Abstract
Copy number variation is a dominant contributor to genomic variation and may frequently underlie an individual’s variable susceptibilities to disease. Here we question our previous proposition that copy number variants (CNVs) are often retained in the human population because of their adaptive benefit. We show that genic biases of CNVs are best explained, not by positive selection, but by reduced efficiency of selection in eliminating deleterious changes from the human population. Of four CNV data sets examined, three exhibit significant increases in protein evolutionary rates. These increases appear to be attributable to the frequent coincidence of CNVs with segmental duplications (SDs) that recombine infrequently. Furthermore, human orthologs of mouse genes, which, when disrupted, result in pre- or postnatal lethality, are unusually depleted in CNVs. Together, these findings support a model of reduced purifying selection (Hill–Robertson interference) within copy number variable regions that are enriched in nonessential genes, allowing both the fixation of slightly deleterious substitutions and increased drift of CNV alleles. Additionally, all four CNV sets exhibited increased rates of interspecies chromosomal rearrangement and nucleotide substitution and an increased gene density. We observe that sequences with high G+C contents are most prone to copy number variation. In particular, frequently duplicated human SD sequence, or CNVs that are large and/or observed frequently, tend to be elevated in G+C content. In contrast, SD sequences that appear fixed in the human population lie more frequently within low G+C sequence. These findings provide an overarching view of how CNVs arise and segregate in the human population.
Copy number variants (CNVs) contribute more than single nucleotide polymorphisms to the number of bases differing between a pair of human genomes (Redon et al. 2006; Shianna and Willard 2006), and as such are expected to contribute substantially to phenotypic variation. CNVs have been identified for a large proportion (up to 12%) (Redon et al. 2006) of the human euchromatic sequence, together encompassing up to 1500 (Wong et al. 2007), 1800 (Sharp et al. 2006a), or 2900 (Redon et al. 2006) genes. Most individual CNVs, however, are observed only rarely (<1%) in the human population (Sharp et al. 2005; Nguyen et al. 2006). Given the large size and abundance of CNVs in each human genome it is unsurprising that copy number variation is associated with disease susceptibility (Singleton et al. 2003; Vissers et al. 2004; de Vries et al. 2005; Gonzalez et al. 2005; Aitman et al. 2006; Koolen et al. 2006; Sharp et al. 2006b; Shaw-Smith et al. 2006; Sleegers et al. 2006; Sebat et al. 2007; Walsh et al. 2008). Nevertheless, if we are to better appreciate the broader relevance of CNVs to human disease, it is essential that we better understand the biases underlying their detection, how they arise and segregate in the human population, as well as the selective processes that act upon them.
Copy number variation has not been observed uniformly within the human genome. It is particularly concentrated near proximally duplicated regions (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005; McCarroll et al. 2006), especially segmental duplications (SDs), defined as duplicated sequences sharing >90% identity over at least 1 kb in the reference human genome assembly (Bailey et al. 2001, 2002). This assembly is a mosaic comprised of sequence from multiple individuals’ genomes. Consequently, it is expected that some SDs in the reference assembly represent duplication alleles. However, owing to the rarity of most CNV alleles, the frequent coincidence of human CNVs and reference assembly SDs is not a trivial result. The nonuniform distribution of CNVs may arise from nearby repetitive sequences, facilitating a duplication or deletion via nonallelic homologous recombination (NAHR) (Stankiewicz and Lupski 2002; Hurles 2005; Lupski and Stankiewicz 2005). CNVs, SDs, and, indeed, other fragile portions of genomes such as synteny breakpoints, are also associated with further mutational biases, such as elevated nucleotide substitution rates (Armengol et al. 2005; Webber and Ponting 2005; Nguyen et al. 2006). Genomic regions both rich in SDs and prone to recombination are expected also to be enriched in CNVs, as allelic homologous recombination and NAHR are intimately related (Lindsay et al. 2006).
We previously presented evidence that positive selection of CNVs has occurred within the modern human population (Nguyen et al. 2006). For that study we considered a set of 627 human CNV regions (CNVRs), collated from a variety of sources that had used diverse experimental protocols. We argued that their significantly increased density of genes, particularly those encoding secreted products and those possessing functions involved in the sensing of the environment, suggested the past action of positive selection. This conclusion was also consistent with elevated rates of protein evolution for these regions, estimated over the long time period spanning the human and mouse lineages. For this we assumed that these rate elevations were caused in great part by episodes of positive selection on amino acid substitution, and that these ancient episodes mirror more recent episodes of positive selection on copy number change (Mouse Genome Sequencing Consortium 2002; Emes et al. 2003). As these effects were most pronounced for the more frequent CNVRs (those observed on at least two occasions), we further argued that CNVs might have been driven to high frequency in the human population as a consequence of their adaptive benefit. In that publication, we did not consider whether selection on human CNVs is dominated instead by reduced purifying selection.
We felt it important to revisit this issue using recently available and genome-wide data sets, each acquired using a different experimental protocol. In addition to a new set of CNVs, we analyzed the properties of three previously described sets of CNVs (Redon et al. 2006; Wong et al. 2007). This allowed us to investigate whether biases associated with different array-based platforms could influence our evolutionary conclusions. The origin and rates of fixation of CNVs were also addressed by analyzing the large numbers of SDs that are apparent from the human genome reference sequence. By exploiting both CNV and SD data, we reconsidered whether nonadaptive processes, rather than positive selection, might underlie our previous observations. We were particularly concerned by four potential confounding factors. First, we were concerned that CNVs’ increased gene densities might have arisen simply because of a positive correlation with the G+C nucleotide content (Lander et al. 2001; Mouse Genome Sequencing Consortium 2002), although our previous study had not revealed such a G+C content bias. We also needed to reconsider the issue of gene density in the light of recent reports from Redon et al. (2006) and Conrad et al. (2006) that some CNV sets are gene poor, not gene rich. Second, there was a need to consider whether selective and mutational forces have acted differentially on small versus large, or rarely versus frequently observed CNVs. Third, CNV genes might have acquired an elevated number of deleterious rather than advantageous amino acid changes if they have often been subject to reduced rates of recombination (“Hill–Robertson interference”) (Hill and Robertson 1966; Charlesworth and Charlesworth 2000; McVean and Charlesworth 2000). Finally, the concentration of CNVs in certain genomic regions, and their enrichments in “environmental” genes, might reflect selective in addition to mutational biases. Specifically, CNV regions may be largely spared from strong constraint on copy number that applies to the remaining bulk of the genome.
Results
We divide our studies between those investigating the mutational processes of how CNVs arise and others seeking to understand the impact of selection on CNVs. We first introduce the CNV data sets we use, and then we describe a nucleotide content-dependent bias on how frequently CNVs arise in a genomic region. Subsequently, we question whether CNVs have been negatively or positively selected in the human population. For these studies we also took advantage of SDs from the reference human genome sequence, considering these to be duplications that either were fixed or remain polymorphic in the human population.
CNV data sets
Analyses were performed on four CNV data sets identified by either Redon et al. (2006) (two sets) or by Wong et al. (2007), or by ourselves (Table 1). For each set, overlapping CNVs were merged (see Methods) in order to obtain nonoverlapping CNVRs (Table 1). In order to consider platform-specific biases, we considered CNVs identified by Redon and colleagues separately by each platform. Our own previously unpublished data set contains 1276 “Nijmegen” CNVs that were identified as a by-product of a study of diagnostic genome profiling in mental retardation (de Vries et al. 2005), and were purged of those variants that arose spontaneously, and thus might be causally linked to disease (see Methods; Supplemental Table 1).
