Abstract
Copy number variants (CNVs) underlie many aspects of human phenotypic diversity and provide the raw material for gene duplication and gene family expansion. However, our understanding of their evolutionary significance remains limited. We performed comparative genomic hybridization on a single human microarray platform to identify CNVs among the genomes of 30 humans and 30 chimpanzees as well as fixed copy number differences between species. We found that human and chimpanzee CNVs occur in orthologous genomic regions far more often than expected by chance and are strongly associated with the presence of highly homologous intrachromosomal segmental duplications. By adapting population genetic analyses for use with copy number data, we identified functional categories of genes that have likely evolved under purifying or positive selection for copy number changes. In particular, duplications and deletions of genes with inflammatory response and cell proliferation functions may have been fixed by positive selection and involved in the adaptive phenotypic differentiation of humans and chimpanzees.
The human genome is structurally dynamic, with thousands of heritable copy number variants (CNVs) among the genomes of individuals with normal phenotypes (Iafrate et al. 2004; Sebat et al. 2004; Redon et al. 2006; Korbel et al. 2007). Despite burgeoning interest, the evolutionary significance of copy number variation remains poorly understood (Conrad and Hurles 2007), and we have relatively few intraspecific CNV data from non-human species with which to make comparisons (e.g., Perry et al. 2006; Dopman and Hartl 2007; Egan et al. 2007; Graubert et al. 2007; Guryev et al. 2008; Lee et al. 2008). In contrast to single-nucleotide polymorphism data, for which population genetic tools were traditionally developed and matured in model organism studies (especially Drosophila), CNV research has thus far focused predominantly on humans. Moreover, we have yet to examine fully patterns of within-species CNVs and between-species copy number differences (CNDs) (e.g., Locke et al. 2003; Fortna et al. 2004; Demuth et al. 2006; Goidts et al. 2006; Wilson et al. 2006; Dumas et al. 2007) together under an evolutionary framework.
In this study, we have used array-based comparative genomic hybridization (aCGH) on a human whole-genome tile-path (WGTP) platform comprised of 28,708 large-insert DNA clones to identify CNVs among the genomes of 30 unrelated chimpanzees (Pan troglodytes) and 30 unrelated humans from Africa. To investigate the mutational mechanisms and forces of natural selection affecting copy number variation and to examine how these processes may have differed in the evolution of our two species, we compared the locations and frequencies of the human and chimpanzee CNVs with each other and with structural and functional features of both genomes. In addition, we used the same platform to identify CNDs between the human and chimpanzee genomes, which facilitated direct comparisons of the rates of copy number fixation and variation. These analyses have identified gene duplications and deletions that may have played important roles in the adaptive phenotypic differentiation of humans and chimpanzees.
Results
Comparative CNV maps in the human and chimpanzee genomes
The human WGTP platform used for this study was an updated version of the aCGH platform that was used in a previous genome-wide CNV study of 270 individuals from four human populations (Fiegler et al. 2006; Redon et al. 2006), with more than 2000 additional clones selected to span gaps in the previous generation of the WGTP array. Of the 30 chimpanzees in our study, all but three were wild-born, with 29 individuals from the Western chimpanzee subspecies (P. troglodytes verus) and one Eastern chimpanzee (P. troglodytes schweinfurthii). For the chimpanzee aCGH experiments, the genomic DNA of each individual was compared with that from Clint, the captive-born donor for the chimpanzee reference genome sequence (The Chimpanzee Sequencing and Analysis Consortium 2005). The 30 human individuals consisted of 10 Yoruba (Ibadan, Nigeria), 10 Biaka rainforest hunter-gatherers (Central African Republic), and 10 Mbuti rainforest hunter-gatherers (Democratic Republic of Congo). A European-American male (NA10851) served as the reference individual for all human aCGH experiments. Copy number gains and losses were detected using CNVfinder, which enables identification of CNVs on the WGTP platform with a false positive rate < 5% per sample (Fiegler et al. 2006); further validation was provided by comparison with human CNVs in the Database of Genomic Variants and by PCR and fluorescence in situ hybridization (FISH) experiments described below.
On average, we found 70 and 80 autosomal CNVs per within-chimpanzee and within-human comparison, respectively. The median sizes of CNVs are also similar for both species (∼250 kb; Supplemental Fig. 1). However, we note that CNV boundaries are likely overestimated on the WGTP platform, because a variant comprising only a portion of a large-insert clone could still be identified as a CNV, but would be reported as if the whole clone was involved. Indeed, using a custom-made aCGH platform with ∼1-kb resolution, 65% of human CNVs detected using the previous generation of the WGTP platform (Redon et al. 2006) were estimated to be less than half of their originally reported sizes, with a total size reduction of ∼33% for all studied CNVs (Perry et al. 2008).
In total, we identified 353 discrete autosomal CNV-containing regions (CNVRs) in humans, compared with 438 in chimpanzees (Fig. 1). There were similar numbers of “common” CNVRs (identified in two or more individuals) in the two species: 223 in humans versus 229 in chimpanzees. Population demography may affect within-species patterns of genetic diversity (Ptak and Przeworski 2002); thus, the higher proportion of “rare” CNVRs (singleton calls) in chimpanzees could be due to differences in population stratification or histories between the human and chimpanzee cohorts included in our study (Supplemental Note). However, this result cannot be explained by the single Eastern chimpanzee in our sample: only seven of the 77 CNVs detected in this individual (9%) were not observed in other individuals.
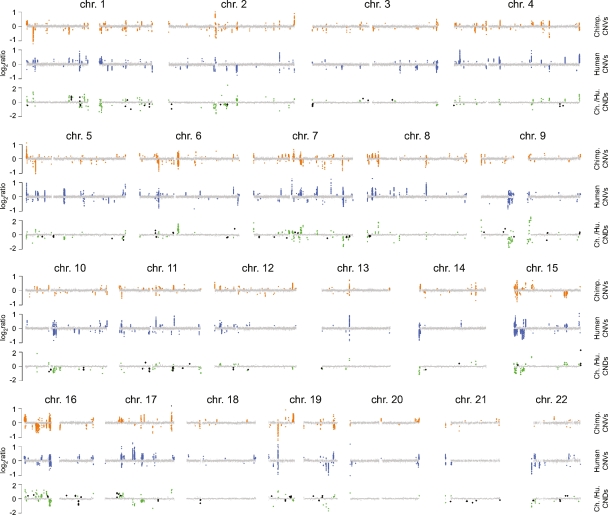
Figure 1.
Whole-genome comparison of human and chimpanzee copy number variation. For each autosome, relative intensity log2 ratios are superimposed for all 30 chimpanzees compared with Clint (top) and all 30 humans compared with NA10851 (middle). Log2 ratios for the interspecies comparison of Clint vs. NA10851 are shown at the bottom. Clones in nonvariable regions are depicted as gray circles (log2 ratios close to 0). Clones reporting copy number change with the CNVfinder algorithm are shown in orange, blue, and green/black for chimpanzees, humans, and the interspecies comparison, respectively. Fixed human–chimpanzee CNDs are indicated by the larger black circles on the interspecies profiles. Large gaps in clone coverage correspond to centromeric regions.
Of the 353 human CNVRs, 313 (89%) overlap copy number variable regions identified in previous human studies, as annotated in the Database of Genomic Variants (version hg18.v3, http://projects.tcag.ca/variation). However, we note that 222 (63%) were expected to overlap at random based on permutation analysis (see Supplemental Note). In contrast, only 53 of our 438 chimpanzee CNVRs (12.1%) overlap those identified in a previous analysis of 20 chimpanzees that used two aCGH platforms with ∼12% genomic coverage (Perry et al. 2006). Because only a portion of chimpanzee individuals were in common between the two studies, we performed a second comparison considering only the 106 CNVs that were identified in multiple individuals in the previous study: 36 of these CNVs (34%) overlapped CNVRs detected by the WGTP platform. This result may reflect relatively lower false-negative and higher false-positive rates in the previous data set, due to the use of less stringent CNV detection parameters (Perry et al. 2006). Nevertheless, 385 of the chimpanzee CNVRs detected by the WGTP platform are newly described in this study.
