Abstract
βγ-Crystallins belong to a superfamily of proteins in prokaryotes and eukaryotes that are based on duplications of a characteristic, highly conserved Greek Key motif. Most members of the superfamily in vertebrates are structural proteins of the eye lens that contain four motifs arranged as two structural domains. Absent in melanoma-1 (AIM1), an unusual member of the superfamily whose expression is associated with suppression of malignancy in melanoma, contains 12 βγ-crystallin motifs in six domains. Some of these motifs diverge considerably from the canonical motif sequence. AIM1g1, the first βγ-crystallin domain of AIM1, is the most variant of βγ-crystallin domains currently known. In order to understand the limits of sequence variation on the structure, we report the crystal structure of AIM1g1 at 1.9Å resolution. In spite of having changes in key residues, the domain retains the overall βγ-crystallin fold. The domain also contains an unusual extended surface loop that significantly alters the shape of the domain and its charge profile. This structure illustrates the resilience of the βγ fold to considerable sequence changes and its remarkable ability to adapt for novel functions.
Keywords: βγ-crystallin, Greek key motif, AIM1g1, Tyr-corner, Trp-corner, Multiple isomorphous replacement using Anomalous scattering (MIRAS)
β- and γ-crystallins are major components of the vertebrate eye lens.1 They share a common all-β fold and form part of a wider βγ-crystallin superfamily.2 They accumulate at very high concentrations in the lens where they contribute to the refractive properties of this transparent tissue. They are highly stable and are among a few proteins having the longest life in the body. Related proteins have been identified in both eukaryotes and prokaryotes and some of these are involved in stress responses or in changes in cell morphology.3
The β- and γ-crystallins are two-domain proteins. Each domain consists of an 8-stranded β-sandwich made up of two variant Greek key motifs with each motif swapping its third strand with the opposite motif thereby rendering the domain more stable.1,4,5 A key feature of the fold is a characteristic β-hairpin motif, which forms a loop between the first two strands of each motif and has a highly conserved signature sequence -Y/FXXXXY/FXG- which interacts with other conserved residues, particularly a serine at the beginning of the fourth strand in each motif.6 This structure is seen in members of the superfamily from Protein S of Myxococcus xanthus to lens crystallins.6, 7 While similar strand topologies have been observed in Williopsis mrakii yeast killer toxin (WmKT) and Streptomyces killer toxin like protein (SKLP),8,9 these proteins lack the conserved motifs and are stabilized by extensive disulfide bridge formation. They are most likely products of convergent evolution with no ancestral link to the βγ-crystallins.10
