Abstract
The endosperm of a sorghum mutant cultivar, with high in vitro uncooked and cooked protein digestibilities, was examined by transmission electron microscopy and α-, β-, and γ-kafirins (storage proteins) were localized within its protein bodies. Transmission electron microscopy micrographs revealed that these protein bodies had a unique microstructure related to high protein digestibility. They were irregular in shape and had numerous invaginations, often reaching to the central area of the protein body. Protein bodies from normal cultivars, such as P721N studied here, with much lower uncooked and cooked digestibilities are spherical and contain no invaginations. Immunocytochemistry results showed that the relative location of α- and β-kafirins within the protein bodies of the highly digestible genotype were similar to the normal cultivar, P721N. γ-Kafirin, however, was concentrated in dark-staining regions at the base of the folds instead of at the protein body periphery, as is typical of normal cultivars. The resulting easy accessibility of digestive enzymes to α-kafirin, the major storage protein, in addition to the increased surface area of the protein bodies of the highly digestible cultivar appear to account for its high in vitro protein digestibility.
Sorghum [Sorghum bicolor (L.) Moench] grain is an important staple food in developing countries of the semiarid tropics and is used as an animal feed in both developed and developing countries. Its nutritional quality is of particular importance in areas where people depend on it as a main source of energy and protein. The nutritional quality of sorghum protein is of concern because, besides being low in the essential amino acid Lys, it has lower digestibility than protein of other cereal grains. Human studies in Peru (1) showed apparent protein digestibility of cooked sorghum to be only 46%, whereas values of 81, 73, and 66% were reported for cooked wheat, maize, and rice, respectively.
Many factors are known to affect the protein digestibility of sorghum. Tannins (secondary metabolites rich in phenolic hydroxyl groups) were initially recognized as being able to complex with proteins and render them unavailable for enzymatic digestion (2). However, even among the low tannin or tannin-free cultivars, low-protein digestibility still exists (3), and in fact, a broad range of uncooked and cooked in vitro digestibilities was found for these types in our laboratory (unpublished data). Low tannin/tannin-free sorghum cultivars are the predominant types grown in the world. Other unpublished data from our group showed that environmental conditions during the growing season also influence sorghum protein digestibility. In addition, processing methods affect digestibility, such as whether sorghum is boiled to a porridge, fermented, decorticated and extruded, or germinated (1, 4–8). Simple cooking of sorghum flour has an uncommon and detrimental effect of decreasing protein digestibility as demonstrated by in vitro (4, 5) and in vivo (9, 10) studies.
Among the proteins of sorghum, kafirins (sorghum prolamins) are the most abundant, making up 70–80% of the total endosperm protein (11). They have been classified according to structure, molecular weight, and solubility characteristics into α- (Mr 25,000 and 20,000), β- (Mr 20,000, 18,000, and 16,000), and γ-kafirins (Mr 28,000) (12). α-Kafirin, the main storage protein, comprises about 80% of total kafirin in the endosperm (13). The amino acid compositions of β- and γ-kafirin are unique because in part to their high content of Cys, which is about 5 and 7 mol%, respectively (14). During development, kafirins are synthesized and deposited inside the rough endoplasmic reticulum to form protein bodies. Within the protein body, the kafirins are distributed in a nonhomogeneous fashion. Immunocytochemistry showed that α-kafirin is located in light-staining areas mainly in the interior of the protein body, and β- and γ-kafirin are found in dark-staining areas inside and at the periphery of the protein body (14).
