Abstract
The folding of chromatin into topologically constrained loop domains is essential for genomic function. We have identified genomic anchors that define the organization of chromatin loop domains across the human major histocompatibility complex (MHC). This locus contains critical genes for immunity and is associated with more diseases than any other region of the genome. Classical MHC genes are expressed in a cell type-specific pattern and can be induced by cytokines such as interferon-gamma (IFNG). Transcriptional activation of the MHC was associated with a reconfiguration of chromatin architecture resulting from the formation of additional genomic anchors. These findings suggest that the dynamic arrangement of genomic anchors and loops plays a role in transcriptional regulation.
After each mitosis in mammalian cells, daughter chromosomes form discrete chromosome territories in the two new nuclei by chromatin decondensation (Cremer and Cremer 2006a, b). The imaging of these chromosome territories indicates that they are each uniquely and stably contained within their own domains. Recent research at both the chromosome and gene level perpetuates the concept of large- and small-scale three-dimensional remodeling of chromatin being a key determinant of genomic regulation. Enhancer–promoter associations, epigenetic modifications, and DNA–protein interactions are all proving to be important control mechanisms in the functioning of our genome. Much, however, has still to be learned.
Several lines of evidence indicate that the folding of higher-order chromatin into loops, mediated by their anchoring to the nuclear matrix, is critically important in genomic regulation (Berezney and Coffey 1977; Berezney et al. 1995; Nickerson 2001; Ottaviani et al. 2008). The nuclear matrix is a collection of proteins that is retained in nuclei treated with high-strength ionic buffers. The identification of some of its molecular constituents, such as lamins, RNA and DNA polymerases, SAFB (scaffold attachment factor-B1), SATB1 (special AT-rich sequence binding protein 1), and RUNX1 transcription factors, provides strong evidence of replication and transcription occurring at the nuclear matrix and has improved our understanding of the mechanisms involved (Luderus et al. 1994; Kimura et al. 1999; Carter et al. 2002; Stein et al. 2003; Townson et al. 2004; Cai et al. 2006).
In interphase cells, chromatin loops are anchored to the nuclear matrix at short genomic sequences called matrix attachment regions (MARs) (Berezney et al. 1995; Linnemann et al. 2007; Ottaviani et al. 2008). These sequences have been called scaffold attachment regions (SARs) in metaphase cells (Paulson and Laemmli 1977). MARs are associated with sequence motifs rich in AT tracts, repetitive sequences, topoisomerase binding sites, and regions of DNA with a propensity for base unpairing (Liebich et al. 2002; Platts et al. 2006; Ottaviani et al. 2008). The Multi-Loop Subcompartment model of chromatin folding predicts rosettes containing loops of 120 kb (Münkel et al. 1999). It is estimated that chromatin loops are formed from 30,000 to 80,000 anchored points across the human genome (Linnemann et al. 2007). Analysis of different eukaryotic organisms has suggested that permanent associations of MARs with the nuclear matrix are important for the maintenance of higher-order chromatin structure while dynamic associations play a prominent role in the regulation of gene expression (Heng et al. 2004; Eivazova et al. 2007). Recent investigations on the TH2 cytokine and beta-globin gene clusters have shown that MARs regulate transcription by functioning as landing platforms for transcription factors and chromatin remodeling enzymes (Cai et al. 2003; Wen et al. 2005). In addition, it is emerging that they can bring together proximal and distal cis-regulatory elements at the nuclear matrix and shield chromatin domains from the effects of flanking regulatory regions (Nabirochkin et al. 1998; Eivazova et al. 2007; Girod et al. 2007). The mapping of MARs directly to or in the vicinity of several origins of replication suggests that they may also participate in DNA replication (Lagarkova et al. 1998; Girard-Reydet et al. 2004).
MARs have classically been identified using in vitro binding affinity assays and polymerase chain reaction (PCR). Due to the limited scale of these methods only a few hundred of these sequences have been experimentally identified. Although computational methods have been developed to determine their positions on a larger scale, progress has been hampered by the limited number of MARs that have been experimentally defined as well as the fact that some MARs may not be permanently attached to the nuclear matrix (Platts et al. 2006; Girod et al. 2007; Linnemann et al. 2007). Microarray technology now provides an opportunity to map MARs at high resolution across the genome and address their role in gene expression.
The human major histocompatibility complex (MHC) was the first genomic region for which higher-order chromatin architecture was shown to reflect gene expression (Volpi et al. 2000; Christova et al. 2007). It is one of the most gene-rich regions of the entire genome, and spans over 4 Mb at band 6p21.31. The gene products of the region were first discovered on the surfaces of leukocytes, and so it is often called the human leukocyte antigen (HLA) region rather than the MHC. The classical MHC is divided into three classes: class I (telomeric), class III, and class II (centromeric) (Trowsdale and Campbell 1992; Horton et al. 2004). The extended MHC (xMHC) includes the classical MHC and the extended class II region (most-centromeric). A high percentage of genes in the MHC play decisive roles in immunity. The products of the classical MHC class I region, HLA-A, HLA-B, and HLA-C, present endogenous antigens to CD8+ T cells, while the classical class II molecules (HLA-DR, HLA-DP, HLA-DQ) present exogenous antigens to CD4+ T cells. Other MHC class I and II genes generate the TAP, HLA-DM, HLA-DO molecules which are involved in antigen processing required for correct cell-surface presentation. Genes in the class III region encode components of the complement system, which is a critical element of innate immunity (Germain 1994; Janeway 2001). The extended class II region contains important nonimmune genes such as the retinoid X receptor (RXR), which is involved in retinoic acid, vitamin D, and thyroid hormone activity.