Reflecting a history of tandem gene duplication, the four motifs of β- and γ-crystallins follow an ABAB pattern with each B type motif containing a conserved Tyrosine corner, a structure believed to play the role of a folding nucleus.11–13 Similar motifs are seen in the microbial Protein S, but arranged in a BABA pattern suggesting that lineages of vertebrate and microbial members of the superfamily may have followed independent evolutionary paths of gene duplication and fusion after diverging from single motif ancestors.12 Other members of the superfamily in micro-organisms, spherulin 3a, Vibrio crystallin and Yersinia crystallin, have motifs that retain the hairpin structure but lack Tyr-corners.12,14 Members of the superfamily also have conserved tryptophan residues that form Trp-corners and make major contributions to the packing of the hydrophobic core of the βγ-crystallin domain.
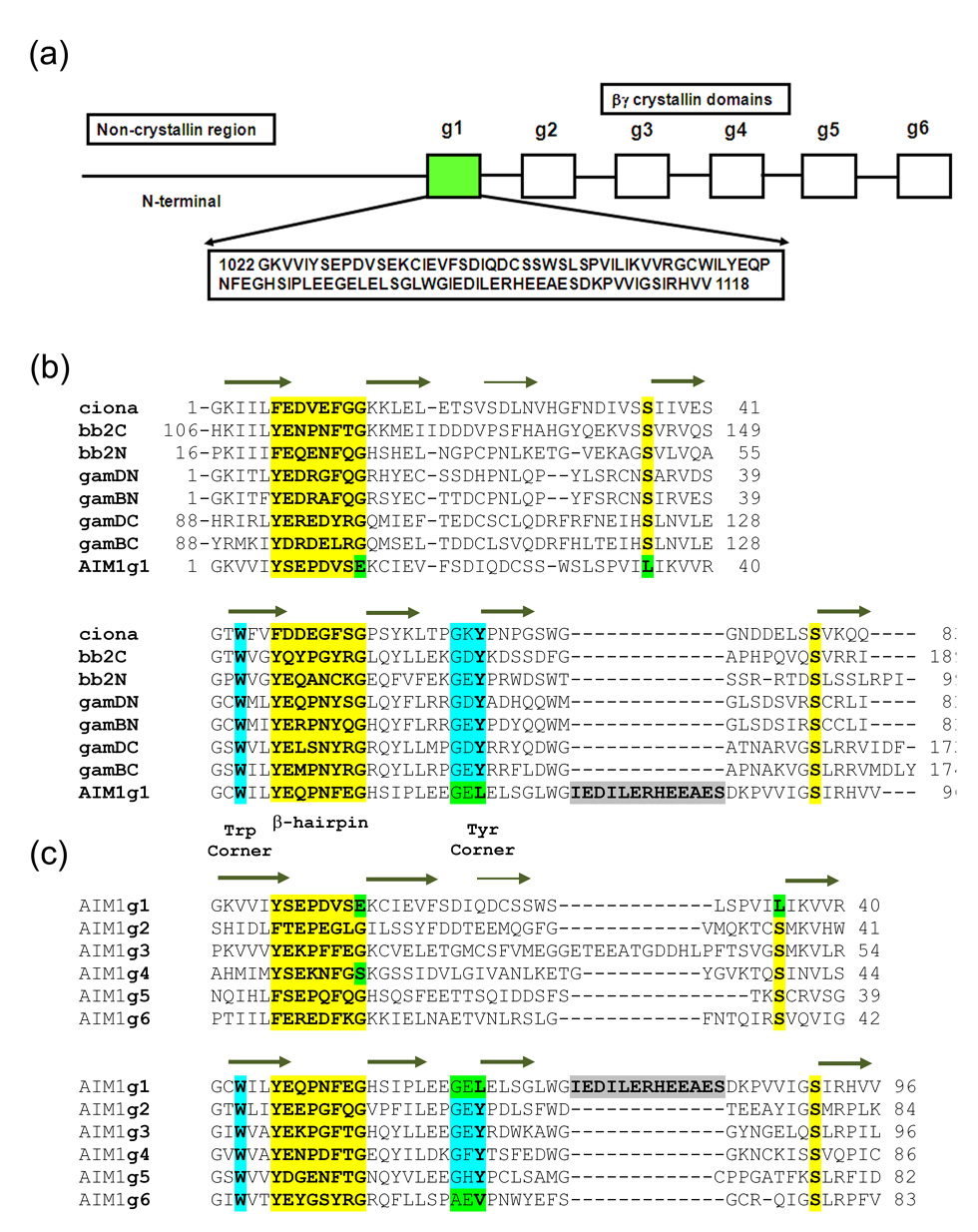
Absent in melanoma 1 (AIM1) was discovered as a transcript that was absent in malignant melanoma cells but was highly expressed in cells in which malignancy was suppressed.15 It is a large protein of over 1700 residues with a poorly defined, possibly filament-like, N-terminal region and a C-terminal Ricin-type beta-trefoil domain. In between is a region containing twelve βγ-crystallin motifs, predicted to form six β-crystallin-like domains.15 Among the six domains of AIM1, the first domain (referred to as AIM1g1) is the most divergent among vertebrate βγ-crystallins.16 In the first motif, it lacks the -YXXXXFXG- signature, replacing the final glycine that is critical for formation of a bend with peptide backbone dihedral angles disallowed for residues with R-groups, with a glutamate (Glu13). In addition, a highly conserved serine is non-conservatively replaced by Leu35. The second motif is more conventional with a typical hairpin motif and a conserved serine but lacks the tyrosine corner. It also has a ten-residue insertion in the loop between third and fourth strands, much longer than in other βγ-crystallin-like domains characterized till date (Fig. 1). The other βγ-crystallin domains of AIM1 (g2–g6) also have some variations and are related amongst themselves and with known βγ-crystallin domains with 24–30% sequence identity. Some of them possess variations like the absence of tyrosine corner (domain g6), presence of an insertion in the domain loop (domain g3) and loss of conserved glycine in the hairpin motif (domain g4), but AIM1g1 is the only domain in which all these variations are collectively incorporated. Such divergence from the template that is so well conserved elsewhere may reflect a specific functional adaptation in this domain of AIM1.15
Figure 1.
(a) βγ-Crystallin domain organization in AIM1 with sequence of AIM1g1. (b) Multiple sequence alignment between AIM1g1 and other vertebrate and urochordate lens βγ-crystallins are shown. g1–g6 represents the six crystallin domains of AIM1. (c) Second panel shows alignment of other βγ-crystallin domains of AIM1 protein. An insertion similar to the 10-residue insertion present in motif B of AIM1g1 is present in the A-motif of the AIM1g3. The first and the last domains lack a Tyr-corner. All crucial structural features of the domains are explicitly identified. Mutations at highly invariant residues are marked in green.
