Abstract
Hypoxia-Inducible Factor 1α (HIF-1α) is critical not only in the regulation of oxygen homeostasis, but also in the regulation of innate and adaptive immune systems. We previously reported that TCR-mediated activation of T cells in mice leads to the expression of an alternative isoform of HIF-1α that inhibits activated T cells in a delayed negative feed-back manner. In this paper, we describe a novel mRNA isoform of human HIF-1α that is upregulated in peripheral T lymphocytes after TCR stimulation. This activation-inducible isoform is expressed using the alternative first exon I.3, and it encodes a protein that is 24 amino acids longer than the ubiquitous HIF-1α isoform. This mRNA isoform I.3 of HIF-1α is expressed in a tissue-specific manner with the highest expression found in peripheral blood leukocytes and the thymus.
Keywords: T cells, TCR, PBMC, HIF-1, hypoxia-inducible factor
INTRODUCTION
Immune cells are exposed to various levels of oxygen tension, including hypoxia as they migrate and function in different tissue microenvironments [1]. Hypoxia-Inducible Factor-1 (HIF-1) is a heterodimeric transcription factor that plays a critical role in mediating the adaptation of cells to low oxygen tension [2]. At higher oxygen tension, the HIF-1α subunit of HIF-1 is degraded due to the activity of prolyl hydroxylase enzymes [3], while in hypoxia HIF-1α is stabilized and translocated into the nucleus [4]. In addition, HIF-1α expression can be upregulated even under normoxic conditions by IL-4 [5], TNF-α [6], IL-1 [7], and other cytokines. The TCR-mediated activation of T cells in mice results in a selective induction of the alternatively spliced mRNA isoform I.1 of HIF-1α [8]. HIF-1α was shown to be critically involved in regulation of antibacterial functions of macrophages [9] and T cells [10]. It was also demonstrated that T-cell specific HIF-1α-deficiency results in enhanced anti-bacterial response in inflamed hypoxic tissues [10], and is associated with the increased expression of NF-κB p50 mRNA in activated T cells [10]. Moreover, genetic deletion of the activation-inducible isoform I.1 of HIF-1α in mice results in significant enhancement of T-cell functions [11]. However, a human isoform of HIF-1α analogous to the mouse activation-inducible isoform has not yet been identified. In this report, we describe the novel activation-inducible mRNA isoform I.3 of human HIF-1α, which is upregulated in activated peripheral T lymphocytes.
MATERIALS AND METHODS
Cells and reagents
PBMCs were isolated from heparinized blood of healthy volunteers (purchased from Research Blood Components, Brighton, MA) by Ficoll–Hypaque density centrifugation. Cells were activated using 1µg/µl plate-bound anti-CD3 and anti-CD28 mAb (BD Biosciences) with 5µg/ml phytohemagglutinin (Sigma) for 24 hours. HeLa cells (ATCC) were maintained in complete RPMI media
Rapid Amplification of cDNA Ends (RACE)
Total RNA from activated PBMCs was extracted using RNA-STAT60 (Tel-test) and subjected to 5'-RACE and 3'-RACE procedures using Marathon cDNA Amplification kit (Clontech) according to the manufacturer's protocol. Gene-specific primers 3.1 and 3.2, complementary to the exon II of human HIF-1α, were described in [12]. PCR products were cloned using Blunt-cloning kit (Clontech) and were subjected to sequencing (service provided by MWG Biotech, Inc). 3'-RACE was performed using TGGTGGTTACTCAGCACTTTTAGA primer complementary to the exon I.3.
RT-PCR analysis
Expression of I.3 isoform of HIF-1α was measured by real-time PCR using comparative Ct analysis (primers for I.3 isoform: TGGTGGTTACTCAGCACTTTTAGA and CTCCGACATTGGGAGCTCAT) with L32 mRNA as a reference (primers: CATCTCCTTCTCGGCATCA and AACCCTGTTGTCAATGCCTC). RT-PCR was performed using SYBR Green mix (Applied Biosystems). cDNA was synthesized from 1 µg of total RNA samples obtained from activated PBMC using First-Strand Amplification kit (Invitrogen). A human tissues cDNA panel was purchased from Clontech.