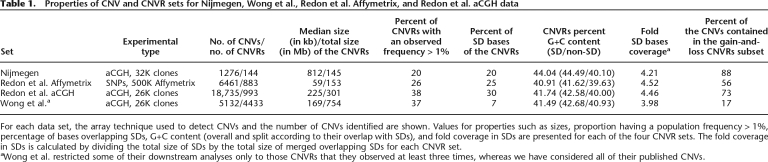
Table 1.
Properties of CNV and CNVR sets for Nijmegen, Wong et al., Redon et al. Affymetrix, and Redon et al. aCGH data
For each data set, the array technique used to detect CNVs and the number of CNVs identified are shown. Values for properties such as sizes, proportion having a population frequency > 1%, percentage of bases overlapping SDs, G+C content (overall and split according to their overlap with SDs), and fold coverage in SDs are presented for each of the four CNVR sets. The fold coverage in SDs is calculated by dividing the total size of SDs by the total size of merged overlapping SDs for each CNVR set.
aWong et al. restricted some of their downstream analyses only to those CNVRs that they observed at least three times, whereas we have considered all of their published CNVs.
As expected, CNVs identified using the Affymetrix SNP array platform tended to be smaller than those found with array-comparative genomic hybridization (aCGH). In contrast, Nijmegen CNVRs were larger and were observed less frequently (Supplemental Fig. 1; Table 1), likely owing to a more stringent CNV calling protocol and to the use of a pooled set of genomes as reference in these aCGH experiments, respectively (see Methods). For three of the four sets, the majority of CNVs lay within CNVRs in which both gain and loss CNVs are observed (Table 1). These “gain-and-loss CNVRs” might represent instances where the reference genomes contain both CNV alleles and where subject genomes are homozygous for one allele. However, many gain-and-loss CNVRs contain at least one CNV pair whose boundaries are not equivalent (94%, 64%, 95%, and 36% of CNVRs for the Nijmegen, Redon et al. aCGH, Redon et al. Affymetrix, and Wong et al. data sets, respectively). Even when the inaccuracies of CNV boundary determination are considered, it appears likely that gain-and-loss CNVRs often reflect recurrent CNV changes, rather than heterozygosity in the reference genomes. We, and others, have demonstrated that CNV changes can be recurrent (White et al. 2007; Turner et al. 2008).
Mutational biases
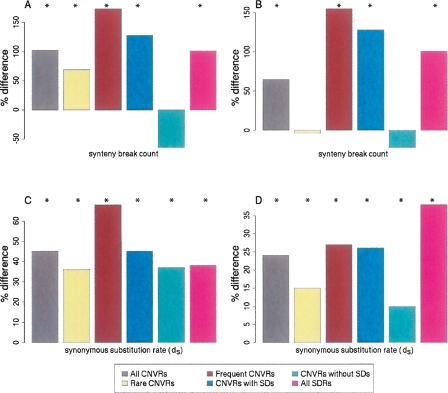
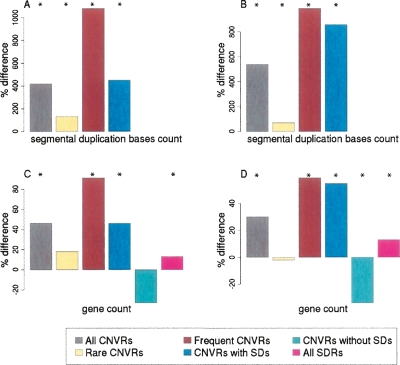
SDs frequently coincide both with CNVs and with breakpoints in conserved synteny for mammalian chromosomes (Bailey et al. 2004; Armengol et al. 2005). Thus, we were interested in whether an increased density of synteny breakpoints would be found within CNVRs. Indeed, all four CNVRs sets contain more breakpoints in synteny between dog and human chromosomes than expected by chance alone (P < 2 × 10−3) (Fig. 1A,B; Supplemental Table 2). (To establish this significance and that for subsequent observations unless otherwise stated, we constructed 500 randomly sampled sets of nonoverlapping genomic segments matched in number and in size to each of the CNVR sets considered; no set of random segments possessed as many breakpoints in synteny as any of the CNVR sets [i.e., P < 2 × 10−3].)
Figure 1.
The departures from expected values for the coincidence of CNVRs with human and dog synteny breakpoints (A,B) and human and dog orthologs’ synonymous substitution rates within CNVRs (C,D), calculated for sets of Nijmegen CNVRs and Redon et al. aCGH CNVRs. Results are shown as percentage differences from values expected by random sampling of the human genome reference assembly (see Methods). (A,C) Nijmegen aCGH CNVRs; (B,D) Redon et al. aCGH CNVRs. For either Nijmegen or Redon et al. aCGH data, the set of genomic regions are: “All CNVRs,” all CNV regions; “Rare CNVRs,” genomic regions in which CNVs were identified in <1% of the individuals sampled; “Frequent CNVRs,” CNVRs in which CNVs were identified in >1% of the individuals sampled; “CNVRs with SDs,” CNV regions overlapping with at least one SD; “CNVRs without SDs,” CNV regions having no overlap with SDs; and “All SDRs,” the set of all 8051 SD regions (shown accompanying each CNVR set). Bars annotated with an asterisk (*) are significant at P < 0.025.
By partitioning the four sets of CNVRs according to their overlap with SDs (see Methods), we found that, as expected, it is only those CNVR subsets that also overlap SDs that are significantly enriched with synteny breakpoints (Fig. 1A,B). The frequent coincidence of CNVRs, SDs, and synteny breakpoints indicate that the most recently fragile regions of the human genome have also been fragile throughout mammalian evolutionary history.
Genomic regions that are prone to copy number variation are known to possess high rates of nucleotide substitution (Armengol et al. 2005; Nguyen et al. 2006; She et al. 2006). Each of the four sets of CNVRs we analyzed exhibit this bias, with median synonymous nucleotide substitution rates being elevated by between 11% and 45% (Fig. 1C,D). Genes present in SDs also exhibit increased synonymous substitution rates (+38% elevation), and similar elevations are observed for orthologous transposable elements (“ancestral repeats”) in CNVRs and in SDs (data not shown). As most of the CNVR sets exhibit unexpectedly high G+C contents (see below), these elevated rates are due, in part, to the well-established positive correlation between G+C or CpG contents and substitution rates (Hardison et al. 2003; She et al. 2006). However, even for CNVRs that do not possess unusual G+C contents, such as those reported from the Redon et al. (2006) Affymetrix platform and even for CNVRs lacking SDs (see below), substitution rates remain significantly elevated. These findings suggest that a common cellular mechanism could underlie the increased rates of substitution and copy number changes in these regions (Nguyen et al. 2006).
G+C content bias
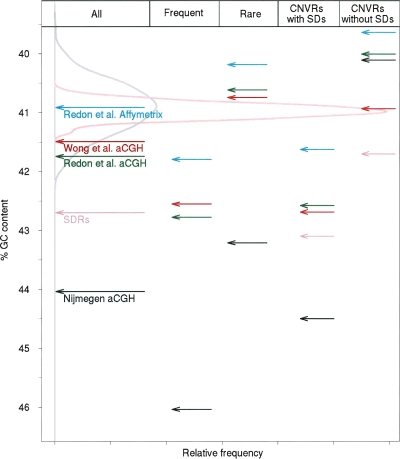
The CNVR set we analyzed previously (Nguyen et al. 2006), and those identified by Redon et al. (2006) or Wong et al. 2007), have each been reported to possess nucleotide contents that reflect the G+C content of the genome as a whole. It came as a surprise, therefore, to observe that each of the three aCGH CNVRs sets is actually enriched in G+C content. To assess the departure of their G+C contents, we performed randomizations as before. The median G+C content for each randomized genomic set was found to be 40.9% (Fig. 2).