The sequence of AIM1g1 is an extreme evolutionary test of the limits of sequence divergence in the βγ-crystallin fold. To determine how the structure is affected by these multiple changes and to classify whether AIM1g1 is indeed a divergent member of βγ-crystallin fold, we solved the crystal structure of AIM1g1 by multiple isomorphous replacement combined with anomalous scattering. The structure shows remarkable conservation of the β-sandwich while the insertion in the second motif produces a large electronegative loop stabilized by a network of water molecules and lattice interactions. In addition to being an extreme variant, this domain structure is the first to be solved for a non-lens vertebrate βγ-crystallin superfamily member.
Domain structure
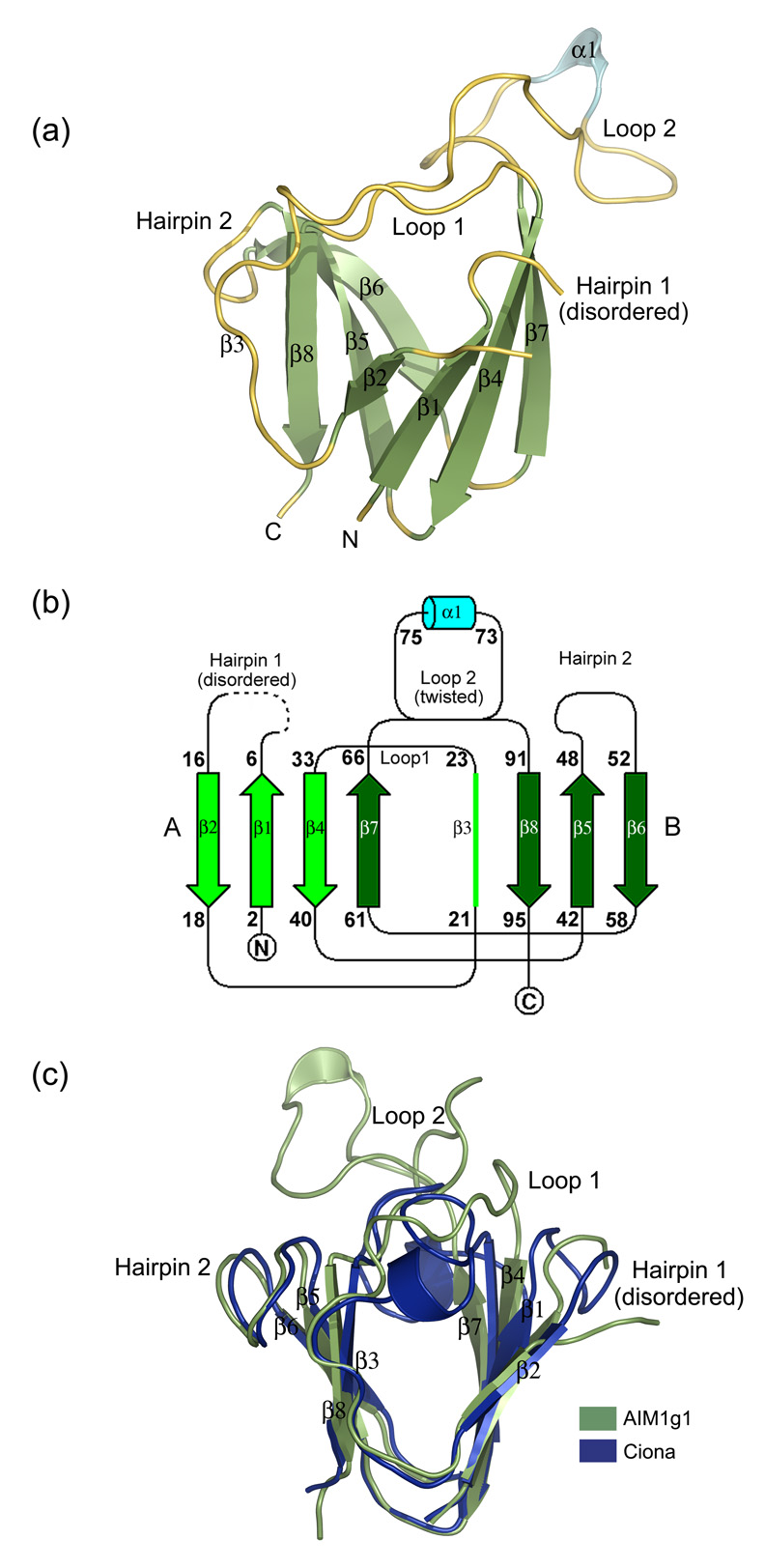
The first βγ-crystallin domain of AIM1 extends from residues 1022 to 1118 (Fig. 1). The sequence details of the domain and its spectral features are described elsewhere.15,16 In the electron density map, 93 residues out of 96 residues could be modeled except residues 10 to 12 and the structure was refined to an Rfactor of 18.14 % and Rfree of 23.2 % (Table 1). Overall, the crystal structure shows a β-sandwich arrangement and a Greek key topology that is similar to other βγ-crystallin domains despite a very low sequence identity and the absence of several key conserved features (Fig. 2a and b).
Table 1.
| Parameter | Native | Iodide (NaI) | Cadmium (CdCl2) |
|---|---|---|---|
| Experimental Details | |||
| X-ray source | CuKα | CuKα | CuKα |
| Heavy atom compound | Sodium iodide | Cadmium Chloride | |
| Soak conc. | 75 mM | 50 mM | |
| Soak duration | 50–70 min | 60–70 min | |
| Crystallographic Data | |||
| Space group | P65 | P65 | P65 |
| Cell Dimensions (Å; °) |
a=b=55.0Å,c=59.6Å α=β=90°; γ=120° |
a=b=55.2Å,c=59.7Å α=β=90°; γ=120° |
a=b=55.0Å,c=59.5Å α=β=90°; γ=120° |
| Unit cell volume (Å3) | 155874 | 157654 | 156204 |
| Resolution range (Å)a | 25.0-1.88 (1.95-1.88) | 25.0-1.9 (1.97-1.9) | 25.0-2.5 (25.0-2.5) |
| Observations | 38569 | 69358 | 13916 |
| Unique reflections | 8382 (810) | 8240 (811) | 3545 (313) |
| Redundancy | 4.6 (3.2) | 8.4 (7.7) | 3.9 (2.6) |
| Completion (%) | 99.6 (96.8) | 99.9 (99.8) | 98.7 (91.8) |
| I/σ(I) | 13.8 (1.9) | 20.4 (4.4) | 7.07 (1.78) |
| Rsym (%)b | 9.7 (44.4) | 9.3 (35.0) | 13.7 (39.0) |
| Molecules/A.U | 1 | 1 | 1 |
| Refinement | |||
| RMSD bonds (Å) | 0.005 | ||
| RMSD angles (°) | 1.27 | ||
| Rcryst (%)c | 18.14 | ||
| Rfree (%) | 23.2 | ||
| No. of residues | 93 | ||
| No. of atoms | 859 | ||
| Protein | 740 | ||
| Water | 113 | ||
| Glycerol | 6 | ||
| Ramachandran plot: | |||
| Residues in: | |||
| Most favored region | 71 (90%) | ||
| Addtnl allowed regions | 6 (7.6%) | ||
| Generously allowed regions | 1(1.3%) | ||
| Disallowed regions | 1 (1.3%) | ||
| Average B-factors (Å2): | |||
| Protein (overall) | 20.38 | ||
| Solvent atoms | 33.15 | ||
| Glycerol atoms | 26.4 | ||
Values in parentheses are for the highest resolution shell
Rsym = ∑|I(h)−<I(h)>|/∑I(h), where I (h) is the observed intensity and <I(h)> is the mean intensity of reflection h over all measurements of I(h).
Rcryst and Rfree = ∑h‖F(h)obs|−|F(h)calc‖/∑h|F(h)obs|.
Throughout refinement, 5% of the total reflections were held aside for Rfree. Refinement was carried out in repeated cycles using the CNS package. Stereochemical quality was assessed using Procheck.
Figure 2.
