Abstract
Approximately one in 500 individuals carries a reciprocal translocation. Balanced translocations are usually associated with a normal phenotype unless the translocation breakpoints disrupt a gene(s) or cause a position effect. We investigated breakpoint junctions at the sequence level in phenotypically normal balanced translocation carriers. Eight breakpoint junctions derived from four nonrelated subjects with apparently balanced translocation t(1;22)(p36;q13) were examined. Additions of nucleotides, deletions, duplications, and a triplication identified at the breakpoints demonstrate high complexity at the breakpoint junctions and indicate involvement of multiple mechanisms in the DNA breakage and repair process during translocation formation. Possible detailed nonhomologous end-joining scenarios for t(1;22) cases are presented. We propose that cryptic imbalances in phenotypically normal, balanced translocation carriers may be more common than currently appreciated.
With the exception of benign copy number variation, it is generally assumed that a normal phenotype is associated only with a balanced genotype. However, a balanced translocation may produce an abnormal phenotype by disruption of a gene at the translocation breakpoint, through the formation of a novel fusion gene product, or by a position effect. Moreover, in apparently balanced translocations with abnormal phenotypes, duplications and deletions at the breakpoints have been identified that impact the phenotype. Baptista et al. (2005) hypothesized that breakpoints of normal individuals with apparently balanced chromosome rearrangements are relatively simple. However, their study was based on molecular cytogenetic methods (fluorescence in situ hybridization [FISH] and microarray-based comparative genomic hybridization [array CGH]) only. Breakpoints in only a few constitutional rearrangements in individuals with normal phenotypes have been examined at the DNA sequence level. These include translocations t(11;22) (Kurahashi et al. 2000, 2007), the most common constitutional translocation in humans, in which junction fragments were identified, and other translocation events including t(1;22)(p21.2;q11.2) (Gotter et al. 2004), t(4;22)(q35.1;q11.2) (Nimmakayalu et al. 2003), and t(8;22)(q24.13;q11.21) (Gotter et al. 2007). We have previously examined apparently balanced translocations in phenotypically normal individuals and found evidence that translocation junctions may show sequence inconsistency at the breakpoints created by addition of nucleotides and duplications (Gajecka et al. 2006b).
The resolution of DNA double-strand breaks (DSBs) can result in translocation formation through a variety of different pathways. In homologous recombination repair, the DNA ends can be aligned and joined using sequence homology. Alternatively, the broken ends can be brought together and rejoined in the absence of long tracks of sequence homology through nonhomologous end-joining (NHEJ). Two DSBs on homologous or nonhomologous chromosomes can be joined, creating derivative chromosomes. During the formation of translocations, it is unknown whether breaks in the participating DNA sequences occur simultaneously and/or are independent events.
Yu and Gabriel (2004) demonstrated that NHEJ-mediated reciprocal translocations can be formed as a consequence of DSB repair in Saccharomyces cerevisiae in the absence of homology. However, little is known about human balanced constitutional translocation formation. In balanced translocations, NHEJ was proposed as the mechanism involved in the rearrangement formation in a limited number of cases (Gajecka et al. 2006b). The key step in NHEJ is the physical juxtaposition of the DNA ends (Hefferin and Tomkinson 2005). Whether the physical juxtaposition of the DNA ends is a random or stimulated event is not known. If the juxtaposed DNA ends can be directly ligated, then the repair event only requires the action of a DNA ligase for completion. However, the majority of DSBs do not have ligatable termini and must be processed prior to ligation (Hefferin and Tomkinson 2005). Whether the additions of new nucleotides, duplications, and/or deletions seen after NHEJ are the consequence of a multistep process of DNA repair is not known.
Of 143 subjects with monosomy 1p36 ascertained by our laboratory, we have identified four subjects who showed unbalanced translocation involving 1p36 and 22q13 chromosomal regions, inherited from phenotypically normal balanced translocation carrier parents (Gajecka et al., in press). Herein, we present the results of molecular investigation of the t(1;22)(p36;q13) translocations in the four phenotypically normal balanced carrier parents. We compared the results to three phenotypically normal cases with six breakpoint junctions derived from balanced translocations t(1;9)(p36;q34) (Gajecka et al. 2006b) and conclude that NHEJ is likely the mechanism that generated all balanced translocations studied. We postulate involvement of multiple mechanisms of NHEJ in translocation formation. Possible detailed NHEJ scenarios for t(1;22) cases are presented. We propose that cryptic imbalances at the breakpoints in phenotypically normal balanced translocation carriers may be more common than simple junctions that result in no net gain or loss of DNA.
Results
Junction analysis
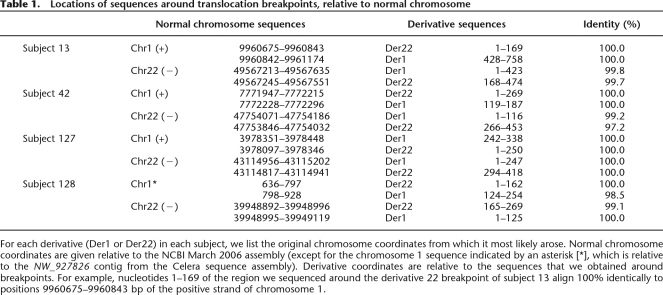
STS marker walking analyses using hybrids containing derivative chromosomes 1 allowed us to define the 1p36 breakpoint in each rearrangement at the sequence level. Using the TOPO Walker protocol and amplification across the junction we were able to clone the junction fragments and perform sequence analyses for the junctions between 1p36 and 22q13 in each derivative chromosome 1. Next, using complementary primers from the regions of the derivative 22 breakpoints, junctions were identified. Breakpoint junctions were sequenced and localized in the human genome as presented in Table 1. The breakpoint regions were unique in all subjects. Sequenced fragments show 98.5%–100% identity to sequences for that respective chromosome.