Figure 2.
Percentage of G+C contents for CNVRs and SDRs compared with size-matched randomized sets for Nijmegen CNVRs (indicated within black arrows), Redon et al. Affymetrix CNVRs (cyan), Redon et al. aCGH (green), Wong et al. CNVRs (red), and SDRs (pink). The first column shows G+C contents for the entire sets. The G+C contents of CNVR sets partitioned according to frequency are shown in the second and third columns (“Frequent,” CNVRs at >1% observed population frequency; “Rare,” CNVRs at <1% observed population frequency). The fourth and fifth columns show G+C contents of CNVR sets partitioned according to their overlap, or not, with SDs. Additionally, the G+C contents of SDRs overlapped, or not, by CNVs, are shown in the fourth and fifth columns, respectively. The frequency distribution of the G+C contents for 500 randomly sampled sets of genomic regions, matched in size to Nijmegen CNVRs (gray density curve) and to SDRs (pink density curve), are also shown.
The elevation of G+C content is significant for each of the three aCGH CNV sets (P < 2 × 10−3). Nijmegen CNVRs exhibit the largest G+C content of 44.0%, ∼3% higher than expected from the genome-wide randomized samples; Wong et al. (2007) and Redon et al. (2006) aCGH CNVRs possess G+C contents of 41.5% and 41.7%, respectively. The increase in G+C for the Nijmegen set is substantial, since only 20.6% of 50-kb windows in the human genome possess a G+C content higher than 44.0%. No significant nucleotide G+C bias was, however, observed for the CNVRs from the Redon et al. Affymetrix set: Their G+C content of 40.9% exactly matches the value expected by random sampling.
The largest G+C content increases are always associated with CNVRs that are observed frequently; hence, rarely (<1%) observed CNVRs possess the lowest G+C contents (Fig. 2). This contrast is particularly pronounced for Nijmegen CNVRs (46.0% for frequent vs. 43.2% for rarely observed CNVRs) (Fig. 2). We also note that for all four data sources, CNVR partitions that overlap SDs exhibit marked increases in G+C content, whereas those outside of SDs exhibit reduced G+C contents (Fig. 2). The four CNVR sets are associated with very different amounts of SDs, but all are significantly higher than the 5.3% of human SDs present genome-wide (P < 2 × 10−3) (International Human Genome Sequencing Consortium 2004). A total of 20%, 25%, 30%, and 7% of Nijmegen, Redon et al. Affymetrix, Redon et al. aCGH, and Wong et al. CNVR bases, respectively, lie within segmentally duplicated sequence (see also Cheng et al. 2005; Sharp et al. 2005). For all CNVR partitions outside of SDs the G+C content is lower than the genome average and is significantly so for each of the two Redon et al. CNVR sets (P < 2 × 10−3). The high G+C content of CNVRs could thus be interpreted as being simply due to the substantial overlap between CNVRs and SDs.
Faster turnover of G+C-rich sequence in SDs
This observation led us to investigate the G+C contents of SDs, despite these sequences not having previously been noted as containing an increased G+C content. For this analysis we constructed segmentally duplicated regions (SDRs) from overlapping SDs exactly as was previously done for CNVs. Compared with the randomized distribution, the G+C content of SDRs is indeed significantly elevated (+1.7%, P < 2 × 10−3) (Fig. 2).
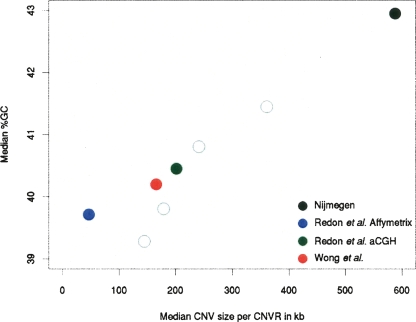
Why might CNVs observed by aCGH exhibit such pronounced increases in G+C content, while CNVRs identified on a single nucleotide polymorphism platform do not? Similarly, why might SD-containing CNVRs possess a high G+C content? We looked to the biases of size that are inherent in experimental designs to explain these differences. We do, indeed, observe that CNVRs of increasingly larger size possess elevated G+C content. Plotting the median size of CNVs underlying these CNVRs against their median G+C content shows that the G+C content increases approximately linearly with CNV size (Fig. 3). This trend does not simply reflect ascertainment biases between platforms, because it is also apparent for data acquired with a single platform (Fig. 3). Thus, across single and multiple data sets there appears to be a strong trend for larger CNVs to contain an unusually high G+C content.
Figure 3.
Relationships between CNVR size and G+C content. We partitioned by CNVR size the Redon et al. aCGH set into four equally populated bins; this data set was chosen, as it provides CNVRs covering the broadest logarithmic range of size variation. Differences in G+C content between each of the two largest bins when compared with either of the two smallest bins are significant (P < 4 × 10−3). Even when discarding segmentally duplicated bases, the two larger CNVR sets exhibit significantly increased G+C content (Nijmegen set, 44.2%; Redon et al. aCGH set, 41.5%; P < 1 × 10−3). For each data set, the median size of CNVs within a CNVR is plotted against the CNVs’ median G+C content (filled circles). Redon et al. aCGH CNVRs were also partitioned into four equally populated bins according to median CNV sizes (open circles).
If the highest G+C portions of the human genome have been particularly susceptible to copy number variation in the human population, then we would expect SDs that are fixed in the human population to be relatively depleted in G+C content compared with SDs that are copy number variable. To examine this proposition, we use as a proxy for fixed SDs all SDs that lie outside of any CNV present among the four data sets we examined. As expected, nonfixed SDs possess a significantly higher G+C content than apparently fixed SDs (43.1% and 41.7%, respectively; P = 1 × 10−7).
We also might expect sequence containing higher proportions of G or C bases to have been more frequently duplicated. To compare genomic regions that have suffered large numbers of SD events with those whose extant duplications have been far less numerous, we computed, for each base pair, the number of overlapping SDs. Next, we calculated the G+C content of sequence that either was overlapped by SDs more than 12 times, or else once only. We argue that if no G+C-dependencies exist on deletion or duplication rates, then the G+C content of these two sets should be equivalent. Instead, the G+C content of >12-fold SD coverage regions is 2.1% higher than that for onefold coverage SD regions (44.5% vs. 42.4%, P < 10−16). We conclude that genomic regions possessing high G+C content have been unusually susceptible to segmental duplication and, more particularly, to copy number variation in the human population.
We observe a positive correlation between G+C content and CNV size (Pearson’s r = 0.18, P = 4 × 10−8; Fig. 3) and a higher G+C content of CNVRs that overlap SDs than of CNVRs that do not. It is thus unsurprising that we also observe a significant correlation between the percentage of SD bases within a CNVR and the median CNV size within each CNVR (r = 0.25, P = 1 × 10−14). However, examining first order partial correlations, we find that the correlation between G+C content and CNV size remains significant even when accounting for SD content (r = 0.16, P < 10−4), as does the correlation between SD content and CNV size when accounting for G+C content (r = 0.24, P < 10−4). Thus, genomic regions high in either G+C content or SD content may be particularly susceptible to the generation of larger CNVs.