We also used the WGTP platform to identify 355 autosomal CNDs between the human and chimpanzee reference individuals by direct comparison in an interspecies aCGH experiment (Fig. 1). One hundred fourteen of these loci (32%; Supplemental Table 1) overlapped previously identified human–chimpanzee CNDs (Wilson et al. 2006; Dumas et al. 2007). Importantly, because we collected both within- and between-species copy number data on the same aCGH platform, for the first time we were able to compare the locations of CNDs with those of human and chimpanzee CNVRs. We found that only 92 of the 355 CNDs (26%) did not overlap any within-species CNVR from our data set, and therefore may be “fixed” between human and chimpanzee (Supplemental Note). This result demonstrates that studies aiming to identify fixed CNDs, potentially critical for our understanding of the genetic basis of interspecific phenotypic differences, should interrogate more than one or a few individual genomes per species.
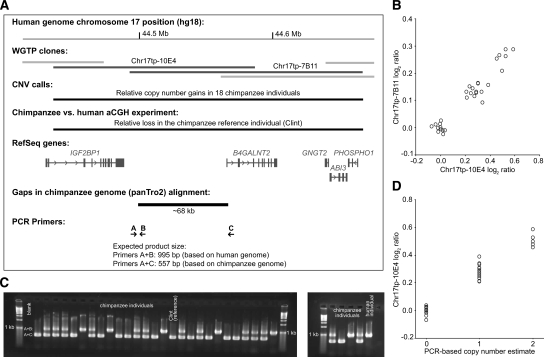
The single donor individual for the chimpanzee reference genome sequence (The Chimpanzee Sequencing and Analysis Consortium 2005), Clint, was also the reference individual for our chimpanzee aCGH experiments. Among the 438 observed chimpanzee CNVRs, we identified nine putative deletion variants in regions with substantial gaps in Clint’s genome (panTro2) compared with the human reference genome (hg18) (Fig. 2A,B). For three of these regions that did not have large repetitive elements at their putative breakpoints, we developed PCR-based assays to confirm and validate the deletions. Two variants were successfully validated with this highly specific approach (Fig. 2C,D; Supplemental Fig. 2) (results from the third CNV were inconclusive), including one that encompassed the promoter region and first exon of the B4GALNT2 gene, whose product is required for synthesis of the Sda and CAD antigens in human erythrocytes and colon mucins (Montiel et al. 2003). In this way, distinguishing among fixed and polymorphic deletions at sequence gap locations—which has implications for evolutionary analyses of patterns of gene gain and loss in primates (Hahn et al. 2007)—and identifying individuals for subsequent recovery of missing sequences is practicable for chimpanzees.
Figure 2.
PCR-based validation of a deletion CNV in chimpanzee. (A) The WGTP clones Chr17tp-10E4 and Chr17tp-7B11 (human chromosome 17q21.32) report a chimpanzee-specific deletion CNV. Based on an alignment of the chimpanzee and human genomes (Karolchik et al. 2003) for this region, the chimpanzee reference sequence (donor: Clint) has a gap of ∼68 kb including the first exon of the B4GALNT2 gene. In the between-species aCGH experiment, we observed a relative loss for these two clones in Clint compared with the human reference individual. (B) Bivariate clustering of Chr17tp-10E4 and Chr17tp-7B11 log2 ratios. The inferred cluster class of Clint, the chimpanzee reference individual (i.e., log2 ratio close to 0 for each clone) corresponds to the lowest copy number state among chimpanzees. (C) Results of a PCR-based genotyping assay using a 1.2% agarose gel with ethidium bromide staining. PCR primer positions are depicted in A. Note that primer combination A+C amplifies only when the intervening sequence is deleted. The 31 chimpanzees (including Clint) are in sample numerical order. One human individual is included as positive control. (D) PCR-based copy number genotype estimates and Chr17tp-10E4 log2 ratio clusters are 100% concordant.
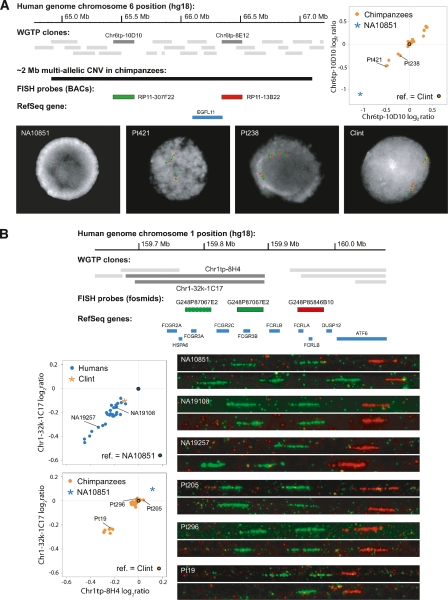
We used FISH on interphase nuclei to validate an extremely large multiallelic chimpanzee CNV (∼2 Mb for each repeated unit, which is considerably larger than any yet-discovered human multiallelic CNV) that encompassed only one RefSeq gene, EGFL11, whose function is still unknown (Fig. 3A). We also performed FISH on stretched DNA fibers to characterize copy number variation at another locus, containing the FCGR3A/B genes. Low copy number of FCGR3B, which codes for a receptor of the IgG Fc fragment, has been associated with systemic autoimmune diseases, including systemic lupus erythematosus in humans (Fanciulli et al. 2007). Based on our experiments (Fig. 3B), more than two-thirds of the human and chimpanzee individuals were estimated to have four diploid FCGR3A/B gene copies; fewer than 10 individuals in each species had three diploid copies, and the human reference individual was found to have five diploid copies. Our observation of similar patterns of copy number diversity at this locus in both species raises the intriguing possibility that FCGR3B copy number also influences autoimmune disease susceptibility in chimpanzees. Finally, we used both interphase- and fiber-FISH approaches to visualize CNV patterns at the CCL3 and CCL3L1 cytokine gene locus (Fig. 4). Confirming our aCGH results, we detected copy number variation of the CCL3L1 gene in humans, but not in chimpanzees, who seem to have only one haploid copy of this gene. We also observed a separation of >1 megabase between the chimpanzee CCL3 and CCL3L1 genes (Fig. 4C,D), a result consistent with the current sequence assembly of the chimpanzee genome (panTro2) (The Chimpanzee Sequencing and Analysis Consortium 2005).
Figure 3.
Validation of CNV loci by fluorescence in situ hybridization. (A) An ∼2-Mb region encompassing the EGFL11 gene (human chromosome region 6q12) is copy-number variable in chimpanzees. The relative intensity log2 ratios for clones throughout this region form four discrete clusters. In addition, there is a relatively large log2 ratio difference between the human and chimpanzee reference individuals (ref., reference). Interphase FISH experiments with two labeled BAC probes (RP11-307F22 in green; RP11-13B22 in red) confirmed the presence of an extremely large, tandem, multi-allelic CNV in chimpanzees. The human reference individual NA10851 was found to have two diploid copies (one copy on each chromosome). The chimpanzee log2 ratio clusters correspond to four (Pt421; 1 + 3), five (Pt238; 2 + 3), six (reference chimpanzee Clint; 3 + 3), and presumably seven copies of this genomic region per diploid cell. (B) The WGTP clones Chr1tp-8H4 and Chr1-32k-1C17 (human chromosome 1q23.3) report CNVs in both humans and chimpanzees. This region includes the FCGR2 and FCGR3 genes, and HSPA6. Fiber FISH experiments with two labeled fosmid probes (G248P87067E2 in green; G248P85846B10 in red) validated the WGTP results and determined absolute copy numbers of the FCGR3 genes, which have been associated with susceptibility to systemic autoimmune diseases (Fanciulli et al. 2007). The human reference individual NA10851 has five diploid FCGR3 copies (3 + 2), while the other humans apparently have either four (e.g., NA19108; 2 + 2) or three (e.g., NA19257; 2 + 1) copies per diploid cell. Chimpanzees have either four (Pt205 and Pt296; 2 + 2) or three (Pt19; 2 + 1) diploid copies of this genomic region.
