane in order to hybridize them to specific REPA and REPB probes. The presence of these repeat elements was confirmed in the three clones, but not in the expected number (Fig. 3B). Both RP11-104J2 and RP11-100B15 have just one REPA and one REPB band (each one was expected to carry two REPA and two REPB instead, or in the alternative RP11-100B15b structure, one REPA and no REPB; Fig. 1). RP11-210H14 showed no REPA bands and two REPB bands, whereas the expected pattern would be one REPA and two REPB bands. We propose a new REPA/B structure that can be seen in Figure 3C (top). The restriction patterns observed for the three clones for both PacI and RsrII enzymes (Fig. 3B) are consistent with those predicted according to that alternative structure (Fig. 3C, bottom). Thus, these BAC experiments present unambiguous evidence that the individual that constitutes the RPCI-11 library carries two distinct structures, likely representations from each chromosome homolog (Fig. 3C).
The final structure, constituted by just one REPA and two REPB, can be achieved after, at least, two NAHR events mediated by the SNORD3 repeats, if each occurred on the Barbouti’s REPA/B type: one inversion mediated by the U3a1 and U3a2 repeats followed by one deletion mediated by the U3b3 and U3b2 (Fig. 3D). The SNORD3 genes are present five times within the REPA/B in the reference genome and are responsible for most of the isochromosome 17q formation (Barbouti et al. 2004). Such a structure is consistent with all results obtained by us for the three clones: the NotI BAC size bands, the digestion patterns obtained by NotI, PacI, and RsrII enzymes, the subsequent REPA and B hybridization unexpected ratios, and the inversion of one of the RP11-210H14 ends (Fig. 3).
Structural analysis of REPA/B using PFGE
To assess the variability of the REPA/B structures within the population, we performed a PFGE assay based on PacI digestion and REPA and B probe hybridizations. We selected two different sample groups to test (1) Smith-Magenis patients carrying 17p11.2 deletions (thereby hemizygous at the REPA/B locus), selected from Stankiewicz et al. (2003) and Shaw et al. (2004), and (2) anonymized random samples whose cell lines were available in our laboratory. The genomic DNA of each sample was digested by PacI followed by membrane transfer and two distinct hybridizations sequentially using REPA and B as probes. We anticipated the presence of two hybridization bands for REPA (76 and 102 kb) and two bands for REPB (76 and 102 kb), for structures similar to the reference genome (Type I) (Fig. 4A). Additionally, the relative intensity ratio of the bands should reflect the copy number of REPA and B present therein. For Type I, the relative intensity between the two REPA 76/102-kb bands should be 1:1, whereas between the REPB 76/102-kb bands such ratio should be ∼1:2 (Fig. 4A). We considered as potential “new structures” all samples whose hybridization patterns presented not only the unexpected sizes of REPA and/or REPB, but also those with unexpected relative intensity ratios (Fig. 4B–E).
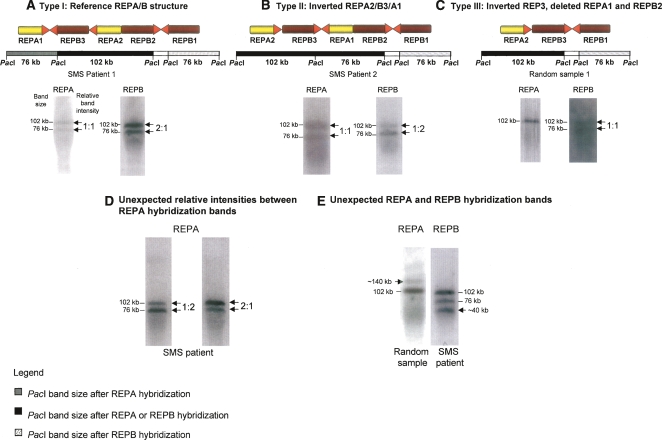
Figure 4.
REPA/REPB copy number polymorphism within the population using PFGE assay. Two different sample groups were used: Smith-Magenis patients carrying 17p11.2 deletions and random samples (refer to text for further details). (A–C) We detected and molecularly described three different REPA/REPB types (Types I, II, and III) based on PacI digestion and both REPA and REPB hybridization profiles of the DNA samples. Band sizes are shown at the left side of the hybridized membrane, whereas relative band intensities are shown on the right. (D,E) Other CNVs detected within these sample groups; each one belongs to a different individual and exemplifies either REPA or REPB variation. Contrary to Type I–III structures, we have no further evidence that allows us to assign a molecular genomic structure for those samples.