Table 1.
Locations of sequences around translocation breakpoints, relative to normal chromosome
For each derivative (Der1 or Der22) in each subject, we list the original chromosome coordinates from which it most likely arose. Normal chromosome coordinates are given relative to the NCBI March 2006 assembly (except for the chromosome 1 sequence indicated by an asterisk [*], which is relative to the NW_927826 contig from the Celera sequence assembly). Derivative coordinates are relative to the sequences that we obtained around breakpoints. For example, nucleotides 1–169 of the region we sequenced around the derivative 22 breakpoint of subject 13 align 100% identically to positions 9960675–9960843 bp of the positive strand of chromosome 1.
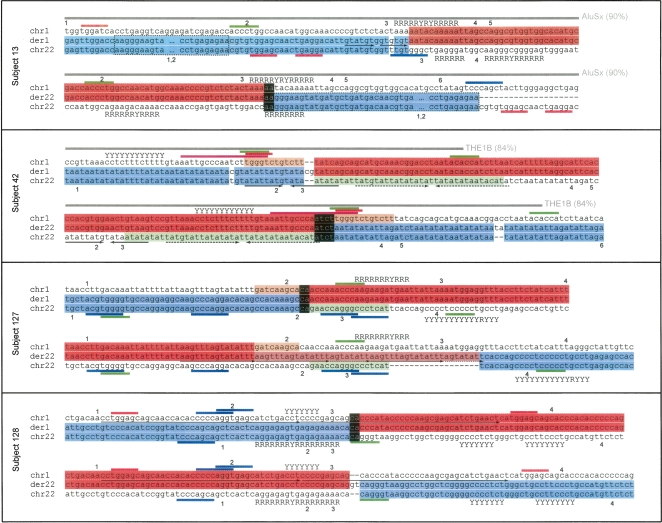
Cryptic imbalances were identified in both junctions at derivatives 1 and junctions at derivatives 22 (Fig. 1). In detail, in subject 13, a simple junction between the 1p36.22 and 22q13.33 was identified on the derivative chromosome 1, whereas a 337-bp duplication of chromosome 22 sequence was evident from comparing both derivative chromosomes (Fig. 1). Additionally, in the sequence originating from chromosome 22, a single nucleotide polymorphism was present within 3 bp of the breakpoint junction of the derivative chromosome 1. In subject 42, insertion of a CG dinucleotide was present at the junction at the derivative 1. Additionally in this case, a 38-bp deletion of sequence derived from chromosome 22 and a 12-bp deletion of sequence derived from chromosome 1 were identified at the derivative breakpoint junctions. In subject 127, a 9-bp deletion of sequence derived from chromosome 1, a 16-bp deletion of sequence derived from chromosome 22, and a 23-bp interstitial duplication with short triplication fragments originating from chromosome 1 on the derivative chromosome 22 were identified. Finally, in subject 128 we identified a simple junction on the derivative 1 and an insertion of new nucleotides (GT) at the derivative 22 junction. To exclude the possibility that the dinucleotide additions identified in the derivative chromosome 1 in case 42 and derivative chromosome 22 in case 128 represent discrepancies in the genome database sequences, the normal chromosomes 1 and 22 were sequenced at these locations in five unrelated control individuals. The normal chromosomes did not show these 2-bp insertions; thus, it is unlikely that these represent errors in the genome database. However, we have not excluded the possibility that these bases represent rare sequence variation.
Figure 1.
Sequence analysis of t(1;22)(p36;q13) breakpoints. For each subject the two alignments show derivatives of chromosome 1 (der1) and chromosome 22 (der22), respectively. Adjacent to these are the original chromosomal sequences, showing contribution to the derivative from chromosome 1 (red) or chromosome 22 (blue). Inverse (white on black) shows regions of derivative chromosome that match both original chromosomes. The area of chromosome 22 in subject 13 outlined by a dashed box indicates a 337-bp duplication present on both derivatives. Segments of chromosomes that are lost (deleted) are shown in light orange (chromosome 1) or light green (chromosome 22). The salmon/light red section of der22 in subject 127 represents a sequence that is generated by duplication from the original chromosome 1 sequence (in red). Motifs are indicated by colored bars adjacent to the alignments: eukaryotic topoisomerase cleavage site consensus (magenta), translin (TSN) target motifs (blue), DNA polymerase frameshift hotspots (green), and deletion hotspot consensus (red). “R” and “Y” adjacent to chromosomal sequences indicate runs of purines and pyrimidines, respectively. Interspersed repeats are represented by a gray bar, together with the repeat type and the similarity of the sequence to the repeat consensus (%). Paired arrows under a sequence indicate direct repeats (arrows in same direction) and inverted repeats (a sequence plus its inverted complement, arrows in opposite directions). The arrow line styles depict which sequences are related. Numbers adjacent to the normal chromosome sequences indicate significant potential DNA structural features, which are shown in the corresponding Supplemental Figure 2.
On the derivative 22 in subject 13, derivative 22 in subject 42, derivative 1 in subject 127, and derivative 1 in subject 128, short (2–4 bp) segments were identified that match both original chromosomes (in-phase) (Fig. 1, inverse presented as white letters on black background). Other than this microhomology, no significant sequence homology was found at the sequence around the breakpoint junctions.