(a) 3D-structure of the AIM1g1 domain. The secondary structural elements have been marked. The third strand (β3) is marked in spite its loop like appearance for greater clarity as it deviates only partly from the strand architecture and is a conserved feature among all vertebrate βγ-crystallins. (b) Topology of AIM1g1 is depicted which shows the presence of the twin Greek key motifs (A & B) with the large twisted loop 2 connecting β7 and β8. (c) Overlap of γB crystallin and βB2 crystallin N-terminal domains with AIM1g1. Grey indicates the backbone of AIM1g1, cyan represents γB crystallin and magenta represents βB2-crystallin domain. The first hairpin of AIM1g1 is splayed due to disorder. Loop 2 clearly shows completely different orientation as compared to the lens βγ-crystallin domains. Extent of overlap is as per values given in Table 2 for structural relatedness which was analyzed by DaliLite program of EBI.31 The purified protein was crystallized using a 2 µl + 2 µl mixture of protein and a reservoir solution containing 1.3–1.4 M Na citrate, 0.1 M HEPES, pH 7.3–7.5 with non-polar solvents like isopropanol (1–3%), DMSO (2%) and salts like ammonium sulphate (0.06 M), sodium chloride (0.4 M) as additives.32 Different heavy atom compounds like thiomersal, cadmium chloride and sodium iodide were initially attempted by soaking crystals in high concentrations (10–50 mM) of these solutions and observing the crystals for morphological changes. Both time of soak and concentration of the soak were altered to get well diffracting isomorphous derivatives. All derivatized crystals were transferred to 15% glycerol solution containing the heavy atom, before being flash frozen at 100K. Data were collected over large sectors to collect the anomalous signal for the heavy atom especially for iodide derivative, as Iodine is known to have high anomalous signal when CuKα X-rays are used.33 Good derivatives and native data were then scaled together and phasing was performed using the program SOLVE. Solve picked up six sites, three each for Iodine derivative and Cadmium derivative, and yielded a Mean figure of merit (<m>) of 0.51 up to a resolution of 2.1 Å with good electron density maps depicting clear β-strands in the map built without density modification. Density modification and automated model building was done by Resolve which built up to ~80% of the main chain with ~60 % of the side chains. Manual model building was performed using the program ‘O’.
Comparison with other βγ-crystallin domain structures
When superimposed on other βγ-crystallin domains, AIM1g1 domain shows close overlap in the β-sheet regions. However, it has distinct differences from the other structures (Fig. 2c). While all other vertebrate domains are related with RMS deviations in the range of 0.6 to 1.5 Å, AIM1g1 is more divergent and shows RMS deviation of 2.0 Å to 2.5 Å for 70–75 Cα residues with conventional βγ-crystallin domains (Table 2). In particular, the β1 and β2 strands are partly splayed apart due to lack of formation of a regular β-hairpin loop while the insertion in the second motif leads to formation of a large loop between strands β7 and β8. The orientation of this loop has very little similarity among the other loops in similar position found in these domains (Fig. 2c).
Table 2.
Vertebrate crystallins: Structural relatedness*
| Rmsd (Å) | Ciona (2bv2) | γD-N (1hk0) | γD-C (1hk0) | γB-N (4gcr) | γB-C (4gcr) | βB2-N (1ytq) | βB2-C (1ytq) | AIM1g1 (3cw3) |
|---|---|---|---|---|---|---|---|---|
| (%) Identity | ||||||||
| Ciona | 0 | 1.1/81 Cα | 1.0/82 Cα | 1.1/81Cα | 1.0/82Cα | 1.1/79Cα | 1.0/82Cα | 2.0/71Cα |
| γD-N | 31% | 0 | 0.9/82 Cα | 0.5/83 Cα | 1.1/82 Cα | 1.2/81 Cα | 0.9/82 Cα | 2.3/72Cα |
| γD-C | 24% | 37% | 0 | 1.6/85 Cα | 0.6/87 Cα | 1.3/83 Cα | 1.5/86 Cα | 2.5/73Cα |
| γB-N | 32% | 83% | 36% | 0 | 1.8/85Cα | 1.2/81Cα | 1.1/85 Cα | 2.2/73Cα |
| γB-C | 23% | 37% | 72% | 17% | 0 | 1.2/82Cα | 1.7/87Cα | 2.4/73Cα |
| βB2-N | 32% | 35% | 29% | 37% | 30% | 0 | 1.5/82Cα | 2.6/75Cα |
| βB2-C | 37% | 40% | 37% | 35% | 40% | 35% | 0 | 2.5/73Cα |
| AIM1g1 | 23% | 31% | 25% | 34% | 26% | 21% | 23% | 0 |
All information obtained by using pair-wise structural alignments of DaliLite.
Cysteines
The domain has a total of three cysteines (Cys15, 25 and 42), which like all other vertebrate βγ-crystallin domains, remain free and are not involved in formation of any disulfide linkages although intra-chain disulfides can form upon oxidation of some γ-crystallins.17,18 The first two cysteines (Cys15 and 25) are in close proximity to aromatic groups Tyr6 and Trp28 respectively. Proximities between polarizable groups are implicated in providing stability to domains 19 and similar interactions were found in the structure of γ-crystallin.5 Interaction between Cys15 and Tyr6 would attain significance in providing local stability in motif A of AIM1g1, where the hairpin is disordered due to mutations in crucial residues as described above. However, these free cysteines do not contribute to the secondary structure and fold stabilization by forming disulfide bridges as seen in the cases of SKLP, WmKT and Streptomyces Metalloproteinase Inhibitor (SMPI). 8,9,20
Hairpin Disorder
The first hairpin motif, usually a well defined structure in this fold, is disordered and three residues (from 10 to 12) could not be modeled in the region due to lack of electron density (Fig. 2a). This could be the result of the replacement of two key residues. The tight bend glycine residue is replaced by glutamate, which cannot adopt the characteristic backbone conformation of the hairpin. The fourth strand serine, which in other examples of the fold, forms side chain H-bonds with backbone thus stabilizing the hairpin, is replaced by Leu35. The second motif of the domain forms a conventional hairpin loop with conserved glycine and serine residues and is well defined in the electron density map.