A nonadaptive explanation for elevated evolutionary rates of CNV genes
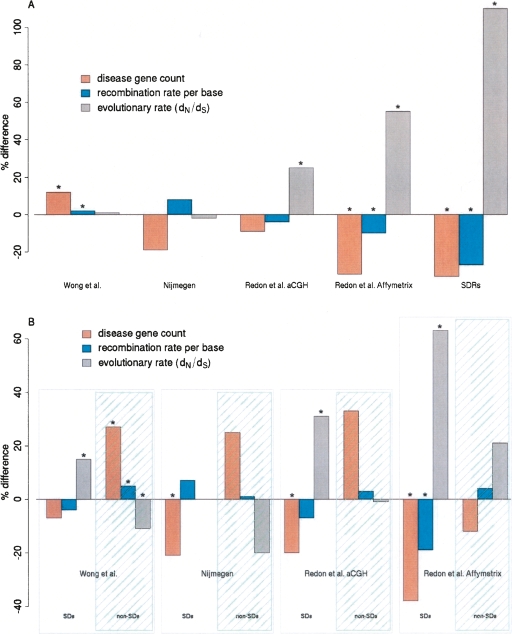
We previously showed that genes located within CNVRs tend to exhibit significantly elevated rates of protein evolution, as measured using dN/dS ratios (Nguyen et al. 2006). For this work we exploited a set of high-quality orthologous gene assignments determined between human and dog Ensembl genes (Hubbard et al. 2002; Birney et al. 2006; Goodstadt and Ponting 2006). Due to the shorter mutational distance between human and dog (median dS = 0.35) compared with that between human and mouse (median dS = 0.56) (Lindblad-Toh et al. 2005), we expect the assignment of orthology between the former species’ pair to be more accurate. By comparing dN/dS distributions for genes from different CNVR sets against dN/dS values for all human genes (see Supplemental Table 2), we showed that the tendency for increased dN/dS values was also exhibited by genes from the two Redon et al. CNVR sets (median dN/dS value increases of +25% and +55% for Affymetrix and aCGH data sets, respectively; P < 10−10; Kolmogorov-Smirnov [KS] test) (Fig. 4A). This tendency, however, was not found for the two remaining sets (Nijmegen CNVRs, −2%; Wong et al. CNVRs, +1%; Fig. 4A).
Figure 4.
The departures from expected values for evolutionary rates, recombination rates, and disease gene count estimated within Nijmegen CNVRs, Redon et al. Affymetrix CNVRs, Redon et al. aCGH CNVRs, Wong et al. CNVRs and SDRs. Evolutionary rates and disease gene counts have been obtained only for genes that entirely lie within these CNVRs. Results are shown as percentage differences from values expected from random sampling of the human genome reference assembly (see Methods). (A) Departures of the rate of protein evolution (dN/dS), recombination rate per base (cM/bp) and disease gene count are shown as percentage differences from the value expected from random sampling of the human genome. Bars annotated with an asterisk (*) are significant at P < 0.025. (B) Departures of the same properties as in A, after splitting the different CNVR data sets according to their overlap with SDs.
To investigate these platform-specific findings we partitioned CNVRs according to their overlap with segmentally duplicated sequence. We chose to do this because human SD genes exhibit a significant and substantial increase in dN/dS, with a median value (0.25) over twice that expected from the genome as a whole (0.12) (Fig. 4A; Supplemental Table 2) (see also Armengol et al. 2005). Dividing the CNVRs from each of the four platforms conditional on their overlap with SDs results in eight partitions. Of these CNVR subsets, six accord with a pattern of elevated dN/dS values in CNVRs overlapping SDs, and decreased dN/dS values in CNVRs lying outside of SDs (Fig. 4B). We will interpret this pattern observed for the six CNVR subsets under a model of reduced purifying selection and will thereafter return to discuss the remaining two subsets.
The elevation in dN/dS ratios for CNV genes that overlap with SDs might have arisen from relaxation of constraints or from positive selection on amino acid substitutions. A third possibility is that the dN/dS elevation is due to an increased rate of fixation of deleterious, rather than adaptive, substitutions because of unusually low rates of recombination (“Hill–Robertson interference”) (Hill and Robertson 1966; Charlesworth and Charlesworth 2000; McVean and Charlesworth 2000). Reduced rates of cross-over could arise mechanistically simply because polymorphic copy number variants interfere with homologous strand invasion (Navarro et al. 1997; Shaw and Lupski 2004; Lupski and Stankiewicz 2005; Erdogan et al. 2006; Lindsay et al. 2006). This third explanation predicts that when median dN/dS values are elevated, human recombination rates are reduced, and vice versa. Indeed, this inverse relationship was found to hold true for all but one of the eight CNVR sets that either overlap, or do not overlap, SDs (Fig. 4B). Hill–Robertson interference thus offers a plausible and nonadaptive explanation for the elevated evolutionary rates of protein-coding genes lying within SD regions. We emphasize that it appears to be the tendency of infrequently recombining regions, SDs, and CNVs to coincide that underlies both the increase in dN/dS ratios of genes lying in CNVs and the decrease in dN/dS ratios of genes outside of CNVs.
As we highlight above, two subsets, namely, Nijmegen CNVRs that overlap SDs and Redon et al. Affymetrix CNVRs that do not overlap SDs, diverge from our proposed model of reduced purifying selection inside SDs, and strong purifying selection outside SDs (Fig. 4B). With regard to Nijmegen CNVRs that overlap SDs, these are distinguished from the other sets not only because of their larger sizes, but because of their significantly lower proportions of segmentally duplicated bases (Nijmegen: 0.8%; Wong et al.: 1.5%; Redon et al. aCGH: 1.6%; Redon et al. Affymetrix 4.8%; P < 2 × 10−4). They are thus expected to be among the least influenced by the effects of low recombination on increasing dN/dS ratios within segmentally duplicated sequence. Finally, we return to the observation that Redon et al. Affymetrix CNVRs that do not overlap SDs encompass genes tending to show higher than expected dN/dS ratios (Fig. 4B). We believe this observation arises from this SNP-based platform’s preference for sampling relatively short CNVRs (Fig. 3) that contain shorter human genes whose protein evolutionary rates tend to be higher. In support of this, we considered all human–dog orthologs and found that the shortest 50% of such genes lying outside of human SD sequences tend to possess significantly higher dN/dS ratios than the longest 50% of such genes (0.09 vs. 0.13, respectively, KS-test P < 1 × 10−16).
Reduced selection explains the functional biases of CNVR genes
CNV genes are enriched in “environmental” functions and encode unexpectedly large numbers of secreted proteins (Gibbs et al. 2004; Feuk et al. 2006; Nguyen et al. 2006; Sharp et al. 2006a). We, and others, previously interpreted these observations as indicating that positive selection on duplications has occurred for these particular gene categories (Mouse Genome Sequencing Consortium 2002; Emes et al. 2003). However, we need to consider whether these enrichments might instead have arisen from nonuniform negative selection on gene copy changes: Duplication or deletion of nonenvironmental genes might be more frequently deleterious than copy number changes of environmental genes.
To distinguish between these two possibilities, we took advantage of phenotypic information from two sources that relate to the deleteriousness of gene disruptions. First, we identified a set of “essential genes,” defined as human orthologs of mouse genes that, when disrupted, result in either pre- or postnatal lethality (Bult et al. 2008). We also considered a set of “disease genes” representing human genes that, when mutated, have been associated with Mendelian disease; as such disruptions less frequently result in pre- or postnatal lethality, it is expected that purifying selection on mutations is considerably stronger on essential genes than it is on disease genes. The two sets are also distinguished by their gene functions: For example, essential genes are unusually depleted (one-third lower) in genes encoding secreted proteins (P < 10−16; Fisher’s exact test), whereas disease genes encode a significantly higher proportion of such proteins (Winter et al. 2004).
