Abstract
We present an overview of current progress and future directions in the molecular genetics of schizophrenia. We review linkage studies, involving the genome-wide scan of chromosomes with closely spaced polymorphic markers, and association studies of candidate genes, identified on the basis of receptors, neurotransmitters and response to certain drugs. The limitations of the research methodology involved in analyzing such a complex disorder are discussed, as are methods to strengthen this methodology with newer statistical and technological advances to give results that are replicable, statistically significant and applicable to a wider population. A greater understanding of the genetic mechanisms and the application of pharmacogenetics would lead to improvements in therapeutic interventions.
Nous présentons un survol des progrès en cours et des orientations futures en génétique moléculaire de la schizophrénie. Nous passons en revue des études de liaison qui recherchent dans tout le génome des chromosomes comportant des marqueurs polymorphes rapprochés, et des études d'association portant sur des gènes candidats, identifiés en fonction des récepteurs, des neurotransmetteurs et de la réponse à certains médicaments. Nous discutons des limites de la méthodologie de recherche utilisée pour analyser un trouble aussi complexe, ainsi que des moyens de la renforcer au moyen de nouvelles percées statistiques et techniques afin de produire des résultats reproductibles, statistiquement significatifs et applicables à une plus grande population. Une meilleure compréhension des mécanismes génétiques et de l'application de la pharmacogénétique permettrait d'améliorer les interventions thérapeutiques.
Medical subject headings: genetic predisposition to disease; genetics, behavioral; linkage (genetics); linkage disequilibrium; schizophrenia
Introduction
Schizophrenia is a chronic disabling disease that affects about 1% of the world's population. Although the causes of schizophrenia remain unknown, evidence from family, twin and adoption studies clearly demonstrates that it aggregates in families, with the clustering being largely attributable to genetic rather than cultural or environmental factors.1,2,3,4
Initial attempts to study the genetic mechanism underlying schizophrenia took the form of segregation analysis or searching for “biologic markers” (such as receptor proteins or neurophysiologic findings from, for example, electroencephalography) that segregate with the disorder.
The advent of molecular genetics was a turning point in schizophrenia research, when it became possible to use both linkage and association methods to study the DNA markers spanning the genome or the candidate gene polymorphism suspected of being related to the disorder. The search began with reports on chromosome 5 by Sherrington et al,5 though it took almost a decade to reach the pace at which studies are being done today in this field.
Identifying the genes, mode of transmission and responsible chromosomes has proved to be a difficult task in view of the imprecise phenotype, the several phenocopies present and the effect of nongenetic risk factors.
Linkage studies and candidate gene association studies have been used in clinical cohorts collected from a variety of populations, and these studies have given us some hint as to the chromosomes and genes involved. The results from these studies also need careful interpretation regarding their statistical significance and applicability to the whole population.
Nevertheless, with the refinement of phenotypes, the use of endophenotypes, reduction of heterogeneity and extensive genetic mapping, especially after the advent of the Human Genome Project, the chances of discovering mechanisms of inheritance as well the susceptible genes have increased substantially.
In this paper, we review recent and important studies in this field, with particular reference to the various molecular methods used to study the genetic mechanism and their limitations; at the same time, we consider potential improvements that could help us develop a model to predict expression of the disease with more precision.
Linkage studies
Linkage analysis seeks to find chromosomal regions within families that tend to be shared among affected relatives but not among unaffected individuals. Conceptually, linkage analysis takes place in 3 steps:
1. A linkage statistic at each of many DNA markers throughout the genome is calculated.
2. The markers that the linkage statistic shows to be transmitted are identified.
3. More markers in that region are tested to locate the gene more precisely.
The statistics are needed in linkage studies to summarize the evidence available from different studies and to help identify the most likely “allele configuration” in case the DNA sample is not available from 1 of the parents. The statistical theory underlying linkage analysis is fairly complex but can be summarized in 2 critical numbers:
1. a statistic whose magnitude increases with the evidence for linkage, and
2. a number between 0 and 1 that indicates the probability of computing the observed statistic in linkage was absent.
The “affected sibling pair” (ASP) method of linkage relies on the ability to identify “identity by descent,” namely, the alleles that were inherited from the same parent. The probability that all ill relative pairs would inherit the same version of the genetic marker is more than that predicted by Mendel's laws or chance. Similarly, sibling pairs discordant for the disease share alleles at the marker locus more often than predicted by Mendel's laws. This affected pedigree method can be used without knowing the mode of inheritance, which makes it very appealing in the study of psychiatric disorders.
The second popular method is to compute a logarithm of the odds ratio (LOD) score. The main drawback of this method is the need to specify the mode of transmission. However, to get around this, when the data are analyzed several times under different modes of inheritance, the highest LOD score is close to the true mode of inheritance. The LOD score is computed by first calculating the maximum likelihood estimate — the rate of a marker being linked or not linked to a disease locus under a specific assumption. In calculating these scores, the genotype of entire families is pooled rather than that of the ill relatives alone. A LOD score of 3 is considered to be evidence for linkage, but lately it has been found that this rule works well for single gene diseases rather than for complex diseases such as psychiatric disorders. To conclude whether apparent linkage is “real,” the concept of “genome-wide significance” has been developed — the probability threshold that declares linkage after testing many DNA markers used in a genome scan. Lander and Kruglyak6 suggest 3 levels of genome-wide significance: suggestive linkage, significant linkage and confirmed linkage, though it is suggested that confirmed linkage only occurs when the results are replicated in an independent study sample.
Association studies
Linkage analysis has been extremely successful for finding the genetic basis of diseases with well-defined modes of inheritance, with the exception of Alzheimer's disease; it has not yet identified genes for psychiatric disorders, hence certain researchers have turned to association studies.
The “population-based association study” considers specific genes believed for theoretical reasons to be involved in the pathogenesis of a disorder. Such genes are called “candidate genes.” The gene frequencies between patients and controls are compared. A weak inferential link when reasoning from association studies is the fact that a positive association finding for a specific gene could occur if the gene was very close to the true disease gene. These genes are said to be in “linkage disequilibrium,” that is, rarely separated by crossover.
Because of this phenomenon, we do not necessarily need a candidate gene for association studies. Population-based association studies are limited by potential ethnic differences between patients and controls. Therefore, “family-based association studies” are carried out in families that have at least 1 affected offspring, and a transmission test of linkage disequilibrium (TDT) is applied. This test essentially compares the number of times the heterozygous parents transmit the associated marker to affected offspring as compared with the other marker. If these probabilities differ from those expected by chance, then linkage disequilibrium exists, that is, that gene is associated with the disease. With this technique there is perfect ethnic matching, because the transmitted and nontransmitted alleles are from the same parent.