Lack of Tyr corner
The domain lacks a tyrosine corner, a common feature of β-sandwich domains,13 which is known to be well conserved in B-type motifs of βγ-crystallins.5,12 In this feature, a tyrosine makes a H-bond with the main chain carbonyl four residues ahead (Fig. 3a), and may act as a nucleus during the initial folding of the Greek key motifs. The presence of tyrosine not only shields the domain core from hydrophobic exposure but also renders the domain flank hydrophilic due to its -OH group. AIM1g1 has Leu63 in place of Tyr in that position. This may create a hydrophobic interaction patch. Interestingly, AIM1g1 and AIM1g6 (the sixth crystallin domain of AIM1) both lack this tyrosine corner (Fig. 1) and one model of possible domain arrangements in AIM1 suggests an interaction between these two domains.15 Although the tyrosine corner is missing, AIMg1 folds into a typical β-sandwich, showing that in vertebrates, as in some microbial members of the superfamily, this feature is not as essential as it was believed to be.
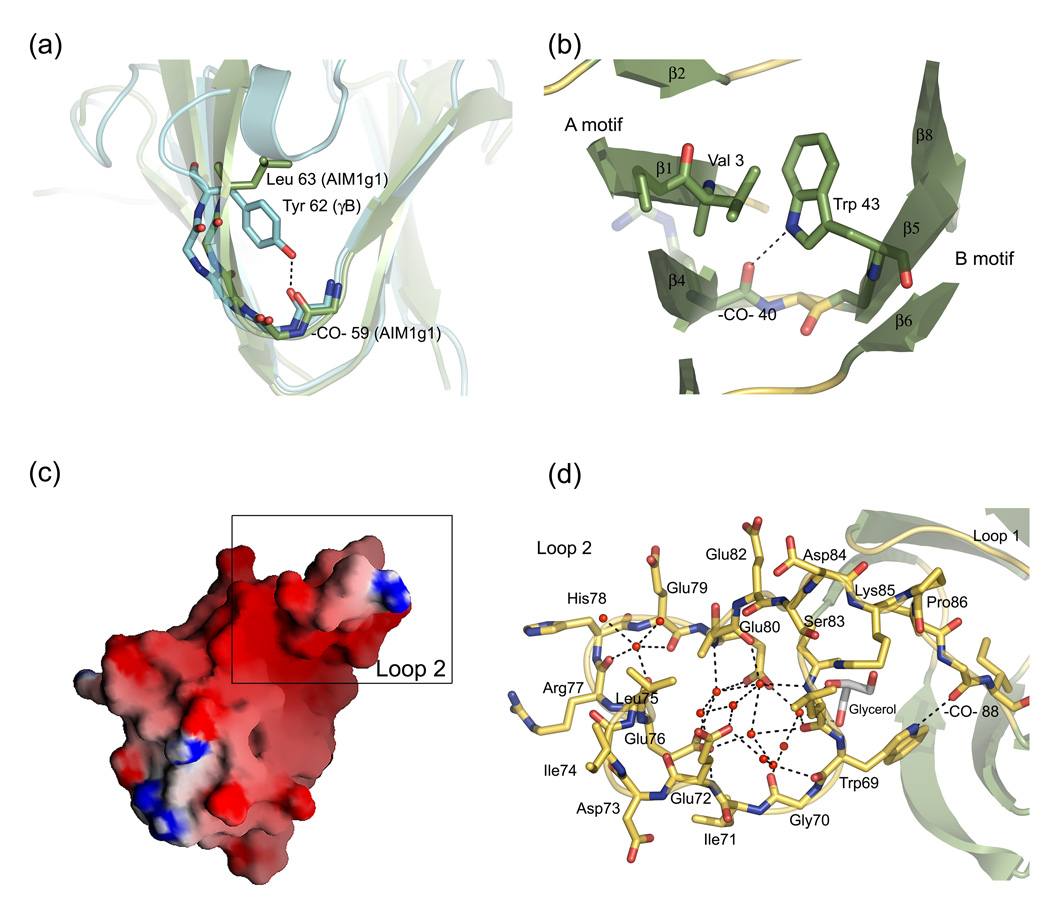
Figure 3.
(a) The Tyr-corner, which is an important feature in other domains, is replaced to Leu63 in AIM1g1. There are no major effects on the fold of the molecule in spite of this mutation. (b) The Trp-corner, one of the features conserved among AIM1g1 and lens crystallins, and its interactions in the hydrophobic core. It acts as a bridge between A and B-motifs. (c) Surface representation of AIM1g1 showing strong electronegative charge distribution in the loop region of the domain. Surface representation was done using GRASP.34 (d) Extensive water networking is observed in the loop region. Water molecules involved in formation of H-bond connectivities are w118, w126, w128 w147, w151, w157, w161, w171, w181, w190, w212 and w216 along with a glycerol molecule found in the vicinity of the loop. These waters bridge the side chains, main chain atoms and help in stabilization. These interactions are in addition to the electrostatic interactions that hold crystal lattice intact.
Trp corner
The Trp corner in the B-Type Greek key motif is a typical feature that is retained in the AIM1g1 domain. Trp43 is located at the beginning of the 2nd Greek key motif between the third and fourth strand (Fig. 3b). The indole -NH- forms a H-bond with a conserved main chain carbonyl (residue 40) and can form strong hydrophobic interactions in the domain core. The Trp-corner seems to make an important contribution to the folding and stability of the domain, but like the Tyr-corner this feature also may not be essential, since it is absent from the microbial Yersinia crystallin.14
Unique surface loop
βγ-crystallin domains contain two long loops that bridge the β-sheets that form the wedge-like sandwich. In AMI1g1, the first loop that connects β3 and β4 appears conventional, similar in length to that of other domains. It possesses Trp28 whose indole ring helps in preventing the exposure of the hydrophobic core at the domain flank. The loop undergoes stabilizing interactions with the loop that connects strands β7 and β8. These include water mediated interactions between main chain carbonyls of Cys25, Trp28 of loop 1 and Val87 and Ile89 of loop 2. H-bond formation takes place between main chain carbonyl of Ser31 and main amide of Val87. The loop 1 ends in a Pro32 that helps the chain reorient to form β4.