Cloning of I.1 and I.3 isoforms of human HIF-1α
The cDNA of I.3 isoform obtained by reverse transcription was cloned into pcNDA3.1(+) by NotI-XbaI sites. The cDNA of I.1 isoform was cut from pCEP4-HIF1a (provided by Dr. Semenza) and cloned into pcDNA3.1(+) by KpnI-XbaI sites.
Luciferase reporter assay
HeLa cells were transfected with plasmid pHRE-Luc (provided by Dr. Blagosklonny) [8] together with plasmids pcDNA3.1(+) encoding either isoform I.1 or I.3 of HIF-1α using Lipofectamine 2000 (Invitrogen) according to the manual. Hypoxia response element-dependent luciferase activity was measured using Luciferase Assay Kit (Promega) as previously described [8].
Western blot
HeLa cells were transfected with plasmids encoding either isoform I.1 or I.3 of HIF-1α using Lipofectamine 2000 (Invitrogen) in 6-well plate according to the manual. After 24h cells were washed with PBS supplemented with 5µM MG132 (inhibitor of 26S proteasome) and solubilized with lysis buffer containing 1% Triton X-100, 50 mM Tris-HCl pH 7.6, 150 mM NaCl, 5mM EDTA, 5µM MG132, and protease inhibitors cocktail (Pierce) on ice for 30 min. Lysate was cleared by centrifugation at 20,000g for 30 min and subjected to 7% NuPage Tris-Acetate gel analysis. Semi-wet transfer to the nitrocellulose membrane (Invitrogen) using XCell II Blot Module (Invitrogen) was used in order to achieve most efficient transfer of high molecular weight proteins. Antibodies against HIF-1α (Novex) at a 1/500 dilution or β-actin (Sigma-Aldrich) at a 1/5000 dilution were used in combination with HRP-conjugated goat anti-mouse Ab (Santa Cruz Biotechnology) at a 1/5000 dilution before detection using SuperSignal Substrate (Pierce, Rockford, IL).
RESULTS
Previously, it was demonstrated that the alternative mRNA isoform I.1 of the mouse HIF-1α is expressed predominantly in lymphoid organs and testis [8, 12] and is significantly induced by TCR activation [8]. Analysis of I.1 gene-deficient mice demonstrated that this activation-inducible isoform of HIF-1α plays an anti-inflammatory role in T cells and acts as a regulator of activated T cells in a delayed negative feed-back manner [11]. These findings prompted the search for a human analogue of the I.1 isoform of HIF-1α. Recently, a novel alternative mRNA isoform of HIF-1α was identified in human testis [13]. This isoform encodes a short dominant-negative HIF-1α protein that lacks fifty nine N-terminal amino acids including DNA-binding bHLH domain [13]. Furthermore, in contrast with the mouse isoform I.1, which is expressed in both testis and in activated T cells [8, 12], this novel human isoform was shown to be only testis-specific.