Genes at the breakpoint junctions
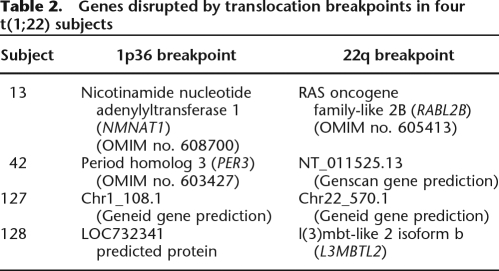
The junction fragment analysis revealed that the breakpoints on 1p36 and 22q13 each interrupt a known or predicted gene (or predicted protein) in all rearrangements (Table 2). To predict whether the subjects’ breakpoint junctions might produce chimeric transcripts, we examined the putative products of each subject’s breakpoint junction.
Table 2.
Genes disrupted by translocation breakpoints in four t(1;22) subjects
In case 13, the chromosome 22 breakpoint interrupts exon 8 (coding) of RABL2B, whereas the chromosome 1 breakpoint lies within an intron of NMNAT1 between exons 3 and 4. Fusions of these genes introduce premature stop codons from both derivatives that would potentially result in truncated protein products. For case 42, the chromosome 1 breakpoint occurs between coding exons 4 and 5 of PER3 whereas the chromosome 22 break occurs in an intron between exons 4 and 5 of a Genscan prediction of a 5-exon gene. Both breakpoints occur within introns, and therefore the translocation potentially can result in two fusion gene products. Because it is unknown if the Genscan gene produces a protein, we cannot determine if the fusion of its 5′ end with PER3 would result in a fusion protein. The chromosome 1 breakpoint location in PER3 would fuse a small portion of its 5′ end with the Genscan prediction (total 21 coding exons). Therefore, translation of the product is possible, but with most of the normal PER3 sequence being lost.
Both breakpoints in case 127 occur within introns of genes predicted by Genscan. Therefore, the translocations of the sequences could result in fusion genes, but it is not known if these would result in aberrant protein products. Finally, in case 128 the chromosome 22 breakpoint occurs between exons 7 and 8 of L3MBTL2, whereas the chromosome 1 breakpoint occurs just downstream from a predicted protein LOC732341. Hence, the most likely outcome of the translocation in this case would be truncation of L3MBTL2, with LOC732341 being retained intact.
Sequence motifs and motif densities
RepeatMasker (http://www.repeatmasker.org) identified interspersed repeats at the breakpoint junctions in subjects 13 and 42. In subject 13, the chromosome 1 breakpoint was located in the poly-A rich central linker region of an AluSx element that is 90% identical to the AluSx family consensus sequence. The chromosome 1 breakpoint of subject 42 is within a THE1B element from the MaLR/LTR family of repeats (84% identical to the family consensus) (Fig. 1). The rest of the breakpoints are located in nonrepetitive DNA. Furthermore, direct repeats (in subject 13), imperfect but long repeats (in subject 128), and inverted repeats (in subject 42) were identified in the breakpoint junctions as indicated in Figure 1.
No putative regions of Z-DNA structure were identified in the breakpoint junctions using Z-Hunt (Clamp et al. 2004; http://gac-web.cgrb.oregonstate.edu/zDNA/). Using Tandem Repeats Finder (Benson 1999), we identified oligopurine/oligopyrimidine tracts in all subjects at the junctions. Using the “fuzznuc” program from EMBOSS (Rice et al. 2000), we identified vertebrate topoisomerase consensus cleavage sites (RNYNNCNNGYNGKTNYNY) (in case 42), translin sites (GCCCWSSW and ATGCAG) (in subjects 13, 127, and 128), DNA polymerase a/b frameshift hotspots (ACCCWR, TGGNGT, GGGGGA, TCCCCC, and CTGGCG) (in all subjects), and deletion hotspot consensus sequences (TGRRKM) (in subjects 13, 42, and 128).
To examine whether the sequences near the breakpoints can form cruciform structures, we used UNAFold (Markham and Zuker 2008) to produce potential configurations. The lowest-energy folding structures for each chromosome are presented in Supplemental Figure 2.
We postulated that the breakage was owing to random damage (e.g., oxidative damage or ionizing radiation) with no particular predisposition caused by the DNA sequence, in which case there would not be a nearby DNA sequence motif. To test this hypothesis, we compared the densities of motifs around the breakpoints to their distribution across chromosomes. The complete chromosome 1 and 22 sequences were split into 1-kb and 5-kb windows, and motifs in each were identified using fuzznuc. We calculated the density of each motif in each window to determine the distributions of densities for both chromosomes. We then found the density of each motif within 1 kb and 5 kb of each breakpoint, and compared these to the densities for whole chromosomes. In each case, the motif density around breakpoints was within two standard deviations of their mean density across chromosomes, indicating that they were not significantly enriched and suggesting that these breaks were random and not caused by any identifiable susceptibility. Next, we performed the motif analysis using a window size of 200 bp, which is comparable in size to the “normal” chromosome sequence regions that we display in Figure 1. We identified motifs in all 200-bp windows across chromosomes 1 and 22, for each type of motif displayed in Figure 1. The results are presented as histograms (Supplemental Fig. 1). Except for translin sites in chromosome 1 of subject 128, none of the breakpoint regions show high numbers of motifs.