The MHC can be used as a model for studying genomic structure–function relationships, since it has cell type-specific and experimentally inducible gene expression. Most genes in the MHC class I and III regions are constitutively expressed in all somatic cell types, whereas the classical MHC class II genes are expressed only in antigen-presenting cells. However, cytokines such as interferon-gamma (IFNG) strongly induce the classical class II genes in a coordinated manner in other cell types including fibroblasts, and also up-regulate genes in the class I and III regions (Rohn et al. 1996; Boehm et al. 1997).
In order to further our understanding of the relationship between higher-order chromatin architecture and gene expression, we identified genomic anchors, or MARs, across the MHC in different cell types using a conventional biochemical isolation method followed by hybridization to a customized MHC tiling path array (Tomazou et al. 2008). We show that genomic anchors are unevenly distributed, and that they can be classified into three classes: constitutive, cell type-specific, and IFNG-dependent. These sequences are mainly positioned in intergenic regions and in close proximity to the MHC class boundaries, subdividing the locus into physical domains. We further show that induction of transcription of the MHC is accompanied by a major recruitment of anchors to the nuclear matrix as well as by extrachromosomal looping, suggesting that the expression of MHC genes is regulated by dynamic chromatin–matrix interactions.
Results
In order to investigate the relationship between the configuration of chromatin loops and gene expression, we mapped genomic anchors, or MARs, across the human MHC in cell types with different expression profiles.
Distribution of MARs across the MHC in human fibroblasts
MARs were identified in MRC5 human fibroblasts using 2 M NaCl extraction followed by competitive hybridization of MAR and loop DNA fractions onto a genomic tiling path array of 2-kb resolution covering the entire 4 Mb MHC region (Tomazou et al. 2008). Real-time PCR reactions were performed to validate the findings. Clones having array enrichment values >0.4 (log2) were consistently confirmed to be MARs by real-time PCR (Supplemental Tables 1, 2). Genomic regions with array enrichment >0.4 were thus designated as MARs.
We found MARs to be present in the MHC on average every 33 kb, consistent with a previous study on the 16q22 locus in HeLa epithelial cells (Shaposhnikov et al. 2007). However, they are nonrandomly distributed across the MHC (Fig. 1). Over 91% of all MARs identified are present within the classical class I and class II regions, while the remaining 9% are within the class III and extended class II regions.
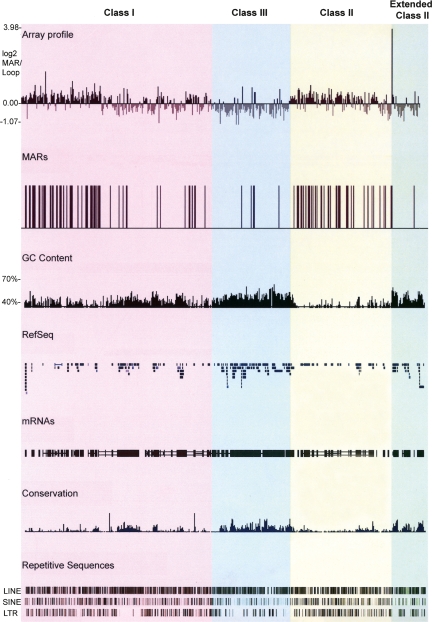
Figure 1.
MARs in the MHC of fibroblasts identified using genomic tiling path arrays. MAR and loop DNA fractions were differentially labeled with Cy3 and Cy5 fluorophores and competitively hybridized onto the MHC microarray platform. The log2 ratios of MAR/loop DNA were then calculated and plotted using the UCSC genome database (http://genome.ucsc.edu/). Datapoints with log2 enrichment values > 0.4 were regarded as MARs, plotted on the UCSC genome database, and represented as vertical black bars. The GC content, the RefSeq genes, the human mRNAs from GeneBank, regions of conservation between 17 vertebrate species, and the repetitive sequences LINE, SINE, and LTR across the MHC are shown below the array data. The different regions of the MHC are highlighted in different colors: classical class I, pink; classical class III, blue; classical class II, yellow; and extended class II, green. This color scheme is used in subsequent figures.
Comparison of the MAR profile with gene density across the MHC reveals an inversely correlated distribution. The classical class I and II regions, which have the lowest gene density in the MHC, have a MAR on average every 29 and 20 kb, respectively. In contrast, the MHC class III and extended class II regions have the highest gene density across the locus and contain a MAR every 148 and 71 kb, respectively. Further analysis revealed that, of the 117 MARs detected in the MHC, 98 are intergenic whereas only 19 are intragenic. Using a χ2 test, we found that the bias of MARs to intergenic regions was statistically significant (P-value < 0.0002; for all statistics a threshold P-value < 0.01 was considered significant). Of the intragenic MARs, five are within introns. Due to the size of DNA fragments generated by our biochemical isolation of MARs and the resolution of the tiling path array, it was not possible to map the remaining 16 intragenic MARs to individual exons or introns.
MARs were also identified at or near class boundaries of the MHC and closely flanking a significant proportion of clustered and individual MHC genes including HLA-A, HLA-C, HLA-E, HLA-G, HLA-DRB5, HLA-DRA, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1, which are essential for the adaptive immune response. These findings suggest that MARs can function as boundary elements defining chromatin domains, which we have tested by comparing their positions with respect to CTCF binding sites and DNase I hypersensitivity sites (see below).
The AT content of MAR and loop regions was compared, as previous studies have shown that MARs are AT-rich (Purbowasito et al. 2004; Shaposhnikov et al. 2007). The average AT content of MAR DNA was found to be 58% while that of the loop DNA was 53% (P-value of 0). Within the MHC classes, which vary in their average AT content, this correlation was also found to be significant (Supplemental Table 3), confirming that MARs are generally AT-rich. Comparative analysis did not reveal any precise organization of AT motifs.