In contrast, the second loop is characterized by the presence of a 10-residue insertion that creates a strikingly novel feature in this domain of AIM1 (Fig. 2c, Fig 3c and 3d). The loop connects strands β7 and β8 and extends from residue 66 to 90. It has an unusual conformation, stabilized by several interactions. The N-terminal part of the loop undergoes a single twist with the Cterminal part of the loop. This is brought about by a strong H-bond between the indole NH of Trp69 and main chain CO of Val88 (fig 3d). As a result, the N-terminal part of the loop faces the second hairpin whereas the C-terminal part of the loop faces the first hairpin of the domain. The C-terminal part of the loop is stabilized by formation of a H-bond between the main chain -NH- of Val87 and the main chain -CO- of Pro31. A short 310 helix observed from residues 73 to 75 is the only secondary structural element found in this extensive loop. This loop has an accessible surface area of ~1200Å2 which is 20% of the total 6009 Å2 accessible surface area of the domain.
A surface charge representation shows that the loop 2 forms a predominantly electronegative region (Fig. 3c), which could play a role in binding cations or interacting partners within the cell. The strong electronegative region is formed due to the presence of carbonyl oxygens 68 and 70 and carboxyl side chains of Glu76 and Glu80 create a spherical region of radius 2.25Å that could potentially bind divalent cations like calcium, which have an average coordination sphere of radius 2.4 Å.21 AIM1g1 and lens βγ-crystallins have earlier been shown to bind Ca2+.16,22,23 Although Ca2+ was present in the crystallization buffer (2–5mM), no bound Ca2+ was observed, perhaps due to the low affinity for Ca2+-binding and presence of high concentrations of sodium citrate (1.3–1.5M) at a pH of 7.5, used as a precipitant during crystallization.
A number of water molecules are bound in this region along with a glycerol molecule in close proximity with the loop. These water molecules form an exquisite network of strong H-bonds along with the glycerol molecule and stabilize this region (Fig. 3d). H-bonded networks mediated through water are known to stabilize charged regions of the protein surfaces and help in improving thermostability. Stabilization through ion-pairs and H-bond networks is a common mechanism for protein stability in thermophilic bacteria, which acts by compensating the loss in entropy incurred during the transition from a disordered to an ordered state.24 The presence of water network in loop 2 could hence be used to maintain the stability of the loop.25, 26
The strong ion-pair formation between Glu76 and 80 with Lys2’ and Arg 40’ of a symmetry related partner leads to a distortion of the main chain dihedral angles (ϕ/ψ=79°/−69°) of His78, which is the only residue that falls outside the Ramachandran plot. Pro86 is involved in reorienting the loop2 at the roof of the wedge-like domain. In spite of the long loop on one side of the domain, that face of the domain is not hidden from solvent and interacts with a symmetry related molecule in the crystal lattice.
Packing analysis
Hexagonal crystal packing is brought about by interactions of the novel surface loop with different regions of the symmetry related partners. The interactions are predominantly electrostatic and water mediated, but some of them are hydrophobic (Table 3). A total of eight symmetry related molecules with four unique interfaces envelop each protomer. All interactions take place in a head to tail fashion so that the lattice is formed by predominantly linear arrangements of these domains. The variant loop is the major component having the lattice forming interactions, summarized in Table 3.
Table 3.
Packing analysis
| Interface | Symmetry operator | Buried Surface area/monomer(Å2) (% Buried surface area) | Major interactions & Interacting residues |
|---|---|---|---|
| Interface 1 | −y,x−y+1,z−1/3 | 740.4 (12.3) |
Electrostatic: Arg 77- Glu 60’ Glu80/Glu76– Lys 2’ Glu 80/76(-CO-) – Arg 40’ His54/70(-NH-) – Glu 17’ Ser55(-NH-) – Cys15 (-CO-) Hydrophobic: Ile 71, Val 39, Phe 19’ |
| Interface 2 | x−y+1,x+1,z−1/6 | 456.8 (7.5) |
Electrostatic: Glu72/w46/81(-CO-)/82(-CO-) – Arg 93’ Ser83/(-NH-)84– Asp 21’ Glu 72 – Tyr 46’ Hydrophobic Leu 68, Leu 75, Ile 74 Phe 51’, Ile 44’ |
| Interface 3 | −x,−y+1,z−1/2 | 309.1 (5.14) |
Electrostatic: 29 (-NH-) – 73 (-CO-)’ 27(-CO-) – 77 (-NH-)’ Ser 26 – His 78’ 26 (-CO-) – Arg 77’ |
| Interface 4 | x−y,x,z−1/6 | 190.9 (3.17) | Water-mediated |
Represents residues from symmetry related protomer; -CO- and -NH- represent main chain carbonyl and amide groups respectively.
In our earlier work, it was reported that AIM1g1 forms a dimeric molecule in solution at a physiological ionic strength.16 Interdomain interactions in lens βγ-crystallins are well documented.5,27 γ-Crystallins which are monomeric 2-domain proteins have their both domains related by a pseudo dyad.5 The interdomain associations formed by H-bonding and hydrophobic interactions are mediated by complimentary residues in strands 5, 6, 8, hairpin 2 and loop2 of both the crystallin domains. This interface formation is facilitated by a short linker, which connects domains 1 and 2 in γ-crystallins. This interface shows a remarkable conservation in domain swapped dimers of β-crystallins wherein the dimer forms through interactions between the N-terminal domain of one protomer and the C-terminal domain of the dimeric partner. Truncated N-terminal domain of βB2-crystallin also dimerizes in a similar fashion and this phenomenon is proposed to be replicative of formation of ancestral homodimers.28 AIM1g1 domain was overlapped onto this homodimeric N-terminal domain of βB2-crystallin. Interestingly, the interface formed did not show any steric clashes, unlike ciona crystallin, which is a monomeric single domain urochordate crystallin.29 Though some of the possible dimeric interactions are distorted by loop 2 in AIM1g1, the residues in the two interfaces (strands 5, 6 and 8) exhibit a lot of similarity with the βB2 N-terminal domain dimers. This could be indicative of the mode of AIM1g1 dimerization in solutions of physiological ionic strength.