Discussion
Complexity at the breakpoint junctions
We observed considerable complexity at the breakpoint junctions in t(1;22) balanced translocations. Each case presented with different genomic alterations and multiple sequence changes. Although we found some similar events across the t(1;22) cases, the complexity at the breakpoint junctions exceeded our expectations given that these subjects are phenotypically normal. Recent reports have suggested the translocation breakpoints in phenotypically normal individuals are relatively simple and not associated with genomic imbalances (Baptista et al. 2005, 2008). However, these studies have examined the translocation breakpoints at the resolution of fosmid clones (∼40 kb) or PCR products (∼10 kb). Only through sequencing the breakpoint junctions can this complexity be uncovered. Previously, in t(1;9) cases, we found similar sequence alterations, including duplications (5 bp) and additions (5 bp) of new nucleotides at the breakpoint junctions (Gajecka et al. 2006a). Considering t(1;22) and t(1;9) together, only one case out of seven [subject 70; with t(1;9)] presented with two “simple” breakpoint junctions, without any sequence imbalances. Although no other data are available, the frequency of complex junctions observed in our subjects (6/7 cases; 85.7%) is surprisingly high. Part of the complexity may be owing to the number of combinations of nicks on DNA strands that result in DSBs. Once the DSB arises, additional complexity could arise due to the variation introduced by the NHEJ process during translocation formation.
Sequence motifs identified at the junctions
Different sequence motifs have been associated with genomic rearrangements and have been identified as factors influencing DNA breakage by non-B-DNA conformations (Chuzhanova et al. 2003; Bacolla and Wells 2004; Bacolla et al. 2004; Raghavan and Lieber 2006). Target motifs of various endonucleolytic enzymes and recombination proteins have also been detected near many chromosomal translocation breakpoints (Abeysinghe et al. 2003). In our t(1;22) subjects, we identified vertebrate topoisomerase consensus cleavage sites, DNA polymerase a/b frameshift hotspots, and a deletion hotspot consensus sequence (TGRRKM). As a comparison, in the t(1;9) subjects previously studied (Gajecka et al. 2006b), we identified a heptamer recombination signal and DNA polymerase a/b frameshift hotspots TGGGGT, which also overlap with immunoglobulin heavy chain class switch repeats TGGGG and GGGGT. Oligopurine/oligopyrimidine tracts were identified in all subjects with t(1;22) or t(1;9). Direct repeats and inverted repeats were observed at the junctions of t(1;22) translocations similar to t(1;9) where breakpoints were also located within short sequence repeats including palindromes, mirror, or direct repeats.
Previously, we reported the presence of the translin consensus GCCCWSW at the t(1;9) breakpoint junctions. With one mismatch allowed, this motif was found in four out of six breakpoints (Gajecka et al. 2006b). In the present study, translin sites GCCCWSSW and ATGCAG were also identified in subjects 13, 127, and 128. Translin binding sites have been found at translocation breakpoints in cancer (Aoki et al. 1995; Chalk et al. 1997; Kanoe et al. 1999). The translin protein is thought to recognize single-stranded DNA ends of staggered breaks that may occur at recombination hotspots (Kasai et al. 1997).
Because the presence of these motifs is associated with genomic rearrangements, we investigated whether the density of the motifs is higher or random at the breakpoint junctions. None of the motifs were enriched in these regions, relative to their average density across the chromosomes, for either 1-kb or 5-kb windows. The analysis using a window size of 200 bp showed similar results. Because the motifs provide a potentially important substrate for specific translocations, it is possible that the motifs may be mechanistically important.
To investigate potentially relevant secondary structures as a cause of DSB, we performed UNAfold analysis for all normal chromosome sequences displayed in Figure 1. In case 128, the imperfect tandem repeats encompass the breakpoint. Also, the tetraplex-forming sequence and inverted repeats were noted in subject 13 and subject 42, respectively. Some of the features could lead to the rearrangement through secondary structure involvement. The t(11;22), the most common constitutional translocation in humans, is thought to be mediated by cruciform formation (Kurahashi and Emanuel 2001). However, those inverted repeats are hundreds of bases long and the translocations occur consistently in the same places (Kurahashi et al. 2006). Shorter cruciforms require a substantial degree of negative supercoiling to form consistently, but on average the human genome is only slightly negatively supercoiled (Sinden et al. 1980). Nevertheless, the 8-bp (and nearby 15-bp) inverted repeats could be a cause of rare-event instability. This instability might occur at such a low level that it would be difficult to find more than one case.
If non-B-DNA structures were directing DNA double-strand breakage, breakpoints would occur more often at such structure-forming elements than at surrounding nonstructure-forming regions. However, breakpoints do not appear to occur consistently at any one structure. In the absence of large numbers of breakpoints consistently around a specific structural element, structure-directed double-strand breakage is indistinguishable from random breakage, e.g., breakage caused by ionizing radiation or reactive oxygen species.
Therefore, based on the presented evidence we postulate that random (sequence-independent) mechanisms for breakage may be most likely such as oxidation damage, ionizing radiation, or replication across a site of DNA damage (Friedberg et al. 2006; Lieber et al. 2006), although we cannot exclude the possibility that sequence features and motifs influenced the rearrangement formation.
Nonhomologous end-joining involvement in translocation formation and evidence of multistep DNA repair process
Previously we proposed that nonhomologous end-joining (NHEJ) resulted in the t(1;9) formation because insertions, duplications, and limited homology at the breakpoints were identified (Gajecka et al. 2006b). Similarly, no long sequence homology (>30 bp) was found at the breakpoints in the t(1;22) translocations. Therefore, the present study further supports NHEJ involvement in balanced translocation formation.
