Abstract
Identification of lineage-specific innovations in genomic control elements is critical for understanding transcriptional regulatory networks and phenotypic heterogeneity. We analyzed, from an evolutionary perspective, the binding regions of seven mammalian transcription factors (ESR1, TP53, MYC, RELA, POU5F1, SOX2, and CTCF) identified on a genome-wide scale by different chromatin immunoprecipitation approaches and found that only a minority of sites appear to be conserved at the sequence level. Instead, we uncovered a pervasive association with genomic repeats by showing that a large fraction of the bona fide binding sites for five of the seven transcription factors (ESR1, TP53, POU5F1, SOX2, and CTCF) are embedded in distinctive families of transposable elements. Using the age of the repeats, we established that these repeat-associated binding sites (RABS) have been associated with significant regulatory expansions throughout the mammalian phylogeny. We validated the functional significance of these RABS by showing that they are over-represented in proximity of regulated genes and that the binding motifs within these repeats have undergone evolutionary selection. Our results demonstrate that transcriptional regulatory networks are highly dynamic in eukaryotic genomes and that transposable elements play an important role in expanding the repertoire of binding sites.
Although cross-species conservation has been successfully used to identify functional regulatory sequences in genomes (Thomas et al. 2003; Boffelli et al. 2004; Wang et al. 2006), there is growing evidence that changes in cis-regulatory elements are important in determining key phenotypic differences, as shown in yeast (Ihmels et al. 2005), pufferfish (Tumpel et al. 2006), Drosophila (Gompel et al. 2005; Marcellini and Simpson 2006), and human (Rockman et al. 2005). Moreover, a number of studies have shown that evolutionary turnover of regulatory elements is a common feature of eukaryotic genomes with examples in yeast (Tanay et al. 2005; Borneman et al. 2007; Tuch et al. 2008), Drosophila (Moses et al. 2006), zebrafish (McGaughey et al. 2008), and mammals (Dermitzakis and Clark 2002; Birney et al. 2007; Chabot et al. 2007; Odom et al. 2007; Jegga et al. 2008).
To gain additional insight into eukaryotic transcriptional regulation and to further quantify the significance of species-specific regulation, we analyzed, from an evolutionary perspective, seven whole-genome occupancy data sets obtained in vivo by chromatin immunoprecipitation (ChIP) (see Table 1). The studied transcription factors (TFs) are: ESR1 (also known as ER), TP53 (also known as p53), MYC (also known as c-MYC), and RELA (also known as the p65 subunit of NFkB) in human and POU5F1-SOX2 (also known as OCT4-SOX2) and CTCF in mouse. These TFs were selected because they play critical roles in a wide spectrum of biological systems. For instance, ESR1 and TP53 are determining factors in cancer while POU5F1 and SOX2 are both required to maintain pluripotency of embryonic stem (ES) cells. CTCF is a methylation-sensitive protein that is important for gene imprinting (Hark et al. 2000) and X chromosome inactivation (Lee 2003). It is also known to act as a chromatin insulator (Bell et al. 1999). Our results extend previous work and show that for these seven TFs, the majority of the genome-wide binding regions do not display signs of sequence conservation even between closely related mammals.
Table 1.
Whole-genome chromatin immunoprecipitation data sets (see Methods)
aThe use of ESR1 by itself will refer to this particular data set.
In looking for a mechanistic explanation for this limited cross-species conservation, we studied the frequency of repeats within the binding regions since repeats are known to account for a large fraction of the sequence differences between human and mouse (Waterston et al. 2002). Although repeats have been hypothesized to play an important role in transcriptional regulation (Davidson and Britten 1979; McClintock 1984; Kidwell and Lisch 1997; Brosius 2003), have been reported to harbor transcription factor binding motifs (Polak and Domany 2006), and have been shown to be bound in a number of cases (Bejerano et al. 2006; Johnson et al. 2006; Laperriere et al. 2007; Wang et al. 2007), the extent of their impact on the evolution of regulation remains elusive. Interestingly, in the current study, we report that hundreds, and in some cases thousands, of binding sites for five of the seven TFs are embedded in distinctive repeat families. Our results quantitatively demonstrate that transposable elements have mediated substantial regulatory expansion throughout the mammalian phylogeny.
Results
Limited evolutionary conservation of transcription factor binding regions
Assessing the conservation of TF binding regions can be challenging, especially with the added complexity that conserved cis-regulatory elements can be locally shuffled which prevents their detection by alignment-based algorithms (Pollard et al. 2006; Sanges et al. 2006). We make a distinction between a binding region, a region that is observed to be bound (typically a few hundred base pairs depending on the detection technique), a binding site, the actual point of contact between the TF and the DNA, and a binding motif, the recognition sequence that mediates this interaction. To evaluate the conservation of the regions detected to be bound, we relied on two metrics: (1) overlap with a phastCons conserved element (Siepel et al. 2005) and (2) presence of conserved binding motifs across multiple species (see Methods). The first metric looks for the presence of a good quality whole-genome alignment of vertebrates within the TF binding region as an indirect measure of the conservation of the binding site. The second metric is more targeted and looks for the presence of binding motifs in the region observed to be bound and in its homologous counterparts in other genomes. Although limited by the quality of the binding motifs used and their applicability across species, this second metric allows for more flexibility within the local alignments in the conservation assessment.