Several studies suggest that it is the nature of the proteins within the protein bodies that is mostly responsible for low-protein digestibility of sorghum grain. It was revealed that kafirins are the last proteins to be digested (5), and that the addition of a reducing agent improved their uncooked and cooked digestibility (15, 16). A developmental study showed that, as the sorghum seed matures, uncooked and cooked in vitro protein digestibilities decreased with a paralleled increase in formation of disulfide bonds among prolamins, most noticeably among β- and γ-kafirins (17). These latter studies (15–17) support the idea that structural features within the proteins, particularly disulfide bonds, negatively influence protein hydrolysis. Also, α-kafirin, although easily digested when isolated from protein bodies in its native unreduced form (B.R.H., unpublished data), was digested only after β- and γ-kafirins when flour was used for in vitro digestion (16). These results plus a scanning electron microscopy study (18) indicate that the breakdown of storage proteins starts on the outside of the protein body and progresses toward the interior. Evidence supports our theory that the poor digestibility of α-kafirin is caused by its interior location and that, at the protein body periphery, disulfide bonds among γ- and possibly β-kafirins form an enzyme-resistant structure that retards digestion of α-kafirin.
We recently discovered a sorghum cultivar whose protein digestibility resembles or surpasses that of other cereals and is not negatively affected by cooking or environment. This may confer a competitive advantage for this sorghum to be used for food and feed. Also, future information on the molecular basis of this trait could be used to improve the digestibility of other agronomically important varieties. The sorghum lines with high protein digestibility were found within a high-Lys population developed from crosses of the high-Lys mutant P721 opaque (Q) and normal cultivars. In vitro protein digestibility was unusually high whether uncooked (about 85%) or cooked (about 80%). Unlike normal sorghum, SDS/PAGE and ELISA time-course analysis of undigested proteins from highly digestible lines showed that the digestion of α-kafirin was rapid and occurred before the digestion of the other kafirins. These findings are reported elsewhere (19). We hypothesized that this unique feature may be reflected in the microstructure of the protein bodies. Likewise, because the relative location of individual kafirins within the protein body appears to influence their rate of digestion, location of kafirins in these unique cultivars was of interest.
The objective of this study was to examine the microstructure of endosperm protein bodies of a highly digestible sorghum line and determine the location of α-, β-, and γ-kafirins within its protein bodies.
Materials and Methods
Plant Material.
A normal sorghum cultivar, P721N, and the highly digestible sorghum line, P851171, were grown in 1994 at the Purdue University Agronomy Research Center, hand harvested at 30 days after half bloom (DAHB), and immediately prepared for microscopic examination. Additionally, P721Q (crop year, 1999) seed at 30 DAHB was used. P851171 was derived from outcrosses of P721Q with agronomically superior lines. P721Q is a single semidominant gene derived by chemical mutagenesis of P721N (20). Mature, field-dried sorghum seeds of P851171 were also collected, decorticated for 2 min in a Tangential Abrasive Dehulling Device (model 4E-110, Venables Machine Works, LTD, Saskatoon, Saskatchewan, Canada), and ground for 5 min at the highest setting on a ball mill (Retsch Vibratory Mill, type MM-2, Brinkmann). Flour was stored in a desiccator.
Western Blot Analysis.
Western blot analysis was performed to demonstrate that the antisera against the individual kafirins were specific for the kafirins of this new cultivar. The antisera used was previously prepared in this laboratory against each of α-, β-, and γ-kafirins (14). Total protein was extracted from flour of mature P851171 seeds (30 mg) by shaking overnight with a 0.0125 M sodium tetraborate buffer (pH 10)/1% (wt/vol) SDS/2% (vol/vol) 2-mercaptoethanol (0.5 ml). Sample solvent [25 μl, 0.06 M Trizma base/2% (wt/vol) SDS/5% (vol/vol) 2-mercaptoethanol/10% (vol/vol) glycerol/0.05% (wt/vol) bromophenol blue] was then added in equal volume for Coomassie blue R-250 staining. The protein extract was diluted 100 times with sample solvent for Western blot analysis. For Coomassie blue and Western blot analysis, the samples (50 μl) were loaded onto a 10–15% gradient SDS/PAGE gel containing 5 M urea and electrophoresed overnight at a constant voltage of 70 V. The presence of urea in the gel decreases the mobility of γ-kafirin, thereby improving its separation from α- and β-kafirins. An LKB Multiphor II Novablot apparatus (Bromma, Sweden) was used to transfer the proteins onto nitrocellulose membranes. A continuous buffer system that had 0.039 M glycine/0.048 mM Trizma base/20% (vol/vol) methanol was used. The transfer time was 2.5 h at a current of 0.8 mA/cm2. Nonspecific binding of the antibodies to the membranes was blocked with 3% (wt/vol) nonfat dry milk in 20 mM Trizma base/500 mM sodium chloride (TBS) and washed twice with TBS-T [TBS with 0.5% (vol/vol) Tween-20]. Membranes were incubated with preimmune serum, anti-α-kafirin, anti-β-kafirin, and anti-γ-kafirin (1:100, 1:1,000, 1:500, and 1:100 dilutions in TBS-T, respectively) after the procedure described by Shull et al. (14). Goat anti-rabbit/horseradish peroxidase conjugate (GAR-HRPR; Bio-Rad) was used as a secondary antibody (1:3,000 dilution in TBS-T). Immunostaining was achieved through reaction with hydrogen peroxide and 4-chloronapthol (Bio-Rad).