Distribution of MARs across the MHC in human IFNG-treated fibroblasts and B-lymphoblastoid cells
We previously showed that giant external chromatin loops containing the entire MHC locus extend outward from the chromosome 6 territory, with higher frequency in B-lymphoblastoid cells and IFNG-induced fibroblasts than in untreated fibroblasts (Volpi et al. 2000). Here, we confirmed that large-scale looping of the MHC is observed with high frequency in PGF B-lymphoblastoid cells (Supplemental Fig. 1).
To determine whether the change in large-scale chromatin organization and expression of MHC genes are accompanied by a rearrangement of genomic anchors, we determined their locations in IFNG-treated fibroblasts and in PGF B-lymphoblastoid cells using the MHC tiling path array (Fig. 2A). A custom track of all the MARs can be obtained by uploading Supplemental Tables 4, 5, and 6 into the UCSC Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway), May 2004 assembly. A detailed profile across the MHC class II region in the different cell types is shown in Figure 3A, and the profile across the entire MHC is shown in Supplemental Figure 2. A striking increase in the number of MARs was found in these two cell types compared to untreated fibroblasts. In IFNG-treated fibroblasts and B-lymphoblastoid cells, MARs within the MHC were identified on average every 17 and 15 kb, respectively, which is approximately twice the frequency of that in untreated fibroblasts.
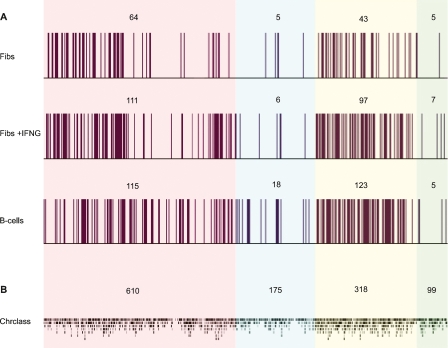
Figure 2.
MARs in the MHC of fibroblasts, IFNG-induced fibroblasts, and B-lymphoblastoid cells. (A) Experimentally identified MARs; (B) predicted MARs using Chrclass. The number of MARs in each region is shown above the data plots. Color scheme as in Figure 1.
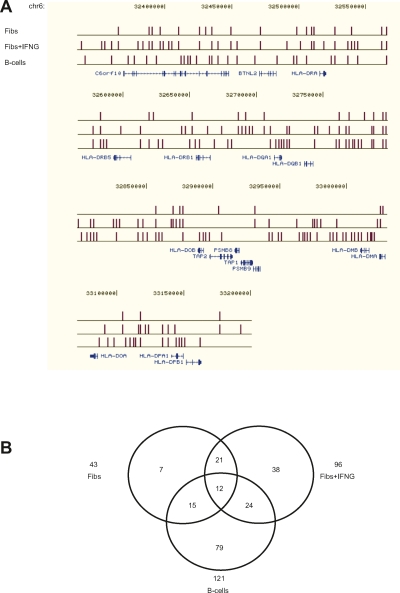
Figure 3.
Comparison of MARs in the MHC class II region in fibroblasts, IFNG-induced fibroblasts, and B-lymphoblastoid cells. (A) MARs are shown relative to RefSeq genes from the UCSC web browser (http://genome.ucsc.edu/). The distribution of MARs across the entire MHC is shown in Supplemental Figure 2. (B) Venn diagram of MARs in the class II region in the different cell types.
The overall distribution of MARs in the MHC is highly conserved in fibroblasts and B-lymphoblastoid cells. Over 90% of the MARs identified in IFNG-treated fibroblasts and B-lymphoblastoid cells are located within the classical class I and class II regions. The increase in the number of MARs in IFNG-treated fibroblasts and B-lymphoblastoid cells, compared to untreated fibroblasts, is mainly within the classical class II region (Fig. 3). In all cell types studied, only a small minority of MARs are contained in the class III and extended class II regions of the MHC.
As observed in untreated fibroblasts, the MARs detected in IFNG-treated fibroblasts and B-lymphoblastoid cells have a higher AT content compared to loop DNA. In addition, in all cell types MAR density tends to anti-correlate with gene density (Supplemental Fig. 3).
Previous studies have used several prediction software programs to define the locations of MARs on a genome-wide scale. One of the best performing programs, Chrclass, was used here to interrogate MARs across the MHC, and the findings were compared with our data (Fig. 2B) (Purbowasito et al. 2004). The Chrclass program maps MARs at base pair resolution. In order to integrate the two sets of data, the sequences identified by Chrclass were organized into the individual probes across the tiling path array, enabling the probes to be classified as MAR or non-MAR. MARs predicted by Chrclass occurred on average every 3 kb and were more evenly distributed across the MHC compared to those identified experimentally. Despite having high sensitivity (0.83), Chrclass was found to have extremely reduced specificity (0.14). Approximately 5% of the probes on the MHC tiling path array consisted entirely of repeat sequences and were excluded from the analysis. Chrclass identified MARs in 88% of clones with repeat regions. This analysis has confirmed previously described limitations of prediction software and shows that further empirical identification of MAR sequences is essential in developing more accurate prediction software programs (Linnemann et al. 2007).
A comparative analysis of MARs between the different cell types identified three main categories: constitutive, cell-specific, and IFNG-dependent (Supplemental Fig. 4). DNA regions attached to the nuclear matrix in all cell types tested account for 17% of MARs. They are defined as constitutive MARs since their tethering function is preserved regardless of the cell type and transcriptional status of the MHC locus. Approximately 45% of MARs are cell type–specific. These are further classified into fibroblast-specific, which include those present in both untreated and treated fibroblasts, and B-lymphoblastoid-specific. IFNG-dependent MARs are transiently associated with the nuclear matrix depending on the transcriptional status of the MHC. They include MARs in untreated fibroblasts that are released from the nuclear matrix in IFNG-treated fibroblasts, those recruited to the nuclear matrix in response to IFNG treatment, and those recruited to the nuclear matrix in both B-lymphoblastoid cells and IFNG-treated fibroblasts. These categories together constitute 34% of all MARs identified. The remaining 4% of MARs were present in both B-lymphoblastoid cells and untreated fibroblasts. The characterization of MARs in other cell types is required to determine whether they are cell type-specific or IFNG-dependent. The exact positions of the MARs in the different subclasses are listed in Supplemental Tables 7–12.