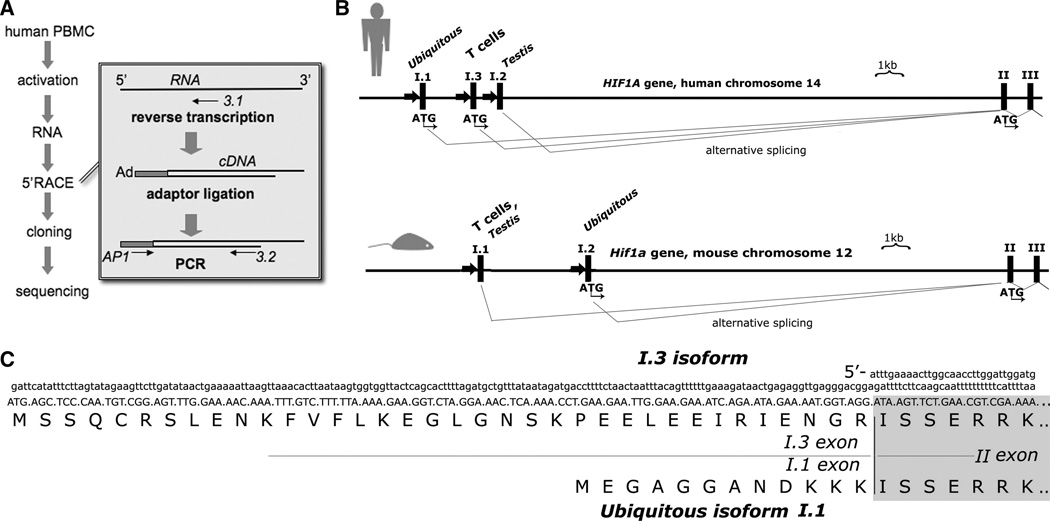
In order to detect an alternative T-cell-specific mRNA isoform of HIF-1α we used a 5'-RACE approach (Figure 1A). In previous mouse studies, it was demonstrated that TCR-activated T cells preferentially upregulate expression of the activation-inducible mRNA isoform I.1 [8]. Therefore, RNA was obtained from human peripheral leukocytes activated with anti-CD3 and anti-CD28 antibodies in combination with mitogen phytohemagglutinin. Using the described 5'-RACE procedure we were able to clone a novel HIF-1α mRNA isoform that contained an alternative first exon that was named I.3. The sequence conformed to the human HIF1A gene (GenBank accession # AL137129) on chromosome 14 between the exon I.1 and exon I.2 (Figure 1B). In addition, we identified an alternative splicing between exons I.3 and II that resulted in the insertion of an additional triplet corresponding to arginine. Such splicing variations were previously reported in other human HIF-1α mRNA isoforms I.1 and I.2 [13]. The subsequent analysis of the nucleotide sequence of exons II-XV of the I.3 mRNA revealed no difference in comparison to the ubiquitous mRNA isoform I.1 of human HIF-1α.
Figure 1. The activation-inducible isoform I.3.
(A) Scheme of experiment designed to identify rapid amplification of cDNA ends (5'-RACE). Ad, adaptor oligonucletoide; AP1, adaptor primer; 3.1 and 3.2, gene-specific primers complementary to the exon II of human HIF-1α mRNA.
(B) Genomic organization of the human HIF1A and mouse Hif1a loci.
(C) Nucleotide sequence of the 5' region of alternative mRNA isoform I.3 and N-terminal sequence of the protein encoded by I.3 mRNA compared to the ubiquitous isoform of human HIF-1α.
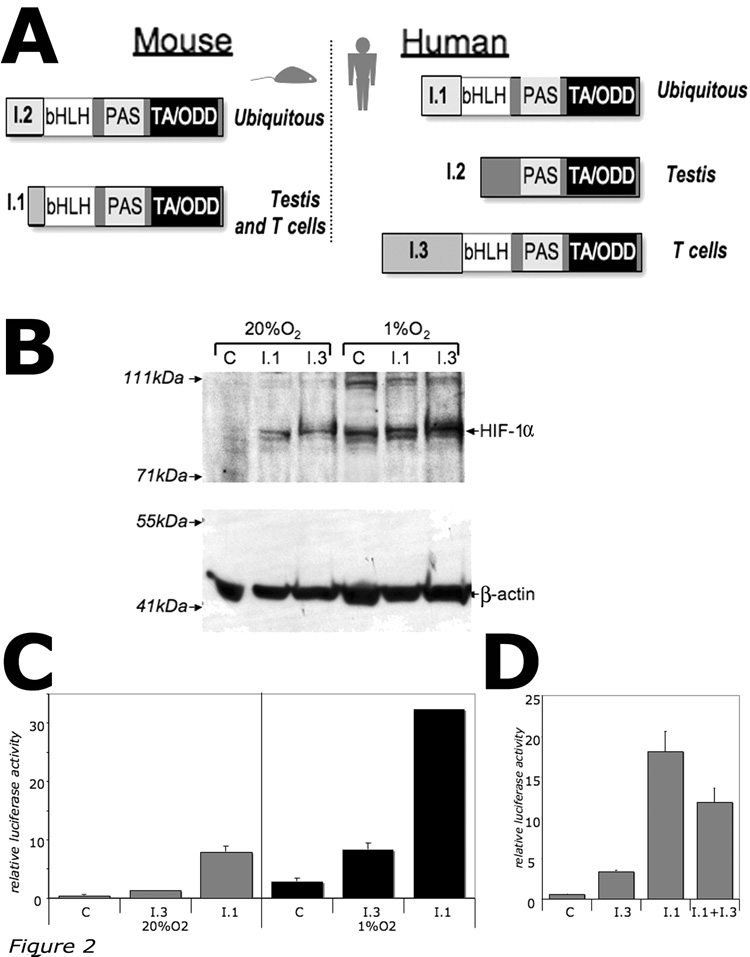
It was shown that the mouse activation-inducible mRNA isoform I.1 produces transcriptionally active HIF-1α protein that is twelve amino acids shorter than the ubiquitous HIF-1α isoform [8, 14]. In contrast, the novel human I.3 mRNA isoform encodes HIF-1α that is twenty four amino acids longer than the ubiquitous isoform (Figure 1C). Thus, in contrast with the mouse HIF-1α, which can be encoded by two alternative mRNA isoforms: T-cell/testis-specific (I.1) and ubiquitous (I.2) [15], the human HIF-1α mRNA isoforms include ubiquitous (I.1), testis-specific (I.2), and the T-cell-specific (I.3) (Figure 2A).