Preparation of Samples for Transmission Electron Microscopy.
Developing seeds from P721N, P851171, and P721Q were sliced transversely into 1–2-mm pieces with a razor blade and fixed in 4% (vol/vol) paraformaldehyde/1% (vol/vol) glutaraldehyde in 0.05 M potassium phosphate buffer (pH 6.8) at 4°C for 16 h. For microstructural analysis, the specimens were stained with 2% osmium tetroxide in 0.05 M potassium phosphate buffer (pH 6.8). After washing with the same buffer, the specimens were dehydrated in a graded ethanol series (15 min each in 10, 30, 50, 70, 90, and 95% aqueous ethanol, and in 100% ethanol). Samples were transferred sequentially to 1:2, 1:1, and 2:1 mixtures of acetone/ethanol for 30 min each, and finally to 100% acetone with three changes every 30 min. Then, they were gradually infiltrated with 50% Spurr's resin in acetone during 3 h, followed by 75% Spurr's resin in acetone overnight. They were further infiltrated twice with 100% resin for 2 h and polymerized at 55°C for 2 days. For immunocytochemical analysis, immature seeds of P851171 were fixed and washed in the same way except that they were not postfixed with osmium tetroxide. After gradual dehydration in the ethanol series, samples were infiltrated for 2 h each in 20, 40, 60, and 80% LR White resin in ethanol, and then for overnight in 100% LR White resin. The infiltration with 100% LR White resin continued for 2 more days with several changes of fresh resin. The samples were placed in plastic molds and polymerized for 2 days under a nitrogen atmosphere in an oven at 55°C.
Prepared specimens were sectioned with a diamond knife on a Richert Om U3 ultramicrotome. Copper grids coated with formvar membrane and carbon were used. Sections from Spurr's resin-embedded tissue were poststained for 5 min with 2.5% (wt/vol) uranyl acetate and for 3 min with 0.1% (wt/vol) lead citrate and examined with a Philips EM 200 transmission electron microscope (Philips–FEI, Eindhoven, The Netherlands) at 60 kV. Sections from LR White-embedded tissue were left to dry overnight and then immunolabeled.
Gold Immunochemical Labeling and Transmission Electron Microscopy.