Distribution of MARs relative to differentially and nondifferentially expressed genes
We compared the expression of MHC genes in untreated fibroblasts with IFNG-treated fibroblasts and B-lymphoblastoid cells using microarray analysis (Fig. 4). A significant proportion of class II genes and a few class I genes are up-regulated in IFNG-treated fibroblasts and B-lymphoblastoid cells. The overall expression of the class III and extended class II genes in fibroblasts does not differ following IFNG treatment, while a proportion of class III genes are up-regulated in B-lymphoblastoid cells relative to untreated fibroblasts. As expected, most of the differentially expressed genes code for classical HLA class I and class II molecules (Supplemental Table 13).
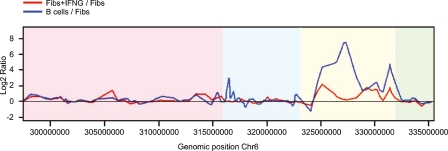
Figure 4.
Comparison of gene expression profiles in the MHC of fibroblasts, IFNG-induced fibroblasts, and B-lymphoblastoid cells. The red line represents the log2 expression ratios of IFNG-induced fibroblast/fibroblast ratios, and the blue line represents the B cell/fibroblast ratios. Color scheme as in Figure 1.
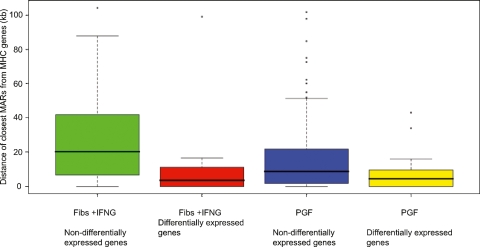
The distances of MARs from differentially and nondifferentially expressed genes in fibroblasts and B-lymphoblastoid cells were calculated to determine how changes in gene expression affect their distribution (Supplemental Fig. 5). In both IFNG-treated fibroblasts and B-lymphoblastoid cells the average distance between MARs and differentially expressed genes was over two times smaller than the distance between MARs and nondifferentially expressed genes (Fig. 5). A Mann-Whitney test showed that the distribution was significant with P-values of 0.003 and 0.009, respectively. The average distance between MARs and nondifferentially expressed genes in IFNG-induced fibroblasts is 26 kb, while that between MARs and differentially expressed genes is 12 kb. In B-lymphoblastoid cells the average distance of MARs from nondifferentially expressed genes is 19 kb while that from differentially expressed genes is 7 kb. This trend is observed for several genes of critical immunological importance including HLA-C, HLA-DPA1, HLA-DPB1, and HLA-DRB1 genes.
Figure 5.
Distances of MARs to differentially and nondifferentially expressed genes in IFNG-induced fibroblasts and B-lymphoblastoid cells. Boxplots show the interquartile range of the data; the horizontal black lines running through each boxplot represent the median values. The whiskers extend as far as 1.5 times the interquartile range. Any values that lie beyond the whiskers are plotted as dots.
MARs and genomic features
In order to further our understanding of the genomic functions of MARs, we compared their positions in the MHC with data available for CTCF binding and DNase I hypersensitive sites (Kim et al. 2007; Boyle et al. 2008). The set of MARs present in uninduced MRC5 fibroblasts and its subset of constitutive MARs were compared to the CTCF binding sites identified in IMR90 fibroblasts. Only a small fraction (<5%) of either set of MARs colocalized with CTCF binding sites, although a high proportion (55%) had CTCF binding sites within 20 kb on either side. The colocalized sites did however occur in the vicinity of the MHC class boundaries. DNase I hypersensitivity data for CD4+ T cells were compared to MARs identified in B cells since they both express classical MHC class I and II genes. A relatively small fraction of MARs (20%) colocalize with DNase I hypersensitive sites, but a high proportion (88%) had DNase I hypersensitive sites within 20 kb on either side.
We also analyzed the position of MARs in the intergenic region between the MHC class II genes HLA-DRB1 and HLA-DQA1 where a regulatory site (XL9) was shown to bind CTCF, class II transactivator (CIITA), and the RFX-CREB-NFY complex, and to be associated with the nuclear matrix (Majumder et al. 2006, 2008). A MAR was found at this locus in IFNG-treated fibroblasts but not in untreated fibroblasts or B cells. The absence of a MAR in B cells was unexpected but might be explained by the stringency of our analysis.
Discussion
The aim of this study was to determine the relationship between the folding of chromatin, mediated by genomic anchors, and gene expression in the MHC. We obtained a detailed map of genomic anchors using an MHC tiling path array to which we competitively hybridized MAR and loop DNA. The findings were then validated by real-time PCR and compared with gene expression data. MHC gene expression is cell-specific and inducible with interferons. In this study, fibroblasts, IFNG-treated fibroblasts, and B-lymphoblastoid cells were examined because of their differential gene expression in the MHC locus.
Despite the overall differences in MHC expression in the different cell types, MARs were found to be positioned mainly in the MHC class I and II regions. These regions have a significantly higher content of AT and repetitive sequences and a lower gene density than the MHC class III and extended class II regions, which contain only 10% of MARs. The chromatin loops, anchored by MARs, correlate positively with gene density and GC composition. Sequence analysis revealed that MARs in the MHC are rich in AT and repetitive sequences, consistent with previous studies (Purbowasito et al. 2004; Shaposhnikov et al. 2007).