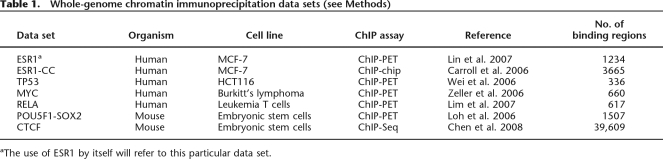
Consistent with previous reports (Borneman et al. 2007; Odom et al. 2007), we found that for these seven TFs, the underlying sequence conservation was limited with evolutionary similarity detected in only 10%–40% of the binding regions (Fig. 1A; Supplemental Table 1). Nonetheless, we also observed that for some TFs (e.g., ESR1 and CTCF), this evolutionary pressure was not limited to sites in proximity to genes (Fig. 1B). Overall and as expected, the assessment that relied on binding motifs rather than sequence alignment was more specific with background levels below 5% in all cases. For TP53, we also note that the targeted approach relying on conserved motifs was able to detect significantly more conserved sites than the untargeted approach relying purely on good quality alignments.
Figure 1.
Limited evolutionary conservation of transcription factor binding regions. (A) Gray bars show the percentage of binding regions that are conserved based on either an overlap with a phastCons conserved element (left panel) or the presence of a conserved binding motif (right panel). ESR1 is the ESR1 ChIP-paired-end diTag (ChIP-PET) data set (Lin et al. 2007) while ESR1-CC is the ESR1 ChIP-chip data set (Carroll et al. 2006). Conservation levels expected by chance are shown in white and are computed from simulated binding data sets (see Methods). (B) Gray bars show the percentage of binding regions for ESR1, MYC, and CTCF that have a conserved binding motif where the regions are further partitioned into four categories: adjacent (within 250 bp of the coding region of a gene), proximal (within 5 kbp of a coding region), distant (intragenic or within 100 kbp of a gene), or desert (>100 kbp from any gene). Conservation levels expected by chance are shown in white. Error bars, 1 SD.
Pervasive association between transcription factor binding regions and repeats
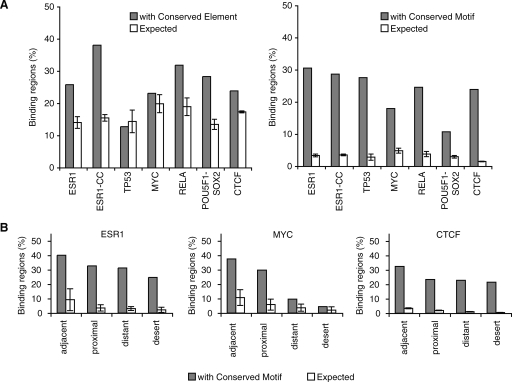
In looking for a mechanistic explanation for this limited cross-species conservation, we tabulated the frequency of the different repeat families within the binding regions for the various TFs. We found that specific families were strongly overrepresented in the experimentally bound regions as compared to randomly selected fragments (Fig. 2A; see Methods). Specifically, ESR1 binding regions overlapped an inordinate number of MIR repeats (19.8% versus the expected 13.3%, P = 1.5 × 10−10), TP53 regions showed an association to ERV1 repeats (39.6% versus the expected 6.1%, P = 1 × 10−70), POU5F1-SOX2 regions showed a predominance of ERVK repeats (23.8% versus the expected 8.7%, P = 1 × 10−68) and B2 repetitive sequences were overrepresented in CTCF binding regions (33.8% versus the expected 12.4%, P < 1 × 10−100). These associations were not due to biases in library preparations since the nonenriched and random immunoprecipitated fragments showed no such preference (Fig. 2A). Two examples of binding regions overlapping repetitive sequences are shown in Figure 2B. We had initially observed that all TFs under study appeared to bind a common set of pericentromeric satellite DNA sequences but since these regions are known to be incompletely assembled, they were removed from downstream analyses to avoid likely mapping artifacts (Supplemental text). The association between ESR1 and MIR repeats would have been challenging to detect in the ChIP-chip data set because of the difficulty in designing probes covering repetitive sequences (Supplemental text).
Figure 2.
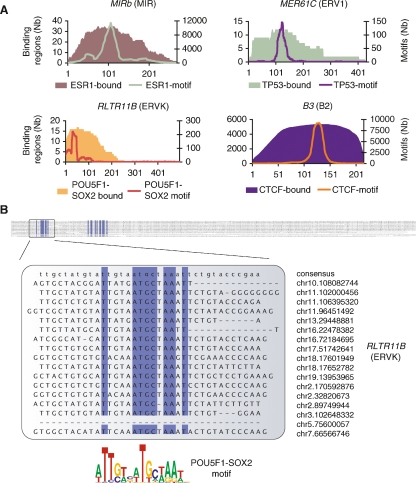
Pervasive association between transcription factor binding regions and transposable elements. (A) Enrichment of specific repeat families in the binding regions of distinct transcription factors. Heatmap shows the percentage of instances of a specific family of repeats that is in excess (yellow) or in deficit (purple) as compared to expected levels. Values were computed for the seven binding data sets but also for background data sets (labeled with “-B”) consisting of only singleton PETs (for ChIP-PET), random selected affymetrix probes (for ChIP-chip), or singleton tags (for ChIP-sequencing [ChIP-Seq]). The specific repeats from the four repeat families showing enrichment are highlighted on the right. These four repeat families are: MIR (mammalian interspersed repeat, a SINE repeat), ERVK (mouse endogenous retrovirus K, an LTR repeat), ERV1 (human endogenous retrovirus 1, an LTR repeat), and B2 (a rodent-specific SINE repeat). (B) Two examples showing ChIP sequencing clusters detecting binding regions in repeat-rich genomic sequences. In the first example, the binding region is identified with three fragments from the POU5F1 ChIP-PET library and four fragments from the SOX2 ChIP-PET library. In the second example, only the tag density is shown for the CTCF ChIP-Seq library.