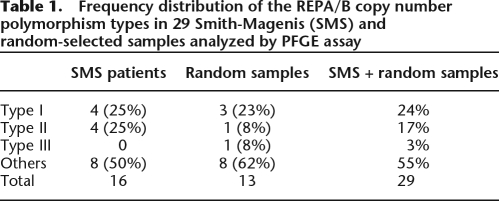
We analyzed 29 samples in total, from which 16 were SMS samples and 13 were random samples. The reference structure (Type I) was detected in 24% of samples; the structure containing an inverted REPB3 (Type II) was observed in 17% of samples and the alternative structure present in the RPCI-11 library (Type III) was detected in 3% of samples (Table 1). Among the SMS patient group, for whom we can assign unambiguously just one REPA/B structure given the hemizygous nature of the locus in such microdeletion patients, we detected four different REPA/B structures (Table 1; Fig. 4B,D,E): Type II (25%), two types with unexpected relative intensities of REPA hybridization bands, and one type with unexpected REPB hybridization band (140 kb). These three “other” last types are present at different frequencies, totaling 50% of the SMS samples. The types classified, as “others” are not structurally described herein, because we could not unambiguously determine the subunit orientation or composition of those structures using the PFGE technique alone. Among the random samples we also detected new different REPA/B structures (Table 1; Fig. 4C,E; data not shown): Type III (8%), one type with an unexpected REPB hybridization band (∼40 kb), and types with unexpected relative intensities of either REPA or REPB (those “other” types totaled 62% of the random samples). As the random samples can be heterozygous for the REPA/B loci, we do not know if the patterns represent the hybridization patterns of one or two different structures, but we can count at least two new structures detected within the random samples: Type III and the one presenting an unexpected 40-kb band. Thus, in total, we detected at least seven REPA/B structures, from which six are novel structures, but as suggested here, this number may be underestimated.
Table 1.
Frequency distribution of the REPA/B copy number polymorphism types in 29 Smith-Magenis (SMS) and random-selected samples analyzed by PFGE assay
Discussion
Barbouti et al. (2004) proposed that the LCRs—REPA and REPB—are important mediators of somatic recombination events leading to the formation of the dicentric isochromosome i(17q). Relatively few examples of mitotic NAHR have been documented (Flores et al. 2007; Bruder et al. 2008) since the description of the proposed mechanism for iso17q. Somatic NAHR may be an important molecular mechanism for other tumor-associated rearrangements.
Here, we propose that REPA and REPB not only lead to the formation of i(17) in somatic cells, but also mediate frequent genomic rearrangements in meiotic cells, producing a highly variable locus in different populations. The paralogous sequences REPA1 and REPA2 as well as REPB1, REPB2, and REPB3 share a significant stretch of identity (∼99.8%) along their 35–48 kb length, making them excellent substrates to undergo strand exchange and ectopic recombination through NAHR. Indeed, the statistical analysis of the log2 intensity ratios of the BAC-array CGH on the four HapMap populations revealed that the CNV average in this region is ∼36%: 19% in CEU, 38% in YRI, 49% in JPT, and 51% in CHB. The frequency of variation is less prominent in the CEU population but this is probably because the control sample used in CGH was from this group. Remarkably, when the four sample groups are compared, the African sample group (YRI) presents higher frequency of REPA/B copy number gain, whereas both Asian sample groups (JBT and CHB) showed a higher frequency of REPA/B copy number loss.