Molecular genetic studies
Linkage studies
After the initial enthusiasm regarding a potential link with chromosome 5 suggested by Bassett,7 subsequent studies have given mixed results. In a genome-wide scan of a nationwide study sample from Finland, the highest LOD scores were found on chromosome arm 5q,8 whereas another study investigating various regions including 5q in 62 pedigrees from Finland found little support for linkage.9 5q has also been implicated in a set of Irish pedigrees and in German and Israeli families.10,11 A recent genome-wide scan suggested that 5q33.2 should be intensively investigated by linkage disequilibrium methods.12 A second region of interest on this chromosome is at the D5S111 locus 5p4.1–p13.1. A significant 2-point LOD score (3.72) and a multipoint score of 4.37 at this locus have been reported in a large Puerto Rico pedigree, using a broad disease definition and dominant inheritance; however, the LOD score has been criticized for not being robust to sensitivity analysis.13 In contrast, studies of 5 multiplex pedigrees from eastern Canada and certain other studies produced evidence against this region.14,15
Another chromosome to be implicated as being associated with schizophrenia has been chromosome 6, because the 6p22–p24 locus emerged as having one of the strongest associations in 265 Irish families using an additive genetic model and an intermediate phenotypic definition, but this evidence substantially declined when either a narrow or a broad disease definition was used.16 Certain other studies have also shown a positive association with certain regions of chromosome 6.17,18,19 A study evaluating 28 genetic markers in 10 moderately large Canadian families using ASP analysis found no evidence for linkage, but the positive symptom scale scores on the Positive and Negative Symptoms Scale for schizophrenia (PANSS) were associated with marker D6S1960, suggesting that the locus might be related to the severity of psychotic symptoms.20 A linkage disequilibrium analysis of 115 ASPs with microsatellite markers also pointed toward a susceptible locus at D6S1960 on 6p.21 Another linkage analysis performed in 186 multiplex families, assuming locus heterogeneity and moderately broad disease definition, revealed a significant LOD score and genome-wide significance of 5%–8%, again supporting a susceptibility locus on chromosome 6 and a model of locus heterogeneity.22 Martinez et al,23 in a follow-up study of a sample that had previously shown suggestive linkage on 6q, found an increase in the LOD score for a 13-cM region after the addition of 43 multiplex pedigrees, and they suggested that a very large sample would be required to narrow down the locus further by ASP linkage methods. There is positive evidence for association with both the long and the short arm of chromosome 6 from genome-wide scans, though there are studies in which linkage could not be replicated; however, interest continues to centre on 6p24–22 and 6q21–22.24,25,26,27,28
Chromosome 6 has several candidate genes, that is, mutations that are hypothesized to predispose individuals to schizophrenia. The spinocerebellar ataxia gene (SCA), LDL-PLA 2, HLA region at 6p21 and, recently, the NOTCH4 gene have been studied; these studies have been primarily negative except for positive linkage disequilibrium between SCA1 CAG repeats and schizophrenia.29,30,31,32,33,34
The implicated region in many studies of chromosome 22 is near the site for the velo-cardio-facial syndrome deletion on 22q, clustering about 4–5 cM around the marker D22S278–9.35 In another study, in 8 Utah multigenerational families, where schizophrenia was phenotyped with P50 auditory evoked potential and ocular motor performance, a genome-wide scan using an autosomal dominant model revealed a significant LOD score (3.55) for marker D22S315, and a nonparametric linkage (NPL) analysis showed evidence for allele sharing over the same broad region (i.e., 3.83).36 There is also evidence from other genome scans, and statistically significant evidence for linkage disequilibrium has been found for polymorphic markers within the catechol-o-methyltransferase (COMT) gene located on this chromosome.37 Several other studies have found negative evidence for linkage in specialized populations such as South African Bantu-speaking families and for allelic association between D22S278 and D22S283 in case–control samples or markers around IL2RB.38,39,40
In a genome-wide scan of 22 extended Canadian families with high rates of schizophrenia, highly significant evidence of linkage to chromosome 1 in the region 1q21–q22, between the markers D1S1653 and D1S1679 under the recessive model of inheritance with parametric analysis, using narrow disease definition, was reported. The power of the study was unusually high, though the authors claim that this was because of the selection of a sample of dense pedigrees.41 Several other linkage studies including genome-wide scans have also implicated regions on chromosome arm 1q especially involving the DISC1 gene,42,12 though a recent report found no linkage at this locus or any heterogeneity in a large multicentre sample.43
Chromosome 15 became a region of interest after initial reports from US schizophrenia pedigrees, the most promising region being 15q13–q15, which is also host to the α-7 nicotinic receptor gene. A significant linkage was found to a physiologic endophenotype (P50 wave of auditory evoked response) of schizophrenia where DI5S1360, containing the gene α-7 nicotinic receptor, was the most positive marker.44 This was further confirmed in the systematic genome scans by the US National Institute of Mental Health (NIMH) Genetics Initiative.45 These scans also found genome-wide significant linkage at 15q14, which maps within 1 cM of the α-7 nicotinic receptor gene.46 Although some studies have failed to confirm linkage at the 15q14 locus, there are significant linkage reports from studies of German, European American, Azorean and Taiwanese families.47,48,49,50
Chromosome 11 includes several possible candidate genes for major psychiatric disorders including tryptophan hydroxylase (TPH), the D4 dopamine receptor gene (DRD4) (both at 11p15) and the D2 dopamine receptor gene (DRD2) at 11q23. Therefore, it has been a site of interest to many molecular geneticists. Interest has been maintained in this region because of a suggestive linkage reported by Maziade et al51 and weak evidence from certain genome-wide scans.52,12
In a genome scan of 43 US nuclear families, using a narrow disease definition, significant evidence for linkage was found at 10p for markers D10S1423 and D10S582 on chromosome 10.53 There is also strong evidence from German, Israeli and Irish pedigrees of a vulnerability locus on 10p for markers other than D10S1423 (D10S674, D10S1714).17,45,54,55
The linkage findings for potentially susceptible genes on chromosome 8 and 13 for schizophrenia are at 8p22–p21 and 13q, respectively, and were first reported in 54 US pedigrees and replicated in 265 multiplex Irish families with a high prevalence of schizophrenia.56,57,58,59,60 In a recent genome scan, strong support of genetic linkage was found for 8p21–p22. Blouin et al58 reported that 13q14–q33 was the only significant region in their genome scan in 54 pedigrees with a maximum NPL score of 4.18 (p < 0.001), satisfying the threshold of a 5% genome-wide significance level.
The hypothesis that a gene in the pseudoautosomal region of the sex chromosomes might confer vulnerability to schizophrenia was based mainly on previous epidemiologic findings of excess anomalous sex chromosomes in psychotic patients, concordance by sex in family studies and sex differences associated with psychosis and underlying brain pathology.61,62,63 The validity of these findings has, however, been questioned, first, because of the only moderate strength of the association in the reverse direction and, second, because of the incomplete and biased ascertainment of ASPs in samples of limited size. Further, molecular genetic studies have yielded no evidence for the association of schizophrenia with the pseudoautosomal region.64,65,66 Even DeLisi et al,67 who initially showed moderate association at Xp11, despite listing a new set of markers and a larger cohort, could not find consistent linkage to the X chromosome and proposed that epigenetic modification rather than variation in the X chromosome should be considered.
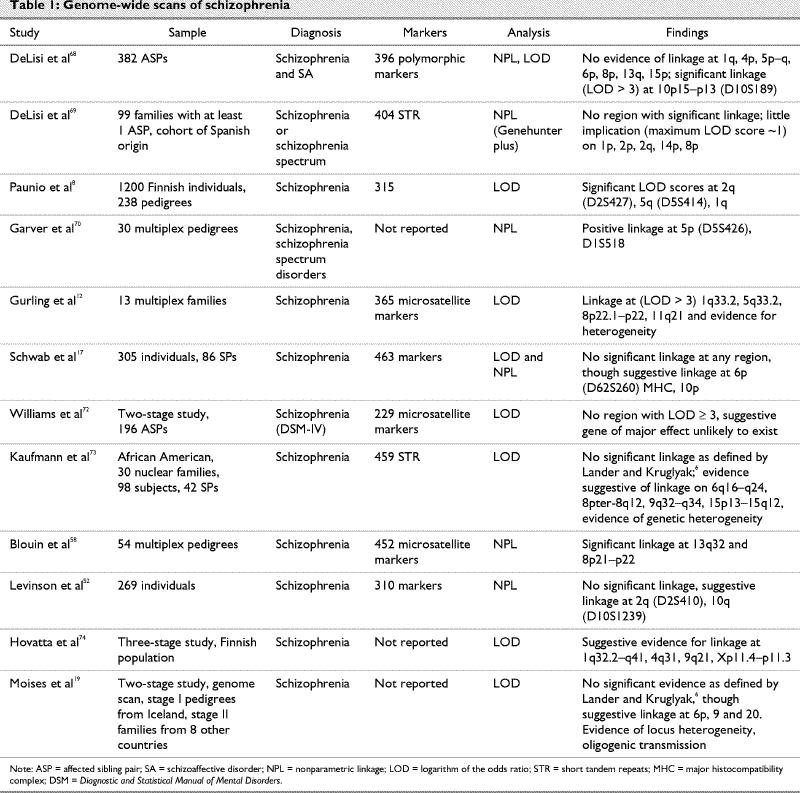
There are several reports of genome-wide scans of linkage studies in schizophrenia.7,12,17,19,28,36,43,45,46,52,58,59,68,69,70,71,72,73,74,75,76 Some of the important features of these scans are listed in Table 1. These reports have again provided statistically significant evidence against a major genetic locus, but weak positive LOD scores were found for as many as 12 chromosomal regions. A meta-analysis of previous samples gave positive results for certain regions, especially on chromosome arms 6p, 6q, 8p and 22q, although drawbacks have been reported in the interpretation of these results.75,76,77
Table 1
Association studies
These studies aim to find the allelic association with a certain disorder. The various mechanisms of this association follow: when a gene is located very close to a true disease gene, for instance at a distance of 1 cM, resulting in linkage disequilibrium, or when a gene has a polymorphism within itself that has functional effect resulting in susceptibility to a disease, or when the phenomenon of population stratification occurs, whereby ethnic differences in allele frequencies contribute to observed differences between affected individuals and controls. Most recent studies focus on the polymorphism in or near candidate genes. These studies have also attempted to do a systematic search through the entire genome for linkage disequilibrium.