CNVR genes fail to randomly sample the set of essential genes: Each of the four CNVR sets is significantly depleted in such genes (P < 2.5 x 10−2; Supplemental Table 3). Deficits of essential genes are most pronounced for both frequently observed (>1%) CNVRs and for gain-and-loss CNVRs (Supplemental Table 3). One explanation of these findings is that fewer essential genes reside within regions that are predisposed to CNV mutations. However, a more likely explanation is that purifying selection on copy number change acts nonuniformly in the genome. Some gene types are poorly represented in CNVs, simply because changes in their copy number tend to be strongly deleterious and thus are often purged from the population. Conversely, other gene types are over-represented in CNVs, either because their changes in copy number are less deleterious, or because delete rious CNV alleles are less frequently purified owing to low recombination rates (Hill–Robertson interference) (see Discussion).
Disease genes, as opposed to essential genes, show no set pattern of enrichment in CNVRs, as they are significantly depleted in the Redon et al. Affymetrix CNVR set and significantly enriched in the Wong et al. CNVR set (Fig. 4A). However, when partitioned further according to whether or not they occur in CNVRs that overlap SDs, three of the four sets exhibit a disease gene surfeit outside of SDs and a deficit inside SDs (Fig. 4B). The deficit of disease genes within CNVRs that overlap SDs is consistent with these regions containing an unusual concentration of genes better able to accept the potentially deleterious effects of variable copy number and, as we propose above, deleterious substitutions.
The enrichment of disease genes within CNVRs lying outside SDs might imply that duplications of such genes are advantageous, perhaps by providing functional compensation when single genes are disrupted. We thus might expect that duplications, in other lineages, of genes that are essential in mouse, would be retained in those lineages due to selection for beneficial redundancy. Instead, we found that essential and disease genes have been preferentially retained as single copies in four lineages (those of human, mouse, dog, and opossum; see Methods) since their last common ancestor. More specifically, we found that among those genes that have remained unduplicated over a total of 485 million years of evolutionary time (Archibald 2003; Springer et al. 2003), there are significant enrichments of disease genes (81% increase) and essential genes (22% increase) when compared with genes that have experienced at least one duplication in these lineages (P < 10−16; Fisher’s exact test).
We thus propose that human disease gene CNVs lying outside of segmentally duplicated sequence will, over sufficient numbers of generations, be preferentially purged from the human population. By way of contrast, CNVs encompassing essential genes, being more deleterious, are purged more rapidly and are thus more rarely observed segregating in the human population.
This proposition appears to be at odds with the surfeit of disease genes observed in three of the four sets of CNVRs that do not overlap SDs. We considered whether disease genes might tend to lie within a sequence that is preferentially sampled by each of these three CNV platforms. Owing to our previous observation that each of the three aCGH platforms show a preference in sampling sequence with high G+C content, we considered whether disease genes also exhibit a preference to lie within high G+C sequence. Indeed, the intronic and flanking sequence (5 kb up- and downstream) of disease genes, after excluding all other coding sequence, was found to be significantly elevated in G+C content compared with genes not known to be involved in Mendelian disorders (median 45.7% vs. 44.0%, respectively; KS-test P = 1.7 × 10−8). The fourth platform, that of Redon et al. Affymetrix, shows a deficit of disease genes (Fig. 4B) that may be explained, at least in part, by its lower G+C content. It may also be explained, however, by this platform’s preference in detecting smaller CNV(R)s (Table 1; Fig. 3): Mendelian disease genes are, on average, twice as large as other genes (median sizes 29.3 kb vs. 16.6 kb, respectively; see Smith and Eyre-Walker 2003) and are thus less likely to be completely overlapped by this set’s CNVRs compared with the other three platforms.
CNVR gene richness and elevated G+C content
If positive selection on human CNVs has occurred frequently, then we might expect an increased number of functional elements to be found within such regions. Previously (Nguyen et al. 2006), we showed that CNVs, especially those found frequently in the human population, are indeed enriched in protein-coding genes, consistent with some CNVs being preferentially retained within the human population because of the benefits accrued from their genes’ copy number changes (Gonzalez et al. 2005; Zhang et al. 2005; She et al. 2006). In our new study, all four CNVRs sets also exhibit a strong and significant enrichment in gene numbers over that expected from their sizes (46%, 12%, 14%, and 30% increases for Nijmegen, Wong et al., Redon et al. Affymetrix, and Redon et al. aCGH CNVRs, respectively; P < 0.02) (Fig. 5A).
Figure 5.
The departures from expected values for the coincidence of CNVRs with segmentally duplicated basepairs (A,B) and with Ensembl protein-coding genes (C,D), calculated for sets of Nijmegen CNVRs and Redon et al. aCGH CNVRs. Results are shown as percentage differences from values expected by random sampling of the human genome reference assembly (see Methods). (A,C) Nijmegen aCGH CNVRs; (B,D) Redon et al. aCGH CNVRs. CNVR sets are described in the legend to Figure 2. Bars annotated with an asterisk (*) are significant at P < 0.025.
Nevertheless, CNVs might contain many genes simply because of a genome-wide tendency for a higher density of genes within G+C-rich sequence (Zoubak et al. 1996), which is more frequently subject to copy number change. The enrichment in genes (Fig. 5C,D) and in G+C content (Fig. 2) within CNVs would be consistent with this alternative and nonadaptive explanation. Other biases are also consistent with this nonadaptive model: CNVRs lying outside of SDs exhibit reduced G+C contents and significantly reduced gene densities (33%, 47%, 34%, and 10% decreases for Nijmegen, Redon et al. Affymetrix, Redon et al. aCGH, and Wong et al. data sets, respectively, P < 0.02) (Fig. 5).
The Redon et al. Affymetrix CNVR data might still be considered to support an adaptive evolution for CNVs, since these data show no overall elevation of G+C content despite a substantial increase in gene content. Nevertheless, here too, a nonadaptive explanation can be provided. The Redon et al. Affymetrix CNVR data appear to be gene rich on account of their high proportion (∼25%) of CNVRs that overlap SDs, yet these SDs possess a lower G+C content than SDs occurring within CNVRs from other data sets (Fig. 2). The Affymetrix SNP platform preferentially samples lower over higher G+C SDs, since higher G+C SD sequence presents greater difficulties when unambiguously identifying markers within commonly structurally variable regions (Wirtenberger et al. 2006) and, as we have argued above, higher G+C sequence is more frequently copy number variable.
The CNVR protein-coding proportion (percentage of exonic base pairs) correlates significantly with both CNVR G+C-content (Pearson’s r = 0.41, P < 10−16) and with SD-fraction (percentage of SD base pairs, r = 0.39, P < 10−16). Examining the first order partial correlations reveals that G+C content remains significantly correlated with CNVR protein-coding content having accounted for SD content (r = 0.39, P < 10−4), as does SD content when accounting for G+C content (r = 0.37, P < 10−4). Thus, in general, we conclude that the gene richness of CNVs is best explained not by positive selection, but instead by platform-specific biases in sampling CNVs with a high G+C-content and/or that overlap SDs.
Discussion
Our study examined 6453 genomic regions with copy number variation that were identified using four different platforms and experimental protocols. Each of these approaches appears to sample CNVs differently according to size and G+C content (Fig. 3). General trends that emerge are that regions showing elevated G+C content give rise to larger CNVs and more frequently overlap with sequence-similar segmental duplications. Such regions also exhibit tendencies to be gene rich and to accumulate more nucleotide substitutions and interspecies synteny breakpoints (Nguyen et al. 2006; Cooper et al. 2007), although whether these tendencies are causally related remains unknown. Human sequence showing high G+C content also tends to experience elevated rates of recombination (Hardison et al. 2003; Duret et al. 2006; Khelifi et al. 2006). NAHR alone, however, cannot explain such sequences’ higher rates of structural rearrangement. This is because, notwithstanding methodological considerations that we discuss below, CNVs that contain SDs in general exhibit lower rather than higher rates of recombination.