In NHEJ, genomic alterations including short deletions, duplications, or insertions at the breakpoint junctions have been reported (Jeggo 1998; Liang et al. 1998; Kanoe et al. 1999; Shaw and Lupski 2004; Yu and Gabriel 2004; Rooms et al. 2007) and confirmed in our studies (Gajecka et al. 2006b; this study). We suspect that broken chromosome ends were incompatible to simply join in all cases except one (subject 70; Gajecka et al. 2006b). NHEJ is synonymous with the Ku- and DNA ligase IV–dependent rejoining mechanisms (Daley and Wilson 2005). Broken DNA strands in derivative chromosome formation must be aligned and, after nucleolytic processing of broken DNA ends, small gaps need to be filled prior to DNA joining. Evidence exists that DNA ligase IV/XRCC4 will ligate one strand when there are gaps and flaps on the opposite strand (Ma et al. 2004). This implies that two strands at the junction can be processed and joined independently (Ma et al. 2004; Hefferin and Tomkinson 2005). Furthermore, Ma et al. (2004) showed that the order of the enzymatic steps in mammalian NHEJ, including nucleolytic, polymerization, and ligation steps can vary not only from one junction to another, but also from one strand to the other in the same junction.
In the case of balanced translocations, we investigated the rearrangements as if they were created as two independent DNA breaks with a total of four broken strands at the breakpoint junctions. We speculate that the complexity at the breakpoint junctions identified in our studies is consistent with the finding that the two strands at the junction were processed and joined independently with the observed additions of nucleotides, deletions, and duplications being the consequence of several events in a multistep DNA repair process.
It has been reported that compatible and blunt DNA ends can be joined using only XRCC4:DNA ligase IV (Grawunder et al. 1997; Wilson et al. 1997). Thus, we speculate that the broken chromosome ends in case 70 (Gajecka et al. 2006b) were compatible and multistep processing prior to ligation was not necessary. Consequently, no sequence alteration (despite a translocation event), or complexity, was observed.
Multiple mechanisms could mediate DNA repair in balanced translocations
Previous junction analysis performed in individuals with translocations allowed us to propose a mechanism of translocation formation similar to the mechanism of transposition in bacteria (Gajecka et al. 2006b). In accordance with this mechanism, DSBs are staggered nicks, and each breakpoint is separated by nucleotide bases that will be duplicated in the rearrangement formation/stabilization. The broken strands ligate, forming a four-ended intermediate, after which the unpaired sequences produced by the staggered nicks are filled in, resulting in the duplicated segments found at both derivative junctions (Gajecka et al. 2006b).
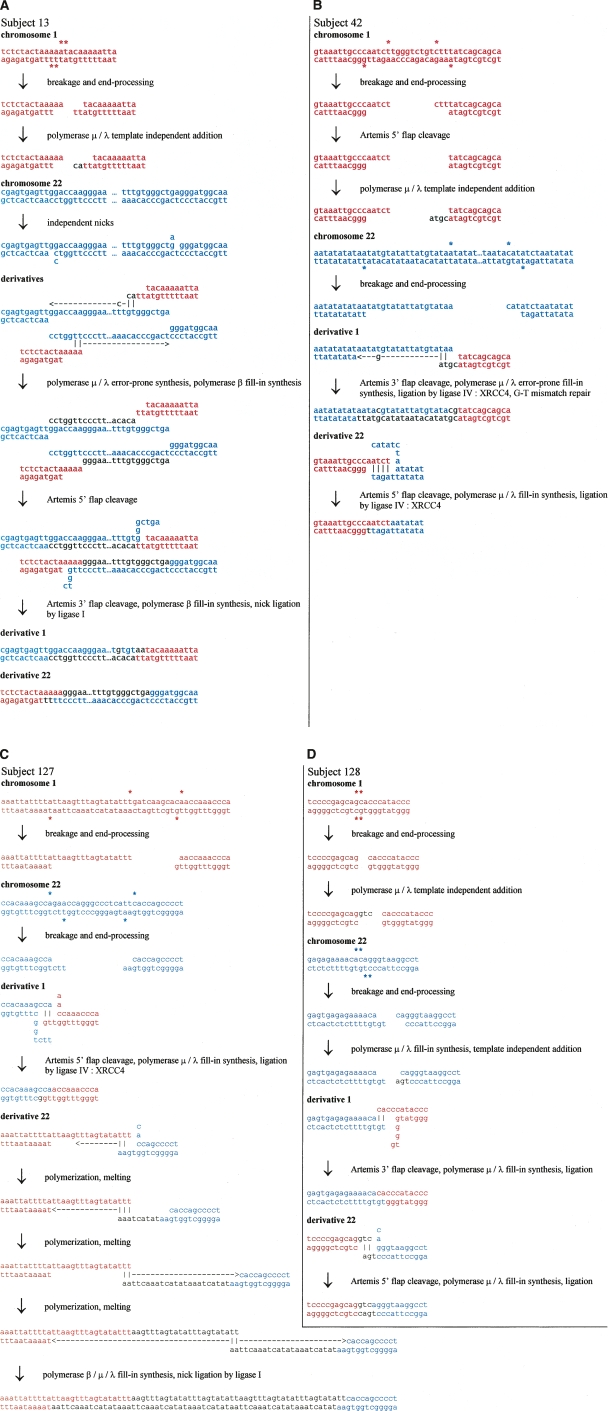
Although we identified short sequence gains in the t(1;22) cases, more complex mechanisms than just repair of staggered nicks are likely involved. In case 13, a 337-bp sequence fragment of chromosome 22 is duplicated and present at both derivative chromosomes, whereas in subject 127, an interstitial duplication with short triplication of fragments originating from chromosome 1 sequence is present only on the derivative chromosome 22. In these cases, multistep, more complex mechanisms are likely to be involved (shown in Fig. 2).
Figure 2.