The dopamine hypothesis of schizophrenia has been accepted for about 3 decades, based on evidence that antipsychotic drugs are D2 receptor antagonists and their efficacy is correlated with affinity for D2 receptors. Defects of the genes in the dopamine system might play an important role in the origin of schizophrenia. Genes for 5 dopamine receptors in the brain (i.e., D1, D2, D3, D4, D5) have been isolated and cloned.78 The chromosomal locations of the dopamine receptor genes are as follows: D1, 5q35.1; D2, 11q22.5; D3, 3q13.3; D4, 11p15.5; D5, 4p15.2.79
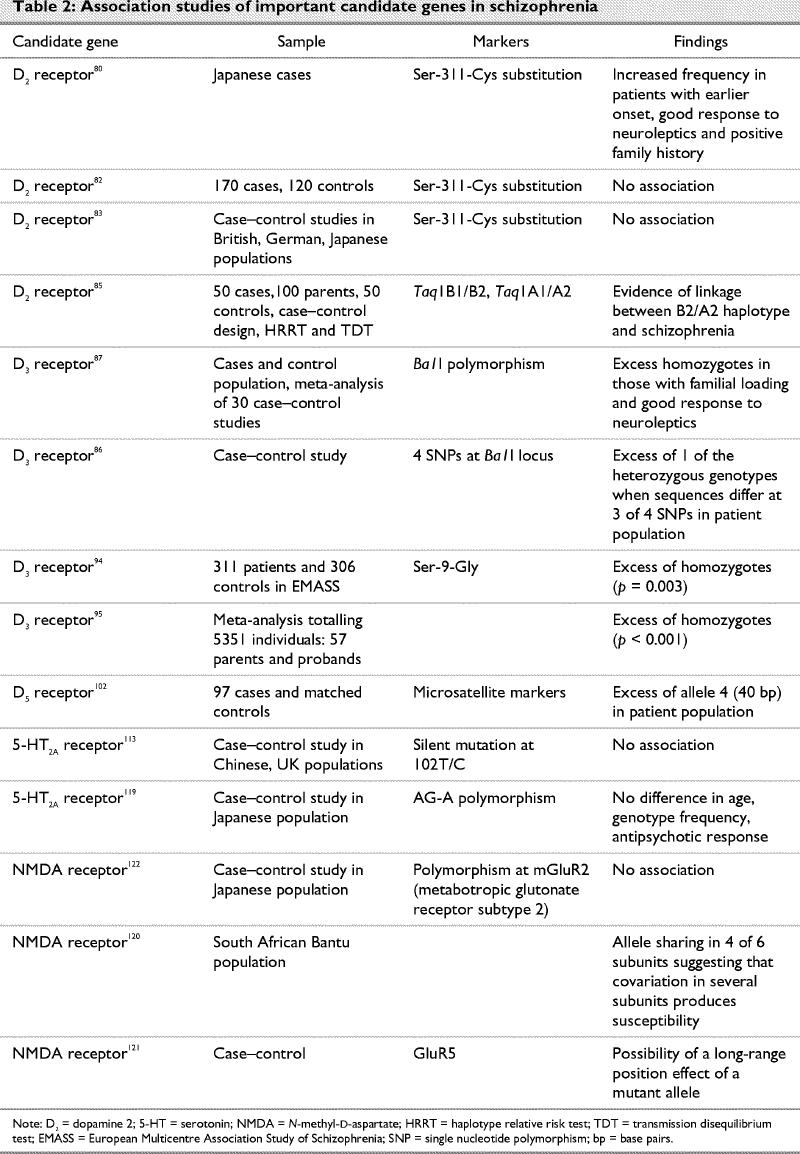
With regard to the genetics of schizophrenia, the crucial question is whether a linkage or association exists between schizophrenia and the genes for dopamine receptors. Since the advent of newer and atypical antipsychotics, several other neurotransmitters and receptors have also been implicated in the pathogenesis of schizophrenia. Some of the association studies related to these are summarized in Table 2.
Table 2
Arinami et al80 found a substitution from serine to cysteine at position 311 of the D2 receptors and reported an increased frequency of the Cys 311 variant in Japanese patients with schizophrenia and earlier onset, good response to medication and a positive family history; this was also reported in another study.81 Conversely, other case–control studies in different populations reported no association.82,83 Arinami et al80 could not replicate their own studies, and several other researchers have found this substitution to be associated to a greater degree with certain clinical subtypes (disorganized schizophrenia) rather than schizophrenia.84 A study using a progressive strategy with 3 different approaches (case–control, haplotype relative risk test and TDT) found evidence for linkage between haplotype and schizophrenia, suggesting the need for further investigation at this receptor.85
The D3 dopamine receptor gene is an important candidate gene for schizophrenia because of its almost exclusive expression in the limbic system. As well, D3 involvement would combine the dopamine receptor system hypothesis with the limbic system hypothesis. Several studies including a meta-analysis of studies of the Bal1 polymorphism in exon 1 of this gene found excess homozygotes in patients with high familial loading and a good response to neuroleptic treatment (Table 2).86,87,88,89,90 There are other studies that have not confirmed this association.91,92,93 However, 2 recent large analyses have provided substantial evidence for an association at the Bal1 site, and it is possible that inconsistent results are the result of a weak association that cannot be detected in every sample.94,95
The D4 receptor may play a major role in schizophrenia given its high density in the frontal cortex and amygdala and its great affinity for an atypical antipsychotic, clozapine.96 However, several groups have failed to find an association between schizophrenia and the D4 dopamine receptor gene.97,98,99 In fact, Serreti et al could not replicate their own finding of association of DRD4 (exon 1 and exon 3) variants with any major psychosis.100
Studies of the D5 dopamine receptor gene have found missense (N351D) and nonsense mutations (C335X). An analysis of combined measures of frontal lobe functions in patients with schizophrenia and controls hints that heterozygotes of C335X might have a vulnerability to mild impairment.101 Certain other studies have reported significant difference (excess) in allele frequency between patients with schizophrenia and controls, though these results were not replicated in an Italian population (Table 2).102,103
Despite its importance as a candidate gene and a 40-base pair polymorphism of a variable number of tandem repeats in the 3' untranslated region of the gene coding for the dopamine transporter (DAT), no linkage has been found between schizophrenia and the different alleles or allele combinations of the dopamine transporter gene.104,105,106
Others have failed to find linkage between schizophrenia and all the dopamine receptor genes.107,108,109,110 Overall, it seems that molecular genetic studies lend only minor support to the dopamine theory of schizophrenia.
Serotonin-2A (5-HT2A) receptors have received much investigative attention in schizophrenia, because several studies have shown a decrease in the concentration of 5-HT2A receptor in the prefrontal cortex, a response to atypical antipsychotics and a positive association between the A and C polymorphism at position 102 of this receptor gene and schizophrenia.111 The various mutations screened in 5-HT receptor variants have been h5-HT1A (Gly-22-Ser, Ile-28-Val, Arg-219-Leu), h5-HT1B (Phe-124-Cys), h5-HT2A (Thr-25-Asn, His-452-Tyr) h5-HT2C (Cys-23-Ser) and h5-HT7 (Thr-92-Lys, Pro-279-Leu).112 The focus of attention has been a silent polymorphism in the 5-HT2A receptor gene, though studies seeking a link with schizophrenia have not yielded positive results consistently (Table 2).111,112,113,114
The serotonin transporter gene is another primary candidate for involvement in major psychosis. A functional polymorphism in its upregulatory region (5HTTLPR) has been recently reported to be associated with a variety of psychopathologic conditions. Serretti et al115 typed patients with schizophrenia for their 5HTTLPR variants and found that the serotonin transporter gene was not a liability factor for symptomatology for schizophrenia; although in another study this region was positively related to individual items on the Brief Psychiatric Rating Scale (BPRS), specifically, intensity of hallucinations, suggesting again a change in presentation according to the allelic variant.116
There is now considerable evidence for glutamatergic system alterations associated with schizophrenia. Reduced expression and regional loss of non-N-methyl-D-aspartate (non-NMDA) glutamate receptors in the temporal lobe of patients with schizophrenia compared with healthy individuals has been reported.117 Other studies also report a disproportionate expression of certain receptor subunits, covariation in several subunits or a long-range position effect to be associated with genetic predisposition to schizophrenia of a mutant allele in the subunit of the NMDA receptor gene.118,119,120,121,122,123
An alteration in γ-aminobutyric acid (GABA) neurotransmission has been indirectly implicated in the pathogenesis of schizophrenia. GABA receptor subunit genes are plausible candidate genes. However, researchers found weak evidence of linkage between this gene and schizophrenia.124 Coon125 found a variant (C–G) in the B1 peptide region of this receptor, which cosegregated in small numbers, and they suggested that this variant might be a predisposing allele in rare instances. In an attempt to study the alteration in markers of GABA neurotransmission, including GABA membrane transporter GAT-1, it was seen that GAT-1 mRNA expression is relatively unaltered in most prefrontal cortex GABA neurons in patients with schizophrenia but is reduced below a detectable level in a subset of GABA neurons.126
The tryptophan hydroxylase gene (TPH), which has been localized to chromosome 11 and codes for the rate-limiting enzymes of serotonin biosynthesis, has also been investigated as a candidate gene along with the COMT gene and dopamine transporter genes (DAT). Studies have reported an association between TPH polymorphism, variation in the region of intron 1 and 7 and negative symptoms of schizophrenia.127,128 COMT inactivates catecholamines, and a common genetic polymorphism in humans is associated with 3- to 4-fold variations in enzyme activity. In a study of 129 Turkish subjects, although no statistically significant difference was found between patients and controls regarding COMT polymorphism, the BPRS scores were associated with COMT polymorphism within the schizophrenia group.129 Associations between schizophrenia and polymorphisms of the 5-HT2A receptor gene, TPH, COMT and DAT were tested in an Indian population, but none were found.130
Recent work has identified 4 novel candidate regions by linkage disequilibrium, namely, neuregulin-1 (chromosome 8p21), G72 (chromosome 13q34), dysbindin (chromosome 6p22.3) and proline dehydrogenase (chromosome 22q11), warranting further investigation. These proteins play an important role in cell-to-cell and intracellular transduction.131,132
Epigenetic mechanisms, defined as nonenvironmental modifications of gene expression, have gained some attention recently. One such mechanism, “anticipation,“ which refers to earlier age of onset and increased severity of disease from parental to offspring generations, has been observed in several neurologic disorders and in schizophrenia.133,134,135 Trinucleotide repeats that cause unstable DNA have been a suggested mechanism for anticipation, and recent studies are searching for trinucleotide repeats in schizophrenia. One such study136 reported correlation between the age of onset of spinocerebellar ataxia and CAG repeats at its gene, which is also seen as a candidate gene for schizophrenia.29 In many other studies, there is evidence both for and against anticipation in schizophrenia.137,138,139,140 Several reports implicate nongenetic factors such as “birth cohort effects” or “adjustment of observation time” (for development of the disorder) rather than anticipation in causing early onset and increased severity of the disorder in offspring.141,142 Imprinting, another epigenetic phenomenon, has been less well studied in schizophrenia. Imprinting refers to the fact that a mutation in the same gene is expressed in different ways depending on whether the defect is transmitted paternally or maternally. It has been examined for the age of onset where no imprinting effect was generally found.143,144,145,146
Discussion
Although most individuals who are first-degree relatives of individuals with schizophrenia do not develop the disease themselves, data from adoption studies are consistent in pointing out that genetic factors rather than intrafamilial culture are responsible for familial aggregation. Twin studies suggest the same conclusion by observing significantly higher concordance rates in monozygotic than in dizygotic twins. It is also evident that it is not schizophrenia per se that is inherited but a susceptibility to it, and environmental factors appear to be necessary for the disease manifestation in many, if not all, cases. Risk factors include obstetric complications, early developmental delays, life events, immigration and drug abuse. But even in this gene–environment model, the heritability is 66%–85%, again emphasizing the high proportion of liability to genetic influence.147
Molecular studies attempt to identify an alteration in genetic expression that causes susceptibility to a disease. The linkage studies reviewed earlier rule out a single major locus but suggest several regions that may be susceptibility loci and warrant replication and investigation in future studies. These regions (especially those identified in the genome-wide scans) include 5p, 5q, 6p, 6q, 8p, 10p, 13q and 15q.