REPA/B genomic structure is certainly different among the worldwide populations, but how exactly they differ is not possible to ascertain by CGH. CGH provides neither positional nor orientation information but rather just changes of copy number for a given interrogated locus relative to the control. Through PFGE assays followed by REPA/B hybridization on Smith-Magenis samples (who are deleted for the SMS region and hemizygous for the REPA/B locus), we could detect four new REPA/REPB structures. The structure described by Barbouti et al. (2004), referred to here as Type I, was observed in 25% of the SMS patients. Type II (containing an A2/B3/A1 inversion) was also detected in 25% of the samples, whereas 50% were represented by three new, previously unrecognized types. Furthermore, through digestion and hybridization analysis of a BAC library of a single individual, we were able to characterize two REPA/REPB structures (Type II and III). In total, we analyzed 29 samples, from which 16 are SMS samples and 13 are random samples. Considering both SMS and random samples, the frequency of samples carrying new structures, classified as Type II, III, and others (Table 1) was surprisingly high and reached up to 75%. The reference structure (Type I) was detected in 24% of samples; Type II was observed in 17% of samples and Type III, detected on the RPCI-11 library, was found in only 3% of samples. Thus, the PFGE assay not only confirmed the high REPA/B locus variability observed on CGH data, but also shows that such variability is far more complex than previously thought.
According to the BAC analysis, as well as the PFGE results, we can conclude that REPA and B can vary either concomitantly or in an independent way. Thus, one can infer that the large log2 ratio variance range observed in all HapMap groups (Fig. 2B,C) is the most probably reflecting different rearrangements within REPA/B region, that is gain and/or losses of either REPA or REPB, as well as both at the same time. By PFGE we were also able to detect a large inversion (A2/B3/A1 Type II; Fig. 4B), not possible to discern by CGH.
The frequency of each type calculated here aims only to illustrate the high frequency of variation of this locus. For instance, Type I frequency should be considered with restrictions because it can represent more than one type. Gao et al. (1997) described an inversion on the REPA/B region (B2/A2/B3), different from the one detected by us (A2/B3/A1, Type II; Fig. 4B). Unfortunately, it is impossible to identify such inversion by PFGE because its profile cannot be distinguished from the Type I haplotype. This has also an implication for the molecular mechanism model proposed in Figure 3D for the generation of the new structure detected in the RPCI-11 library as it can also be generated by the Gao’s structure. In this case, the intrachromosomal or intrachromatid/interchromatid misparing, followed by NAHR, would occur between the two REPA elements (now in direct orientation) instead of between the REPB elements, as suggested here (Fig. 3D). Further research is needed to understand the structural variation underlying the REPA/B region.
Although the REPA and REPB copy number changes probably contribute to the CGH diversity (Fig. 2C), the CNV of the REPA/B in CGH assays cannot be simply analyzed as a dimorphic complete deletion (log2 ratio = −1) or a complete duplication (log2 ratio = 0.6) of the whole region. Actually those values are barely observed in the CGH results (Fig. 2B,C). As the log2 results cannot be easily interpreted, the region was classified as a complex locus, and hence one cannot reliably infer the underlying molecular basis of the CNV by CGH alone (Redon et al. 2006). Thus, such region, along with several other genomic regions, cannot be assayed by most of the current commercial array-CGH platforms. According to Redon et al. (2006), up to 12% or 131 CNVs detected by the WGTP platform were classified as complex loci and most of them (70.2%) are constituted by or are associated with segmental duplications. Recently, Hollox et al. (2008) reported an association between risk of psoriasis and increased copy number in a multiallelic CNV locus on 8p23, introducing an important technical breakthrough for typing multiallelic variants in a large cohort. However, despite the important contribution of the technique developed, it still does not provide us detailed structural information of how those regions are organized along the genome. Such structural characteristics have been emerging as very important for the understanding of several genomic disorders as well as susceptibility to genomic instability and the genomic rearrangements causing them. For example, in Williams Beuren, 17q21.31 microdeletion syndrome, and Sotos syndrome, each can be caused by deletions generated by NAHR during meiosis in heterozygous carriers of polymorphic inversions of the interval between flanking LCRs (Osborne et al. 2001; Visser et al. 2005; Koolen et al. 2006; Lupski 2006; Sharp et al. 2006; Shaw-Smith et al. 2006). Duplication/triplication rearrangement at 8p23.2 caused by NAHR in heterozygous carriers of a polymorphic 8p23.1 inversion was also recently reported (Giorda et al. 2007). Barbouti’s model for the iso17q formation relies on an interchromatid misparing of direct repeats, especially stimulated by the potential cruciform structure formed because of the presence of the REB1 and REPB2 inversions (Barbouti et al. 2004). As the REPA/B region is a highly polymorphic and dynamic region, it is an intriguing question whether any of the different REPA/B structures identified are more prone than others to undergo disease-causing rearrangements, such as those responsible for the iso17q formation.