Goat anti-rabbit colloidal gold conjugates (10 nm) were prepared by the method of Slot and Geuze (21). Colloidal gold particles were prepared by reduction of gold by a mixture of tannic acid/trisodium citrate. Affinity-purified goat anti-rabbit IgG (R-2004, Sigma) was rehydrated and dialyzed against 2 mM sodium borate buffer (pH 9.0) at 4°C. After centrifugation of the dialyzate to remove precipitated protein, the supernatant was conjugated with colloidal gold (21) and centrifuged in a 10–30% glycerol gradient at 65,000 × g until a band was formed in the middle section of the tube. Aliquots of the conjugated gold particles were collected (1 ml) and stored at 4°C. Nonspecific sites on the sections were blocked by floating the grids on 50-μl drops of TBS-T(0.3%) [TBS with 0.3% (vol/vol) Tween-20] containing 1% (wt/vol) BSA, and 10% (vol/vol) goat serum [TBS-T(0.3%)-B-G] for 15 min at room temperature. Sections were incubated at 4°C overnight in 50 μl of the anti-α-kafirin, anti-β-kafirin, anti-γ-kafirin, or preimmune serum (1:2,000, 1:500, 1:500, and 1:500 dilutions in TBS-B-G, respectively). After washing for 10 min in 2 ml of TBS-T(0.3%), the grids were blocked for 15 min with TBS-T(0.3%)-B-G. This was followed by incubation on a 50-μl drop of goat anti-rabbit colloidal gold conjugate [1:50 dilution in TBS-T(0.3%)] for 1 h and two washes with two 100-μl drops of water. The grids were allowed to dry for 2 h before poststaining with 2.5% (wt/vol) uranyl acetate for 5 min followed by 0.1% (wt/vol) lead citrate for 20 s. Sections were examined with a Philips EM 200 transmission electron microscope at 60 kV.
Results
Microstructure of Protein Bodies.
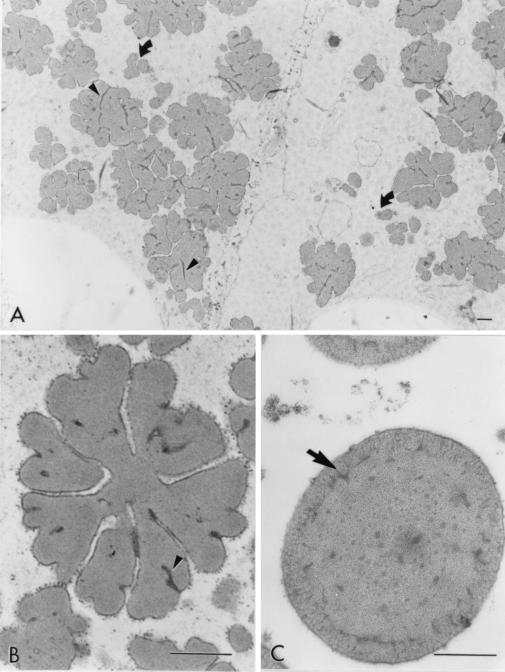
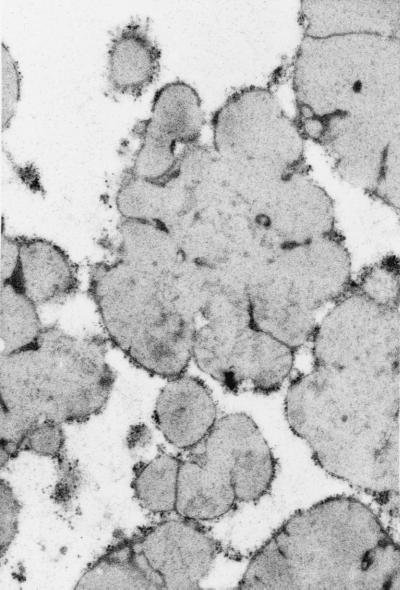
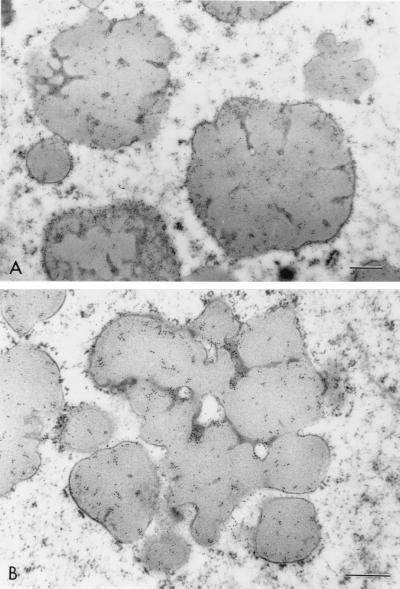
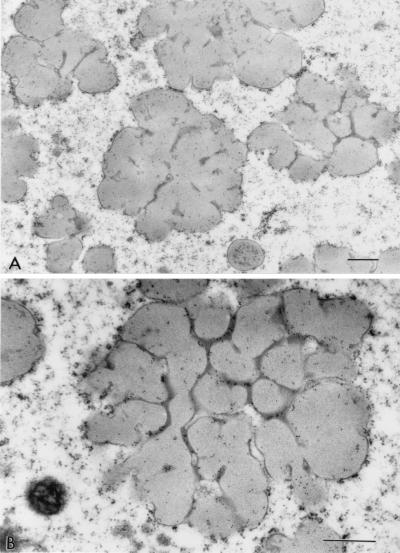
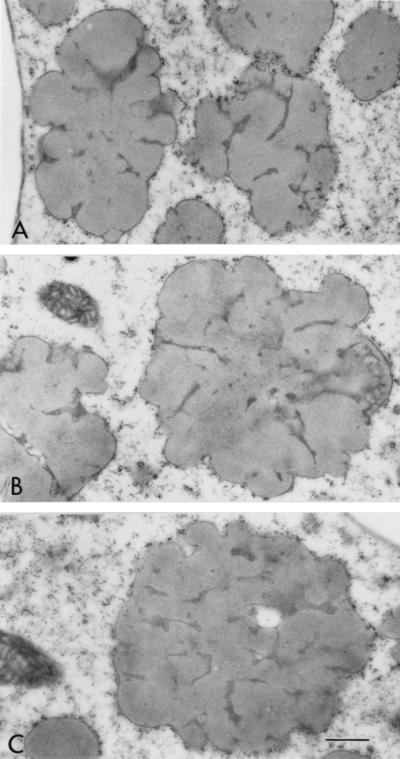