The issue still remains as to whether these linkages are “real” or false positives. Determining an appropriate statistical threshold at which to accept or reject these findings is important, but to expect a “highly significant finding” according to Lander and Kruglyak's criteria,6 as discussed earlier, is realistically difficult in view of the uncertainties in the phenotype and model of transmission. It is, therefore, important that the “highly suggestive” leads are vigorously pursued. Moreover, the model proposed by Lander and Kruglyak assumes a single multipoint analysis of a very dense map, whereas most studies compute multipoint statistical analysis, which is likely to give inflated significance levels and make comparison across various studies difficult. The findings for 13q formally meet the criteria for replicated linkage and the other strongest finding is for chromosome 6p,58,24 but it would be discouraging to assume that the other results generated in different genome scans are all false positive. Suraez et al148 suggest that in a disorder caused by a set of interactive loci of small effect, the regions that produce the most positive results in multiple scans should be considered strong “candidate genes,” despite their nonsignificant p values.
The association methods give good results for a gene of modest effect for which a very large sample size is required by the linkage studies. Although the interpretation of these studies is particularly difficult because of problems of repeat analysis, multicentre analyses still support an association between schizophrenia and polymorphism in loci for the D3 dopamine receptor gene and the 5-HT2A receptor gene.
A number of other limitations have been pointed out that lead to a wide discrepancy in results and failure identify the genes associated with schizophrenia. There is wide variability, from acute psychosis to the schizophrenia spectrum of personality disorders, in what has been included as “cases” in genetic studies. The importance of precise “phenotype selection” is highlighted by reports of overlap of candidate region genes of schizophrenia with those of disorders as distinct as bipolar disorder.149 The current suggestion is that one should classify subjects using affected, unaffected and unknown as categories, which would make the studies statistically more robust.
Considering the multidimensional phenotype of schizophrenia, studies have suggested that multiple symptomatic dimensions such as the different clinical subtypes or biologic endophenotypes may be more closely related to underlying genetic vulnerability. The use of quantitative measures rather than discrete phenotypes may increase the power of a study to detect linkage. The 2 neurobiologic dysfunctions associated with familial schizophrenia: impaired gating of the auditory evoked response (identified by measuring P50 of the auditory evoked potential) and ocular motor dysfunctions (e.g., smooth pursuit eye movement) are the 2 biologic endophenotypes being used for linkage studies.
Another important issue, overlooked in the past, but important in the current context of gene–environment interaction, is ascertainment and sequential extension of pedigrees studied. Initially, pedigrees were ascertained only on the basis of size and number of affected individuals (opportunistic ascertainment) in contrast to “systematic ascertainment.” The assumption then was that the informativeness for linkage was all that mattered, even if there was a nonreplicable sample frame. The most advanced and most demanding designs for combined linkage and association studies initiated to date are those by the NIMH Genetics Initiative on Schizophrenia. This design employs explicit rules for the systematic ascertainment and extension of pedigrees to permit both segregation and linkage analysis, replication, and interpretation of nonlinear genetic and environmental interaction. Future simulation studies are also being carried out to support proper linkage, because present simulation studies have shown that linkage or association may not be detected because of incomplete penetrance, small samples and allele frequencies at disease and marker loci.
The current evidence is more compatible with “multigenic transmission” in schizophrenia. Certain studies postulate that genetic loci vary across different genetic populations, whereas others point out that in the absence of familial clinical subtypes it is quite possible that there are common multigenic mechanisms.150,151
Risch and Teng151 conclude that when considering relative risk of schizophrenia in relatives of affected patients, the “Gottesman and Shields model of multigenic inheritance,” with 3–5 interacting loci, none of which increases the susceptibility by more than 2–3 times, fits best.
Future directions in this area include the following:
· Collection of sufficiently large samples and better methods of pedigree ascertainment to obtain power to detect linkage and then loci to chromosomal regions.
· Application of large-scale family-based association tests across the whole genome.
· Consideration of the interaction between different loci, because we know that linkage and association studies treat these loci independently of each other, and there is a suggestion that these loci might interact to produce other disorders. New methods have been developed that have the power to detect genes of small effect and interaction between loci.
· Potential combination of path, segregation and linkage analysis to increase the power of linkage and association studies to detect genes of small effect.
· Resolution of genetic heterogeneity requires further work on phenotypic classification to identify clinical characteristics that can delineate genetically distinct groups.
· Linkage analysis that is reported, for validity reasons, should be replicable by an independent group of investigators and should hold retain their validity even after sample augmentation (increase in families, subjects).
· Joint application of linkage and association studies as a 2-step strategy. Initial genome-wide scanning using linkage analysis should be followed by high-density association studies.
· Use of state-of-the-art techniques for molecular and quantitative analysis, including nonparametric methods for multipoint linkage, microchip DNA arrays and new genetic markers to identify the source of variance and newer mutations.
Conclusion
Several linkage and association studies have produced positive results, mostly at a suggestive level, for chromosomal regions 1q, 5p, 5q, 6p, 6q, 8p, 10p, 13q, 15q and 22q, but in each case there are reports that were either negative or researchers were unable to replicate the original findings. However, as with other complex diseases, this should not discourage researchers, especially given the novel methods and new statistical tools available. It is apparent that no single locus could cause anything other than a modest effect, though that might be the first to be identified, and that intensive efforts will be required to identify multiple genes of minor effect. At the same time, it is difficult to ignore the possibility that multiple nonlinear interactions between genes and environmental factors might cause such a heterogeneous disorder.
Footnotes
Competing interests: None declared.