The relationships between G+C content and both CNVR size and frequency highlight an antagonistic relationship between, on the one hand, increased mutation and, on the other, increased purifying selection, within high G+C CNVs that are likely to be enriched in functional elements. The amount of functional sequence within high G+C CNVs is expected to rise as a result of increased duplication rates; however, it will also fall because of purifying selection on deleterious copy number increases. Nevertheless, as recombination tends to occur at an unusually low rate within human CNVs, even deleterious copy number changes are more likely to be fixed here than elsewhere in the genome. This model has previously been invoked to explain regional enrichments of repetitive sequence (Charlesworth et al. 1994). Regions of the human genome that are not observed to be copy number variable, on the other hand, tend to contain a lower G+C content and a higher efficacy of purifying selection on copy number change, in part owing to a higher rate of recombination.
Selection on CNVs
Previously, we presented two lines of evidence that CNVs have been subject to positive selection in the human population (Nguyen et al. 2006). The first of these was that CNVs contain an unusually elevated gene density, but do not exhibit a concomitant increase in G+C content, which would otherwise have represented a confounding factor. Our previous report that CNVs were not G+C-rich sequence might have arisen from some of the earlier studies biasing their detection of structural variation either away from SDs (Lucito et al. 2003) or toward smaller, presumably lower G+C (Fig. 3) variants (Tuzun et al. 2005). In this study we again found a significantly increased gene density in all CNVR sets (Fig. 5A), particularly within frequently observed CNVRs (Fig. 2). However, on this occasion we found that G+C contents were elevated in three of the four CNVRs sets. The high gene densities of some CNVs, therefore, can be explained by their frequent coincidence with high G+C content SD sequence.
We note that Redon et al. (2006) previously reported their CNVR sets to be gene poor, rather than gene rich. However, whereas Redon et al. investigated the number of CNVRs overlapping genes, in our study we considered the converse, namely the number of genes that are overlapped by CNVs. These findings can be reconciled if fewer than expected CNVs overlap genes, yet those CNVs that do overlap contain many more genes than expected (M.E. Hurles, pers. comm.). This is indeed what we observe: CNVs outside of SDs more rarely contain genes, whereas CNVs overlapping SDs frequently contain many. CNVs that do not overlap SDs may be gene poor simply because regions outside of SDs are more likely to harbor genes whose copy numbers are more strongly constrained. This is due in part because of increased recombination outside of SDs, and therefore more efficient selection, but also because mutations involving genes outside of SDs are more likely to be strongly deleterious.
Our previous second line of evidence for positive selection on CNVs was that copy number variable genes exhibit higher than expected protein evolutionary (dN/dS) rates. We argued then that genes that are more likely to have accumulated beneficial amino acid changes during mammalian evolution have also a greater susceptibility to more contemporary events of positive selection on gene duplicates (Nguyen et al. 2006). Here, we observed significantly elevated evolutionary rates for genes located within CNVRs from both Redon et al. data sets (Redon et al. 2006) (see Fig. 4A and Results).
Our previous approach to interpreting these data, however, was not comprehensive. The observed increases in dN/dS values could have arisen not by positive selection, but by nonadaptive processes. These might include reduced purifying selection or increased fixation of deleterious substitutions in regions with low recombination rates (Charlesworth et al. 1995; McVean and Charlesworth 2000; Haddrill et al. 2007). An inverse relationship between recombination rate and dN/dS values was, indeed, observed for SDRs. Conditional on their overlap with SDs, CNVR data that show significantly high recombination rates also exhibited unexpectedly low dN/dS values, and vice versa (Fig. 4B; Supplemental Table 2). A similar doubling of dN/dS rates has also been observed for low- or nonrecombining regions of the Drosophila genome (Haddrill et al. 2007).
It is possible that the measurement of recombination rates across CNV-rich sequence may be less accurate than elsewhere in the genome, due both to a lower density of SNP markers and increased structural variation that would alter physical distances between markers. Nevertheless, recombination rates do remain substantially (−26%) and significantly (P < 2 × 10−3) lower than expected for those SDs that are not seen to be copy number variable within our four data sets. This argues that the lower recombination rates in SDs are not artifacts of the SNP ascertainment scheme.
This reduction of recombination in SDRs would need to have been sustained for the extended periods of time since the last common ancestor of human and dog in order for the accumulation of deleterious substitutions to be apparent in increased dN/dS values. We present evidence in a separate manuscript that, indeed, recombination rates are an ancestral trait of metatherian and eutherian mammals (C. Webber and C.P. Ponting, in prep.).
A model of inefficient purifying selection acting on CNV genes explains the previously reported surfeit of so-called “environmental” genes within CNVRs (Nguyen et al. 2006). Instead of environmental gene copy number changes being of frequent benefit, we argue that they accumulate simply because they are substantially less deleterious than copy number changes in other genes. Even if the duplication of such genes is mildly deleterious, rather than being neutral or beneficial, they may persist in the population because of inefficient purifying selection. A recent study of CNVs within Drosophila melanogaster reported that genes overlapped by CNVs have fewer network interactions, reduced lethality, and increased evolutionary rates (Dopman and Hartl 2007). These findings are also consistent with a model of reduced selective constraint and, despite the very different population dynamics between human and Drosophila, may illustrate common features of copy number variants across diverse species.
Conclusions
We conclude that a model of reduced recombination and reduced purifying selection in G+C-rich and highly duplicating sequence is able to account for the unusual evolutionary properties of most CNVRs. An alternative argument that these properties have arisen because of positive selection on gene copy number (within the human population) and amino acid substitution (over mammalian evolution) is disfavored because it cannot account for the inverse relationship observed between gene evolutionary rates and recombination rates. Strong selection may account for the segregation of CNV alleles in specific instances (Gonzalez et al. 2005; Perry et al. 2007), but our results imply that such examples are exceptions rather than the rule. Our findings indicate that copy number changes are most likely to be deleterious, and thus lead to human disease when involving genes lying outside of the segmentally duplicated portion of the human genome. This should be most evident for unduplicated human genes whose orthologs have previously been observed to elicit a deleterious phenotype when disrupted in mouse. We hope that our findings of ascertainment, mutational, and selective biases will now enable improved discrimination of neutral, deleterious, and beneficial CNVs in the human population.
Methods
Identification of Nijmegen CNVs
A 32-k tiling resolution genomic microarray consisting of 32,447 overlapping BAC clones, selected to cover the entire human genome, was used to generate genomic copy number profiles for 494 samples using methods that have been described previously (de Vries et al. 2005). These samples were originally analyzed by this method within a diagnostic setting with the aim of identifying copy number changes causally related to mental retardation. The samples therefore consisted of patients with unexplained mental retardation (n = 405, of which 102 were run in replicate with dye-reversal) as well as unaffected parents (n = 89, 38 complete trios). All BAC array data have been deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession no. GSE7391; CNV intervals are given in Supplemental Table 1. In all cases, genomic DNA was isolated from uncultured blood leukocytes, thereby excluding culture-induced rearrangements and aneuploidies, as described by Redon et al. (2006).