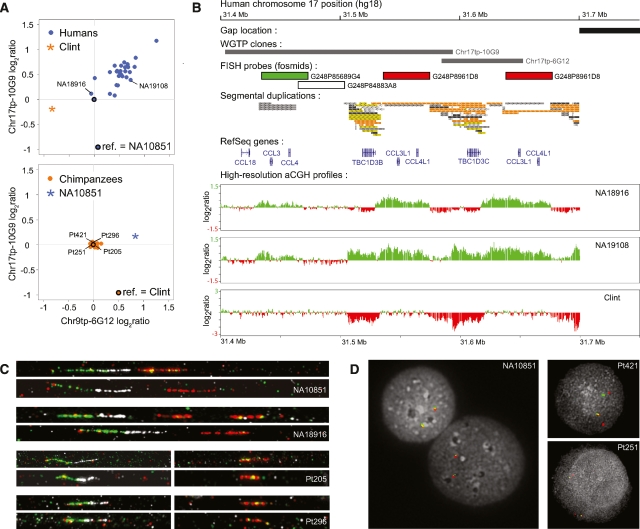
Figure 4.
Genome architecture of the CCL3/CCL3L locus in humans and chimpanzees. (A) Log2 ratio distributions of the WGTP clones Chr17tp-10G9 and Chr17tp-6G12 from the human 17q12 locus for humans (top) and chimpanzees (bottom; ref., reference). (B) Sequence annotation of this locus based on the human reference genome (hg18), with locations of the WGTP clones and fosmids used in this study. We compared two human samples and Clint to NA10851 by aCGH with a human oligonucleotide platform covering the 17q12 locus with a median spacing of 50 bp. The high-resolution profiles are concordant with the WGTP results displayed in A: NA18916 shows an increase in CCL3L1 copy number together with a decrease in TBC1D3 copy number compared with NA10851, while NA19108 shows relative copy number gains for both genes. In contrast, no difference in CCL3L1 copy number can be detected between Clint and NA10851. Instead, a high-fold relative copy number loss of the TBC1D3 gene is identified in Clint, thus explaining the CND loss detected with the WGTP platform. (C) Absolute CCL3L1 copy number measurement by fiber-FISH, using probes containing CCL3 (green), CCL3L1 (red), and a DNA segment between these two genes (in white). NA10851 carries a single copy of CCL3L1 per chromosome, while NA18916 carries one copy of CCL3L1 on one chromosome, but two copies on the other chromosome. For Pt205 and Pt296, no DNA fiber shows green and red signals together, suggesting that CCL3 and CCL3L1 are not adjacent genes in chimpanzee. In addition, we observed only single red signals for these two chimpanzee individuals, with no evidence of tandem CCL3L1 duplication. (D) Interphase-FISH with probes containing CCL3 (green) and CCL3L1 (red). In NA10851 cells, the signals corresponding to CCL3 and CCL3L1 cannot be discriminated spatially. In contrast, gaps between the red and green signals can be observed in chimpanzee nuclei, confirming major structural differences in the architecture of the CCL3L1 locus between human and chimpanzee. These results are concordant with the published sequence of chimpanzee chromosome 17 (panTro2 assembly), where a single copy of CCL3L1 is present at 19.41 Mb and CCL3 is mapped at 21.07 Mb.
Evolutionary origins of copy number variation
By comparing the respective locations of human and chimpanzee CNVs to each other and with particular features of our genomes, we may gain considerable insight into the evolution of genomic instability, and specifically, copy number variation. We found that 144 of the 353 human CNVRs (42%) overlapped chimpanzee CNVRs, versus the random expectation of only 39 CNVRs (11%; permutation test; P < 1 × 10−15). With the exception of variants under extreme and long-term balancing selection pressures (e.g., the MHC locus) (Lawlor et al. 1988), it is unlikely that genetic polymorphisms present in the human–chimpanzee common ancestor would have been maintained to our extant species (for discussion, see Perry et al. 2006). This theoretical expectation is supported by empirical studies of variants with relatively low likelihoods of multiple hits (recurrent mutation at the same site) on this timescale (Hacia et al. 1999; Weber et al. 2002; Asthana et al. 2005). For example, Hacia et al. (1999) found that none of the 271 human single nucleotide polymorphisms (SNPs) they examined were also polymorphic in 23 chimpanzees. Therefore, the presence of both human and chimpanzee CNVs in orthologous genomic regions likely reflects recurrent CNV genesis rather than maintenance of ancestral polymorphisms, suggesting that sequence motifs or architectures shared between the human and chimpanzee genomes may predispose certain chromosomal regions to structural instability in both extant species.
Previous studies in humans (Redon et al. 2006), chimpanzees (Perry et al. 2006), rhesus macaques (Macaca mulatta) (Lee et al. 2008), and mice (Mus musculus) (Graubert et al. 2007) have reported that CNVs are significantly enriched for segmental duplications (SDs; low-copy repeats ≥1 kb and ≥90% similarity) (Bailey et al. 2002), suggesting that nonallelic homologous recombination (NAHR) mechanisms may play important roles in the formation of CNVs (see Cooper et al. 2007) in diverse mammalian lineages. To examine in more depth these genomic patterns using our multispecies CNV data set, we identified the locations of SDs in the human and chimpanzee genomes, applying size and similarity cutoffs (≥10 kb and ≥94% identity, respectively) to achieve roughly similar resolution in our databases of human and chimpanzee SDs (i.e., accounting for the more limited power to identify SDs of smaller size and lower similarity in the chimpanzee genome). We then compared the locations of the human and chimpanzee CNVs with those of SDs and used permutations to gauge levels of enrichment relative to random expectations.
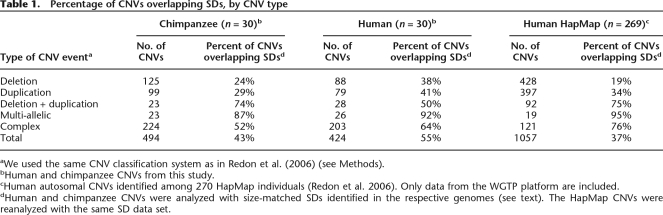
We found that 182 of the 353 human CNVRs (51.6%) overlapped segmental duplications in the human genome, compared with 47 (13.4%) expected by chance alone (permutation test; 3.9-fold enrichment; P < 1 × 10−15). A similar level of enrichment was observed for chimpanzees: 171 of 453 CNVRs (39.0%) overlapped SDs in the chimpanzee genome versus only 41 (9.3%) expected (4.2-fold enrichment; P < 1 × 10−15). While levels of enrichment were highest for common CNVRs, rare (singleton) CNVRs were still threefold or more enriched for SDs in each species (Supplemental Table 2). When we isolated and classified individual CNV events (i.e., a given CNVR may be comprised of two or more overlapping CNV events, distinguished by log2 ratio profiles), we found that the frequency of SD overlap is considerably higher for certain CNV types—especially multiallelic and deletion + duplication CNVs—in humans as well as in chimpanzees (Table 1). Such CNVs appear to occur almost without exception in genomic regions that are hotspots for SD-mediated NAHR.
Table 1.
Percentage of CNVs overlapping SDs, by CNV type
aWe used the same CNV classification system as in Redon et al. (2006) (see Methods).
bHuman and chimpanzee CNVs from this study.
cHuman autosomal CNVs identified among 270 HapMap individuals (Redon et al. 2006). Only data from the WGTP platform are included.
dHuman and chimpanzee CNVs were analyzed with size-matched SDs identified in the respective genomes (see text). The HapMap CNVs were reanalyzed with the same SD data set.