Correspondence to: Dr. Neeraj Berry, Department of Psychiatry, All India Institute of Medical Sciences (AIIMS), New Delhi-110029, India; fax 91-11-26588663; berry_neeraj@hotmail.com
Submitted May 17, 2002 Revised May 5, 2003 Accepted May 27, 2003
References
- 1.Kendler KS, McGuire M, Gruenberg AM, Walsh D. An epidemiological, clinical and family study of simple schizophrenia in County Roscommon, Ireland. Am J Psychiatry 1994; 151: 27-34. [DOI] [PubMed]
- 2.Tsuang MT, Gilbertson MW, Faraone SV. The genetics of schizophrenia. Current knowledge and future directions. Schizophr Res 1991;4:157-71. [DOI] [PubMed]
- 3.Kendler KS. Overview: a current perspective on twin studies of schizophrenia. Am J Psychiatry 1983;140:1413-25. [DOI] [PubMed]
- 4.Kety SS. Schizophrenic illness in the families of schizophrenic adoptees: findings from the Danish national sample. Schizophr Bull 1988;14:217-22. [DOI] [PubMed]
- 5.Sherrington R, Brynjolfsson J, Petursson H, Potter M, Dudleston K, Barraclough B, et al. Localization of a susceptibility locus for schizophrenia on chromosome 5. Nature 1988;336:164-7. [DOI] [PubMed]
- 6.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995;11(3):241-7. [DOI] [PubMed]
- 7.Bassett AS. Chromosome 5 and schizophrenia: implications for genetic linkage studies. Schizophr Bull 1989;15:393-402. [DOI] [PubMed]
- 8.Paunio T, Ekelund J, Varilo T, Parker A, Hovatta I, Turunen JA, et al. Genome-wide scan in a nation wide study sample of schizophrenia families in Finland reveals susceptibility loci on chromosomes 2q and 5q. Hum Mol Genet 2001;10:3037-48. [DOI] [PubMed]
- 9.Hovatta I, Lichtermann D, Juvonen, H, Suvissari J, Terwilliger JD, Ara Jarvi R, et al. Linkage analysis of putative schizophrenia gene candidate regions on chromosomes 3p, 5q, 6p, 8p, 20p and 22q in population based sampled Finnish family set. Mol Psychiatry 1998;3:452-7. [DOI] [PubMed]
- 10.Straub RE, MacLean CJ, O'Neill FA, Walsh D, Kendler KS. Support for a possible schizophrenia vulnerability locus in region 5q22-31 in Irish families. Mol Psychiatry 1997;2:148-55. [DOI] [PubMed]
- 11.Schwab SG, Eckstein GN, Hallmayer J, Lerer B, Albus M, Ertl MA, et al. Evidence suggestive of a locus on chromosome 5q31 contributing to the susceptibility to schizophrenia in German and Israeli families by multipoint affected sib-pair linkage analysis. Mol Psychiatry 1997;2:156-60. [DOI] [PubMed]
- 12.Gurling H, Kalsi G, Brynjolfsson J, Sigmundsson T, Sherrington R, Baljinder S, et al. Genome wide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q 32.2, 5q 33.2 and 8p 21-22 and provides support for linkage to schizophrenia, on chromosomes 11q 23.3 – 24 and 20q 12.1 – 11.23. Am J Hum Genet 2001;68:661-73. [DOI] [PMC free article] [PubMed]
- 13.Silverman JM, Greenberg DA, Altstiel LD, Davis KL, Siever IJ, Moks RC, et al. Evidence of a locus for schizophrenia and related disorders on the short arm of chromosome 5 in a large pedigree. Am J Med Genet 1996;67;162-71. [DOI] [PubMed]
- 14.King N, Bassett AS, Honer WG, Masellis M, Kennedy JL. Absence of linkage for schizophrenia on short arm of chromosome 5 in multiplex Canadian families. Am J Med Genet 1997; 74; 472-4. [PMC free article] [PubMed]
- 15.Crowe RR, Vieland W. Report of the chromosome 5 workshop of the Sixth World Congress on Psychiatric Genetics. Am J Med Genet 1999;88:229-32. [PubMed]
- 16.Straub RE, Maclean CJ, O'Neill FA, Burke J, Murphy B, Duke F, et al. A potential vulnerability locus for schizophrenia on chromosome 6p 24-22: evidence for genetic heterogeneity. Nat Genet 1995;11;287-93. [DOI] [PubMed]
- 17.Schwab SG, Hallmayer J, Albus M, Borrmann M, Segman RH, Hanses C. A genome-wide autosomal screen for schizophrenia susceptibility loci in 71 families with affected siblings: support for loci on chromosome 10p and 6. Mol Psychiatry 2000;5:638-49. [DOI] [PubMed]
- 18.Schwab SG, Albus M, Hallmayer J, Honig S, Borrmann M, Lichtermann D, et al. Evaluation of a susceptibility gene for schizophrenia on chromosome 6p by multi point affected sib-pair linkage analysis. Nat Genet 1995;11:325-7. [DOI] [PubMed]
- 19.Moises HW, Yang L, Kristbjarnarson H, Wiese C, Byerley W, Macciardi F, et al. An international two stage genome-wide search for schizophrenia susceptibility genes. Nat Genet 1995; 11: 321-4. [DOI] [PubMed]
- 20.Bruzustowicz LM, Honer WG, Chow EW, Hogan J, Hodgkinson K, Bassett AS. Use of a quantitative trait to map a locus associated with severity of positive symptoms in familial schizophrenia to chromosome 6p. Am J Hum Genet 1997;61:1388-96. [DOI] [PMC free article] [PubMed]
- 21.Deng H, Liu X, Cai G, Terwedow H, Wang Z, Xu X. Linkage disequilibrium study of microsatellite markers on chromosome 6 and schizophrenia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2002;19:6-9. [PubMed]
- 22.Wang S, Sun CE, Walczak CA, Ziegle JS, Kipps BR, Goldin LR. Evidence for a susceptibility locus for schizophrenia on chromosome 6pter p22. Nat Genet 1995;10:41-6. [DOI] [PubMed]
- 23.Martinez M, Goldin LR, Cao Q, Zhang J, Sander AR, Nancarrow DJ, et al. Follow up study on a susceptibility locus for schizophrenia on chromosome 6q. Am J Med Genet 1999;88: 337-43 [PubMed]
- 24.Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U, et al. A schizophrenia susceptibility locus at 6q 25 in one of the world's largest reported pedigrees. Am J Hum Genet 2001;69:96-105. [DOI] [PMC free article] [PubMed]
- 25.Cao Q, Martinez M, Zhang J, Sanders AR, Badner JA, Cravchi A, et al. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics 1997;43:1-8. [DOI] [PubMed]
- 26.Gurling H, Kalsi G, Chen A, Green M, Butler R, Read T, et al. Schizophrenia susceptibility and chromosome 6p 24-22. Nat Genet 1995;11:234-5. [DOI] [PubMed]
- 27.Mowry BJ, Nancarrow DJ, Lennon DP, Crow RR, Sandkuijl LA, Silverman JM, et al. Schizophrenia susceptibility and chromosome 6p24-22. Nat Genet 1995;11;233-4. [DOI] [PubMed]
- 28.Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, et al. A genome wide search for schizophrenia susceptibility genes. Am J Med Genet 1998;81:364-76. [DOI] [PubMed]
- 29.Joo EJ, Cannon TD, Lee JH, Price CA. Possible association between schizophrenia and a CAG repeat polymorphism in the spinocerebellar ataxia type 1 (SCA1) gene on human chromosome 6p23. Psychiatr Genet 1999;9:7-11. [DOI] [PubMed]
- 30.Pujana MA, Martorell L, Volpini V, Valero J, Labad A, Viella E, et al. Analysis of amino acid and nucleotide variants in spinocerebellar ataxia type 1 [SCAI] gene in schizophrenic patients. Hum Genet 1997;99:772-5. [DOI] [PubMed]
- 31.Bell R, Collier DA, Kerwin RW, Gloger IS, Rice SQ, Roberts GW, et al. Systematic screening of the LDL-PLA2 gene for polymorphic variants and case control analysis in schizophrenia. Biochem Biophys Res Commun 1997;241:630-5 [DOI] [PubMed]
- 32.Sklar P, Schwab SG, Williams NM, Daly M, Maier W, Albus M, et al. Association analysis of NOTCH4 loci in schizophrenia using family and population based controls. Nat Genet 2001; 28:126-8. [DOI] [PubMed]
- 33.Blackwood DH, Muir WJ, Stephenson A, Wentzel J, Clair DM, Robets DF, et al. Reduced expression of HLA-B35 in schizophrenia. Psychiatr Genet 1996;6:51-9. [DOI] [PubMed]
- 34.Fan JB, Tang JX, Gu NF, Feng GY, Zou FG, Xing YL, et al. A family based and case-control association study of the NOTCH4 gene and schizophrenia. Mol Psychiatry 2002;7:100-3. [DOI] [PubMed]
- 35.Murphy KC. Schizophrenia and velo-cardio-facial syndrome. Lancet 2002;359:426-30. [DOI] [PubMed]
- 36.Myles-Worsley M, Coon H, McDowell J, Brenner C, Hoff M, Lind B, et al. Linkage of a composite inhibitory phenotype to a chromosome 22q locus in eight Utah families. Am J Med Genet 1999; 88:544-50. [PubMed]
- 37.Kunugi H, Vallada HP, Sham PC, Hoda F, Arrang MJ, Li T, et al. Catechol-O-methyl transferase polymorphisms and schizophrenia: a transmission disequilibrium study in multiply affected families. Psychiatr Genet 1997;7:97-101. [DOI] [PubMed]