However, none of the interactions in the crystal lattice are indicative of a dimeric AIM1g1, although this may be a result of the high ionic strength used for crystal formation. Since the unique surface loop is extensively involved in lattice contacts, its structure in solution may be more dynamic from that seen in the crystal. To our knowledge none of the other βγ-crystallin crystal lattices are formed by head to tail juxtaposition of domains. This is a unique case in AIM1g1 lattice which leads to linear agglomerates of domains making up the crystalline lattice. It could, in part, be a result of the open flanks of the domain unlike the other crystallins whose hydrophobic cores are protected by tyrosine corners and interactions mediated by the loops found on the wedge like domains.
Conclusions
The structure of AIM1g1 illustrates how the βγ-crystallin fold can accommodate significant variations in its sequence signature. In spite of non-conservative changes in key residues and the addition of a large insertion, the basic shape of the ancestral fold is retained. In particular, the residues of the characteristic hairpin structure are missing from the first motif and yet the motif follows the familiar topology, albeit with some deviations. It is possible that the retention of the correctly folded hairpin in the second motif allows that motif to form a template that helps to direct the folding of the more variant first motif.
Insertions and deletions that take place during evolution allow domains to evolve new functions while maintaining a basic structural scaffold.30 The insertion in AIM1g1, forming an extensive novel loop, shows how variations can be engineered by nature into the template of a conserved protein fold. Such changes could give rise to new functional roles while maintaining at least some of the basic stability of the ancestral fold. This structure suggests that AIM1 has indeed acquired a new role distinct from that of the well characterized βγ-crystallins of the eye-lens. Thus, the work provides a structural frame work for one of the domains (AIM1g1) of AIM1 protein to further explore not only the βγ-crystallin fold in detail but also the role of the other domains in AIM1 protein.
Acknowledgements
PA acknowledges Council of Scientific and Industrial Research (CSIR) for senior research fellowship. This work is supported by a Department of Science and Technology (DST), Govt of India grant (YS) and by a Wellcome Trust International fellowship (RS). GW is supported by the Intramural program of the National Eye Institute. Thanks to Drs. Orval Bateman of Birkbeck College, Jeff Trent and Paul Meltzer of NHGRI and Caroline Graham of NEI for the clone of AIM1g1.
Abbreviations used
- AIM1
Absent in Melanoma-1
- AIM1g1
First crystallin domain of AIM1
- SKLP
Streptomyces killer toxin like protein
- WmKT
Williopsis mrakii yeast killer toxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Deposition: Coordinates and structure factors are deposited in the protein data bank with an identity code 3CW3.
References
- 1.Wistow GJ, Piatigorsky J. Lens Crystallins: The Evolution and Expression of Proteins for a Highly Specialized Tissue. Ann. Rev. Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 2.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Prog. Biophys. Mol. Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Jaenicke R, Slingsby C. Lens Crystallins and their microbial homologs: Structure, stability, and function. Crit. Rev. Biochem. Mol. Biol. 2001;36:435–499. doi: 10.1080/20014091074237. [DOI] [PubMed] [Google Scholar]
- 4.Blundell TL, Lindley P, Miller L, Moss D, Slingsby C, Tickle I, Turnell B, Wistow G. The molecular structure and stability of the eye lens: X-ray analysis of gamma-crystallin II. Nature. 1981;289:771–777. doi: 10.1038/289771a0. [DOI] [PubMed] [Google Scholar]
- 5.Wistow G, Turnell B, Summers L, Slingsby C, Moss D, Miller L, Lindley P, Blundell T. X-ray analysis of the eye lens protein gamma-II crystallin at 1.9 A resolution. J. Mol. Biol. 1983;170:175–202. doi: 10.1016/s0022-2836(83)80232-0. [DOI] [PubMed] [Google Scholar]
- 6.Slingsby C, Norledge B, Simpson A, Bateman OA, Wright G, Driessen HPC, Lindley PF, Moss DS, Bax B. X-ray diffraction and structure of crystallins. Prog. Retinal Eye Res. 1997;16:3–29. [Google Scholar]
- 7.Bagby S, Go S, Inouye S, Ikura M, Chakrabartty A. Equilibrium folding intermediates of a Greek key beta-barrel protein. J. Mol. Biol. 1998;276:669–681. doi: 10.1006/jmbi.1997.1563. [DOI] [PubMed] [Google Scholar]
- 8.Antuch W, Guntert P, Wuthrich K. Ancestral βγ crystallin precursor structure in a killer toxin. Nat. Struct. Biol. 1996;3:662–665. doi: 10.1038/nsb0896-662. [DOI] [PubMed] [Google Scholar]
- 9.Ohki SY, Kariya E, Hiraga K, Wakamiya A, Isobe T, Oda K, Kainosho M. NMR structure of Streptomyces killer toxin-like protein, SKLP: further evidence for the wide distribution of single-domain betagamma-crystallin superfamily proteins. J. Mol. Biol. 2001;305:109–120. doi: 10.1006/jmbi.2000.4244. [DOI] [PubMed] [Google Scholar]
- 10.Clout NJ, Wistow G, Slingsby C. An eye on crystallins. Nat. Struct. Biol. 1997;4:685. doi: 10.1038/nsb0997-685. [DOI] [PubMed] [Google Scholar]
- 11.Wistow G. Evolution of a protein superfamily: relationships between vertebrate lens crystallins and microorganism dormancy proteins. J. Mol. Evol. 1990;30:140–145. doi: 10.1007/BF02099940. [DOI] [PubMed] [Google Scholar]