Our data show that there are approximately twice as many MARs in IFNG-treated fibroblasts than untreated fibroblasts, and these MARs are ∼50% closer to MHC genes, representing a reduction in the average size of chromatin loops. Most of the anchored sequences are situated in the classical class I and II regions and are, with few exceptions, attached outside of genes. The same pattern of anchoring was observed in B-lymphoblastoid cells which constitutively express the MHC genes that are up-regulated in fibroblasts by IFNG. A recent investigation of loop organization in 300 kb of the MHC class I region in T cells, with and without IFNG, also shows the recruitment of additional MARs upon transcriptional induction (Kumar et al. 2007). Furthermore, PML was shown to interact with SATB1 to organize the chromatin into loops. We have previously shown that PML nuclear bodies are positioned in close spatial proximity to the MHC (Shiels et al. 2001; Wang et al. 2004). An earlier study on the human IFNB1 locus also showed correlation between the recruitment of DNA sequences to the nuclear matrix and transcription (Heng et al. 2004).
Independent of the transcriptional status of the MHC, MARs are positioned closer to genes which are differentially expressed. Our analysis of MAR-gene distances showed that differentially expressed genes are on average half the distance away from their closest MAR compared to those which are constitutively expressed. These observations suggest that MARs may function as dynamic cis-regulatory elements facilitating transcriptional regulation (Heng et al. 2004; Eivazova et al. 2007).
Previous studies have proposed insulator and boundary element roles for MARs in regulating gene expression (Dunn et al. 2003; Yusufzai and Felsenfeld 2004). We found that MARs predominantly occupy intergenic regions, closely flank the differentially expressed classical class I and class II genes, and cluster near the MHC class boundaries. Comparative analysis of our data with high resolution CTCF and DNase I hypersensitivity sites suggests that MARs may have diverse roles in genome organization beyond topologically constraining chromatin loops (Kim et al. 2007; Boyle et al. 2008). Additionally, MARs could delineate regions of open chromatin containing regulatory sequences which are DNase I hypersensitive (Crawford et al. 2006). Our work further suggests that a complex network of intergenic looping events occurs in the MHC upon activation consistent with that observed by others in the HLA-DRB1 and HLA-DQA1 region (Majumder et al. 2006, 2008).
To improve our understanding of the roles of MARs in the MHC, the MARs in each cell type were compared and classified into three groups: constitutive, cell-specific, and IFNG-dependent (Jackson et al. 1992; Heng et al. 2004; Shaposhnikov et al. 2007). Constitutive MARs are attached to the nuclear matrix in all cell types regardless of the transcriptional status of the MHC (Heng et al. 2001, 2004). Their conserved matrix binding property across these different cell types suggests that these DNA sequences have an important role in maintaining the higher-order chromatin structure of the MHC, which may be facilitated by a particular, so far unidentified, group of proteins. Cell type-specific tethering of the MHC is likely to reflect interactions between chromatin and cell type-specific nuclear matrix proteins (Jackson et al. 1992). Studies on SATB1 have shown that the interaction of cell type-specific matrix proteins at MARs is critical for tissue-specific regulation of gene expression (Cai et al. 2006; Galande et al. 2007). Therefore, cell-specific MAR–protein interactions could be involved in the distinct MHC expression profiles of fibroblasts and B-lymphoblastoid cells. Those MARs, which are recruited or released based on the transcriptional status of a given genomic domain, account for the third group of anchors (Heng et al. 2004). They too may be associated with a subclass of MAR-binding proteins that contributes to the regulation of MHC genes dynamically in IFNG-treated fibroblasts and constitutively in B-lymphoblastoid cells.
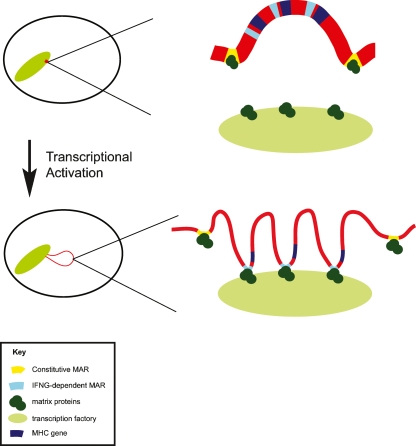
Our findings need to be examined in the context of our previous studies which showed that IFNG-treatment of fibroblasts was accompanied by higher-order chromatin reorganization in the MHC, resulting in an increase in giant external chromatin loops, compared to untreated fibroblasts (Volpi et al. 2000). Here we show that the number of giant loops in B-lymphoblastoid cells is equivalent to that observed in IFNG-treated fibroblasts. We showed recently that these giant loops represent an increase in decondensation of local chromatin, providing a more “open” chromatin environment (Christova et al. 2007). Decondensed chromatin could facilitate chromatin–matrix interactions by increasing accessibility for nuclear matrix proteins and transcription machinery (Hart and Laemmli 1998; Kimura et al. 1999). Furthermore, giant external chromatin loops may place the MHC in distinct nuclear microenvironments containing specific matrix proteins required for the regulation of MHC genes. Taken together our findings indicate that there is an intimate relationship between gene expression, higher-order chromatin structure, and the association of MARs with the nuclear matrix (Fig. 6).
Figure 6.
Model showing dynamic changes in chromatin organization of the MHC locus upon transcriptional up-regulation. In cells that are not expressing MHC class II genes, such as uninduced fibroblasts, the 4 Mb MHC locus (red) is positioned within the chromosome 6 territory anchored by constitutive MARs (yellow) to nuclear matrix proteins (dark green), forming stable chromatin loops. Upon up-regulation of gene expression the entire MHC locus adopts a more open, decondensed chromatin structure, represented by the thinning and lengthening of the chromatin line, and the entire MHC region is repositioned outside of the chromosome territory. IFNG-dependent MARs (light blue) are recruited to the nuclear matrix, folding chromatin into smaller loops and facilitating the interaction of chromatin and transcription factors (light green). Many IFNG-dependent MARs are positioned immediately adjacent to genes. The constitutive MARs remain attached to nuclear matrix proteins following transcriptional up-regulation, and some may have an insulatory function.