The use of SMS patients enabled us to interpret the PFGE results, because we could unambiguously assign just one structure, otherwise confounded by the presence of digestion/hybridization allele profiles resulting from the presence of both chromosome homologs. Thus, such an approach seems to us fundamental to ascertain the genomic structure of this region. If one seeks to test the hypothesis that different REPA/B structures can lead to different rates of iso17q formation, it may be necessary to generate somatic cell hybrids to analyze the REPA/B hemizygous state. The REPA/B structure of the tumor tissue should be determined and, if possible, compared to the normal tissue (fibroblast, for example) from the same patient, enabling the molecular characterization of the altered chromosome 17. Other techniques to carry out the analysis can also be considered in order to complement the PFGE results, such as fiber-FISH (Heiskanen et al. 1994). In this case the fiber-FISH should be done using specific probes for REPA and REPB (Barbouti et al. 2004) instead of BACs that can produce cross-hybridization confounding the final result.
This work exemplifies how large complex genomic regions still represent a technical challenge both for determining precise genome structural organization in an individual and assaying its variation in different world populations. Such regions have been poorly analyzed even though new structural variation techniques have been recently developed. Precise knowledge of how REPA/B structure varies in populations, and how this variation is distributed worldwide is essential to investigating whether different REPA/B structures may play a role in susceptibility to neoplasia by inducing the iso(17q) formation.
Methods
Downloading log2 intensity ratios from REPA/B spanning clones and statistical data analysis
We downloaded the log2 probe ratio intensities available on the Copy Number Variation project (http://www.sanger.ac.uk/humgen/cnv/data/) for the 270 HapMap samples of each spanning REPA/B clone: RP11-368L23, RP11-104J2, RP11-160E2, RP11-744A16, and RP11-135L13. The data were compared by χ2 test and Bonferroni adjusting for multiple test comparisons and Mann-Whitney pairwise comparison using software Past version 1.74 downloaded from the website http://folk.uio.no/ohammer/past (Hammer et al. 2001). Box plots were employed to graphically represent the log2 ratio distribution of each HapMap group; it was drawn using the SSP statistical package (SSP, Smith's Statistical Package, http://www.economics.pomona.edu/StatSite/SSP.html). The HapMap collection comprises four populations: 30 parent-offspring trios of European descent from Utah, USA (CEU), 45 unrelated Japanese individuals from Tokyo, Japan (JPT), 30 parent-offspring trios of the Yoruba from Nigeria (YRI), and 45 unrelated Han Chinese subjects from Beijing, China (CHB).
BAC clone selection from the RP11 library
The RPCI-11 BAC clones were selected from the Genome Browser Gateway at the University of California Santa Cruz (http://genome.ucsc.edu/cgi-bin/hgGateway, NCBI build 35), either fully or just end-sequenced, which overlapped the REPA/B region (chr17: 18869254–19081415, NCBI 35). We also included BAC clones once identified by hybridization screening of the RPCI-11 library in our previous work, which were mapped at the REPA/B region (Park et al. 2002). Selected clones used in our analysis include: RP11-788I14, RP11-358A04, RP11-330C20, RP11-766J18, RP11-977H13, RP11-368L23, RP11-816G17, RP11-73E4, RP11-48J10, RP11-970O14, RP11-181I17, RP11-251B7, RP11-210H14, RP11-104J2, RP11-100B15, RP11-160E2, RP11-744A16, and RP11-135L13. We also included clone hCIT.457_L_16, although it is from a different library, because we sought to include all BAC clones used by Barbouti et al. (2004) in our analysis.
Genomic DNA samples
We selected two different sample groups to analyze for variation at the iso(17q) breakpoint cluster region, (1) 16 Smith-Magenis patients carrying 17p11.2 deletions and thus hemizygous for the REPA/B locus, selected from Stankiewicz et al. (2003) and Shaw et al. (2004), and (2) 13 random samples. Both the SMS and random samples were selected on the basis of the availability of stored high-molecular-weight DNA agarose plugs isolated from peripheral blood samples or lymphoblastoid cell lines, established by standard methods. Peripheral blood samples from patients and family members were obtained after informed consent.