- 12.Clout NJ, Kretschmar M, Jaenicke R, Slingsby C. Crystal structure of the calcium-loaded spherulin 3a dimer sheds light on the evolution of the eye lens betagamma-crystallin domain fold. Structure. 2001;9:115–124. doi: 10.1016/s0969-2126(01)00573-1. [DOI] [PubMed] [Google Scholar]
- 13.Hamill JH, Cota E, Chothia C, Clarke J. Conservation of folding and stability within a protein family: The tyrosine corner as an evolutionary cul-de-sac. J. Mol. Biol. 2000;295:641–649. doi: 10.1006/jmbi.1999.3360. [DOI] [PubMed] [Google Scholar]
- 14.Jobby MK, Sharma Y. Calcium-binding crystallins from Yersinia pestis: Characterization of two single beta gamma-crystallin domains of a putative exported protein. J. Biol. Chem. 2005;280:1209–1216. doi: 10.1074/jbc.M409253200. [DOI] [PubMed] [Google Scholar]
- 15.Ray ME, Wistow G, Su YA, Meltzer PS, Trent JM. AIM1, a novel non-lens member of the ββ-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanoma. Proc. Natl. Acad. Sci (USA) 1997;94:3229–3234. doi: 10.1073/pnas.94.7.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajini B, Graham C, Wistow G, Sharma Y. Stability, homodimerization, and calcium-binding properties of a single, variant ββ-crystallin domain of the protein absent in melanoma 1 (AIM1) Biochemistry. 2003;42:4552–4559. doi: 10.1021/bi027384l. [DOI] [PubMed] [Google Scholar]
- 17.Hanson SR, Smith DL, Smith JB. Deamidation and disulfide bonding in human lens gamma-crystallins. Exp. Eye Res. 1998;67:301–312. doi: 10.1006/exer.1998.0530. [DOI] [PubMed] [Google Scholar]
- 18.Skouri-Panet F, Bonneté F, Prat K, Bateman OA, Lubsen NH, Tardieu A. Lens crystallins and oxidation: the special case of gammaS. Biophys. Chem. 2001;89:65–76. doi: 10.1016/s0301-4622(00)00216-7. [DOI] [PubMed] [Google Scholar]
- 19.Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ. Evidence for a strong sulfur-aromatic interaction derived from crystallographic data. Biopolymers. 2000;53:233–248. doi: 10.1002/(SICI)1097-0282(200003)53:3<233::AID-BIP3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Ohno A, Tate S, Seeram SS, Hiraga K, Swindells MB, Oda K, Kainosho M. NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gammacrystallin precursor structure. J. Mol. Biol. 1998;282:421–433. doi: 10.1006/jmbi.1998.2022. [DOI] [PubMed] [Google Scholar]
- 21.Strynadka NC, James MN. Crystal structures of the helix-loop-helix calcium-binding proteins. Ann. Rev. Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 22.Rajini B, Shridas P, Sundari CS, Muralidhar D, Chandani S, Thomas F, Sharma Y. Calcium binding properties of gamma-crystallin: calcium ion binds at the Greek key beta gamma-crystallin fold. J. Biol. Chem. 2001;276:38464–38471. doi: 10.1074/jbc.M102164200. [DOI] [PubMed] [Google Scholar]
- 23.Jobby MK, Sharma Y. Calcium-binding to lens βB2- and βA3-crystallins suggests that all β-crystallins are calcium-binding proteins. FEBS J. 2007;274:4135–4147. doi: 10.1111/j.1742-4658.2007.05941.x. [DOI] [PubMed] [Google Scholar]
- 24.Scandurra R, Consalvi V, Chiaraluce R, Politi L, Engel PC. Protein stability in extremophilic archaea. Front. Biosci. 2000;5:787–795. doi: 10.2741/A551. [DOI] [PubMed] [Google Scholar]
- 25.Vogt G, Woell S, Argos P. Protein thermostability, hydrogen bonds, and ion pairs. J. Mol. Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 26.Cooper A. Heat capacity of hydrogen-bonded networks: an alternative view of protein folding thermodynamics. Biophys. Chem. 2000;85:25–39. doi: 10.1016/s0301-4622(00)00136-8. [DOI] [PubMed] [Google Scholar]
- 27.Lapatto R, Nalini V, Bax B, Driessen H, Lindley PF, Blundell TL, Slingsby C. High resolution structure of an oligomeric eye lens beta-crystallin. Loops, arches, linkers and interfaces in beta B2 dimer compared to a monomeric gammacrystallin. J. Mol. Biol. 1991;222:1067–1083. doi: 10.1016/0022-2836(91)90594-v. [DOI] [PubMed] [Google Scholar]
- 28.Clout NJ, Basak A, Wieligmann K, Bateman OA, Jaenicke R, Slingsby C. The N-terminal domain of βB2-crystallin resembles the putative ancestral homodimer. J. Mol. Biol. 2000;304:253–257. doi: 10.1006/jmbi.2000.4197. [DOI] [PubMed] [Google Scholar]
- 29.Shimeld SM, Purkiss AG, Dirks RP, Bateman OA, Slingsby C, Lubsen NH. Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr. Biol. 2005;15:1684–1689. doi: 10.1016/j.cub.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 30.Grishin NV. Fold change in evolution of protein structures. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Park J. Dalilite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 32.Aravind P, Rajini B, Sharma Y, Sankaranarayanan R. Crystallization and preliminary X-ray crystallographic investigations on a βγ-Crystallin domain of Absent In Melanoma 1 (AIM1), a protein from Homo sapiens. Acta. Cryst. Sect. F. 2006;62:282–284. doi: 10.1107/S1744309106005380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yogavel M, Gill J, Mishra PC, Sharma A. SAD phasing of a structure based on co-crystallized iodides using an in-house Cu Kα X-ray source: effects of data redundancy and completeness on structure solution. Acta. Cryst. Sect. D. 2007;63:931–934. doi: 10.1107/S0907444907029174. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struc. Func. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]