The MHC is the most important immunological region of the genome and is associated with more diseases than any other from diabetes to rheumatoid arthritis. Understanding the regulation of the MHC beyond its linear DNA sequence could provide novel targets for therapy. Our study has shown that the roles of the genomic anchors of the MHC are linked to its expression; thus, abnormal associations of MARs with the nuclear matrix might play a role in certain diseases associated with the MHC. The identification and characterization of novel components of the nuclear matrix in combination with high throughput studies of MARs of different species will be decisive in our understanding of how DNA tethering contributes to the spatial and temporal regulation of genomic function.
Methods
Cell culture
The human B-lymphoblastoid cell line, PGF, and MRC5 lung fibroblasts were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum and L-glutamine at 37°C in a 5% CO2 atmosphere. Transcriptional up-regulation of MHC genes in MRC5 cells was induced by adding 200 IU/mL of IFNG (recombinant human IFNG, R&D Systems) to the culture medium for 24 h (Darley et al. 1993).
Isolation of MAR and loop DNA fractions
The preparation of MAR and loop enriched DNA samples was carried out essentially as described previously (Krawetz et al. 2005). Optimal extraction time was determined to be 12 min for PGF and MRC5 cells ± IFNG. The isolation of MAR and loop DNA fractions was repeated five times for each cell line to obtain a total of five biological replicates. Two of these replicates were used for array hybridizations, and the other three for RT-PCR.
Construction of a tiling path array for the MHC
A tiling path array across the MHC was constructed and optimized essentially as described elsewhere (Tomazou et al. 2008). Briefly, 1747 PCR fragments of average size 2 kb covering 99.67% of the MHC were generated and printed in quadruplicate onto activated glass slides. The final array comprised 7832 features of which 97% can be expected to be informative based on quality control data.
Array hybridization
MAR and loop DNA fractions were labeled with Cy3- and Cy5-labeled dCTPs (Amersham Biosciences) using random prime labeling (Bioprime DNA Labeling System, Invitrogen) as described previously (Mulholland et al. 2006). Unincorporated nucleotides were removed from the labeled DNA using Micro-spin G50 columns. Pre-blocking of the slide and probe preparation were performed as described previously (Fiegler et al. 2003). Equal amounts (200 pmol) of Cy3- and Cy5-labeled probe were combined with 100 μg of Cot-1 DNA followed by ethanol precipitation. Resuspension of the pellet was done in hybridization buffer (50% formamide, 10% dextrane sulfate, 0.1% Tween 20, 2× SSC, and 10 mM Tris-HCl at pH 7.5). The slide was blocked with 50 μg of Cot-1 DNA and 300 μg of herring sperm DNA dissolved in 60 μL of hybridization buffer. Blocking solution and probe mixture were denaturated for 10 min at 80°C. The blocking solution was placed onto the MHC tiling path array, which was incubated in a humid chamber saturated with 20% formamide and 2× SSC for 1 h at 37°C. Meanwhile, the probe was placed at 37°C for pre-annealing. The probe was then placed on the array, which was incubated and placed in a humid chamber saturated with 20% formamide and 2× SSC. Hybridization was allowed to take place for 48 h at 37°C. The array was removed from the chamber and placed in a Coplin jar containing 2× SSC/0.03% SDS to wash off any excess hybridization solution. The array was then transferred to another Coplin jar and washed in 2× SSC/0.03% SDS for 5 min at 65°C, three times in 0.2× SSC for 20 min at room temperature and in PBS/0.05% Tween 20 for 10 min at room temperature. Finally, the array was spin-dried and placed in a dark chamber at room temperature.
Image and data analysis
Arrays were scanned at 532 nm (Cy3) and 635 nm (Cy5) using the confocal scanner ScanArray 4000 (GSI Lumonics). For each clone, a ratio of Cy5 to Cy3 fluorescent intensity was calculated by processing the array images in TIGR Spotfinder. For each channel the median signals were subtracted from the median foreground signals. Log2 ratios of chA/chB were created and subsequently normalized by subtracting the weighted median for each array. Intensities were normalized between arrays using the “quantile” method of “NormaliseBetweenArrays” function from the Bioconductor package limma. For each clone, a mean log2 was calculated using the four replicates within each array. Pearson correlation coefficients were calculated between all arrays and found to be strongest between duplicates. Summarized log2 ratios were calculated by taking a mean across duplicate arrays. Dye-swap hybridization for each pair of arrays was performed to normalize for any dye bias. The genomic position of the BACs used to generate the MHC tile path arrays was established using the NCBI build 34 assembly of the human genome.
Real-time PCR
PCR primers (Supplemental Table S2) were designed using Primer 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Real-time PCR was performed using SYBR Green Jumpstart Taq ReadyMix kit (Sigma Aldrich), 50 ng DNA, and 2 μM primers using Opticon2 (MJ Research). Standard curves were constructed and used for product quantitation. The log2 fold enrichment in the MAR fraction compared to its corresponding loop fraction was calculated and a value of +1.00 was considered significant to validate a MAR. Enrichment values less than +1.00 validated a loop.
In order to determine a threshold to distinguish between MAR and loop-enriched sequences, MHC regions with different array enrichments were randomly selected and tested with RT-PCR. Regions with array enrichments greater than log2 value of 0.4 were consistently validated and were therefore defined as MARs. Those with a log2 value of <0.4 were defined as loops. This threshold corresponds to 3.6 standard deviations of the average of genomic versus genomic hybridization experiments. Different subtypes of MARs, constitutive, cell-specific, and IFNG-dependent, as well as negative control regions (loops) were also randomly selected and validated (Supplemental Table S1). All RT-PCR reactions were performed in triplicate using three biologically independent samples.