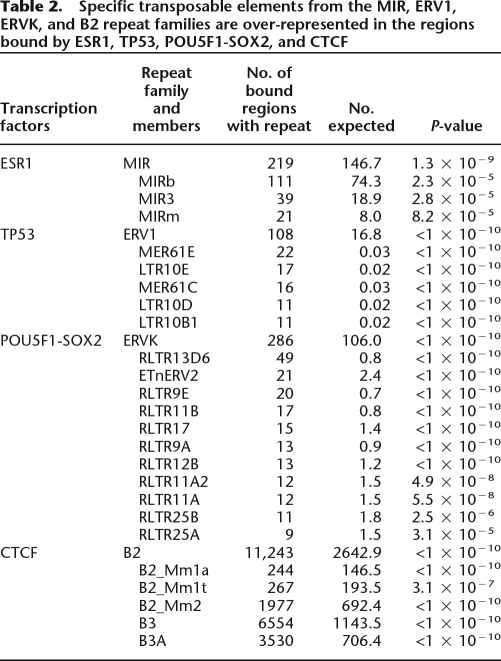
The enrichment for ESR1, TP53, POU5F1-SOX2, and CTCF binding in repeats was even more pronounced within the subtypes of the major repeat families. In the case of CTCF, for instance, there were 10,084 binding regions overlapping B3 and B3A repeats (two subclasses of the B2 family) corresponding to 8234 more than expected by chance (Table 2). To this point, all binding regions that have been described were ascertained by digital counts of sequenced fragments. Previous work showed a high validation rate (∼95%) for precise sites within regions identified by these genomic approaches. We further validated the TF binding on the repetitive sequence fragments described here using quantitative PCR and confirmed that between 86%–100% of designated sites could be confirmed (Supplemental Fig. 1; see Methods). This verifies that a large fraction of the binding sites of some TFs are embedded in highly restricted classes of repetitive sequences. Collectively, we call these sites repeat-associated binding sites (RABS).
Table 2.
Specific transposable elements from the MIR, ERV1, ERVK, and B2 repeat families are over-represented in the regions bound by ESR1, TP53, POU5F1-SOX2, and CTCF
Transposable elements harbor progenitor sequences for ESR1, TP53, POU5F1-SOX2, and CTCF binding motifs
The specificity of TFs to be found within distinct repetitive sequence families suggests that there may be intrinsic sequence signatures embedded in these transposable elements that can predispose them to becoming TF binding sites. Given that a majority of the RABS had sequence binding motifs associated with ESR1, TP53, POU5F1-SOX2, and CTCF (Supplemental Table 2), we scanned all the MIR, ERV1, ERVK, and B2 repeats in the genome and looked for instances of the associated motifs. We observed that specific regions of the repeat consensus consistently harbor the TF binding motifs (Fig. 3A; Supplemental Fig. 2). Given that one of the strengths of the ChIP sequencing approaches is that the precise TF binding site is frequently found in the area of maximal overlap of the sequenced clones (Lin et al. 2007; Chen et al. 2008), we were interested in mapping the section of the repeat consensus actually observed to be bound by the TF. This analysis confirmed a surprising specificity of binding to the same regions of the repeat consensus (Fig. 3A; Supplemental Fig. 2). This validates that these TF-repeat associations are mediated by sequence binding motifs and are highly targeted. Figure 3B shows an alignment of the 17 bound instances of the RLTR11B repeat verifying the presence of the POU5F1-SOX2 motif.
Figure 3.
Transposable elements harbor progenitor sequences for ESR1, TP53, POU5F1-SOX2, and CTCF binding motifs. (A) The same regions of the repeats harbor sequence binding motifs and are observed to be bound by the transcription factor. Filled areas represent the number of instances, at a given position relative to the consensus sequence, observed to be bound by ESR1, TP53, POU5F1-SOX2, and CTCF, respectively. Similarly, the green, purple, red, and orange curves show the number of instances of the ESR1, TP53, POU5F1-SOX2, and CTCF motifs at a given position across all instances of that repeat in the genome. (B) Multiple sequence alignment of the 17 bound instances of the RLTR11B repeat. Columns with >90% identity are in blue and highlight two regions of high sequence similarity. The first region is where the POU5F1-SOX2 motif (Loh et al. 2006) is detectable. Genomic positions of the repeat instances are shown on the right.
To evaluate further whether MIR, ERV1, ERVK, and B2 repeats represent a good source of binding motifs, we assessed the similarity between their ancestral sequence (as estimated by the consensus sequence) and motifs for ESR1, TP53, POU5F1-SOX2, and CTCF, respectively. We then compared this similarity to the one between random promoter sequences and the same binding motifs. For all four families of repeats, we found that, in a majority of cases, a good binding motif was embedded in the ancestral repeat sequence, a property present in less than 10% of the random promoter sequences for ESR1 and POU5F1-SOX2 and less than 4% for TP53 and CTCF (Supplemental Table 3; see Methods). This confirms that the cognate ancestral repeats were poised to generate the appropriate binding motifs more readily than if random promoter fragments were used as an alternative starting template. Also consistent with the idea of sequence drift in transposable elements toward functional binding sites, we found that repeat instances empirically observed to be bound were more likely to harbor a strong TF binding motif than repeats of the same class that were not observed to be bound. Specifically, for TP53, POU5F1-SOX2 and CTCF, the difference in the motif scores between the bound repeats and the whole population of repeats was statistically significant (P < 0.001 for ERV1, P = 0.005 for ERVK, and P < 0.001 for B2; Supplemental Fig. 3; see Methods).