Figure 2. Comparison of the T-cell specific isoform I.3 and the ubiquitous isoform I.1.
(A) Domain structure of the tissue-specific alternative HIF-1α isoforms. bHLH, DNA-binding basic helix-loop-helix domain; PAS, Per-Arnt-Sim domain; TA/ODD, transactivational oxygen-dependent degradation domain.
(B) Western blot analysis of expression of HIF-1a isoforms I.1 and I.3. HeLa cells were transfected with pcDNA3.1(+) plasmid (C, control) or with plasmids expressing either I.1 or I.3 isoforms of HIF-1α, then incubated at 20%O2 and 1%O2 for 24h.
(C) Hypoxia-response element (HRE)-dependent transcriptional activity of I.1 and I.3 isoforms as measured by luciferase reporter gene assay. HeLa cells were transfected with HRE-Luciferase reporter plasmid alone (C, control) or in combination with plasmids expressing either I.1 or I.3 isoforms of HIF-1α, then incubated at 20%O2 and 1%O2 for 24h.
(D) Dominant-negative effect of I.3 isoform on transcriptional activity of I.1 isoform. HeLa cells were transfected with HRE-Luciferase reporter plasmid alone or in combination with plasmids expressing I.1 or I.3 isoforms of HIF-1α, then incubated at 20%O2 for 24h.
Analysis of the nucleotide sequence of the I.3 mRNA isoform revealed that the translation initiation codon located in I.3 exon is surrounded by weak Kozak consensus sequence, therefore the ribosomal scanning mechanism may not be effective in consistently recognizing this suboptimal translation initiation codon. In this case, the translation would start using the next available AUG codon located in exon II, therefore producing shorter HIF-1α protein like testis-specific HIF-1α isoform I.2 [13]. To estimate a size of the I.3 isoform of HIF-1α, the recombinant HIF-1α isoforms I.3 and I.1 were compared by Western blot. It was observed that I.3 isoform produces a large HIF-1α protein as compared with the ubiquitous isoform I.1 (Figure 2B).
The transcriptional activity of I.3 isoform was estimated by its ability to transactivate the Hypoxia-Response Element (HRE)-dependent transcription using luciferase reporter assay. It was demonstrated that the I.3 isoform of HIF-1α is transcriptionally active but in less extent when compared to the ubiquitous HIF-1α isoform I.1 (Figure 2C). The following co-transfection luciferase assay revealed a dominant negative effect of I.3 isoform on the activity of the I.1 isoform (Figure 2D), suggesting that I.3 isoform is capable to compete for dimerization and/or DNA-binding, but has a reduced transcriptional activity.