Gene expression analysis
The gene expression of PGF and MRC5 cells ± IFNG was determined using Affymetrix U133 plus 2.0 GeneChips (Affymetrix). Total RNA was extracted using the QIAGEN RNeasy Mini kit. RNA quality was assessed using an Agilent 2100 Bioanalyser. Samples were prepared and hybridized according to the manufacturers instructions. Statistical analysis was performed using the open source statistical environment R (www.r-project.org) and Bioconductor packages (Smyth 2004). Probe set expression measures were calculated using the “Affy” package's robust multi-chip average (RMA) default method. Probe sets exhibiting a >2-fold change were called differentially expressed. Where multiple probe sets mapped to the same gene, a mean fold change was taken.
Fluorescence in situ hybridization (FISH) and image analysis
Cells were harvested by standard techniques using methanol:acetic acid fixation. The PAC RP1-172K2 probe was selected for the MHC class II gene HLA-DRA and obtained from the Wellcome Trust Sanger Institute. Probe DNA was labeled with digoxigenin-11-dUTP (Roche) by nick-translation with the BioNick kit (Invitrogen) and cohybridized with FITC-labeled chromosome 6 paint (Cambio) to fixed nuclear preparations. Signal was detected with rhodamine-conjugated anti-digoxigenin (Vector). The nuclei were counterstained with DAPI (200 ng/mL) and mounted in Cityfluor antifade solution. Slides were examined with a Zeiss Axiophot epifluorescence microscope. Separate grayscale images were recorded with a cooled CCD-camera (Hamamatsu). Images were pseudocolored and merged using SmartCapture X software (Digital Scientific). The position of signal from the HLA-DRA probe was analyzed in relation to the domain defined by the chromosome 6–specific paint. Each chromosome homolog was counted separately and a score derived for the percentage of loci that were located on an external chromatin loop. Experiments were repeated at least twice on cells from independent cultures.
In silico predictions of MARs
Chrclass (ftp.ncbi.nih.gov/pub/kondrashov/MAR/Chrclass.zip) was used to predict the position of MARs across the MHC using default parameters.
Acknowledgments
We thank our colleagues for helpful discussions, especially Frank Uhlmann, Petros Takousis, and Amelia Linnemann. D.O. and R.M. were supported by the Cancer Research UK London Research Institute; and E.L., T.J., T.F., R.C., B.S., and D.S. by Cancer Research UK Programme grant C5321/A8318. S.A.K. and A.E.P. were supported by NICHD grant HD36512. E.T. was supported by a Ph.D. Fellowship from the Wellcome Trust Sanger Institute. V.K.R. was supported by the Barts and The London Charitable Trust and a C.J. Martin Fellowship from the NHMRC, Australia. S.B. was supported by Wellcome Trust Programme Grant WT084071.
Footnotes
[Supplemental material is available online at www.genome.org. The data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession no. E-MEXP-1793 and NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE10880.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.082313.108.
References
- Berezney R., Coffey D.S. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J. Cell Biol. 1977;73:616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Mortillaro M.J., Ma H., Wei X., Samarabandu J. The nuclear matrix: A structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Boehm U., Klamp T., Groot M., Howard J.C. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Han H.J., Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Christova R., Jones T., Wu P.J., Bolzer A., Costa-Pereira A.P., Watling D., Kerr I.M., Sheer D. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNγ. J. Cell Sci. 2007;120:3262–3270. doi: 10.1242/jcs.012328. [DOI] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C. Rise, fall and resurrection of chromosome territories: A historical perspective. Part I. The rise of chromosome territories. Eur. J. Histochem. 2006a;50:161–176. [PubMed] [Google Scholar]
- Cremer T., Cremer C. Rise, fall and resurrection of chromosome territories: A historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: Experiments and models from the 1990s to the present. Eur. J. Histochem. 2006b;50:223–272. [PubMed] [Google Scholar]
- Darley R., Morris A., Passas J., Bateman W. Interactions between interferon γ and retinoic acid with transforming growth factor β in the induction of immune recognition molecules. Cancer Immunol. Immunother. 1993;37:112–118. doi: 10.1007/BF01517043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.L., Zhao H., Davie J.R. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 2003;288:218–223. doi: 10.1016/s0014-4827(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Eivazova E.R., Vassetzky Y.S., Aune T.M. Selective matrix attachment regions in T helper cell subsets support loop conformation in the Ifng gene. Genes Immun. 2007;8:35–43. doi: 10.1038/sj.gene.6364349. [DOI] [PubMed] [Google Scholar]
- Fiegler H., Carr P., Douglas E.J., Burford D.C., Hunt S., Scott C.E., Smith J., Vetrie D., Gorman P., Tomlinson I.P., et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Galande S., Purbey P.K., Notani D., Kumar P.P. The third dimension of gene regulation: Organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Germain R.N. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- Girard-Reydet C., Gregoire D., Vassetzky Y., Mechali M. DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the Xenopus and mouse c-myc promoter. Gene. 2004;332:129–138. doi: 10.1016/j.gene.2004.02.031. [DOI] [PubMed] [Google Scholar]
- Girod P.A., Nguyen D.Q., Calabrese D., Puttini S., Grandjean M., Martinet D., Regamey A., Saugy D., Beckmann J.S., Bucher P., et al. Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat. Methods. 2007;4:747–753. doi: 10.1038/nmeth1076. [DOI] [PubMed] [Google Scholar]