Transposable elements have facilitated species-specific binding sites
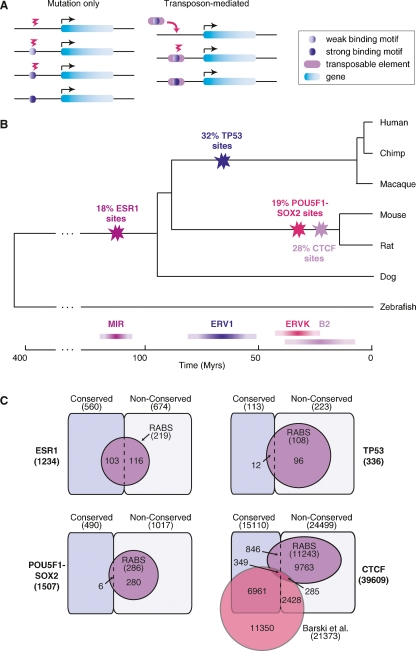
Novel TF binding sites can be acquired directly by point mutations or, when good binding motif precursors exist within transposable elements, they can be facilitated by the insertion of such elements (Fig. 4A). Having demonstrated that a significant fraction of the observed TF binding sites are directly embedded into special classes of repeats, we determined an upper bound for the emergence of these RABS by timing the expansion of the repeat families themselves. By measuring the average divergence from the consensus and consistent with previous reports (Waterston et al. 2002), we find that the expansion of MIR repeats took place ∼130 million years ago (Mya), for the ERV1s it occurred between 56 and 81 Mya, for the ERVKs it is between 32 and 53 Mya, and for the B2s it is between 13 and 57 Mya (Supplemental Table 4; see Methods). Overlaying the emergence of these repeats onto the species tree demonstrates that the RABS identified for ESR1 are likely to be ancestral to mammals, the ones for TP53 are primate-specific, the ones for POU5F1-SOX2 are rodent-specific, and the ones for CTCF are either rodent-specific or even mouse-specific (Fig. 4B).
Figure 4.
Evolution of the mammalian transcription factor binding repertoire via transposable elements. (A) Two evolutionary models for the gain of transcription factor binding sites: (1) via point mutations only or (2) by the insertion of a transposable element in which the seed of a binding motif is embedded. (B) Overlaying the age of the repeats on the species tree determines the age of the RABS. The time scale is in millions of years and divergence times are from Murphy et al. (2007). (C) RABS constitute a large fraction of the nonconserved binding regions of TP53, POU5F1-SOX2, and CTCF. Venn diagrams show the number of conserved and nonconserved binding regions that also correspond to RABS. The CTCF binding regions, which were detected in mouse embryonic stem cells, are also compared to a set of CTCF binding regions detected in human T cells (Barski et al. 2007).
When we initially examined the proportion of bona fide binding regions that did not appear to be evolutionarily conserved at the sequence level under either metric (Fig. 1; Supplemental Table 1), we surmised that a major contributing factor for evolutionary TF binding site diversity could be retrotranspositional dispersion (Fig. 4A). By calculating the proportion of nonconserved regions that are now identified as RABS, we find that 43% (96 out of 223), 28% (280 out of 1017), and 41% (10,048 out of 24,499) of the evolutionary variance in TP53, POU5F1-SOX2, and CTCF binding regions, respectively, can be explained by this mechanism (Fig. 4C). For ESR1, this effect is offset by the fact that the MIR repeat expansion predates the divergence time of the species used in the current study to assess conservation.
The availability of a genome-wide binding map of CTCF in human T cells (Barski et al. 2007) allowed further validation of the sequence-based conservation classification of the binding regions (Fig. 1; Supplemental Table 1) and, more importantly, that RABS are implicated in lineage-specific regulatory expansions. Indeed, we find that 48% (7310 out of 15,110) of the regions bound in mouse and predicted to be conserved have a homologous region that was also observed to be bound experimentally in human (Fig. 4C; see Methods). In contrast, only 6% (634 out of 11,243) of the mouse CTCF RABS were observed to be bound in human.
Binding motifs within repeats are under selection
We have established that repeats harbor better binding motif progenitor sequences compared to typical promoters and that many RABS are lineage-specific. This, however, still leaves unanswered whether there has been selection in the use of these repeats as a template for generating binding motifs. To answer this, we measured the enrichment of binding motifs observed among the repeat families as compared to the expected number should the repeat instances be mutated randomly. We performed a series of simulations generating artificial repeat instances using comparable rates of mutations and counted the number of binding motifs found among them (Supplemental Table 5; see Methods). Our results showed that four out of the five repeat subclasses for TP53 and nine out of 11 for POU5F1-SOX2 contained significantly more binding motifs than expected by chance (P < 0.001). The fact that mutations within these repeat families are not distributed randomly is also illustrated in the alignment of the instances of the RLTR11B repeat that were functionally bound by POU5F1-SOX2 (Fig. 3B). In this alignment, the region associated with the binding motif is highly preserved compared to other parts of the repeat.