Pulsed-field gel electrophoresis
We selected three restriction enzymes for the restriction map analysis. NotI is a rare-cutter enzyme and does not have restriction sites within the REPA/B region. It was selected because it has one restriction endonuclease recognition site per clone (within the bacterial sequence), thus we would be able to estimate their overall size. PacI and RsrII are also rare-cutter enzymes but, according to the reference genome, they may cut at spaced region within the REPA/B region. We digested 1 μg of BAC clones with NotI, PacI, and RsrII (New England Biolabs) at 37°C DNA overnight followed by a separation, ranging from 20 to 400 kb on a 1% agarose gel (Bio-Rad) in 0.5× Tris–Borate–EDTA buffer using a Bio-Rad CHEF Mapper. All samples co-migrated with lambda chromosome size marker (New England Biolabs). We visualized separated DNA by ethidium bromide staining. Agarose plugs containing human DNA were digested with NotI and PacI restriction enzymes for 2 d at 37°C. PFGE conditions were similar to those described previously.
Probe design and Southern hybridization
The BLAST 2 browser (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) and Sequencher software (Gene codes Corporation) were used to search for REPA- and REPB-specific targets. Repeat sequence analysis was performed with RepeatMasker (http://www.repeatmasker.org). The primers were designed using Primer3 website (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). The REPA and REPB probes, 602 bp and 488 bp length respectively, were generated by PCR product (REPA–REPAF: GGGTAGGTTTTGCCAGAGTTG and REPAR: GTCTC TAGACCCAAGTGTGGTGTT; REPB–REPBF: GTCTCCAGTCTCT CAGATTGAGGT and REPBR: AGGGAACAAGAGACCCAGAAG) of the REPA and REPB present in the RP11-160E2 clone.
Following the separation of either BAC clones or human DNA samples by PFGE, we transferred DNA to positively charged Sure Blot Nylon Membranes (Intergen Company) by standard methods for 2 d. We labeled ∼200–250 ng of probe DNA with 32P-dCTP (MP Biomedicals) by random priming for at least 2 h at 37°C (Roche). We prehybridized membranes with salmon sperm DNA (Sigma) for 4 h and hybridized overnight at 65°C with labeled probes preassociated with human placental DNA for 1.5–2 h. Subsequently, we analyzed hybridized blots by autoradiography for the presence of bands of the expected size and also for bands of varying sizes. The radioisotope quantitative analysis was done after the membrane being exposed for 1–2 d to a Molecular Dynamics PhosphorImager plate or after autoradiography scanning; data analysis was managed through the Molecular Dynamics software package, ImageQuant. We stripped and rehybridized blots with different probes, as necessary.
Bioinformatic and sequence analysis
The human reference genome sequence for the REPA/B region (chr17:18869254–19081415, NCBI 35) was downloaded from the Genome Browser Gateway at the University of California Santa Cruz (http://genome.ucsc.edu/cgi-bin/hgGateway). The genomic architecture was defined using BLAST 2 browser with default parameters (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) and Sequencher software version 4.1.4 (Gene Codes Corporation). The restriction map of the whole REPA/B region, as well as of the overlapping BACs previously selected, were based on Sequencher software.
Acknowledgments
We thank Matt Hurles and Pawel Stankiewicz for critical reviews. This work was supported in part by NICHD P01 HD39420.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.080697.108.