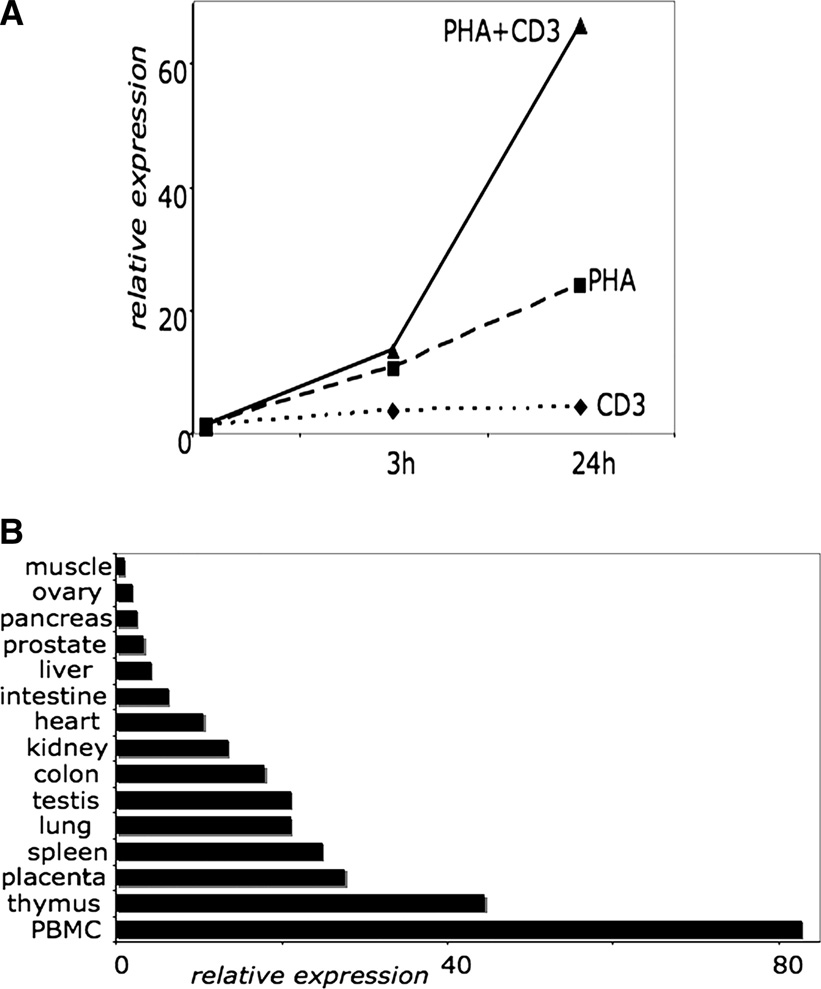
We then tested whether the expression of the I.3 isoform is dependent on TCR-activation as it was shown for the activation-inducible isoform I.1 of mouse HIF-1α [8]. Indeed, the analysis of I.3 mRNA levels by real-time PCR demonstrated that expression of I.3 mRNA is significantly induced by T-cell activation (Figure 3A). Interestingly, the upregulation of I.3 mRNA expression was most prominent after combined activation by anti-CD3/anti-CD28 antibodies and mitogenic phytohemagglutinin. In addition, human tissue-specific expression of I.3 mRNA was analyzed by real-time PCR. It was demonstrated that I.3 mRNA is expressed in a variety of human tissues, while being most abundant in PBMCs, thymus, placenta, and spleen (Figure 3B).
Figure 3. Differential and tissue-specific expression of HIF-1α isoform I.3.
(A) Relative expression of I.3 mRNA in activated human PBMCs measured by real-time PCR. PBMCs were activated by 1µg/ml plate-bound anti-CD3 and anti-CD28 antibodies (CD3), 5µg/ml phytohemagglutinin (PHA), or both (PHA+CD3) for indicated time.
(B) Relative expression of I.3 mRNA in cDNA panel of various human tissues measured by real-time PCR.
DISCUSSION
We have described a novel isoform of human HIF-1α that is upregulated in activated human T lymphocytes. This report suggests future possibilities of development of the new HIF-1α targeting drugs that may specifically inhibit HIF-1α in activated T cells. It may also lead to development of monoclonal antibodies, since the novel isoform I.3 of HIF-1α is different from the ubiquitous HIF-1α thereby providing unique twenty four amino acid sequence as an antigenic determinant to be recognized by selective antibody.
The novel activation-inducible mRNA isoform I.3 of HIF-1α encodes a functional transcriptional factor, although the transcriptional activity of the I.3 HIF-1α isoform is weaker in comparison to ubiquitous HIF-1α. Future development of diagnostic antibodies that are reactive with the unique N-terminal sequence of I.3 isoform will enable studies of its protein expression and functions. It will be interesting to test whether the activation-inducible isoform I.3 of human HIF-1α plays an anti-inflammatory role in regulation of activated T cells as it was shown in previous mouse studies [10, 11].