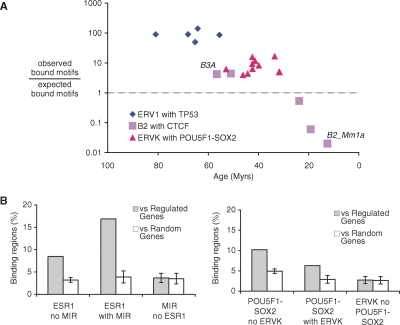
Next, we looked at the fraction of functionally bound motifs within the enriched repeat subfamilies and compared it to the fraction of bound motifs from randomly selected fragments from the genome. Interestingly, we found that the older the repeat, the higher the enrichment in the fraction of bound motifs within the subfamily (Fig. 5A). For CTCF, for instance, while 7.9% of randomly selected motifs were observed to be bound, 34.2% of the motifs within the B3A subfamily were observed to be bound. In contrast, in the younger B2_Mm1a subfamily, this percentage drops to 0.2% (Supplemental Table 6). Together, these results strongly suggested that the repeat instances were subjected to evolutionary selection toward good binding motifs and that, as expected, this selection is most visible in the older repeats which had more time to evolve to bona fide binding sites.
Figure 5.
Transposable elements are enriched for bound motifs and are associated with regulated genes. (A) Ratio between the fraction of motifs within a given repeat subfamily that is observed to be bound and the fraction of motifs that is expected to be bound. The x-axis represents the estimated age of the repeat subfamily (in millions of years). Two subfamilies of the B2 repeat associated with CTCF are highlighted: B3A and B2_Mm1a. (B) Gray bars indicate the percentage of ESR1 and POU5F1-SOX2 binding regions with and without repeats that are within 10 kb of a regulated gene. Expected levels based on a random set of genes are shown in white. An additional control is shown using a random sample of instances from the same repeat families. Error bars, 1 SD.
RABS are associated with regulated genes
Finally, though it is difficult to globally ascertain whether RABS are directly controlling transcriptional regulation, we sought to address this by assessing the association of RABS to genes regulated by the specific TFs. The hypothesis is that if RABS are involved in gene regulation, then they should be more likely to be in the proximity of genes regulated by their cognate TF. To evaluate this possibility, we sought coordinated binding and gene expression data sets that used the same cell lines and similar treatments. Of the experimental data sets, the ones for ESR1 and POU5F1-SOX2 had detailed and well-defined expression matching the binding data. In this analysis we found that, for both TFs, RABS were more likely to be adjacent to regulated genes than to randomly selected genes: for ESR1, the fold enrichment is approximately fourfold, and for POU5F1-SOX2, the enrichment is approximately twofold (Fig. 5B; Supplemental Tables 7, 8). Interestingly, for POU5F1-SOX2, the level of association was comparable, and in the case of ESR1, superior, to that of other nonrepeat associated binding regions. Looking at randomly selected but unbound instances from the same family of repeats showed no specific proximity to regulated genes (see Methods). Taken together, these results suggest that RABS are functional at the level of TF binding and are likely to regulate many associated genes.
Discussion
Although cross-species conservation can be a powerful tool to identify regulatory sequences in genomes (Thomas et al. 2003; Boffelli et al. 2004; Wang et al. 2006), we have now shown using seven whole-genome in vivo occupancy data sets that only a minority of the TF binding regions appears to be evolutionarily conserved (Fig. 1). This result extends on previous reports (Dermitzakis and Clark 2002; Birney et al. 2007; Chabot et al. 2007; Odom et al. 2007; Jegga et al. 2008) and implies that a significant proportion of true TF binding regions would have been missed by in silico approaches relying exclusively on cross-species sequence conservation.
By leveraging on the ability of the ChIP sequencing platforms to detect TF binding even in repeat-rich regions, we showed that, for five of the seven TFs, a significant proportion of the binding sites are embedded in specific families of repeats (17.7% within MIR for ESR1, 32.1% within ERV1 for TP53, 19.0% within ERVK for POU5F1-SOX2, and 28.4% within B2 for CTCF). We demonstrated that progenitor sequences for the motifs were not only included in these repeats but that in many cases binding motifs were overrepresented within the repeat instances suggesting some evolutionary selection. Interestingly, MIR repeats have recently been reported to be under strong selection (Kamal et al. 2006), their association to ESR1 binding could now explain in part this selection. Also supporting a functional role of RABS in transcriptional regulation was the trend that the older the repeat, the higher the enrichment in the fraction of motifs observed to be bound (Fig. 5A) and their overrepresentation in proximity to regulated genes (Fig. 5B). Methodologically, our findings also suggest that approaches detecting TF occupancy that mask repeats (e.g., ChIP-chip) could miss a significant subset of bona fide functional elements for at least some of the TFs.
Why some transcription factor binding sites (ESR1, TP53, POU5F1-SOX2, and CTCF) reside on repeats, whereas others (e.g., MYC and RELA in this study) do not, is unclear. One possibility is that no repeat class harbors MYC or RELA progenitor sequences. Alternatively, we noted that ESR1, TP53, POU5F1-SOX2, and CTCF involve complex recognition motifs that span 10–20 base pairs (bp). In contrast, MYC and RELA binding motifs used shorter 6–11mer recognition sequences. Thus, though binding site diversity enhances robustness, the mechanism for binding site dispersion may be dependent on constraints imposed by the relative sequence complexity of the response elements. Whereas simple binding motifs (as in MYC and RELA) can be generated by mutation of random fragments, such de novo motif construction is not as probable for complex motifs. In the latter case, an alternative mechanism, retrotranspositional dispersion, was linked to the creation of new binding sites.