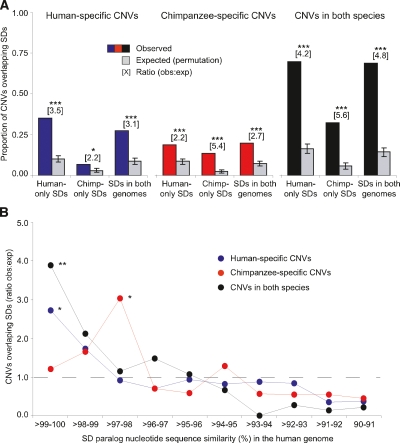
We were particularly interested in comparing relationships between CNVs and SDs while considering whether they were observed in one or both species. For these analyses, we isolated subsets of SDs found only in the human genome, only in the chimpanzee genome, or in orthologous regions in both species (shared SDs), and compared these with similar groupings of CNVRs (Fig. 5A). We found that “human-specific CNVRs” (i.e., human CNVRs that do not overlap any chimpanzee CNVRs in our data set) were enriched 3.5-fold for human-only SDs versus only 2.2-fold for chimpanzee-only SDs mapped onto the human genome. Similarly, “chimpanzee-specific CNVRs” (i.e., chimpanzee CNVRs that do not overlap any human CNVRs in our data set) were enriched 5.4-fold for chimpanzee-only SDs compared with 2.2-fold for human-only SDs. Here, it is important to note that since SD identifications for any given genomic region are based largely on the sequence of a single individual, all human-only and chimpanzee-only SDs do not necessarily reflect species-specific duplications (i.e., there may be a subsequent deletion polymorphism in the region in one species). Still, these results strongly suggest that species-specific CNV patterns are directly associated with architectural differences between the human and chimpanzee genomes.
Figure 5.
Colocalization of CNVs and SDs in the human and chimpanzee genomes. (A) Observed and expected proportion of human-specific CNVs (i.e., human CNVs that do not overlap any chimpanzee CNVs), chimpanzee-specific CNVs, and CNVs that were observed in orthologous regions in both species that overlap human-only, chimpanzee-only, and shared SDs. A given CNV may intersect more than one type of SD. Expected values are based on 10,000 randomized permutations. (B) Ratio of observed (obs) versus expected (exp) number of common CNVs (observed in two or more individuals) overlapping SDs in the human genome, binned by nucleotide sequence similarity between intrachromosomal SD paralogs. For CNVs overlapping multiple SDs, the SD with maximum (similarity x length) is reported. Expected values are based on 1000 randomized permutations. *P < 0.01; **P < 0.001; ***P < 1 × 10−8.
Therefore, it is reasonable to ask whether the observed excess of CNVs in orthologous genomic regions in both species might have been driven by recurrent NAHR of shared SDs, which are inferred to have been present in the genome of the human–chimpanzee common ancestor and maintained in both lineages. In fact, we found that the existence of a large majority of such CNVs could be explained by this mechanism: 96 of 140 human–chimpanzee CNVRs (69%) overlapped shared SDs, compared with 20 CNVRs that were expected to overlap shared SDs based on chance alone (14%; 4.8-fold enrichment; P < 1 × 10−15).
To explore in more detail the evolutionary relationships between CNVs and SDs, we estimated nucleotide sequence similarities for intra- and interchromosomal SD paralogs in the human genome and examined their intersection with the above-classified CNVRs. For CNVRs that overlapped more than one SD, we assigned one intra- and one interchromosomal SD percent similarity value, based on the overlapping SD paralog with the greatest product of percent identity and length. Although similar patterns were observed for rare CNVRs (Supplemental Table 3), the observed enrichments were greatest for CNVRs observed in two or more individuals (Fig. 5B). For human-specific CNVRs, a significant 2.7-fold enrichment was observed for SDs with >99% intrachromosomal nucleotide sequence identity (P = 0.007), with generally concomitantly decreasing enrichments for lower SD percent similarity bins. This pattern for human-specific CNVRs provides several evolutionary insights. First, given that human–chimpanzee nucleotide sequence divergence is ∼1.23% (The Chimpanzee Sequencing and Analysis Consortium 2005), a subset of SDs in the >99%–100% similarity frequency bin may be human specific and thus unavailable as a substrate for NAHR-mediated CNV genesis in chimpanzees. In addition, NAHR is more likely to occur among sequences with higher nucleotide similarity (Bailey and Eichler 2006); therefore, duplication and deletion mutation rates and CNV diversity levels may be relatively high in regions containing nearly identical SD paralogs. Finally, some human SD paralogs with the very highest similarities (near 100%) may not even represent previously fixed duplications, instead reflecting duplication CNVs for which the donor for the human genome reference sequence carried the derived allele.
In contrast to human-specific CNVRs, chimpanzee-specific CNVRs are not significantly enriched for human SDs in the two highest percent similarity bins, but are threefold enriched for SDs with >97%–98% similarity in the human genome (P = 0.009) (Fig. 5B). The observed enrichment with the 97%–98% similarity human SD paralogs for chimpanzee- but not human-specific CNVRs could reflect divergence in human and chimpanzee NAHR mutation rates among a subset of shared SD regions. One possible source for this divergence could be species-specific gene conversion events that may homogenize SDs (Chen et al. 2007) and maintain higher NAHR mutation rates. Alternatively, or in parallel, higher NAHR mutation rates may facilitate multiple rounds of subsequent duplication (Johnson et al. 2006), thereby creating new 100% identical SD paralogs only in one species (some of these duplications may be relatively recent polymorphisms that were ascertained as SDs). Regardless of the mechanism, this hypothesis is consistent with the SD-enrichment pattern for the CNVRs that were observed in orthologous regions in both species (Fig. 5B); these CNVRs have a 3.9-fold enrichment for SDs with >99%–100% similarity in the human genome (P < 0.001). That is, strikingly, the highest level of enrichment for these CNVRs was with SD paralogs that are less diverged than our a priori expectations for shared SDs (a minimum of ∼1.23%; the level of human–chimpanzee nucleotide sequence divergence (The Chimpanzee Sequencing and Analysis Consortium 2005) in the absence of subsequent gene conversions or duplications or both.
We expected to find CNVRs and interchromosomal SD paralogs to intersect no more than would be expected by chance. However, surprisingly, the enrichment patterns we observed for CNVRs with interchromosomal SDs were broadly similar to those with intrachromosomal SDs (Supplemental Table 3). To examine this interesting result in more detail, we compared the locations of human CNVRs with both intra- and interchromosomal SD paralogs in a single analysis, as many SD loci are organized in mosaic architectures of juxtaposed duplicated segments, where intra- and interchromosomal SDs overlap within and around core duplications (Jiang et al. 2007). Of the 139 common CNVRs that overlapped SDs, 126 (91%) were associated with intrachromosomal SD paralogs, including 87 CNVRs (69%) that also overlapped interchromosomal SD paralogs. Therefore, there are relatively very few CNVRs that overlapped interchromosomal but not intrachromosomal SDs (13 of 139, or 9%; Supplemental Fig. 3). Thus, we believe that the observed association between CNVRs and interchromosomal SDs is predominantly a shadowing effect from intrachromosomal SD paralogs, which, via NAHR, are likely responsible for the structural instability of the majority of these genomic regions.
CNV frequency distribution analyses
It has been noted that genes with sensory perception and immune response functions are over-represented in human CNV regions (Redon et al. 2006; Cooper et al. 2007). This enrichment has been interpreted to reflect positive selection for copy number variation of these genes (Nguyen et al. 2006). However, such a pattern could also arise if CNV mutation rates were relatively higher for sensory perception and immune response genes, or if copy number changes for genes with other functions (i.e., nonsensory and nonimmune response) were relatively more deleterious (Nguyen et al. 2008; Young et al. 2008). For example, in a recent study, Nozawa et al. (2007) observed that a similar proportion of functional olfactory receptor genes and nonfunctional olfactory pseudogenes overlapped CNVs that were identified among 270 HapMap individuals (Redon et al. 2006), leading them to suggest that CNV patterns for functional olfactory receptors may largely reflect neutral evolution (i.e., genetic drift), rather than positive or purifying selection.