- 38.Parsian A, Suarez BK, Isenberg K, Hampe CL, Fisher L, Chakraverty S, et al. No evidence of schizophrenia susceptibility gene in the vicinity of IL2RB on chromosome 22. Am J Med Genet 1997;74:361-4. [DOI] [PubMed]
- 39.Williams NM, Jones LA, Murphy KC, Cardno AG, Asherson P, Williams J, et al. No evidence for an allelic association between schizophrenia and markers D22 S278 and D22 S283. Am J Med Genet 1997;74:37-9. [DOI] [PubMed]
- 40.Riley B, Mogudi-Carter M, Jenkins T, Williamson R. No evidence for linkage of chromosome 22 markers to schizophrenia in southern African Bantu-speaking families. Am J Med Genet 1996;67:515-22. [DOI] [PubMed]
- 41.Brzustowicz LM, Hodgkinson KA, Chow EWC, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-22. Science 2000;288:678-82. [DOI] [PMC free article] [PubMed]
- 42.Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, et al. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet 2001;10:1611-7. [DOI] [PubMed]
- 43.Levinson DF, Holmans PA, Laurent C, Riley B, Pulver AF, Gejman PV, et al. No major schizophrenia locus detected on chromosome 1q in a large multicentre sample. Science 2002; 296: 739-41. [DOI] [PubMed]
- 44.Freedman R, Leonard S, Gault J. Linkage disequilibrium for schizophrenia at the chromosome 15q13-14 locus of the alpha- 7-nicotinic acetylcholine receptor subunit gene [CHRNA 7]. Am J Med Genet 2001;105:20-2. [PubMed]
- 45.Leonard S, Gault J, Moore T, Hopkins J, Robinson M, Oliney A, et al. Further investigation of a chromosome 15 locus in schizophrenia: analysis of affected sibpairs from the NIMH Genetics Initiative. Am J Med Genet 1998;81:308-12. [DOI] [PubMed]
- 46.Freedman R, Leonard S, Olincy A, Kaufmann CA, Malaspina D, Cloninger CR, et al. Evidence for the multigenic inheritance of schizophrenia. Am J Med Genet 2001;105:794-800. [DOI] [PubMed]
- 47.Gejman PV, Sanders AR, Cao Q, Zhang J. Linkage analysis of schizophrenia to chromosome 15. Am J Med Genet 2001; 105: 789-93. [DOI] [PubMed]
- 48.Tsuang DW, Skol A, Faraone SV, Bingham S, Young KA, Prabhudesai S, et al. Genetic linkage of the CHRNA7 region on chromosome 15 to schizophrenia in a large Veterans Affairs cooperative study sample. Am J Med Genet 2001;105:662-8. [PubMed]
- 49.Xu J, Pato MT, Torre CD, Medeiros H, Carvalho C, Basile VS, et al. Evidence for linkage disequilibrium between the alpha 7-nicotinic receptor gene [CHRNA7] locus and schizophrenia in Azorean families. Am J Med Genet 2001;105:669-74. [DOI] [PubMed]
- 50.Liu CM, Hwu HG, Lin MW, Ou-Yang WC, Lee SF, Fann CS, et al. Suggestive evidence for linkage of schizophrenia to markers at chromosome 15q13-14 in Taiwanese families. Am J Med Genet 2001;105:658-61. [DOI] [PubMed]
- 51.Maziade M, Raymond V, Cliche D, Fournier JR, Caron C, Garneau Y, et al. Linkage results on 11q 21-22 in Eastern Quebec pedigrees densely affected by schizophrenia. Am J Med Genet 1995;60:522-8. [DOI] [PubMed]
- 52.Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Mowry BJ, et al. Genome scan of schizophrenia. Am J Psychiatry 1998;155;741-50. [DOI] [PubMed]
- 53.Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suraez B, et al. Genome scan of European-American Schizophrenic pedigrees: results of the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 1998;81:290-5. [PubMed]
- 54.Schwab SG, Hallmayer J, Albus M, Lerer B, Hanses C, Kanyas K, et al. Further evidence for a susceptibility locus on chromosome 10p14-11 in 72 families with schizophrenia by nonparametric linkage analysis. Am J Med Genet 1998;81:302-7. [PubMed]
- 55.Straub RE, MacLean CJ, Martin RB, Ma Y, Myakishe MV, Harris-Kerr C, et al. A schizophrenia locus may be located in region 10p15-p11. Am J Med Genet 1998;81:296-301. [DOI] [PubMed]
- 56.Bruzustowicz LM, Honer WG, Chow EWC, Little D, Hogan J, Bassett AS. Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 1999;65:1096-103. [DOI] [PMC free article] [PubMed]
- 57.Kendler KS, MacLean CJ, O'Neill FA, Burke J, Murphy B, Duke F, et al. Evidence of a schizophrenia vulnerability locus on chromosome 8p in the Irish Study of High density Schizophrenia Families. Am J Psychiatry 1996;153:1534-40. [DOI] [PubMed]
- 58.Blouin JC, Dombroski BA, Nath SK, Lasseter VK, Neufeld K, Pulver AE, et al. Schizophrenia susceptibility loci on chromosomes 13q 22 and 8p21. Nat Genet 1998;20:70-3. [DOI] [PubMed]
- 59.Faraone SV, Skol AD, Tsuang DW, Bingham S, Young KA, Prabhudesai S, et al. Linkage of chromosome 13q32 to schizophrenia in a large veteran affairs cooperative study sample. Am J Med Genet 2002;114:598-604. [DOI] [PubMed]
- 60.Lin MW, Curtis D, Williams N, Arranz M, Nanko S, Collier D, et al. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13q14.1-q32. Psychiatr Genet 1995;5: 117-26. [DOI] [PubMed]
- 61.Sturt E, Shur E. Sex concordance for schizophrenia in proband-relative pairs. Br J Psychiatry 1985;147:44-7. [DOI] [PubMed]
- 62.Crow TJ. Sex chromosome and psychosis: the case for a pseudoautosomal locus. Br J Psychiatry 1998;153:675-83. [DOI] [PubMed]
- 63.Delisi LE, Devoto M, Lofthouse R, Poulter M, Smith A, Shields G, et al. Search for linkage to schizophrenia on the X and Y chromosomes. Am J Med Genet 1994;54:113-21. [DOI] [PubMed]
- 64.Kalsi G, Curtis D, Brynjolfsson J, Butler R, Sharma T, Murphy P, et al. Investigation by linkage analysis of the XY pseudoautosomal region in the genetic susceptibility to schizophrenia. Br J Psychiatry 1995;167:390-3. [DOI] [PubMed]
- 65.Lichtermann D, Hovatta I, Terwilliger JD, Peltonen L, Lonnqvist J. Concordance for sex and the pseudoautosomal gene hypothesis revisited: no evidence of increased sex concordance in a nationwide Finnish sample of siblings with paternally derived schizophrenia. Am J Psychiatry 1998;155:1365-75. [DOI] [PubMed]
- 66.Kalsi G, Gamble D, Curtis D, Butler R, Read T, Murphy P, et al. No evidence for linkage of schizophrenia to DXS7 at chromosome Xp 11. Psychiatr Genet 1999;9:197-9. [DOI] [PubMed]
- 67.DeLisi LE, Shaw S, Sherington R, Nanthaa Kumar B, Sheilds G, Smith AB, et al. Failure to establish linkage on the X chromosome in 301 families with schizophrenia or schizoaffective disorder. Am J Med Genet 2000;96:335-41. [DOI] [PubMed]
- 68.DeLisi LE, Shaw SH, Crow TJ, Shields G, Smith AB, Larach VW, et al. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am J Psychiatry 2002;159:803-12. [DOI] [PubMed]
- 69.DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LalPrade B, Llach M, et al. Genome-wide scan for linkage to schizophrenia in a Spanish origin cohort from Costa Rica. Am J Med Genet 2002;114:497-508. [DOI] [PubMed]
- 70.Garver Dl, Holocomb J, Mapua FM, Wilson R, Barnes B. Schizophrenia spectrum disorders: an autosomal-wide scan in multiplex pedigrees. Schizophr Res 2001;52:145-60. [DOI] [PubMed]
- 71.Badner JA, Gershon ES. Meta-analysis of whole genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 2002;7(4):405-11 [DOI] [PubMed]
- 72.Williams NM, Rees MI, Holmans P, Norton N, Cardno A, Jones LA, et al. A two-stage genome scan for schizophrenia susceptibility genes in 196 affected sibling pairs. Hum Mol Genet 1999;8:1729-39. [DOI] [PubMed]
- 73.Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, et al. NIMH Genetics Initiative Millennium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet 1998;81:282-9. [PubMed]
- 74.Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajarvi R, et al. A genome wide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 1999;65:1114-24. [DOI] [PMC free article] [PubMed]
- 75.Straub RE, MacLean CJ, O'Neill FA, Walsh D, Kendler KS. Genome scans for schizophrenia genes: a detailed progress report in an Irish cohort. Am J Med Genet 1997;74:558.
- 76.Paunio T, Ekelund J, Hovatta I, Varilo T, Terwilliger JD, Meyer J, et al. Genome wide scan of an extended Finnish schizophrenia study sample. Am J Med Genet 2000;96:460.