Finally, using the age of the repeat families, we showed that RABS have been associated with significant regulatory expansion throughout the mammalian phylogeny (Fig. 4) with, for instance, an example predating the mammalian radiation (ESR1 on MIR) and a more recent example on a class of transposable elements currently active in rodents (CTCF on B2). Our findings raise important questions about the similarity of the transcriptional regulatory networks between human and mouse in central biological processes ranging from cancer to stem cell. For instance, the fact that a large fraction of the bona fide CTCF binding sites in mouse are associated with rodent-specific transposable elements is likely to have profound functional implications. We note that at the Xist locus, a repeat element bound by CTCF was recently shown to have also descended from an ancient retrotransposon (albeit different from B2). At this particular locus, the bound repeat was also shown to directly regulate random and imprinted X inactivation (Cohen et al. 2007). In another well-studied locus where CTCF acts as a chromatin insulator, the H19/Igf2 locus (Hark et al. 2000), it was recently reported that the developmentally regulated expression of the B2 repeat itself acts as a chromatin domain boundary in organogenesis (Lunyak et al. 2007). These examples combined with the fact that we now report thousands of repeat elements bound by transcription factors help confirm the hypothesis that repeats act as critical “control elements” in eukaryotic genomes (Davidson and Britten 1979; McClintock 1984; Kidwell and Lisch 1997; Brosius 2003; Gentles et al. 2007).
Changes in regulatory elements can have important phenotypic effects across species (Gompel et al. 2005; Ihmels et al. 2005; Rockman et al. 2005; Marcellini and Simpson 2006; Tumpel et al. 2006) and within populations with examples from various human diseases, such as Alzheimer (Theuns et al. 2000), obesity (Esterbauer et al. 2001), and cancer (Bond et al. 2004). Using genome-wide in vivo TF binding data, our study quantitatively substantiate the link between repeat expansions and regulatory evolution in mammalian genomes.
Methods
Whole-genome chromatin immunoprecipitation data sets
We used seven whole-genome occupancy data sets: five used a ChIP-paired-end diTag (ChIP-PET) assay, one used a ChIP-chip assay and one used a ChIP-sequencing (ChIP-Seq) assay (Table 1; Supplemental Tables 9–15). Only the POU5F1-SOX2 data set required some additional experiments and processing different from the ones reported in the original publications. Because POU5F1 and SOX2 function as a heterodimer based on precise juxtaposition of the two binding sites (Loh et al. 2006), we treated these two transcription factors as one regulatory unit. Specifically, an additional ChIP-PET library for SOX2 binding sites also in mouse embryonic stem cells was generated under the same conditions reported in (Loh et al. 2006). Real time PCR validation showed that clusters with more than four PET overlaps have more than 95% validation rate (data not shown). All clusters with at least two overlapping PETs from both libraries were used as binding regions. In effect, this increased the stringency of the binding motif analysis because of the greater specificity of the dual recognition sequences.
Observed and expected overlap with conserved elements
To assess the evolutionary conservation of the binding regions we first looked for the presence of conserved elements identified from global alignments of vertebrate genomes (Siepel et al. 2005). PhastCons conserved element files were downloaded from the UCSC Genome Browser (Kent et al. 2002) for both human (hg17) and mouse (mm5). For this analysis, we only report the results of overlapping the conserved elements with 200-bp windows centered around the middle of the binding regions since larger windows lead to higher background levels and fold enrichments below two for all TFs (data not shown). To account for the conservation bias associated with proximity to coding regions, expected levels were estimated independently for each library using 1000 Monte Carlo simulations where the number of simulated regions falling into four categories with respect to the proximity to genes was fixed to the actual number of binding regions falling into the same categories. The four mutually exclusive categories are: adjacent—within 250 bp of the coding region of a known gene (KG), proximal—within 5 kbp of a coding region, distant—intragenic or within 100 kbp of a KG, and desert—more than 100 kbp from any KG. The analysis for CTCF was similar but based on the mm8 assembly.
Observed and expected binding motifs in multiple species
The binding regions (using centered windows of size 200, 500, 1000, and 2000 bp) were scanned for the motifs reported in the original publications: ERE consensus motif (GGTCAnnnTGACC) with up to two mutations for ESR1 (Lin et al. 2007), position weight matrix for TP53 (Wei et al. 2006), RELA (Lim et al. 2007), POU5F1-SOX2 (Loh et al. 2006), and CTCF (Chen et al. 2008), and finally perfect Ebox consensus motif (CACGTG) for MYC (Zeller et al. 2006). Homologous binding regions were identified using liftOver, a tool that relies on BLASTZ whole-genome alignments available through the UCSC Genome Browser (Kent et al. 2002; Schwartz et al. 2003), and scanned for motifs. Specifically, for the human TF ChIP experiments, windows observed to be bound in human (hg17) were converted into homologous regions in chimpanzee (panTro1), macaque (rheMac2), mouse (mm5), and dog (canFam2) using a 10% base pairs match cutoff and the multiple hits option. A region was said to contain a cross-species conserved motif if a motif was found in two out of the three primates and either mouse or dog. Similarly, for the mouse TF ChIP experiments, windows observed to be bound in mouse (mm5) were converted into rat (rn3), human (hg17), and dog (canFam1) and the motif was required to be found in both rodents and either human or dog. Expected levels were measured using similar distribution-matched simulated data sets as described above. Numbers reported in Figure 1 and Supplemental Table 1 are from 2 kbp windows (except for the ESR1 data sets where they are from 500 bp windows) because significant motif enrichment was observed to extend to these homologous neighborhoods (Supplemental text). The reference assemblies used for the analysis with CTCF were mm8, rn4, hg17, and canFam2.