Possible NHEJ scenarios for four balanced translocations t(1;22)(p36;q13). Sequences presented in red and blue correspond to chromosome 1 and 22 sequence, respectively. (A) Scenario for subject 13. A DSB on chromosome 1 results in a pair of 3′ overhangs—ends with unpaired bases on the 3′ strand. These overhangs invade into separate nicks on chromosome 22 that have opened a little. Polymerase μ or λ polymerizes a few bases on one of the ends, template-independently, and this finds some homology in one of the opened nicks. The polymerase then adds template-dependently, accidentally creating a T–C mismatch in the process. Polymerase β (POLB) fills in the gap. The other 3′ overhang has some microhomology with the other nick on chromosome 22, and polymerase μ or λ polymerizes a few bases template-dependently before ceding to polymerase β. The polymerases continue until they reach the original position of the other nick, and the original chromosome 22 DNA strands are separated along the way. The remaining flaps from the microhomology search are cleaved by ARTEMIS and the nicks sealed by ligase I. The result is a long duplication of chromosome 22 sequence. (B) Scenario for subject 42. A DSB on chromosome 22 results in a 3′ overhang end—an end with unpaired bases on the 3′ strand—and a 5′ overhang end—an end with unpaired bases on the 5′ strand. Chromosome 1 also has a DSB which results in a 3′ overhang end and a 5′ overhang end. The 5′ overhang end is resected by ARTEMIS and extended by polymerase μ or λ template-independently, creating a 3′ overhang end. This end has some microhomology with the 3′ overhang end of chromosome 22, and the microhomology is extended by polymerase μ or λ, which accidently polymerizes a G–T mismatch during the process. The mismatch is aberrantly repaired, resulting in a one-base mutation. The remaining 3′ overhang end of chromosome 1 has some microhomology in the 5′ overhang end of chromosome 22 after some opening. Any remaining flaps are cleaved by ARTEMIS, single-stranded DNA gaps are filled by polymerase μ or λ, and nicks are sealed by ligase IV:XRCC4. (C) Scenario for subject 127. A DSB on chromosome 1 generates two 3′ overhang ends—ends with unpaired bases on the 3′ strand—while a DSB on chromosome 22 results in a 3′ overhang end and a 5′ overhang end. One of the chromosome 1 ends has some microhomology with the 3′ overhang end from chromosome 22 after it has opened a little. The 5′ flaps are resected by ARTEMIS and the single-stranded DNA gaps filled by polymerase μ or λ and sealed by ligase IV:XRCC4. The remaining chromosome 1 3′ overhang end has some microhomology on the remaining chromosome 22 end, which is extended by polymerase μ or λ. The ends undergo several rounds of melting and microhomology-mediated polymerization. Any remaining single-stranded DNA gaps are filled by polymerases μ or λ and β, and the nicks sealed by ligase IV:XRCC4 or ligase I. (D) Scenario for subject 128. A DSB on chromosome 1 results in two blunt ends, one of which is extended template-independently by polymerase μ or λ to result in a 3′ overhang end. Another DSB on chromosome 22 results in two 5′ overhang ends. One of these ends is filled template-dependently by polymerase μ or λ, and then an extra base added template-independently, resulting in a 3′ overhang end. The 5′ overhang end of chromosome 22 has some microhomology with the 5′ overhang of chromosome 1 after both have opened a little. The 5′ flap is cleaved by ARTEMIS, the single-stranded DNA gaps filled by polymerase μ or λ, and the nicks sealed by ligase IV:XRCC4. The remaining chromosome 1 3′ overhang end has some microhomology with the remaining chromosome 22 5′ overhang end after it has opened, and the flap cleavage, gap-filling, and sealing proceed as before.
In contrast to the breakpoint junctions in t(1;9) (Gajecka et al. 2006b), short deletions were identified on both derivatives 1 and 22 in cases 42 and 127 in each case. However, deletions were not observed in cases 13 and 128. Based on this evidence, we conclude that the complexity found at the translocation breakpoints indicates involvement of multiple mechanisms in the DNA breakage/repair process. At one extreme, we observed perfect ligation with simple junctions in case 70 (Gajecka et al. 2006b), although this was the rarest event. At the other extreme, we found complexities such as those seen in case 127 with two different small deletions, an interstitial duplication with short triplication fragments and addition of nucleotides. Thus, complexity in the breakpoints/junctions varies from case to case.
Possible detailed NHEJ scenarios for these cases
Based on the biochemical properties of the nuclease, polymerases, and ligase involved in human NHEJ, we propose a reasonable series of steps that may account for the translocation junctions seen in the four cases here. The mechanisms involve fill-in synthesis (template-dependent addition) by polymerase μ (POLM) and polymerase λ (POLL), as well as some instances of template-independent addition by polymerase μ. Polymerase μ is capable of template-independent addition under physiologic conditions during NHEJ (Gu et al. 2007). The mechanisms also require removal of unpaired overhangs or flaps of DNA by the NHEJ nuclease ARTEMIS:DNA-PKcs (ARTEMIS also known as DCLRE1C). ARTEMIS:DNA-PKcs has been shown to be capable of removing flaps at gapped structures or removing overhangs at DNA ends (Ma et al. 2002, 2005). The ligase of NHEJ, XRCC4:DNA ligase IV, can ligate one strand independent of the other (Ma et al. 2004). Possible detailed NHEJ scenarios for our cases are presented in Figure 2. Because case 13 has a >300-bp duplication at both derivative chromosomal junctions, we propose one of the simpler mechanisms to achieve duplication after staggered nicks at the edges of the region of duplication (Fig. 2).