The protein bodies of the highly digestible sorghum mutant cultivar had a remarkably different and irregular shape compared with protein bodies of the normal cultivar, P721N (Fig. 1). Nearly all were characterized by deep invaginations or folds that in many cases reached the central area of the protein body forming irregularly shaped lobes. Dark inclusions also could be seen mainly at the base of the folds (Fig. 1 A and B; see small arrows). What appears to be small protein bodies are most likely cross-sectioned lobes that are adjacent to the central plane dissecting protein bodies (Fig. 1A; see curved arrows). A membrane structure surrounding the protein bodies was distinguishable. Normal protein bodies usually are characterized by a dark-stained periphery or ring (Fig. 1C; arrow) that was not present in protein bodies of this new genotype. The isogenic counterpart of P721N, P721Q, was found to have the same altered protein body structure (Fig. 2).
Figure 1.
Micrographs of peripheral endosperm of sorghum cultivars P851171 and P721N (30 DAHB). (A) Low magnification of P851171 highly digestible cultivar. Curved arrows point to lobes of protein bodies; small arrows point to dark-staining base folds (B) High magnification of a protein body from P851171; small arrow points at dark-staining folds. (C) High magnification of a protein body from P721N; the arrow points to a dark-staining peripheral ring. (Bar = 0.5 mm.)
Figure 2.
Micrograph of protein bodies of sorghum cultivar P721Q.
Immunolocalization of Kafirins.
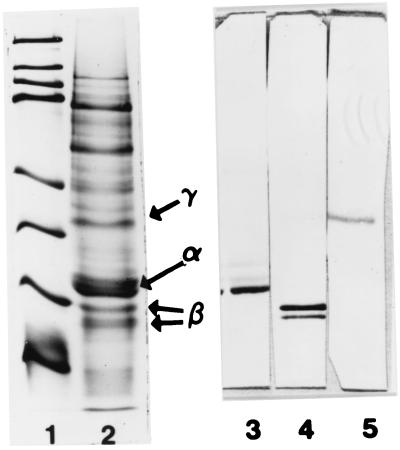
Western blots against individual kafirins showed that anti-α-, anti-β-, and anti-γ-kafirin sera reacted only with α-, β-, and γ-kafirin, respectively, from the highly digestible cultivar (Fig. 3, lanes 3–5). The high specificity of the antisera demonstrated the appropriateness for their use in the immunolocalization study.