Association with repeat elements
RepeatMasker data files (A.F.A. Smit, R. Hubley, and P. Green, http://www.repeatmasker.org) were downloaded from UCSC for both human (hg17) and mouse (mm5). Repeat content of the binding regions was measured in windows centered on the middle of the overlap. In all cases, the size of the windows used was 500 bp and the proportion of windows overlapping a specific type of repeat was reported and compared to the expected proportion observed in one million random locations selected on the appropriate genome from which gaps in the assembly had been removed. ChIP-PET background was estimated by intersecting repeat elements with centered windows for all singleton PETs (i.e., clusters of size one) of the individual TF libraries. ChIP-chip background was estimated using one million randomly selected probe locations based on the array design and obtained from Affymetrix (www.affymetrix.com). The ChIP-Seq background was estimated using singleton tags mapped outside binding clusters. The analysis for CTCF was based on the mm8 assembly. P-values were computed using a one-sided binomial test.
Validation of RABS using quantitative PCR
Quantitative PCR values of RABS for TP53 and ESR1 were extracted from the original publications (Wei et al. 2006; Lin et al. 2007). Additional RABS for POU5F1-SOX2 were tested by POU5F1 ChIP and quantified by real time PCR as described previously (Loh et al. 2006).
Repeat sequences as binding motif progenitors
The susceptibility of a piece of DNA sequence to generate a good binding motif solely through a series of random single nucleotide mutations can be approximated by the minimum Hamming distance (minHD) between any of its substrings and a good binding motif. For each repeat, we computed the minHD of its consensus sequence (as defined in RepBase; Jurka 2000) to a good binding motif of the associated transcription factor. Following that, we extracted all promoter sequences (and matched the length to the repeat consensus sequence) in the genome, based on the UCSC Genome Browser knownGene database, and similarly computed the minHD of each promoter to a good binding motif. A repeat consensus sequence whose minHD fell within the extreme lower tail of the promoter-based minHD distribution can create a good binding motif with fewer point mutations than most promoters—and can probably act as a motif progenitor.
Estimating the age of repeat families
The RepeatMasker output and align files for the human genome sequence (hg17) and mouse genome sequence (mm5) were downloaded from the UCSC Genome Browser. We calculated the age of the repeats using the formula: age = divergence/substitution rate. We used the substitution rates: 2.2 × 10−9 for the human genome and 4.5 × 10−9 for the mouse genome (Lander et al. 2001; Waterston et al. 2002). We computed the age of the repeat subfamily using three methods: (1) Jukes Cantor method, (2) Kimura two-distance, and (3) PAML. For the Jukes-Cantor method, we used the divergence rate (number of mismatches) from the RepeatMasker output file, while in the Kimura two-distance, we extracted the transition and transversion rates from the align files. We followed a similar approach to the one described by Pace and Feschotte (2007) to calculate the sequence divergence using PAML (Yang 1997). We generated a single concatenated sequence for each chromosomal repeat with the corresponding consensus sequence. The process was repeated with and without masking the CG dinucleotides (for + strand) and GC dinucleotides (for – strand) and as well as all non-ATGC characters removed. The combined sequences were analyzed using PAML version 3.15 using the REV model (Tavare 1986) with the global clock option. The corrected divergences (with and without GC masked) were extracted to calculate the age of the MIR, ERV1, ERVK, and B2 repeats (Supplemental Table 5).
Comparison between mouse and human CTCF binding regions
We used the top 21,373 CTCF binding regions detected in a study of human T cells (Barski et al. 2007). As before, 2 kb windows associated with CTCF binding regions in mouse were converted into human homologous regions using the tool liftOver (Kent et al. 2002). Converted regions falling within 1 kb of regions bound in human were said to be bound in both mouse and human.
Enrichment of binding motifs within the repeat families
The enrichment of good binding motifs in the repeats was estimated by comparing the number of good binding motifs found in the repeat instances in the genome to the expected number of good binding motifs had the repeat instances undergone random single nucleotide mutations uniformly across the instance. Sequences of the repeat instances were extracted from the genome and then scanned for motifs. A series of Monte Carlo simulations was run to estimate the expected number of good motifs. In each Monte Carlo iteration, we reconstructed the generation of each repeat instance by (1) extracting the aligned portion of consensus sequence as a seed sequence and (2) mutating its base pairs with the probability r, the mismatch rate reported in the RepeatMasker output file. Following which, the artificial repeat instances were scanned for good motifs as before and the total number of good motifs found was noted. The average number of good motifs was reported as the expected count of good motifs and the fraction of time artificial repeat instances contained as many or more good motifs than observed was reported as the P-value. We note that the effectiveness of this test will be limited by the fraction of functional sites within the repeat family that is directly under selection for the cognate TF binding motif.
Enrichment of bound binding motifs within the repeat families
The enrichment of bound motifs in the repeats was estimated by comparing the fraction of the motif observed to be bound in the different repeat subfamily to the fraction of motifs observed to be bound in one million 100 bp fragments randomly extracted from the respective genomes.
Association with regulated genes
A list of 1638 affy probes corresponding to 1187 differentially regulated genes following E2 treatment was extracted from Lin et al. (2007). A similar list of 1847 affy probes corresponding to 1719 differentially regulated genes following POU5F1 or SOX2 knockdown was extracted from Ivanova et al. (2006). Binding regions were partitioned into two groups: the ones with and without the cognate repeat. A binding region was said to be associated with a regulated gene if it was within 10 kb or internal to this gene. Expected levels were measured in 100 Monte Carlo simulations using the same procedure but where the set of regulated probes was replaced by a random set of the same size. In the final control, the set of bound repeat instances was replaced by random samples of instances coming from the same repeat family.