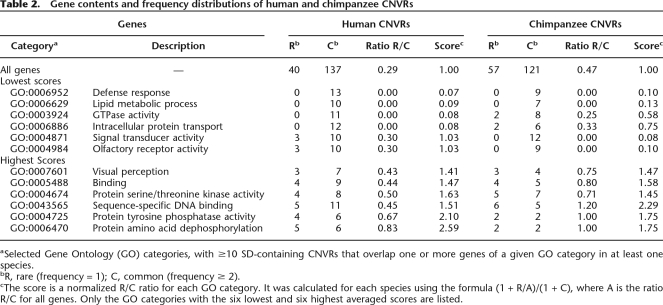
To address these issues, we examined CNVR frequency distributions for Gene Ontology functional categories (Ashburner et al. 2000), based on the number of rare versus common variants containing one or more genes (for each category). Purifying selection may prevent deleterious variants from reaching intermediate frequencies. Therefore, by identifying Gene Ontology functional categories with relatively high proportions of rare variants, we can identify classes of genes for which copy number variation is the most likely to be deleterious (Ohta 1973; Akashi 1999; Williamson et al. 2005). We focused our analyses on SD-overlapping CNVRs, because CNV mutation rates (and thereby neutral frequency distributions) may differ considerably between SD and non-SD regions, and in humans, considerably more gene-containing CNVRs overlap SDs (see Supplemental Note).
Compared with the ratios for all gene-containing CNVRs, in both species there were relatively high proportions of rare CNVRs that overlap genes with sequence-specific DNA-binding and protein phosphatase activity functions (Table 2). In contrast, there were no rare CNVRs that overlap genes with defense response or lipid metabolic process functions in either species. These results provide evidence that levels of evolutionary constraint on gene copy number variation may vary considerably among functional categories. Specifically, CNVRs containing genes with sequence-specific DNA-binding and protein phosphatase activity functions are most likely to be under purifying selection. These CNVRs are intriguing candidates for disease association studies. Furthermore, defense response and lipid metabolic process genes appear relatively tolerant to copy number change in both humans and chimpanzees, although at present we are unable to distinguish between neutral evolution and positive or balancing selection hypotheses for these CNVRs (Supplemental Note).
Table 2.
Gene contents and frequency distributions of human and chimpanzee CNVRs
aSelected Gene Ontology (GO) categories, with ≥10 SD-containing CNVRs that overlap one or more genes of a given GO category in at least one species.
bR, rare (frequency = 1); C, common (frequency ≥ 2).
cThe score is a normalized R/C ratio for each GO category. It was calculated for each species using the formula (1 + R/A)/(1 + C), where A is the ratio R/C for all genes. Only the GO categories with the six lowest and six highest averaged scores are listed.
Comparisons of copy number fixation and polymorphism
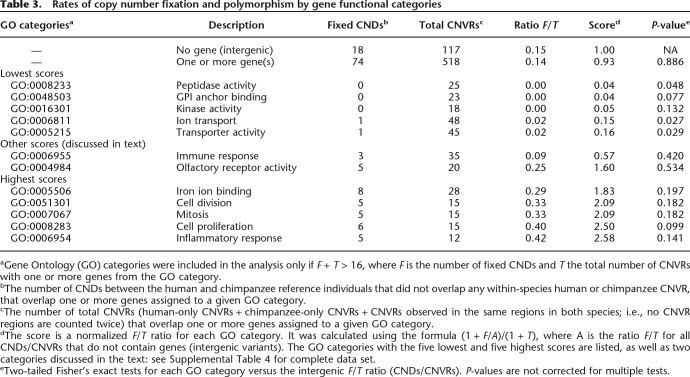
Recent genome comparison studies have discovered an elevated rate of gene-containing duplications in the human lineage (Fortna et al. 2004; Cheng et al. 2005; Demuth et al. 2006), raising the possibility that some of these duplications may have been fixed by positive selection. Therefore, identifying the specific genes or gene families affected may help us to better understand hominin evolution. In the McDonald-Kreitman (MK) test (1991), the ratio of the number of between-species fixed differences to the number of intraspecific polymorphisms for a putatively functional class of variation is compared with that for a neutral class of variation. These ratios will be similar under neutrality, whereas a relative excess of fixed differences for the functional class would suggest that some functional variants may have been fixed by positive selection (McDonald and Kreitman 1991). The MK test was originally developed to identify signatures of selection in gene coding regions. Here, we have adapted this framework for use with CNV data.
For this analysis, we determined the number of fixed CNDs between human and chimpanzee and the number of CNVs within each species that contained one or more genes with a given Gene Ontology function, compared with a similar ratio for intergenic CNDs and CNVs that did not overlap any genes. Although some intergenic variants may be of functional importance (for example, by influencing the expression levels of nearby genes; Stranger et al. 2007), as a whole, this group likely better reflects neutral evolution than gene-containing CNDs and CNVs.
A subset of the MK results is provided in Table 3. Due to the small number of variants representing any one functional category, we lacked statistical power to identify significant outliers (the P-values reported in Table 3 are not corrected for multiple tests)—therefore, these findings should be considered preliminary. Relative to fixation and polymorphism in intergenic regions, there were lower proportions of fixed differences overlapping genes with peptidase activity, ion and intracellular protein transport, and carbohydrate metabolic process functions. This pattern may be consistent with either purifying selection against the fixation of copy number changes or selection for the maintenance of CNVs (i.e., balancing selection). Ratios for immune response and olfactory receptor genes—often discussed in terms of CNVs and natural selection—were similar to those for intergenic regions, which is consistent with neutrality (see also Zhang 2007). In contrast, inflammatory response and cell proliferation function categories were both characterized by a relative excess of fixed CNDs, suggesting that some of these duplications or deletions may have been fixed by positive selection. Therefore, the affected genes will be of particular interest for subsequent studies focusing on the evolution of adaptive phenotypic-level differences between humans and chimpanzees.
Table 3.
Rates of copy number fixation and polymorphism by gene functional categories
aGene Ontology (GO) categories were included in the analysis only if F + T > 16, where F is the number of fixed CNDs and T the total number of CNVRs with one or more genes from the GO category.
bThe number of CNDs between the human and chimpanzee reference individuals that did not overlap any within-species human or chimpanzee CNVR, that overlap one or more genes assigned to a given GO category.
cThe number of total CNVRs (human-only CNVRs + chimpanzee-only CNVRs + CNVRs observed in the same regions in both species; i.e., no CNVR regions are counted twice) that overlap one or more genes assigned to a given GO category.
dThe score is a normalized F/T ratio for each GO category. It was calculated using the formula (1 + F/A)/(1 + T), where A is the ratio F/T for all CNDs/CNVRs that do not contain genes (intergenic variants). The GO categories with the five lowest and five highest scores are listed, as well as two categories discussed in the text: see Supplemental Table 4 for complete data set.
eTwo-tailed Fisher’s exact tests for each GO category versus the intergenic F/T ratio (CNDs/CNVRs). P-values are not corrected for multiple tests.
Interestingly, every fixed CND that overlaps inflammatory response genes is a copy number loss in chimpanzees relative to humans. Based on an alignment of the two reference genome sequences (Karolchik et al. 2003), the APOL1, APOL4, CARD18 (previously known as ICEBERG, RefSeq accession no. NM_021571), IL1F7, and IL1F8 genes have been completely deleted in chimpanzees. While these deletions were also identified and discussed in the initial description of the chimpanzee genome sequence (The Chimpanzee Sequencing and Analysis Consortium 2005), our analyses in the present study have further shown that (1) these gene deletions are likely fixed in chimpanzees, unlike the B4GALNT2 first exon deletion depicted in Figure 2 (see also Supplemental Table 5, for additional examples of nonfixed gene disruptions previously identified in Clint’s genome), and (2) they may have been driven to fixation by positive selection.