Hybridization was performed in a two-color experiment against a reference pool containing equal amounts of genomic DNA from 10 healthy blood donors, and genomic copy numbers were estimated using a highly conservative hidden Markov model (Rabiner 1989; de Vries et al. 2005). Other studies such as Redon et al. (2006) and Wong et al. (2007) use single individuals for the reference, thus enabling CNVs to be detected in either subject or reference individuals. However, for experiments that seek to diagnose copy number variation in the subject only, it is preferable to use a reference pool of multiple unrelated individuals; this disfavors the detection of CNVs in the reference, whilst enjoying a low false-positive rate (de Vries et al. 2005). Use of a reference pool also allows for an accurate estimation of the frequency of each CNV as well as the type of CNV (i.e., loss or gain). In a previous study (White et al. 2007), we used multiplex ligation-dependent probe amplification (MLPA) (Schouten et al. 2002; White et al. 2004) and validated the presence and frequency of six recurrent CNVs. This showed an excellent correlation between these different approaches for CNVs identified using our BAC arrays. Because of the use of large-insert clones for the detection of CNVs, we decided to include only CNVs larger than 100 kb and required a CNV to be covered by a minimum of three BAC clones (see also Hehir-Kwa et al. 2007). These CNVs are much larger than virtually all SDs; hence, strong signals arising from cross-hybridization between highly identical sequences (the “shadowing effect”) are unlikely. As with all array-CGH experiments, the sizes of CNVs and overlapping CNV regions we report are upper-bound estimates.
In this study we focus on inherited CNVs without a known clinical relevance. In order to distinguish clinically relevant copy number alterations from normal copy number variations, we performed several additional steps (see also de Vries et al. 2005). First, we analyzed 89 unaffected parents using the same tiling resolution microarrays. These parental samples served as a control population but, in addition, provided valuable information on the inheritance of specific copy number changes. From this first analysis we identified a large set of inherited CNVs; of these, many were detected in multiple unrelated patients as well as unaffected parents. Secondly, all nonrecurrent CNVs were validated both in the patient as well as in the parents by either MLPA using specifically designed synthetic probe sets and/or fluorescence in situ hybridization (FISH). All de novo copy number alterations in the patient were excluded from the list of CNVs used in this study, as were CNVs on the sex chromosomes. For the frequency analysis, parental samples were excluded, leading to inherited CNVs being counted once only per family.
Other CNV and SD data sets
We obtained all 5132 and 25,196 CNVs identified by Wong et al. (2007) and Redon et al. (2006), respectively, and CNVs identified by Redon et al. were considered separately by platform. The 6461 CNVs identified by Redon et al. (2006) on the Affymetrix SNP array platform were herein termed “Redon et al. Affymetrix” CNVs, while the 18,735 CNVs identified using the array-CGH platform were herein termed “Redon et al. aCGH” CNVs.
In their study, Wong et al. (2007) considered for further analysis only those CNVRs that were observed for three or more (out of 95) individuals. For our analyses, however, we considered all Wong et al. CNVs for the reason that their properties are broadly consistent with those of the other three sets (Figs. 2, 3, 4, 5; Supplemental Table 2). The genomic properties of those Wong et al. CNVRs that encompass CNVs from three or more individuals appear much as would be expected for frequent CNVs: They possess proportions of segmentally duplicated bases (19%), G+C content (43%), and protein-coding gene content (+53%) that are significantly higher than expected from genome-wide distributions.
Segmental duplications, as identified using the method described by Bailey et al. (2001), were obtained from the University of California Santa Cruz (UCSC) genome browser (Kent et al. 2002) (http://genome.cse.ucsc.edu, human: hg17, segmental dups track). Segmental duplications and CNVs located either on X or on Y chromosomes were excluded from our analyses in order to remove any evolutionary biases related to sex chromosomes.
Partitioning and merging of the data sets
We wished to examine the regions of the genome that give rise to observed copy number variation or segmental duplication. Thus, within their respective sets, overlapping segments were either merged together if they overlapped by ≥50% or else trimmed equally so as not to overlap in order to produce a set of nonoverlapping regions. The procedure for merging and trimming of overlapping CNVs was applied to the different portions of all four CNVs sets (rare, frequent, gain-and-loss, and [non]overlapping with SDs) and to SDs.
As CNVs often overlap, properties averaged over all CNVRs will differ from those calculated from all CNVs. If independent copy number variations frequently coincide, as they certainly do for gain-and-loss CNVRs, then rarely observed CNVRs will contribute disproportionately to our calculations. Nevertheless, as we find that frequently observed CNVRs are associated with the greatest departures of properties from the genomic average, our approach is conservative, particularly since results (Figs. 1, 2, 3, 4, 5) then represent lower-bound values.
Each of the four CNVR sets was partitioned according to CNV observation frequency, their overlap with segmental duplications, and whether they contain both copy number gains and losses. This resulted in five partitions: (1) CNVRs with ≤1% observed frequency, termed “rare”; (2) CNVRs observed at higher frequencies (>1%), termed “frequent”; (3) CNVRs that overlap with at least one SD, termed “CNVRs with SDs”; (4) CNVRs having no overlap with SDs, termed “CNVRs non-SDs”; and, (5) CNVRs containing both gain CNVs and loss CNVs, termed “gain-and-loss CNVRs.”
Genomic data sets
Simple tandem repeats (from Tandem Repeats Finder; Benson 1999) and genomic sequence were obtained from the University of California Santa Cruz (UCSC) genome browser (Kent et al. 2002), and gene predictions and annotations were assigned to CNVs according to Ensembl (Hubbard et al. 2002) (Ensembl mart version 37). Ensembl genes annotated as OMIM morbid map genes were used to define the set of disease genes (Hamosh et al. 2005).
Recombination rates across the human genome were obtained from Myers et al. (2005) and recast to the NCBI35 assembly using the UCSC NCBI34 → NCBI35 chained alignment (Hinrichs et al. 2006). Rates were calculated as the average recombination rate per base pair. Such rate predictions are expected to be less accurate in repetitive sequence, simply because marker densities are lower, and also in copy number variable sequence, owing to the uncertainty in physical distances separating markers (see Discussion).
Orthologs between human and dog were predicted using PhyOP (Goodstadt and Ponting 2006). We argue that these orthologs will be more reliable than, for example, those between human and mouse, because of their lower degree of divergence. These orthologs were those that have remained unduplicated between the species’ last common ancestor, as well as those arising from lineage-specific gene duplication events. dN and dS values and their ratios were calculated for orthologs using the codeml program from the PAML package (Yang and Nielsen 2000). Ancestral repeats were defined as those RepeatMasker annotated repeats (Jurka 2000) that were aligned between the genomes of dog and human within the UCSC chained alignment, and therefore we infer to have been present in their last common ancestor (Hardison et al. 2003). The substitution rate between those aligned ancestral repeats that extended over 150 bp was calculated using the REV model in the BASEML program from the PAML package (Yang 1997).
Unduplicated 1:1:1:1 human:mouse:dog:opossum orthologs were obtained from the OPTIC database (Heger and Ponting 2008). Similar to PhyOP, OPTIC is an automated orthology assignment procedure that defines phylogenetic relationships on the basis of dS trees calculated through the codeml program from the PAML package (Yang and Nielsen 2000).
Synteny breakpoints between dog and human genomes were defined as the gaps between the 100-kb synteny blocks obtained from the Dog Genome Sequencing Consortium (Lindblad-Toh et al. 2005). The dog genome sequence is a high-quality and high-coverage sequence and is used here in preference to other available genome sequences to be consistent with other evolutionary analyses in this work. As centromeres are always annotated as representing synteny breakpoints, these were excluded from our analyses.