References
- Babicka L., Zemanova Z., Pavlistova L., Brezinova J., Ransdorfova S., Houskova L., Moravcova J., Klamova H., Michalova K. Complex chromosomal rearrangements in patients with chronic myeloid leukemia. Cancer Genet. Cytogenet. 2006;168:22–29. doi: 10.1016/j.cancergencyto.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Barbouti A., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Höglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F., et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Park S.-S., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel J.A. Genetics of pediatric central nervous system tumors. J. Pediatr. Hematol. Oncol. 1997;19:492–501. doi: 10.1097/00043426-199711000-00002. [DOI] [PubMed] [Google Scholar]
- Bown N., Cotterill S., Lastowska M., O'Neill S., Pearson A.D., Plantaz D., Meddeb M., Danglot G., Brinkschmidt C., Christiansen H., et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- Bruder C.E., Piotrowski A., Gijsbers A.A., Andersson R., Erickson S., de Stahl T.D., Menzel U., Sandgren J., von Tell D., Poplawski A., et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.-S., Manian P., Koeuth T., Potocki L., Zhao Q., Chinault A.C., Lee C.C., Lupski J.R. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Fioretos T., Strömbeck B., Sandberg T., Johansson B., Billström R., Borg A., Nilsson P.G., Van Den Berghe H., Hagemeijer A., Mitelman F., et al. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood. 1999;94:225–232. [PubMed] [Google Scholar]
- Flores M., Morales L., Gonzaga-Jauregui C., Domínguez-Vidaña R., Zepeda C., Yañez O., Gutiérrez M., Lemus T., Valle D., Avila M.C., et al. Recurrent DNA inversion rearrangements in the human genome. Proc. Natl. Acad. Sci. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Frey M.R., Matera A.G. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorda R., Ciccone R., Gimelli G., Pramparo T., Beri S., Bonaglia M.C., Giglio S., Genuardi M., Argente J., Rocchi M., et al. Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum. Mutat. 2007;28:459–468. doi: 10.1002/humu.20465. [DOI] [PubMed] [Google Scholar]
- Hammer Ø., Harper D.A.T., Ryan P.D. Palaeontologia Electronica. Vol. 4. 2001. PAST: Paleontological statistics software package for education and data analysis.http://palaeo-electronica.org/2001_1/past/past.pdf [Google Scholar]
- Heiskanen M., Karhu R., Hellsten E., Peltonen L., Kallioniemi O.P., Palotie A. High resolution mapping using fluorescence in situ hybridization to extended DNA fibers prepared from agarose-embedded cells. Biotechniques. 1994;17:928–934. [PubMed] [Google Scholar]
- Hollox E.J., Huffmeier U., Zeeuwen P.L., Palla R., Lascorz J., Rodijk-Olthuis D., van de Kerkhof P.C., Traupe H., de Jongh G., den Heijer M., et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Lupski J.R. Genome structural variation and sporadic disease traits. Nat. Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- Mendrzyk F., Korshunov A., Toedt G., Schwarz F., Korn B., Joos S., Hochhaus A., Schoch C., Lichter P., Radlwimmer B. Isochromosome breakpoints on 17p in medulloblastoma are flanked by different classes of DNA sequence repeats. Genes Chromosomes Cancer. 2006;45:401–410. doi: 10.1002/gcc.20304. [DOI] [PubMed] [Google Scholar]
- Osborne L.R., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K., Mammoser A.G., Wu C., Frengen E., Zeng C., Catanese J.J., de Jong P.J. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.S., Stankiewicz P., Bi W., Shaw C., Lehoczky J., Dewar K., Birren B., Lupski J.R. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L., Chen K.-S., Park S.-S., Osterholm D.E., Withers M.A., Kimonis V., Summers A.M., Meschino W.S., Anyane-Yeboa K., Kashork C.D., et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurlen W.G., Schwabe G.C., Seranski P., Joos S., Harbott J., Metzke S., Döhner H., Poustka A., Wilgenbus K., Haas O.A. Mapping of the breakpoints on the short arm of chromosome 17 in neoplasms with an i(17q) Genes Chromosomes Cancer. 1999;25:230–240. doi: 10.1002/(sici)1098-2264(199907)25:3<230::aid-gcc5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Bi W., Lupski J.R. Genetic proof of unequal crossovers in reciprocal deletion and duplication of 17p11.2. Am. J. Hum. Genet. 2002;71:1072–1081. doi: 10.1086/344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C.J., Withers M.A., Lupski J.R. Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am. J. Hum. Genet. 2004;75:75–81. doi: 10.1086/422016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P., Shaw C.J., Dapper J.D., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Holt B., Breems D.A., Berna Beverloo H., van den Berg E., Burnett A.K., Sonneveld P., Löwenberg B. Various distinctive cytogenetic abnormalities in patients with acute myeloid leukaemia aged 60 years and older express adverse prognostic value: Results from a prospective clinical trial. Br. J. Haematol. 2007;136:96–105. doi: 10.1111/j.1365-2141.2006.06403.x. [DOI] [PubMed] [Google Scholar]
- Visser R., Shimokawa O., Harada N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am. J. Hum. Genet. 2005;76:52–67. doi: 10.1086/426950. [DOI] [PMC free article] [PubMed] [Google Scholar]