Summary
We have identified sequence alterations at the breakpoint junctions in phenotypically normal individuals with apparently balanced translocations. The frequency of complex junctions observed in our studies was surprisingly high. We speculate that the sequence complexity found at balanced translocation breakpoints has been underestimated. The complexity included short deletions, duplications, triplication, and insertions. No substantial homology was found in the junctions. Sequence motifs were found frequently at the breakpoints; however, except for translin sites in one subject, none of the breakpoint regions show high numbers of motifs. These features correspond with the multistep repair process in NHEJ mechanisms present.
Methods
Generation of somatic cell hybrids
To identify the 1p36 breakpoints at the sequence level, somatic cell hybrids from the unbalanced subject 13 and 42 were generated as previously described (Page and Shaffer 1997). Somatic cell hybrids containing either the derivative chromosome 1 or the normal chromosome 1 were identified for those subjects. Additionally, hybrids were constructed from transformed lymphoblast cells from the father of subject 127 and mother of subject 128. Hybrids containing the derivative chromosome 1 and derivative chromosome 22 were identified after screening with microsatellite markers and FISH as previously described (Gajecka et al. 2006a).
STS marker walking
To identify the 1p36 breakpoints in each subject, STS markers were designed and PCR analyses were performed using hybrid DNA containing the derivative chromosome 1 from each subject, as described previously (Gajecka et al. 2006a). DNA sequences were obtained for analysis from GenBank (www.ncbi.nlm.nih.gov), corresponding to the genomic region containing the 1p36 breakpoint of each subject. Repetitive elements were masked from the sequence using RepeatMasker (http://www.repeatmasker.org/), and 200–400 bp STS markers were designed from the unmasked unique sequence using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Breakpoints were narrowed using markers spaced ∼20 kb apart along the length of the chromosome followed by ∼5 kb apart and finally ∼1 kb apart. To prepare for the subsequent TOPO Walker protocol, the breakpoint for each subject was narrowed to 200–300 bp. PCR was performed as described previously (Gajecka et al. 2006a).
TOPO Walker protocol and junction identification in subjects 13, 42, and 127
The TOPO Walker Kit (Invitrogen, Version E, for isolating and sequencing unknown BAC DNA) was used to amplify junction fragments at derivative chromosomes 1 in subjects 13, 42, and 127, as described previously (Gajecka et al. 2006a). We used restriction enzymes that are predicted to leave a 3′ overhang based on the sequence near each breakpoint: SphI (subject 13), PstI (subject 42), SacI (subject 127).
Using complementary primers from the regions of interest, derivative junctions could be analyzed after amplifying across the junctions. For each case, derivative chromosome 1 junctions were confirmed using PCR with forward primers derived from chromosome 22 sequence, and reverse primers derived from chromosome 1 sequence, at the derivative 1 breakpoint location. Similarly, derivative 22 junctions were confirmed by PCR using forward primers derived from chromosome 1 sequence, and reverse primers derived from chromosome 22 sequence, in the breakpoint region.
Amplification across the junctions in subject 128
In subject 128, the breakpoint region at derivative 1 was narrowed by STS marker walking to a sequence fragment consisting of >2 kb of repetitive elements. Breakpoint locations must be determined to within 200–300 bp for the TOPO Walker protocol. Therefore, the breakpoint for the derivative 1 could not be attempted. The breakpoint for the derivative 22 was narrowed to 200–300 bp, and amplification across the junction was performed. To accomplish this we used a unique-sequence reverse primer located 200–300 bp from the putative 22q13 breakpoint and a forward primer from the putative 1p36 breakpoint. The sequence junction at derivative 22 was then amplified. Similarly, we used a unique-sequence forward primer located 200–300 bp from the putative 22q13 breakpoint and reverse primer from the putative 1p36 breakpoint to identify the sequence junction at derivative 1 chromosome.
Breakpoint junction sequencing
Gel-purified PCR products from the TOPO Walker protocol or PCR products after amplification across the junctions were sequenced using an ABI PRISM 3100-Avant genetic analyzer and Big Dye reagents (Applied Biosystems). Sequencher 4.6 (Gene Codes) was used to analyze sequence at least 200 bp on either side of the breakpoint junctions to predict intron/exon boundaries and potential transcripts.
Junction DNA analysis
Original chromosomal sequences from which derivatives originate were identified using BLAT (Kent 2002) from the UCSC Genome Browser Web site (http://genome.ucsc.edu). Sequences were downloaded, including 200 bp adjacent flanking sequence. For subject 128, derivatives could not be assigned to the chromosome 1 consensus assembly. In this case, we searched Celera human genome contigs using NCBI BLASTN (http://ncbi.nlm.nih.gov/blast). The matching unassigned contig (NW927826.1) was downloaded for further analysis.
Interspersed repeats were identified from the RepeatMasker (http://www.repeatmasker.org) track of the UCSC Genome Browser. Alignments between chromosomal sequences and derivatives were generated using the LINSI module of MAFFT (Katoh et al. 2005), and edited manually with Jalview (Clamp et al. 2004). Putative regions of Z-DNA structure were searched for using Z-Hunt (Clamp et al. 2004; http://gac-web.cgrb.oregonstate.edu/zDNA/). We searched for sequence motifs which could be involved in translocation formation process using the “fuzznuc” program from EMBOSS (Rice et al. 2000). Potentially relevant motifs were considered (vertebrate topoisomerase consensus cleavage sites [RNYNNCNNGYNGKTNYNY], translin sites [GC CCWSSW and ATGCAG], DNA polymerase a/b frameshift hotspots [ACCCWR, TGGNGT, GGGGGA, TCCCCC, and CTG GCG], and deletion hotspot consensus sequences [TGRRKM]). We searched both strands using the “-complementary” option of fuzznuc. For topoisomerase consensus and the longer translin motif, one mismatch was permitted; for DNA polymerase and deletion hotspots no mismatches were allowed. Inverted repeats in sequences were found using the “palindrome” program of EMBOSS. Direct repeats were identified with Tandem Repeats Finder (Benson 1999) with default parameters. Oligopurine (R) and oligopyrimidine (Y) tracts were identified by translating the nucleotide sequences into the R/Y alphabet, and locating the longest tracts occurring in the breakpoint regions. Tracts of R or Y were concatenated if only one or two mismatches would be introduced.