Figure 3.
SDS/PAGE of molecular mass standards (lane 1; 14.4, 21.5, 31.0, 45.0, 66.2, 97.4, 116.2, and 200.0 kDa), extracted proteins of highly digestible sorghum cultivar P851171 (lane 2), and Western blots against α-, β-, and γ-kafirin antiserum (lanes 3, 4, and 5, respectively); α-, β-, and γ-kafirin are shown.
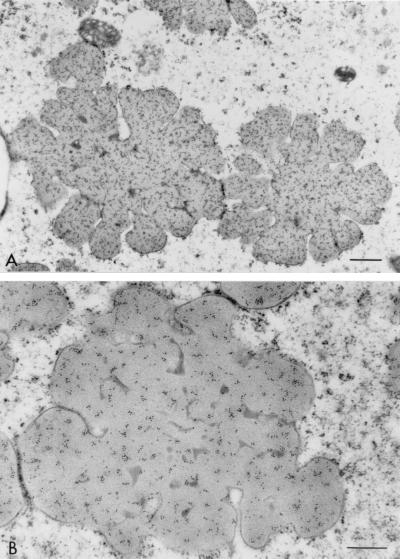
The incubation of sections with preimmune sera showed minimum nonspecific reaction (Fig. 4). α-Kafirin was localized homogeneously throughout the light-staining regions of the interior of the protein body (Fig. 5). Dark-staining areas showed little binding of the anti-α-kafirin serum and there appeared to be little anti-α-kafirin binding present at the protein body periphery. Examination of anti-β-kafirin-incubated sections revealed that β-kafirin was predominantly associated with the dark areas and at the periphery of the protein body lobes (Fig. 6). γ-Kafirin was strongly associated with the dark-staining inclusion bodies seen at the base of the invaginations (Fig. 7). A small proportion of the γ-kafirin was also located in the peripheral regions. Some of the β- and γ-kafirins were present as well in the light-staining areas but to a much lesser extent.
Figure 4.
Low magnification of sections of highly digestible sorghum cultivar P851171 incubated with preimmune serum from the three New Zealand rabbits used for α-kafirin (A), β-kafirin (B), and γ-kafirin (C) antibody production. (Bar = 0.5 mm.)
Figure 5.
Low (A) and high (B) magnification of sections of highly digestible sorghum cultivar P851171 (30 DAHB) peripheral endosperm incubated with antiserum against α-kafirin. (Bar = 0.5 mm.)
Figure 6.
Low (A) and high (B) magnification of sections of highly digestible sorghum cultivar P851171 (30 DAHB) peripheral endosperm incubated with antiserum against β-kafirin. (Bar = 0.5 mm.)
Figure 7.
Low (A) and high (B) magnification of sections of highly digestible sorghum cultivar P851171 (30 DAHB) peripheral endosperm incubated with antiserum against γ-kafirin. (Bar = 0.5 mm.)
Discussion
The numerous invaginations of the irregularly shaped protein bodies of this novel, highly digestible sorghum cultivar are unique and have not been reported before in sorghum. In normal cultivars, evidence supports that the peripheral location of the highly disulfide cross-linked γ-kafirin within the protein bodies impedes the digestion of other kafirins in the protein body (14, 16, 17). It appears that, in this new cultivar, the altered structure of the protein bodies and the change in location of γ-kafirin to dark-staining inclusions at the base of the folds are directly related to its substantially higher in vitro protein digestibility compared with normal cultivars (as shown in ref. 19). We believe that the related improved accessibility of proteases to the major storage protein α-kafirin, and the increased surface area caused by abnormal morphology of its protein bodies are the main causes of the high-protein digestibility of this cultivar. This is supported by data showing that, after 30 min of pepsin digestion, digestibility of α-kafirin in the highly digestible cultivar was 2.5 times higher than in the normal cultivar (19).