Mouse genome informatics (MGI) phenotype data
Information on human NCBI genes whose mouse orthologs’ disruption had been assayed were obtained from MGI 3.54 (Bult et al. 2008). Two phenotypes, “lethality-embryonic/perinatal” (MP:0005374) and “lethality-postnatal” (MP:0005373), were selected to provide two sets of genes whose disruptions were strongly deleterious. Of 4509 human NCBI genes whose mouse orthologs’ disruption had been assayed, 739 were classed as postnatally lethal, while 1545 were classed as embryonic or perinatally lethal.
Statistical tests
To test the null hypothesis that a property is higher or lower within a set of regions than elsewhere in the genome, we performed a randomization test. For this, 500 sets of regions were sampled randomly from the genome assembly; these regions were matched in both number and size to the set of regions under consideration. We calculated the fraction P of such randomly chosen regions that contained higher or lower values of the property. Values of P > 0.025 were generally considered to indicate that the CNV data were not significantly different from the genome data taken as a whole. The probability that two sets of dN, dS, or dN/dS values sample an equivalent distribution was calculated using the two-sided Kolmogorov-Smirnov test (Feller 1948). Partial correlations were performed using the service provided at http://faculty.vassar.edu/lowry/par.html.
Acknowledgments
We thank Andreas Heger for his advice and guidance with the OPTIC database. We also thank members of the Oxford and Nijmegen research groups for valuable discussions, the UK Medical Research Council (C.W., C.P.P.), the Berrow Lord Florey scholarship (D.Q.N.), and the Swiss National Science Foundation (D.Q.N.) for funding.
Footnotes
[Supplemental material is available online at www.genome.org. The BAC array data from this study have been submitted to Gene Expression Omnibus [GEO] (www.ncbi.nlm.nih.gov/geo) under accession no. GSE7391.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.077289.108.
References
- Aitman T.J., Dong R., Vyse T.J., Norsworthy P.J., Johnson M.D., Smith J., Mangion J., Roberton-Lowe C., Marshall A.J., Petretto E., et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Archibald D.J. Timing and biogeography of the eutherian radiation: Fossils and molecules compared. Mol. Phylogenet. Evol. 2003;28:350–359. doi: 10.1016/s1055-7903(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Armengol L., Marques-Bonet T., Cheung J., Khaja R., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X. Murine segmental duplications are hot spots for chromosome and gene evolution. Genomics. 2005;86:692–700. doi: 10.1016/j.ygeno.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E.2004Hotspots of mammalian chromosomal evolution Genome Biol. 5R23 . http://genomebiology.com/2004/5/4/R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Blake J.A. The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Morgan M.T. The pattern of neutral molecular variation under the background selection model. Genetics. 1995;141:1619–1632. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Nickerson D.A., Eichler E.E. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- de Vries B.B., Pfundt R., Leisink M., Koolen D.A., Vissers L.E., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H., Leeuw N., et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman E.B., Hartl D.L. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Eyre-Walker A., Galtier N. A new perspective on isochore evolution. Gene. 2006;385:71–74. doi: 10.1016/j.gene.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Emes R.D., Goodstadt L., Winter E.E., Ponting C.P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 2003;12:701–709. doi: 10.1093/hmg/ddg078. [DOI] [PubMed] [Google Scholar]
- Erdogan F., Chen W., Kirchhoff M., Kalscheuer V.M., Hultschig C., Muller I., Schulz R., Menzel C., Bryndorf T., Ropers H.H., et al. Impact of low copy repeats on the generation of balanced and unbalanced chromosomal aberrations in mental retardation. Cytogenet. Genome Res. 2006;115:247–253. doi: 10.1159/000095921. [DOI] [PubMed] [Google Scholar]
- Feller W. On the Kolmogorov-Smirnov limit theorums for empirical distributions. Ann. Math. Stat. 1948;19:177–189. [Google Scholar]
- Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Goodstadt L., Ponting C.P. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R., Halligan D.L., Tomaras D., Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R.C., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A., Ponting C.P. OPTIC: Orthologous and paralogous transcripts in clades. Nucleic Acids Res. 2008;36:D267–D270. doi: 10.1093/nar/gkm852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa J.Y., Egmont-Petersen M., Janssen I.M., Smeets D., van Kessel A.G., Veltman J.A. Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: A platform comparison based on statistical power analysis. DNA Res. 2007;14:1–11. doi: 10.1093/dnares/dsm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G., Robertson A. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M. How homologous recombination generates a mutable genome. Hum. Genomics. 2005;2:179–186. doi: 10.1186/1479-7364-2-3-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A., Meunier J., Duret L., Mouchiroud D. GC content evolution of the human and mouse genomes: Insights from the study of processed pseudogenes in regions of different recombination rates. J. Mol. Evol. 2006;62:745–752. doi: 10.1007/s00239-005-0186-0. [DOI] [PubMed] [Google Scholar]
- Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lindsay S.J., Khajavi M., Lupski J.R., Hurles M.E. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J.R., Stankiewicz P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- McVean G.A., Charlesworth B. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Navarro A., Betran E., Barbadilla A., Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.Q., Webber C., Ponting C.P. Bias of selection on human copy-number variants. PLoS Genet. 2006;2:e20. doi: 10.1371/journal.pgen.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R., et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L.R. A tutorial on hidden Markov models and selected applications inspeech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Cheng Z., Eichler E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006a;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006b;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- She X., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shianna K.V., Willard H.F. Human genomics: In search of normality. Nature. 2006;444:428–429. doi: 10.1038/444428a. [DOI] [PubMed] [Google Scholar]
- Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Sleegers K., Brouwers N., Gijselinck I., Theuns J., Goossens D., Wauters J., Del-Favero J., Cruts M., van Duijn C.M., Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Smith N.G., Eyre-Walker A. Human disease genes: Patterns and predictions. Gene. 2003;318:169–175. doi: 10.1016/s0378-1119(03)00772-8. [DOI] [PubMed] [Google Scholar]
- Springer M.S., Murphy W.J., Eizirik E., O'Brien S.J. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Turner D.J., Miretti M., Rajan D., Fiegler H., Carter N.P., Blayney M.L., Beck S., Hurles M.E. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A., et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Webber C., Ponting C.P. Hotspots of mutation and breakage in dog and human chromosomes. Genome Res. 2005;15:1787–1797. doi: 10.1101/gr.3896805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.J., Vink G.R., Kriek M., Wuyts W., Schouten J., Bakker B., Breuning M.H., den Dunnen J.T. Two-color multiplex ligation-dependent probe amplification: Detecting genomic rearrangements in hereditary multiple exostoses. Hum. Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- White S.J., Vissers L.E., Geurts van Kessel A., de Menezes R.X., Kalay E., Lehesjoki A.E., Giordano P.C., van de Vosse E., Breuning M.H., Brunner H.G., et al. Variation of CNV distribution in five different ethnic populations. Cytogenet. Genome Res. 2007;118:19–30. doi: 10.1159/000106437. [DOI] [PubMed] [Google Scholar]
- Winter E.E., Goodstadt L., Ponting C.P. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtenberger M., Hemminki K., Burwinkel B. Identification of frequent chromosome copy-number polymorphisms by use of high-resolution single-nucleotide-polymorphism arrays. Am. J. Hum. Genet. 2006;78:520–522. doi: 10.1086/500793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.K., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lu H.H., Chung W.Y., Yang J., Li W.H. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 2005;22:135–141. doi: 10.1093/molbev/msh262. [DOI] [PubMed] [Google Scholar]
- Zoubak S., Clay O., Bernardi G. The gene distribution of the human genome. Gene. 1996;174:95–102. doi: 10.1016/0378-1119(96)00393-9. [DOI] [PubMed] [Google Scholar]