Discussion
We have constructed a genome-wide map of copy number variation in humans and chimpanzees using a single human aCGH platform. These data represent the first comprehensive resource to examine the evolutionary significance of copy number variation in both the human and chimpanzee genomes. Human and chimpanzee CNVs occurred in orthologous genomic regions far more often than would be expected by chance and were strongly associated with the presence of highly homologous intrachromosomal SD paralogs at these loci. This result seems to reflect, in large part, recurrent NAHR involving duplicated DNA segments that probably originated in the human–chimpanzee common ancestor and have been retained in the genomes of both extant species. This hypothesis is supported further by observations of several other genomic instabilities shared between humans and other non-human primates that also occur in regions of ancestral duplication (Fortna et al. 2004; Babcock et al. 2007; Dumas et al. 2007; Lee et al. 2008).
Additionally, we have analyzed our data in an evolutionary population genetic framework, considering CNV data by Gene Ontology category to perform tests of neutrality. These analyses have identified specific genes, particularly those with inflammatory response functions, which may have faced exceptional natural selection pressures at the copy number level during human and chimpanzee evolution. The specific functional roles of these genes, such as APOL1, APOL4, CARD18, IL1F7, and IL1F8 that are completely deleted in chimpanzees, are therefore of great interest. In humans, the APOL1 gene (Apolipoprotein L-1) is involved in resistance to protozoan trypanosome parasites that cause sleeping sickness (Vanhollebeke et al. 2007). Potential selective advantages of APOL1 gene deletion for chimpanzees are not immediately apparent; however, we may be able to generate testable hypotheses once we understand better the other functions of this gene in humans (Pays et al. 2006). The interleukin-1 family member 7 (IL1F7) protein and CARD18 both interact with caspase 1 (Humke et al. 2000; Kumar et al. 2002), which plays critical roles in innate immunity and inflammation (Kersse et al. 2007). For example, CARD18 binds to and inhibits the function of caspase 1 (Humke et al. 2000), thereby inhibiting production of the interleukin-1-beta inflammatory cytokine. Therefore, this inflammatory pathway is likely regulated differently in chimpanzees than in humans. Interestingly, a mutation that inactivates the human CASP12 gene has been driven to near fixation by positive selection (Wang et al. 2006; Xue et al. 2006), likely because loss-of-function of caspase 12 confers resistance to sepsis (Saleh et al. 2004). Thus, it is particularly striking that gene-disrupting mutations affecting similar inflammatory response pathways have now been associated with potential signatures of positive selection in both humans and chimpanzees. Together, these observations provide support for Olson’s “less-is-more” hypothesis (Olson 1999)—which proposes that gene losses can be adaptive—and highlight the potentially significant role played by turnover of inflammatory response genes in hominoid evolution.
We also investigated in detail the pattern of copy number variation at another gene with inflammatory response functions, CCL3L1 (chemokine C-C motif ligand 3-like 1). Gonzalez et al. (2005) found that in human populations, lower CCL3L1 copy numbers were associated with increased susceptibility to HIV infection and progression to AIDS. We were particularly interested in this locus because Gonzalez et al. (2005) reported extensive CCL3L1 copy number variation in chimpanzees, with an average copy number that was substantially higher than that observed in any human population. This finding is intriguing because HIV has only recently been a human disease and is therefore unlikely to have driven any adaptive changes in human CCL3L1 copy number. In contrast, chimpanzees were likely exposed much earlier to an HIV-like virus (Keele et al. 2006). Thus, one could hypothesize that HIV-related positive selection has led to a relatively increased chimpanzee CCL3L1 copy number.
However, unexpectedly, our results showed no CND for the CCL3L1 gene between the human and chimpanzee reference individuals, and no within-species copy number variation at the 17q12 locus among the 30 chimpanzees examined in our study (Fig. 4A,B). Instead, we identified a relative copy number loss of the nearby oncogene TBC1D3 in the chimpanzee compared with the human reference individual (Fig. 4B). In humans, eight paralogous copies of the TBC1D3 gene (named TBC1D3 and TBC1D3B to TBC1D3H) have been described at 17q12, many with different expression patterns (Hodzic et al. 2006). In contrast, only one copy of TBC1D3 is found in the chimpanzee genome reference sequence assembly (panTro2) (The Chimpanzee Sequencing and Analysis Consortium 2005). While future studies will be required to understand better the relative evolutionary roles of CCL3L1 and TBC1D3 in shaping the complex chromosome 17q12 region, our results raise the possibility that the human–chimpanzee difference and the unusually high level of human population differentiation observed at this locus (Gonzalez et al. 2005; Redon et al. 2006) were driven by positive selection for TBC1D3, rather than CCL3L1, duplications. Related to this hypothesis, it was recently demonstrated that TBC1D3 strongly influences cell proliferation (Wainszelbaum et al. 2008), a functional category with one of the highest relative rates of CND fixation (Table 3). Moreover, TBC1D3 is located within one of the 14 “core duplicons” that have been identified in the human genome (Jiang et al. 2007). The rapid expansion of these gene-enriched cores, which are the focal points for large blocks of human lineage-specific duplications, may reflect positive selection for copy number increases in human evolution (Jiang et al. 2007).
In summary, our analysis reveals the power of comparing within- and between-species patterns of variation at the copy number level for providing insights into mutational mechanisms and selective forces acting on this important class of genetic diversity. This framework can be expanded to CNV studies in other species, and augmented for future studies using next-generation technology platforms to investigate more complete size and class ranges of structural variation.
Methods
Samples
Human DNAs were obtained from the Coriell Institute for Medical Research and the HGDP-CEPH Human Genome Diversity Cell Line Panel. We selected African individuals for this study because nucleotide sequence genetic diversity in humans is regularly observed to be highest in sub-Saharan Africa, at a level generally comparable or slightly higher than that of Western chimpanzees (e.g., Fischer et al. 2006). A sample of only one or a few individuals from many different worldwide populations, while possibly facilitating the identification of a larger number of human CNVs, would have been less comparable in population genetic analyses to our chimpanzee sample that is comprised primarily of one subspecies. Chimpanzee B-lymphoblast cell lines were obtained from the Coriell Institute for Medical Research and Integrated Primate Biomaterials and Information Resource. Chimpanzee whole blood samples were collected at the New Iberia Research Center and the Primate Foundation of Arizona during routine veterinary appointments. DNA was isolated from cell lines and whole blood using the PureGene DNA Isolation Kit (Gentra Systems). Subspecies identification for wild-born individuals was based on mitochondrial DNA hypervariable region sequencing and comparison to individuals from known capture location (Stone et al. 2002). Estimated subspecies ancestry for captive-born individuals was based on mitochondrial DNA and Y chromosome sequencing (Stone et al. 2002) and pedigree analysis. The captive-born reference chimpanzee, Clint, is primarily of Western subspecies origin (The Chimpanzee Sequencing and Analysis Consortium 2005).