- 77.Rice JP. The role of meta-analysis in linkage studies of complex traits. Am J Med Genet 1997;74:112-4. [DOI] [PubMed]
- 78.Seeman P, Van Tol HH. Dopamine receptor pharmacology. Trends Pharmacol Sci 1994;15:264-70. [DOI] [PubMed]
- 79.Hartman DS, Civelli O. Molecular attributes of dopamine receptors: new potential for antipsychotic drug development. Ann Med 1996;28:211-9. [DOI] [PubMed]
- 80.Arinami T, Itokawa M, Enguchi H, Tagaya H, Yano S, Shimizu H, et al. Association of dopamine D2 receptor molecular variant with schizophrenia. Lancet 1994;343:703-4. [DOI] [PubMed]
- 81.Ohara K, Nagai M, Tani K, Nakamura Y, Ino A, Ohara K. Functional polymorphism of 141 Ins/Del in the dopamine D2 receptor gene promoter and schizophrenia. Psychiatry Res 1998; 81:117-23. [DOI] [PubMed]
- 82.Kaneshima M, Higa T, Nakamoto H, Nagamine M. An association study between the Cys 311 variant of dopamine D2 receptor gene and schizophrenia in the Okinawan population. Psychiatry Clin Neurosci 1997;51;379-81. [DOI] [PubMed]
- 83.Tallerico T, Ulpian C, Liu IS. Dopamine D2 receptor promoter polymorphism: no association with schizophrenia. Psychiatry Res 1999;85:215-9. [DOI] [PubMed]
- 84.Serretti A, Lilli R, Lorenzi C, Smeraldi E. Further evidence supporting the association between the dopamine receptor D2 Ser / Cys 311 variant and disorganized symptomatology of schizophrenia. Schizophr Res 2000;43:161-2. [PubMed]
- 85.Dubertret C, Gorwood P, Gouya L, Deybach JC, Ades J. Association and excess of transmission of a DRD2 haplotype in a sample of French schizophrenia patients. Schizophr Res 2001; 49: 203-12. [DOI] [PubMed]
- 86.Crocq MA, Mant R, Asherson P, William J, Hode Y, Mayerova A, et al. Association between schizophrenia and homozygosity at the dopamine D3 receptor gene. J Med Genet 1992;29:858-60. [DOI] [PMC free article] [PubMed]
- 87.Mant R, Williams J, Asherson P, Parfitt E, Mcguaffin P, Owen MJ. Relationship between homozygosity at the dopamine D3 receptor gene and schizophrenia. Am J Med Genet 1994;54:21-6. [DOI] [PubMed]
- 88.Kennedy JL, Billett EA, Macciardi FM, Verga M, Parsons TJ, Meltzer HY, et al. Association study of dopamine D3 receptor gene and schizophrenia. Am J Med Genet 1995;60:558-62. [DOI] [PubMed]
- 89.Williams J, Spurlock G, Holmans P, Mant R, Murphy K, Jones L, et al. A meta-analysis and transmission disequilibrium study of association between the dopamine D3 receptor gene and schizophrenia. Mol Psychiatry 1998;3:458. [DOI] [PubMed]
- 90.Sivagnanasundaram S, Morris AG, Gaitonde EJ, McKenna PJ, Mollon JD, Hunt DM. A cluster of single nucleotide polymorphisms in the 5'-leader of the human dopamine D3 receptor gene [DRD3] and its relationship to schizophrenia. Neurosci Lett 2000;279:13-6. [DOI] [PubMed]
- 91.Johnson E, Lanfelt L, Sokoloff P, Schwartz JC, Sedvall G. Lack of association between schizophrenia and alleles in the dopamine D3 receptor gene. Acta Psychiatr Scand 1993;87:345-9. [DOI] [PubMed]
- 92.Inada T, Sugita T, Dobashi I, Inagaki A, Kitao Y, Matsuda G, et al. Dopamine D3 receptor gene polymorphism and the psychiatric symptoms seen in first break schizophrenic patients. Psychiatr Genet 1995;5:113-6. [DOI] [PubMed]
- 93.Hawi Z, McCabe U, Straub RE, O'Neill A, Kendler KS, Walsh D, et al. Examination of new and reported data of the DRD3/MSCI polymorphism: no support for the proposed association with schizophrenia. Mol Psychiatry 1998;3:150-5. [DOI] [PubMed]
- 94.Spurlock G, Williams J, McGuffin P, Aschauer HN, Lenzinger E, Fuchs K, et al. European Multicentre Association study of schizophrenia : a study of the DRD2 and Ser311Cys and DRD3 Ser9Gly polymorphisms. Am J Med Genet 1998;81(1):24-8. [DOI] [PubMed]
- 95.Williams J, Spurlock G, Holmans P, Mant R, Murphy K, Jones L, et al. A meta-analysis and transmission disequilibrium study of association between the dopamine D3 receptor gene and schizophrenia. Mol Psychiatry 1998;3(2):141-9. [DOI] [PubMed]
- 96.Van Tol HH, Bunzow JR, Guan HC, Seeman P, Niznik HB, Civelli O. Cloning of the gene for a human dopamine D4 receptor with high affinity for the antipsychotic clozapine. Nature 1991;350:610-4. [DOI] [PubMed]
- 97.Maier W, Schwab S, Hallmayer J, Ertl MA, Minges J, Ackenhil M, et al. Absence of linkage between schizophrenia and the dopamine D4 receptor gene. Psychiatry Res 1994;53:77-86. [DOI] [PubMed]
- 98.Shaikh S, Gill M, Owen M, Asherson P, McGuffin P, Nanko S, et al. Failure to find linkage between a functional polymorphism in the dopamine D4 receptor gene and schizophrenia. Am J Med Genet 1994;54:8-11. [DOI] [PubMed]
- 99.Jonsson EG, Ivo R, Forslund K, Mattila-Evenden M, Rylander G, Cichon S, et al. No association between a promoter dopamine D4 receptor gene variant and schizophrenia. Am J Med Genet 2001;105:525-8. [DOI] [PubMed]
- 100.Serretti A, Lilli R, Di Bella D, Bertelli S, Nobile M, Nouelli E, et al. Dopamine receptor D4 gene is not associated with major psychosis. Am J Med Genet 1999;88:486-91. [PubMed]
- 101.Sobell JL, Lind TJ, Sigurdson DC, Somner SS, Zald DH, Snitz BE, et al. The D5 dopamine receptor gene in schizophrenia: identification of a nonsense change and multiple missense changes but lack of association with disease. Hum Mol Genet 1995; 4:507-14. [DOI] [PubMed]
- 102.Williams NM, Cardino AG, Murphy KC, Jones LA, Asherson P, McGuffin P, et al. Association between schizophrenia and a microsatellite polymorphism at the dopamine D5 receptor gene. Psychiatr Genet 1997;7:83-5. [DOI] [PubMed]
- 103.Muir WJ, Thomson ML, McKeon P, Mynett-Johnson L, Whitton C, Evans KL, et al. Markers close to the dopamine D5 receptor gene [DRD5] show significant association with schizophrenia but not bipolar disorder. Am J Med Genet 2001; 105:152-8. [DOI] [PubMed]
- 104.Li T, Yang L, Wiese C, Xu CT, Zeng Z, Giros B, et al. No association between alleles or genotypes at the dopamine transporter gene and schizophrenia. Psychiatry Res 1994;52:17-23. [DOI] [PubMed]
- 105.Martinez D, Gelernter J, Ali-Dargham A, van Dyck CH, Kegeles L, Inmis RB, et al. The variable number of tandem repeats polymorphism of the dopamine transporter gene is not associated with significant change in dopamine transporter phenotype in humans. Neuropsychopharmacology 2001;24:553-60. [DOI] [PubMed]
- 106.Byerley W, Coon H, Hoff M, Holik J, Waldo M, Freedman R, et al. Human dopamine transporter gene not linked to schizophrenia in multigenerational pedigrees. Hum Hered 1993; 43 : 319-22. [DOI] [PubMed]
- 107.Coon H, Byerley W, Holik J, Hoff M, Myles-Worsley M, Lannfelt L, et al. Linkage analysis of schizophrenia with five dopamine receptor genes in nine pedigrees. Am J Hum Genet 1993; 52:327-34. [PMC free article] [PubMed]
- 108.Serretti A, Lattuada E, Cusin C, Lilli R, Lorenzi C. Dopamine D3 receptor gene not associated with symptomatology of major psychosis. Am J Med Genet 1999;88:476-80. [PubMed]
- 109.Moises HW, Gelernter J, Giuffra LA, Zarcone V, Wetterberg L, Civelli O, et al. No linkage between D2 dopamine receptor gene region and schizophrenia. Arch Gen Psychiatry 1991;48: 643-7. [DOI] [PubMed]
- 110.Su Y, Burke J, O'Neill FA, Murphy B, Nie L, Kipps B, et al. Exclusion of linkage between schizophrenia and D2 dopamine receptor gene region of chromosome 11 q in 112 Irish multiplex families. Arch Gen Psychiatry 1993;50:205-11. [DOI] [PubMed]
- 111.Ohara K, Nagai M, Tani K, Tsukamoto T, Ohara K. Schizophrenia and the serotonin 2A receptor promoter polymorphism. Psychiatry Res 1999;85:221-4. [DOI] [PubMed]
- 112.Gothert M, Propping P, Bonisch H, Bruss M, Nothen MM. Genetic variation in human 5-HT receptors: potential pathogenetic and pharmacological role. Ann N Y Acad Sci 1998; 861: 26-30. [DOI] [PubMed]