Acknowledgments
We thank C. Feschotte for help in estimating the age of repeats, and N. Clarke, L. Lipovich, S. Prabhakar, and S. Pott for comments on the manuscript. This work was supported by the Agency for Science, Technology and Research (A*STAR) of Singapore.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.080663.108.
References
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., Nobrega M.A., Rubin E.M. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Bond G.L., Hu W., Bond E.E., Robins H., Lutzker S.G., Arva N.C., Bargonetti J., Bartel F., Taubert H., Wuerl P., et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Borneman A.R., Gianoulis T.A., Zhang Z.D., Yu H., Rozowsky J., Seringhaus M.R., Wang L.Y., Gerstein M., Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chabot A., Shrit R.A., Blekhman R., Gilad Y. Using reporter gene assays to identify cis regulatory differences between humans and chimpanzees. Genetics. 2007;176:2069–2076. doi: 10.1534/genetics.107.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cohen D.E., Davidow L.S., Erwin J.A., Xu N., Warshawsky D., Lee J.T. The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell. 2007;12:57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., Britten R.J. Regulation of gene expression: Possible role of repetitive sequences. Science. 1979;204:1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Clark A.G. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Esterbauer H., Schneitler C., Oberkofler H., Ebenbichler C., Paulweber B., Sandhofer F., Ladurner G., Hell E., Strosberg A.D., Patsch J.R., et al. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat. Genet. 2001;28:178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- Gentles A.J., Wakefield M.J., Kohany O., Gu W., Batzer M.A., Pollock D.D., Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P.J., Kassner V.A., Carroll S.B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Ihmels J., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., Berman J., Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jegga A.G., Inga A., Menendez D., Aronow B.J., Resnick M.A. Functional evolution of the p53 regulatory network through its target response elements. Proc. Natl. Acad. Sci. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Gamblin R.J., Ooi L., Bruce A.W., Donaldson I.J., Westhead D.R., Wood I.C., Jackson R.M., Buckley N.J. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kamal M., Xie X., Lander E.S. A large family of ancient repeat elements in the human genome is under strong selection. Proc. Natl. Acad. Sci. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M.G., Lisch D. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laperriere D., Wang T.T., White J.H., Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 2003;13:R242–R254. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- Lim C.A., Yao F., Wong J.J., George J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol. Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Vega V.B., Thomsen J.S., Zhang T., Kong S.L., Xie M., Chiu K.P., Lipovich L., Barnett D.H., Stossi F., et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lunyak V.V., Prefontaine G.G., Nunez E., Cramer T., Ju B.G., Ohgi K.A., Hutt K., Roy R., Garcia-Diaz A., Zhu X., et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Marcellini S., Simpson P. Two or four bristles: Functional evolution of an enhancer of scute in Drosophilidae. PLoS Biol. 2006;4:e386. doi: 10.1371/journal.pbio.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- McGaughey D.M., Vinton R.M., Huynh J., Al-Saif A., Beer M.A., McCallion A.S. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Pollard D.A., Nix D.A., Iyer V.N., Li X.Y., Biggin M.D., Eisen M.B. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Dowell R.D., Jacobsen E.S., Gordon W., Danford T.W., MacIsaac K.D., Rolfe P.A., Conboy C.M., Gifford D.K., Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J.K., Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P., Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard D.A., Moses A.M., Iyer V.N., Eisen M.B. Detecting the limits of regulatory element conservation and divergence estimation using pairwise and multiple alignments. BMC Bioinformatics. 2006;7:376. doi: 10.1186/1471-2105-7-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M.V., Hahn M.W., Soranzo N., Zimprich F., Goldstein D.B., Wray G.A. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanges R., Kalmar E., Claudiani P., D’Amato M., Muller F., Stupka E. Shuffling of cis-regulatory elements is a pervasive feature of the vertebrate lineage. Genome Biol. 2006;7:R56. doi: 10.1186/gb-2006-7-7-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A., Regev A., Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavare S. Lectures in mathematics in the life sciences. American Mathematical Society; Providence, RI: 1986. Some probabilistic and statistical problems on the analysis of DNA sequences; pp. 57–86. [Google Scholar]
- Theuns J., Del-Favero J., Dermaut B., van Duijn C.M., Backhovens H., Van den Broeck M.V., Serneels S., Corsmit E., Van Broeckhoven C.V., Cruts M. Genetic variability in the regulatory region of presenilin 1 associated with risk for Alzheimer’s disease and variable expression. Hum. Mol. Genet. 2000;9:325–331. doi: 10.1093/hmg/9.3.325. [DOI] [PubMed] [Google Scholar]
- Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Tuch B.B., Galgoczy D.J., Hernday A.D., Li H., Johnson A.D. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpel S., Cambronero F., Wiedemann L.M., Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes) Proc. Natl. Acad. Sci. 2006;103:5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Cheng Y., Zhou Y., King D.C., Taylor J., Chiaromonte F., Kasturi J., Petrykowska H., Gibb B., et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zeng J., Lowe C.B., Sellers R.G., Salama S.R., Yang M., Burgess S.M., Brachmann R.K., Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zeller K.I., Zhao X., Lee C.W., Chiu K.P., Yao F., Yustein J.T., Ooi H.S., Orlov Y.L., Shahab A., Yong H.C., et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]