aCGH experiments
The aCGH platform used in this study was the human Whole-Genome TilePath (WGTP) array, previously used to identify CNVs among the genomes of 270 individuals from four human populations of the International HapMap Project (Fiegler et al. 2006; Redon et al. 2006). After addition of new clones to fill coverage gaps, the array includes 28,708 large-insert clones, covering ∼97% of the euchromatic portion of the human genome reference sequence. The clone set and mapping information can be accessed and downloaded using the Ensembl genome browser (www.ensembl.org/Homo_sapiens/index.html) by activating the “30K TPA clones” decoration within the graphical overview. Microarray hybridizations were performed as previously described (Fiegler et al. 2007). All experiments were performed in duplicate with DNA labeling color reversal (dye swap). Array images were acquired using a 5-μm resolution Agilent Technologies G2505A laser scanner. Fluorescence intensities and log2 ratio values were extracted using BlueFuse software (BlueGnome Ltd). Array information, experimental design, raw intensities and processed log2 ratio values are all available through ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-TABM-479. Fusion of dye-swap results and subsequent analyses were performed using custom Perl scripts as previously described (Fiegler et al. 2006), with one additional step: After image quantification, log2 ratio calculation and block median normalization, we have introduced a G+C correction, which consists of normalizing the log2 ratios of each clone using the content in G+C percent of that clone. This correction was performed by linear regression using the module LineFit in Perl (http://search.cpan.org/~randerson/Statistics-LineFit-0.07) and applied on each individual profile before fusion of dye-swap results. Copy-number variable segments were automatically detected using the CNVfinder algorithm (Fiegler et al. 2006). The full set of CNV calls per individual is available in Supplemental Table 6. Because human–chimpanzee nucleotide sequence divergence is on average ∼1.23% (The Chimpanzee Sequencing and Analysis Consortium 2005), there is an expected sequence mismatch effect for the between-species Clint versus NA10851 aCGH experiment. To address this issue, this experiment was carried out following identical procedures, but with addition of one normalization step to account for chimpanzee vs. human nucleotide divergence (which varies among clones; based on an alignment of the human and chimpanzee genome sequences) and with slight modifications of the CNVfinder algorithm (R. Redon, pers. comm.). However, we note that we would be unable to identify deletions that have been fixed in the human lineage, because these sequences would not be represented on the array; the same limitation does not exist for chimpanzee-lineage deletions. We also note that the use of a human platform comprised of large insert clones is not expected to have a large effect on the quality of the within-species chimpanzee experiments, because both the test and reference individuals for these experiments would have similar numbers of nucleotide sequence mismatches from the clones on the array. High-resolution aCGH experiments were performed using a custom oligonucleotide array (NimbleGen Systems) that cover the hg18 genome interval chr17:31Mb-34Mb with 40,000 isothermal probes.
CNV validation by FISH and PCR
For fiber-FISH analyses, DNA fibers were stretched on slides as previously described (Korbel et al. 2007). Large-insert clone DNAs were labeled with digoxigenin-11-dUTP (Roche), fluorescein-12-dUTP (Roche) or biotin-16dUTP (Roche) by using a modified whole-genome amplification kit (WGA3; Sigma). Approximately 100 ng of each labeled probe was used to carry out FISH experiments following previously published protocols (Korbel et al. 2007). Digoxigenin-labeled probes were detected using a 1:500 dilution of monoclonal mouse anti-dig antibody (Sigma) and a 1:200 of Texas Red-X-conjugated goat anti-mouse IgG (Invitrogen); fluorescein-labeled probes using with a 1:200 dilution of Alexa 488-conjugated rabbit anti-fluorescein IgG and Alexa 488-conjugated donkey anti-rabbit IgG (Invitrogen); biotin-labeled probes were detected with one layer Cy3-avidin (final concentration at 2 μg/mL). After detection, slides were mounted with SlowFade Gol mounting solution containing 4′,6-diamidino-2-phenylindole (Invitrogen). Images were captured on a Zeiss Axioplan fluorescent microscope and processed with the SmartCapture software (Digital Scientific).
For EGFL11 interphase FISH analyses, probes were labeled with SpectrumOrange dUTP and Spectrum Green dUTP using a nick translation kit (Abbott Molecular Laboratories). B-lymphoblast cell cultures were subjected to the hypotonic treatment (Ohnuki’s hypotonic solution: five parts 55 mM sodium nitrate, two parts 55 mM sodium acetate, and 10 parts 55 mM potassium chloride) for 1 h at 37°C, followed by three fixations using methanol and glacial acetic acid. Following overnight hybridization and washes, DAPI was applied under a glass coverslip and hybridization signals were viewed on an Olympus BX-51 fluorescent microscope. Images were captured and processed with Cytovision software.
The primer sequences used for genotyping the chromosome 17 deletion were (all 5′–3′ [F] forward; [R] reverse): F flanking, TAGCCAATCAAACAATGGTGTC; R flanking, TCCTCTATT CAACGTGTGTTGC; R internal, TATCCCATTAGGTTTGGTC CAG. Primer sequences for the chromosome 8 deletion were: F flanking, CAGAGAACAGGGTCACAGACAC; R flanking, CTCCT GAAAGGCTGCTAGTGAT; R internal, TGGCCTAGGTTTTGCT CATAAT. PCR assays were performed with 25 ng DNA in 25-μL reaction volumes and HotMaster Taq polymerase (Eppendorf). Cycling conditions were 93°C for 3 min, followed by 40 cycles of 93°C for 30 sec, 60°C for 30 sec, and 70°C for 2 min, using a DNA Engine Thermal Cycler (Bio-Rad). PCR products were visualized by electrophoresis on a 1.2% agarose gel, followed by ethidium bromide staining.
CNV data analyses
CNVRs for each species were generated by merging all individual CNV calls into a single list of nonredundant CNV regions independently to the size of the overlap and the frequency of the calls. Human and chimpanzee CNVRs were further merged—using the same method—in a single list of regions comprising three classes: human-only CNVRs, chimpanzee-only CNVRs, and CNVRs found in both species. All CNVRs for both species are listed in Supplemental Table 7. We defined CNV events (Table 1) by applying more stringent merging criteria to separate juxtaposed CNVs, as previously described (Redon et al. 2006). Duplication and deletion CNV types are designated based on the minor copy number state. Deletion + duplication variants are special cases of multi-allelic CNVs, with low-frequency losses and gains relative to the majority of individuals. Multi-allelic CNVs have log2 ratio distributions that are more evenly distributed across four or more discrete clusters. Complex CNVs could not be classified otherwise: These CNVs may (1) reflect the masking of true CNV type by experimental noise, (2) comprise multiple smaller CNV events that cannot be distinguished with the resolution of our aCGH platform, or (3) be truly complex with different breakpoints, juxtaposed gains and losses, or smaller CNVs contained within larger ones. For the SD-overlap analysis reported in Table 1, we only considered 200 kb at both extremities for CNV events with a size >400 kb.
SD analyses
We downloaded the positions of human and chimpanzee SDs from the Segmental Duplication Database (http://humanparalogy.gs.washington.edu/; http://chimpparalogy.gs.washington.edu/). For this database, SDs were detected by Whole-genome Shotgun Sequence Detection (WSSD) (Bailey et al. 2001; Cheng et al. 2005) by mapping 31.3 million chimpanzee and 27.4 million human sequence reads against the human reference genome (hg17) to identify regions with significantly deeper read coverage depth compared with known unique regions. Regions showing an excess of both chimpanzee and human reads were considered to be duplicated in both species (shared SDs). The proportions of CNVRs (transferred to hg17 positions based on clone end-sequence coordinates) expected to overlap SDs by chance alone were estimated using permutation tests of 10,000 randomized trials. In each permutation, the locations of CNVRs were randomized based on the midpoint coordinates of all autosomal clones on the WGTP platform (the sizes of the randomized CNVRs were maintained) and assessed for SD overlap.
For each SD-overlapping CNVR, we estimated maximum nucleotide sequence percent identities of human genome intra- and interchromosomal SD paralogs. For this analysis, we retrieved Whole-Genome Assembly Comparison (WGAC) (Bailey et al. 2002) SD alignments from the human Segmental Duplication Database. Percent identity calculations include indels. For CNVRs that overlapped more than one SD, we assigned a single SD percent identity value based on the SD with the greatest product of percent identity and length. This weighted approach is based on the resolution of the WGTP platform; we are more likely to have identified the CNVs associated with slightly larger SDs, rather than internal, smaller SDs. The proportions of CNVRs expected to overlap SDs were estimated using permutation tests of 1000 randomized trials. The same procedure for calculating maximum SD paralog percent identities was applied for the randomized CNVRs.
Acknowledgments
We thank Gloria Tam, Diana Rajan, Beiyuan Fu, Stephen Clayton, Stephen Montgomery, Richard Smith, and Heike Fiegler for their help and advice; Jan Korbel for the permutation script used for CNV-SD overlap analyses; and Don Conrad, Stephen Scherer, Brian Verrelli, and Gary Schwartz for helpful discussions regarding this study. We thank the Human Genome Diversity Project, the Coriell Institute for Medical Research, the Integrated Primate Biomaterials and Information Resource, New Iberia Research Center, and the Primate Foundation of Arizona for samples. This work was funded by grants from the L.S.B. Leakey Foundation (to G.H.P.), the Wenner-Gren Foundation for Anthropological Research (to G.H.P.), the National Institutes of Health (C.L., HG004221; The University of Louisiana at Lafayette-New Iberia Research Center, RR015087, RR014491 and RR016483), the Howard Hughes Medical Institute (E.E.E.), and the Wellcome Trust (F.Y., T.F., M.E.H., N.P.C., C.T.-S., and R.R.).