- 113.He L, Li T, Melville C, Liu S, Fang GY, Collier D, et al. 102 T/C polymorphism of serotonin receptor type 2A gene is not associated with schizophrenia in either Chinese or British populations. Am J Med Genet 1999;88:95-8. [PubMed]
- 114.Chen CH, Lee YR, Wei FC, Koong FJ, Hure HG, Hsiao KJ. Lack of allelic association between 102 T/C polymorphism of serotonin receptor type 2A gene and schizophrenia in Chinese. Psychiatr Genet 1997;7:35-8. [DOI] [PubMed]
- 115.Serretti A, Lilli R, Lorenzi C, Lattuada E, Cusin C, Smeraldi E. Serotonin transporter gene [5HTTLPR] and major psychosis. Mol Psychiatry 2002;7:95-9. [DOI] [PubMed]
- 116.Malhotra AK, Goldman D, Mazzanti C, Clifton A, Brier A, Pickar D. A functional serotonin transporter [5-HTT] polymorphism is associated with psychosis in neuroleptic free schizophrenics. Mol Psychiatry 1998;3:328-32. [DOI] [PubMed]
- 117.Eastwood SL, McDonald B, Burnet PW, Beckwith JP, Kerwin RW, Harrison PJ. Decreased expression of mRNAs encoding non-NMDA glutamate receptors GluR1 and GluR2 in medial temporal lobe neurons in schizophrenia. Brain Res Mol Brain Res 1995;29:211-23. [DOI] [PubMed]
- 118.Dracheva S, Marras SA, Elhakem SL, Kramer FR, Davis KL, Haroutunian V. N-methyl-D-aspartic acid receptor expression in the dorsolateral prefrontal cortex of elderly patients with schizophrenia. Am J Psychiatry 2001;158:1400-10. [DOI] [PubMed]
- 119.Ohnuma T, Augood SJ, Emson PC, Arai H, Mckenna PJ. Expression of the human excitatory amino acid transporter 2 and metabotropic glutamate receptors 3 and 5 in the pre-frontal cortex from normal individuals and patients with schizophrenia. Brain Res Mol Brain Res 1998;56;207-17. [DOI] [PubMed]
- 120.Riley BP, Tahil E, Rajagopalan S, Mogudi-Carter M, Jenkins T, Williamson R, et al. A linkage study of the N-methyl D aspartate receptor subunit gene loci and schizophrenia in Southern African Bantu Speaking families. Psychiatr Genet 1997;7:57-74. [DOI] [PubMed]
- 121.Devson RS, Porteus DJ. Physical mapping of glutamate receptor gene in relation to a balanced translocation associated with schizophrenia in a large Scottish family. Psychiatr Genet 1997; 7: 165-9. [PubMed]
- 122.Joo A, Shibata H, Ninomiya H, Kawasaki H, Tashiro N, Fukumaki Y. Structure and polymorphism of the human metabotropic glutamate receptor type 2 gene [mGluR2]: analysis of association with schizophrenia. Mol Psychiatry 2001; 6:186-92. [DOI] [PubMed]
- 123.Semple CA, Devson RS, Le Hellard S, Porteous DJ. Identification of genes from schizophrenia–linked translocation breakpoint region. Genomics 2001;73:123-6. [DOI] [PubMed]
- 124.Papadimitriou G, Dikeos D, Daskalopoulou E, Kardima G, Avramopoulos D, Contis C, et al. Association between GABA-A receptor alpha 5 subunit gene locus and schizophrenia of a later age of onset. Neuropsychobiology 2001;43:141-4. [DOI] [PubMed]
- 125.Coon H, Sobell J, Heston L, Sommer S, Hoff M, Holik J, et al. Search for mutations in the beta 1 GABAA receptor subunit gene in patients with schizophrenia. Am J Med Genet 1994;54:12-20. [DOI] [PubMed]
- 126.Volk D, Austin M, Pierri J, Sampson A, Lewis D. GABA transporter-1 mRNA in the prefrontal cortex in schizophrenia: decreased expression in a subset of neurons. Am J Psychiatry 2001; 158:256-65. [DOI] [PubMed]
- 127.Shinkai T, Ohmori O, Suzuki T, Kojima H, Hori H, Terao T, et al. Polymorphisms of tryptophan hydroxylase case gene and the symptomatology of schizophrenia: an association study. Psychiatr Genet 2000;10:165-71. [DOI] [PubMed]
- 128.Hong CJ, Tsai SJ, Wang YC. Association between tryptophan hydroxylase gene polymorphism [A218C] and schizophrenic disorders. Schizophr Res 2001;49:59-63. [DOI] [PubMed]
- 129.Herken H, Erdal ME. Catechol-O-methyltransferase gene polymorphism in schizophrenia: evidence for association between symptomatology and prognosis. Psychiatr Genet 2001; 11: 105-9. [DOI] [PubMed]
- 130.Semwal P, Prasad S, Bhatia T, Deshpande SN, Wood J, Nimgaonkar VL, et al. Family based association studies of monoaminergic gene polymorphism among North Indians with schizophrenia. Mol Psychiatry 2001;6:220-4. [DOI] [PubMed]
- 131.McGuffin P, Tandon K, Curisco A. Linkage and association studies of schizophrenia. Curr Psychiatry Rep 2003;5(2):121-7 [DOI] [PubMed]
- 132.Straub RE, Jiang Y, Maclean CJ, Ma Y, Webb BT, Myakishev MV, et al. Genetic variation in the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 2002;71(2):337-48. [DOI] [PMC free article] [PubMed]
- 133.Petronis A, Paterson AD, Kennedy JL. Schizophrenia: an epigenetic puzzle? Schizophr Bull 1999;25:639-55. [DOI] [PubMed]
- 134.Heiden A, Willinger U, Scharfetter J, Maszaros K, Kasper S, Aschauer HN. Anticipation in schizophrenia. Schizophr Res 1999; 35:25-32. [DOI] [PubMed]
- 135.McInnis MG, Mc Mohan FJ, Crow T, Ross CA, De Lisi LE. Anticipation in schizophrenia: a review and reconsideration. Am J Med Genet 1999;88:686-93. [DOI] [PubMed]
- 136.Joo EJ, Lee JH, Canon TD, Price RA. Possible association between schizophrenia and a CAG repeat polymorphism in the spinocerebellar ataxia type 1 [SCA 1] gene on human chromosome 6p23. Psychiatr Genet 1999;9:7-11. [DOI] [PubMed]
- 137.Valero J, Martorell L, Marine J, Vilella E, Labad A. Anticipation and imprinting in Spanish families with schizophrenia. Acta Psychiatr Scand 1998;97:343-50. [DOI] [PubMed]
- 138.Vincent JB, Kalsi G, Klempan T, Tatuch Y, Sherrington RP, McInnis MG, et al. No evidence of expansion CAG or GAA repeats in schizophrenia families and monozygotic twins. Hum Genet 1998;103;41-7. [DOI] [PubMed]
- 139.Ohara K, Ikeuchi T, Suzuki Y, Ohtani M, Ohara K, Tsuji S. A CAG trinucleotide repeat expansion and familial schizophrenia. Psychiatry Res 2000;94:257-62. [DOI] [PubMed]
- 140.Joober R, Benkelfat C, Brisebois K, Toulouse A, Ronald GA, Lal S, et al. Lack of association between the hSkCa3 channel gene CAG polymorphism and schizophrenia. Am J Med Genet 1999; 88:154-7. [PubMed]
- 141.DiMaggio C, Martinez M, Menard JF, Petit M, Thibaut F. Evidence of a cohort effect for age at onset of schizophrenia. Am J Psychiatry 2001;158:489-92. [DOI] [PubMed]
- 142.Picco MF, Goodman S, Reed J, Bayless TM. Methodologic pitfalls in the determination of genetic anticipation: the case of Crohn's disease. Ann Intern Med 2001;134:1124-9. [DOI] [PubMed]
- 143.Husted J, Scutt LE, Basset AS. Parental transmission and anticipation in schizophrenia. Am J Med Genet 1998;81:156-62. [PMC free article] [PubMed]
- 144.Ohara K, Xu HD, Mori N, Suzuki Y, Xu DS, Wang ZC. Anticipation and imprinting in schizophrenia. Biol Psychiatry 1997; 42: 760-6. [DOI] [PubMed]
- 145.Imamura A, Honela S, Nakane Y, Okazaki Y. Anticipation in Japanese families with schizophrenia. J Hum Genet 1998;43: 217-23. [DOI] [PubMed]
- 146.Asherson P, Walsh C, Williams J, Sargeant M, Taylor C, Clements A, et al. Imprinting and anticipation. Are they relevant to genetic studies of schizophrenia? Br J Psychiatry 1994; 164: 619-24. [DOI] [PubMed]
- 147.Murray MR, Castle DJ. Aetiology: genetic and environmental risk factors for schizophrenia. In: Gelder GM, Lopez-Ibor JJ, Andreason N, editors. New Oxford Textbook of Psychiatry. 1st ed. Oxford: Oxford University Press; 2000. p. 603.
- 148.Suraez B, Hampe CL, Van Eedewlgh P. Problems replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR, editors. New genetic approaches to mental disorders. Washington: American Psychiatric Press; 1995.
- 149.Nurnberger JI Jr, Foroud T. Genetics of bipolar disorder. Curr PsychiatryRep 2000;2:147-57. [DOI] [PubMed]
- 150.Weiss KM. Genetic variation and human disease: principles and evolutionary approaches. Cambridge: Cambridge University Press; 1993.
- 151.Risch N, Teng JR. The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases I. DNA pooling. Genome Res 1998; 8(12):1273-88. [DOI] [PubMed]