Mfold analysis
We used UNAFold (Markham and Zuker 2008) to predict foldings of normal chromosome sequences given in Figure 1. The lowest-energy foldings obtained with the “DNA” option are displayed in Supplemental Figure 2.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.077453.108.
References
- Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Aoki K., Suzuki K., Sugano T., Tasaka T., Nakahara K., Kuge O., Omori A., Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J., Prigmore E., Gribble S.M., Jacobs P.A., Carter N.P., Crolla J.A. Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur. J. Hum. Genet. 2005;13:1205–1212. doi: 10.1038/sj.ejhg.5201488. [DOI] [PubMed] [Google Scholar]
- Baptista J., Mercer C., Prigmore E., Gribble S.M., Carter N.P., Maloney V., Thomas N.S., Jacobs P.A., Crolla J.A. Breakpoint mapping and array CGH in translocations: Comparison of a phenotypically normal and an abnormal cohort. Am. J. Hum. Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalk J.G., Barr F.G., Mitchell C.D. Translin recognition site sequences flank chromosome translocation breakpoints in alveolar rhabdomyosarcoma cell lines. Oncogene. 1997;15:1199–1205. doi: 10.1038/sj.onc.1201285. [DOI] [PubMed] [Google Scholar]
- Chuzhanova N., Abeysinghe S.S., Krawczak M., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer II: Potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 2003;22:245–251. doi: 10.1002/humu.10253. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Daley J.M., Wilson T.E. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA repair and mutagenesis. ASM Press; Washington, DC: 2006. [Google Scholar]
- Gajecka M., Glotzbach C.D., Jarmuz M., Ballif B.C., Shaffer L.G. Identification of cryptic imbalance in phenotypically normal and abnormal translocation carriers. Eur. J. Hum. Genet. 2006a;14:1255–1262. doi: 10.1038/sj.ejhg.5201710. [DOI] [PubMed] [Google Scholar]
- Gajecka M., Pavlicek A., Glotzbach C.D., Ballif B.C., Jarmuz M., Jurka J., Shaffer L.G. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum. Genet. 2006b;120:519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- Gajecka M., Saadeh R., Mackay K.L., Glotzbach C.D., Spodar K., Chitayat D., Shaffer L.G. Clinical and molecular cytogenetic characterization of four patients with unbalanced translocation der(1)t(1;22)(p36;q13) Am. J. Med. Genet. A. 2008 doi: 10.1002/ajmg.a.32427. in press. [DOI] [PubMed] [Google Scholar]
- Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L., Nimmakayalu M.A., Jalali G.R., Hacker A.M., Vorstman J., Conforto Duffy D., Medne L., Emanuel B.S. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T.E., Mann M., Lieber M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Gu J., Lu H., Tippin B., Shimazaki N., Goodman M.F., Lieber M.R. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefferin M.L., Tomkinson A.E. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. DNA breakage and repair. Adv. Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- Kanoe H., Nakayama T., Hosaka T., Murakami H., Yamamoto H., Nakashima Y., Tsuboyama T., Nakamura T., Ron D., Sasaki M.S., et al. Characteristics of genomic breakpoints in TLS-CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene. 1999;18:721–729. doi: 10.1038/sj.onc.1202364. [DOI] [PubMed] [Google Scholar]
- Kasai M., Matsuzaki T., Katayanagi K., Omori A., Maziarz R.T., Strominger J.L., Aoki K., Suzuki K. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J. Biol. Chem. 1997;272:11402–11407. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Ohye T., Kogo H., Kato T., Emanuel B.S. Palindrome-mediated chromosomal translocations in humans. DNA Repair. 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Hosoba E., Kato T., Ohye T., Kogo H., Emanuel B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Han M., Romanienko P.J., Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R., Yu K., Raghavan S.C. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.L., Schwarz K., Lieber M.R. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Ma Y., Lu H., Schwarz K., Lieber M.R. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: The iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- Markham N.R., Zuker M. UNAFold: Software for nucleic acid folding and hybriziation. In: Keith J.M., editor. Bioinformatics: Structure, functions and applications. Vol. 2. Humana Press; Totowa, NJ: 2008. pp. 1–31. [Google Scholar]
- Nimmakayalu M.A., Gotter A.L., Shaikh T.H., Emanuel B.S. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- Page S.L., Shaffer L.G. Nonhomologous Robertsonian translocations form predominantly during female meiosis. Nat. Genet. 1997;15:231–232. doi: 10.1038/ng0397-231. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Lieber M.R. DNA structures at chromosomal translocation sites. BioEssays. 2006;28:480–494. doi: 10.1002/bies.20353. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rooms L., Reyniers E., Kooy R.F. Diverse chromosome breakage mechanisms underlie subtelomeric rearrangements, a common cause of mental retardation. Hum. Mutat. 2007;28:177–182. doi: 10.1002/humu.20421. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Sinden R.R., Carlson J.O., Pettijohn D.E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: Analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Wilson T.E., Grawunder U., Lieber M.R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- Yu X., Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1534/genetics.166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]