Footnotes
[Supplemental material is available online at www.genome.org. The array data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession no. E-TABM-479.]
Article published online before print. Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.082016.108.
References
- Akashi H. Within- and between-species DNA sequence variation and the 'footprint' of natural selection. Gene. 1999;238:39–51. doi: 10.1016/s0378-1119(99)00294-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S., Schmidt S., Sunyaev S. A limited role for balancing selection. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Babcock M., Yatsenko S., Hopkins J., Brenton M., Cao Q., de Jong P., Stankiewicz P., Lupski J.R., Sikela J.M., Morrow B.E. Hominoid lineage specific amplification of low-copy repeats on 22q11.2 (LCR22s) associated with velo-cardio-facial/digeorge syndrome. Hum. Mol. Genet. 2007;16:2560–2571. doi: 10.1093/hmg/ddm197. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Chen J.M., Cooper D.N., Chuzhanova N., Ferec C., Patrinos G.P. Gene conversion: Mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Hurles M.E. The population genetics of structural variation. Nat. Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.M., Nickerson D.A., Eichler E.E. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- Demuth J.P., De Bie T., Stajich J.E., Cristianini N., Hahn M.W. The evolution of mammalian gene families. PLoS One. 2006;1:e85. doi: 10.1371/journal.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman E.B., Hartl D.L. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L., Kim Y.H., Karimpour-Fard A., Cox M., Hopkins J., Pollack J.R., Sikela J.M. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C.M., Sridhar S., Wigler M., Hall I.M. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 2007;39:1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- Fanciulli M., Norsworthy P.J., Petretto E., Dong R., Harper L., Kamesh L., Heward J.M., Gough S.C., de Smith A., Blakemore A.I., et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H., Redon R., Andrews D., Scott C., Andrews R., Carder C., Clark R., Dovey O., Ellis P., Feuk L., et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H., Redon R., Carter N.P. Construction and use of spotted large-insert clone DNA microarrays for the detection of genomic copy number changes. Nat. Protocols. 2007;2:577–587. doi: 10.1038/nprot.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Pollack J., Thalmann O., Nickel B., Paabo S. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:e207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., et al. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum. Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Saar K., Adamovic T., Verheul M., van Heesch S.A., Cook S., Pravenec M., Aitman T., Jacob H., Shull J.D., et al. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- Hacia J.G., Fan J.B., Ryder O., Jin L., Edgemon K., Ghandour G., Mayer R.A., Sun B., Hsie L., Robbins C.M., et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- Hahn M.W., Demuth J.P., Han S.G. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177:1941–1949. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic D., Kong C., Wainszelbaum M.J., Charron A.J., Su X., Stahl P.D. TBC1D3, a hominoid oncoprotein, is encoded by a cluster of paralogues located on chromosome 17q12. Genomics. 2006;88:731–736. doi: 10.1016/j.ygeno.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Humke E.W., Shriver S.K., Starovasnik M.A., Fairbrother W.J., Dixit V.M. ICEBERG: A novel inhibitor of interleukin-1beta generation. Cell. 2000;103:99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- Johnson M.E., Cheng Z., Morrison V.A., Scherer S., Ventura M., Gibbs R.A., Green E.D., Eichler E.E. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc. Natl. Acad. Sci. 2006;103:17626–17631. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., Van Heuverswyn F., Li Y., Bailes E., Takehisa J., Santiago M.L., Bibollet-Ruche F., Chen Y., Wain L.V., Liegeois F., et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersse K., Vanden Berghe T., Lamkanfi M., Vandenabeele P. A phylogenetic and functional overview of inflammatory caspases and caspase 1-related CARD-only proteins. Biochem. Soc. Trans. 2007;35:1508–1511. doi: 10.1042/BST0351508. [DOI] [PubMed] [Google Scholar]
- Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Hanning C.R., Brigham-Burke M.R., Rieman D.J., Lehr R., Khandekar S., Kirkpatrick R.B., Scott G.F., Lee J.C., Lynch F.J., et al. Interleukin-1F7B (IL-1H4/IL-1F7) is processed by caspase 1 and mature IL-1F7B binds to the IL-18 receptor but does not induce IFN-gamma production. Cytokine. 2002;18:61–71. doi: 10.1006/cyto.2002.0873. [DOI] [PubMed] [Google Scholar]
- Lawlor D.A., Ward F.E., Ennis P.D., Jackson A.P., Parham P. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature. 1988;335:268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- Lee A.S., Gutierrez-Arcelus M., Perry G.H., Vallender E.J., Johnson W.E., Miller G.M., Korbel J.O., Lee C. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum. Mol. Genet. 2008;17:1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Segraves R., Carbone L., Archidiacono N., Albertson D.G., Pinkel D., Eichler E.E. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.H., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Montiel M.D., Krzewinski-Recchi M.A., Delannoy P., Harduin-Lepers A. Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Acα2-3Galβ-R β1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: Evidence for an unusual extended cytoplasmic domain. Biochem. J. 2003;373:369–379. doi: 10.1042/BJ20021892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.Q., Webber C., Ponting C.P. Bias of selection on human copy number variants. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.Q., Webber C.P., Hehir-Kwa J., Pfundt R., Veltman J., Ponting C.P.2008Reduced purifying selection prevails over positive selection in human copy number variant evolution Genome Res. (this issue) 10.1101/gr.077289.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Kawahara Y., Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc. Natl. Acad. Sci. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Olson M.V. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E., Vanhollebeke B., Vanhamme L., Paturiaux-Hanocq F., Nolan D.P., Perez-Morga D. The trypanolytic factor of human serum. Nat. Rev. Microbiol. 2006;4:477–486. doi: 10.1038/nrmicro1428. [DOI] [PubMed] [Google Scholar]
- Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak S.E., Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M., Vaillancourt J.P., Graham R.K., Huyck M., Srinivasula S.M., Alnemri E.S., Steinberg M.H., Nolan V., Baldwin C.T., Hotchkiss R.S., et al. Differential modulation of endotoxin responsiveness by human caspase 12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Stone A.C., Griffiths R.C., Zegura S.L., Hammer M.F. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc. Natl. Acad. Sci. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhollebeke B., Nielsen M.J., Watanabe Y., Truc P., Vanhamme L., Nakajima K., Moestrup S.K., Pays E. Distinct roles of haptoglobin-related protein and apolipoprotein L-I in trypanolysis by human serum. Proc. Natl. Acad. Sci. 2007;104:4118–4123. doi: 10.1073/pnas.0609902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainszelbaum M.J., Charron A.J., Kong C., Kirkpatrick D.S., Srikanth P., Barbieri M.A., Gygi S.P., Stahl P.D.2008The hominoid-specific oncogene TBC1D3 activates Ras and modulates EGF receptor signaling and trafficking J. Biol. Chem. .28313233–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Grus W.E., Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:e52. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.L., David D., Heil J., Fan Y., Zhao C., Marth G. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S.H., Hernandez R., Fledel-Alon A., Zhu L., Nielsen R., Bustamante C.D. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.M., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Daly A., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., et al. Spread of an inactive form of caspase 12 in humans is due to recent positive selection. Am. J. Hum. Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.M., Endicott R.M., Parghi S.S., Walker M., Kidd J.M., Trask B.J. Extensive copy-number variation of the human olfactory receptor gene family. Am. J. Hum. Genet. 2008;83:228–242. doi: 10.1016/j.ajhg.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. The drifting human genome. Proc. Natl. Acad. Sci. 2007;104:20147–20148. doi: 10.1073/pnas.0710524105. [DOI] [PMC free article] [PubMed] [Google Scholar]