ACKNOWLEDGEMENTS
The authors thank Susan Ohman for help in preparation of the manuscript.
Abbreviations
- HIF-1α
hypoxia-inducible factor 1α
- TCR
T-cell receptor
- PBMC
peripheral blood mononuclear cells
- 5'-RACE
rapid amplification of 5' cDNA ends
- bHLH
basic-helix-loop-helix
- HRE
hypoxia-response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sitkovsky M, Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1alpha and adenosine receptors. Nat Rev Immunol. 2005;5(9):712. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell. Dev. Biol. 1999;15:551. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 3.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279(37):38458. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001;107(1):1. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 5.Scharte M, Jurk K, Kehrel B, Zarbock A, Van Aken H, Singbartl K. IL-4 enhances hypoxia induced HIF-1alpha protein levels in human transformed intestinal cells. FEBS Lett. 2006;580(27):6399. doi: 10.1016/j.febslet.2006.10.053. [DOI] [PubMed] [Google Scholar]
- 6.Haddad JJ, Land SC. A non-hypoxic, ROS-sensitive pathway mediates TNF-alpha-dependent regulation of HIF-1alpha. FEBS Lett. 2001;505(2):269. doi: 10.1016/s0014-5793(01)02833-2. [DOI] [PubMed] [Google Scholar]
- 7.Frede S, Freitag P, Otto T, Heilmaier C, Fandrey J. The proinflammatory cytokine interleukin 1beta and hypoxia cooperatively induce the expression of adrenomedullin in ovarian carcinoma cells through hypoxia inducible factor 1 activation. Cancer Res. 2005;65(11):4690. doi: 10.1158/0008-5472.CAN-04-3877. [DOI] [PubMed] [Google Scholar]
- 8.Lukashev D, Caldwell C, Ohta A, Chen P, Sitkovsky M. Differential regulation of two alternatively spliced isoforms of hypoxia-inducible factor-1 alpha in activated T lymphocytes. J Biol Chem. 2001;276(52):48754. doi: 10.1074/jbc.M104782200. [DOI] [PubMed] [Google Scholar]
- 9.Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112(5):645. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel M, Caldwell CC, Kreth S, Kuboki S, Chen P, Smith P, Ohta A, Lentsch AB, Lukashev D, Sitkovsky MV. Targeted deletion of HIF-1alpha gene in T cells prevents their inhibition in hypoxic inflamed tissues and improves septic mice survival. PLoS ONE. 2007;2(9):e853. doi: 10.1371/journal.pone.0000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, Wenger RH, Sitkovsky M. Cutting edge: hypoxia-inducible factor 1alpha and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J Immunol. 2006;177(8):4962. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- 12.Marti H, Katschinski D, Wagner K, Schäffer L, Stier B, Wenger RH. Isoform-specific expression of hypoxia-inducible factor-1alpha during the late stages of mouse spermiogenesis. Mol Endocrinol. 2002;16(2):234. doi: 10.1210/mend.16.2.0786. [DOI] [PubMed] [Google Scholar]
- 13.Depping R, Hagele S, Wagner KF, Wiesner RJ, Camenisch G, Wenger RH, Katschinski DM. A dominant-negative isoform of hypoxia-inducible factor-1 alpha specifically expressed in human testis. Biol Reprod. 2004;71(1):331. doi: 10.1095/biolreprod.104.027797. [DOI] [PubMed] [Google Scholar]
- 14.Gorlach A, Camenisch G, Kvietikova I, Vogt L, Wenger RH, Gassmann M. Efficient translation of mouse hypoxia-inducible factor-1alpha under normoxic and hypoxic conditions. Biochim Biophys Acta. 2000;1493(1–2):125. doi: 10.1016/s0167-4781(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 15.Wenger RH, Rolfs A, Spielmann P, Zimmermann DR, Gassmann M. Mouse hypoxia-inducible factor-1 is encoded by two different mRNA isoforms - expression from a tissue-specific and a housekeeping-type promoter. Blood. 1998;91:3471. [PubMed] [Google Scholar]