- Hart C.M., Laemmli U.K. Facilitation of chromatin dynamics by SARs. Curr. Opin. Genet. Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- Heng H.H., Krawetz S.A., Lu W., Bremer S., Liu G., Ye C.J. Re-defining the chromatin loop domain. Cytogenet. Cell Genet. 2001;93:155–161. doi: 10.1159/000056977. [DOI] [PubMed] [Google Scholar]
- Heng H.H., Goetze S., Ye C.J., Liu G., Stevens J.B., Bremer S.W., Wykes S.M., Bode J., Krawetz S.A. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Wright M.W., et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Dolle A., Robertson G., Cook P.R. The attachments of chromatin loops to the nucleoskeleton. Cell Biol. Int. Rep. 1992;16:687–696. doi: 10.1016/s0309-1651(05)80013-x. [DOI] [PubMed] [Google Scholar]
- Janeway C.A. How the immune system protects the host from infection. Microbes Infect. 2001;3:1167–1171. doi: 10.1016/s1286-4579(01)01477-0. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R.G., Cook P.R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: Little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz S.A., Draghici S., Goodrich R., Liu Z., Ostermeier G.C. In silico and wet-bench identification of nuclear matrix attachment regions. Methods Mol. Med. 2005;108:439–458. doi: 10.1385/1-59259-850-1:439. [DOI] [PubMed] [Google Scholar]
- Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- Lagarkova M.A., Svetlova E., Giacca M., Falaschi A., Razin S.V. DNA loop anchorage region colocalizes with the replication origin located downstream to the human gene encoding lamin B2. J. Cell. Biochem. 1998;69:13–18. doi: 10.1002/(sici)1097-4644(19980401)69:1<13::aid-jcb2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Liebich I., Bode J., Reuter I., Wingender E. Evaluation of sequence motifs found in scaffold/matrix-attached regions (S/MARs) Nucleic Acids Res. 2002;30:3433–3442. doi: 10.1093/nar/gkf446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann A.K., Platts A.E., Doggett N., Gluch A., Bode J., Krawetz S.A. Genomewide identification of nuclear matrix attachment regions: An analysis of methods. Biochem. Soc. Trans. 2007;35:612–617. doi: 10.1042/BST0350612. [DOI] [PubMed] [Google Scholar]
- Luderus M.E., den Blaauwen J.L., de Smit O.J., Compton D.A., van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P., Gomez J.A., Boss J.M. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J. Biol. Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- Majumder P., Gomez J.A., Chadwick B.P., Boss J.M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland P.J., Fiegler H., Mazzanti C., Gorman P., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–791. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- Münkel C., Eils R., Dietzel S., Zink D., Mehring C., Wedemann G., Cremer T., Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- Nabirochkin S., Ossokina M., Heidmann T. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 1998;273:2473–2479. doi: 10.1074/jbc.273.4.2473. [DOI] [PubMed] [Google Scholar]
- Nickerson J. Experimental observations of a nuclear matrix. J. Cell Sci. 2001;114:463–474. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- Ottaviani D., Lever E., Takousis P., Sheer D. Anchoring the genome. Genome Biol. 2008;9:201. doi: 10.1186/gb-2008-9-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.R., Laemmli U.K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Platts A.E., Quayle A.K., Krawetz S.A. In-silico prediction and observations of nuclear matrix attachment. Cell. Mol. Biol. Lett. 2006;11:191–213. doi: 10.2478/s11658-006-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbowasito W., Suda C., Yokomine T., Zubair M., Sado T., Tsutsui K., Sasaki H. Large-scale identification and mapping of nuclear matrix-attachment regions in the distal imprinted domain of mouse chromosome 7. DNA Res. 2004;11:391–407. doi: 10.1093/dnares/11.6.391. [DOI] [PubMed] [Google Scholar]
- Rohn W.M., Lee Y.J., Benveniste E.N. Regulation of class II MHC expression. Crit. Rev. Immunol. 1996;16:311–330. doi: 10.1615/critrevimmunol.v16.i3.40. [DOI] [PubMed] [Google Scholar]
- Shaposhnikov S.A., Akopov S.B., Chernov I.P., Thomsen P.D., Joergensen C., Collins A.R., Frengen E., Nikolaev L.G. A map of nuclear matrix attachment regions within the breast cancer loss-of-heterozygosity region on human chromosome 16q22.1. Genomics. 2007;89:354–361. doi: 10.1016/j.ygeno.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Shiels C., Islam S.A., Vatcheva R., Sasieni P., Sternberg M.J., Freemont P.S., Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. http://www.statsci.org/smyth/pubs/ebayes.pdf. [DOI] [PubMed] [Google Scholar]
- Stein G.S., Zaidi S.K., Braastad C.D., Montecino M., van Wijnen A.J., Choi J.Y., Stein J.L., Lian J.B., Javed A. Functional architecture of the nucleus: Organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Tomazou E.M., Rakyan V.K., Lefebvre G., Andrews R., Ellis P., Jackson D.K., Langford C., Francis M.D., Bäckdahl L., Miretti M., et al. Generation of a genomic tiling array of the human Major Histocompatibility Complex (MHC) and its application for DNA methylation analysis. BMC Med. Genomics. 2008;1:19. doi: 10.1186/1755-8794-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson S.M., Kang K., Lee A.V., Oesterreich S. Structure-function analysis of the estrogen receptor alpha corepressor scaffold attachment factor-B1: Identification of a potent transcriptional repression domain. J. Biol. Chem. 2004;279:26074–26081. doi: 10.1074/jbc.M313726200. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Campbell R.D. Complexity in the major histocompatibility complex. Eur. J. Immunogenet. 1992;19:45–55. doi: 10.1111/j.1744-313x.1992.tb00047.x. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P.J., Islam S.A., Freemont P.S., Freemont P.S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Huang S., Rogers H., Dickinson L.A., Kohwi-Shigematsu T., Noguchi C.T. SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood. 2005;105:3330–3339. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- Yusufzai T.M., Felsenfeld G. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]