It might be argued that the higher protein digestibility of P851171 compared with the normal genotype P721N is in part the result of damage of the seemingly more fragile, irregularly shaped protein bodies during grinding to flour or from cooking. To investigate this possibility, P851171 mature grain was ground as for the digestibility assay (19). Cooked and uncooked flour were prepared and examined by transmission electron microscopy (not shown). The protein bodies remained intact and we concluded that the high-protein digestibility characteristic of this cultivar is not caused by disruption of protein bodies during grinding or cooking, but to the nature of the protein bodies themselves.
The mechanism of formation of protein bodies has not been well studied in sorghum, although it is understood to be similar to that of maize. The formation of protein bodies in maize is speculated to be partly related to the intrinsic structure of the prolamins. α-Zein expressed in Xenopus oocytes was shown to be assembled into protein body-like structures (22). However, the expression of only α-zein resulted in the formation of low-density protein bodies, which suggested that all prolamins are necessary for synthesis of normal protein bodies. Prolamins are deposited in the protein body with β- and γ-zeins located at the periphery and α-zein accumulating in the central region (23). Recently, it has been discovered that certain domains of γ-zeins are essential for the formation of normal protein bodies (24). In this study, various mutant γ-zeins were constructed and it was demonstrated that a proline-rich domain at the N terminus was one of the critical domains for normal protein body development. Also, by using transmission electron microscopy, it was shown that abnormal protein bodies with an amorphous structure were formed when a Cys-rich domain was deleted from γ-zein.
In the naturally occurring maize floury2 mutant, protein bodies exhibit a unique morphology consisting of an amorphous structure (25). Lopes et al. (25) found an unusual α-zein of 24 kDa in this mutant, and hypothesized that the unique features of floury2 may be a result of the accumulation of a defective α-zein outside of the endoplasmic reticulum. Subsequently, they found that there was a 1-bp substitution in the signal peptide region of the gene encoding the 24-kDa α-zein, resulting in its retention (26). They concluded that accumulation of some of the α-zein outside the endoplasmic reticulum results in, among other phenotypic effects, irregularly shaped protein bodies. In support of our findings in sorghum, we found that the maize Oh43 floury2 mutant had somewhat higher protein digestibility (15% higher titration volume by using the pH stat in vitro protein digestibility method) than its normal Oh43 counterpart (unpublished data). Given the similarities between zeins and kafirins (12, 14), and the nucleotide and aa sequence homologies between γ-zein and γ-kafirin (27), it is possible that there is a similar basis, originating in the α- or γ-kafirin proteins, for the distinctly invaginated sorghum protein bodies found in this study.
In conclusion, the cause of high-protein digestibility of this cultivar appears to be caused by the change in location of γ-kafirin, exposure of α-kafirin to digestive proteases, and structural alteration of the protein bodies resulting in greater surface area compared with normal bodies. These factors cause the rapid digestion of sorghum α-kafirin. Research to determine compositional and structural differences in the various kafirins needs to be performed to clarify the mechanism that causes the altered protein body structure.
Acknowledgments
We thank the Electron Microscope Center at Purdue University for the use of the facilities, and particularly the help and numerous suggestions and recommendations of Prof. Charles Bracker and Debbie Sherman throughout the microscopy study. This study was supported in part by the Agency for International Development INTSORMIL Grant LAG-1254-G-00–6009-00.
Abbreviations
- Q
(P721) opaque
- DAHB
days after half bloom
- TBS
20 mM Trizma base/500 mM sodium chloride
- TBS-T
TBS/0.5% Tween-20
- TBS-T(0.3%)
TBS/0.3% Tween 20
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080076297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080076297
References
- 1.MacLean W C, Lopez de Romanna G, Placko R P, Graham G G. J Nutr. 1981;111:1928–1936. doi: 10.1093/jn/111.11.1928. [DOI] [PubMed] [Google Scholar]
- 2.Butler L G, Riedl D J, Lebryk D G, Blytt H J. J Am Oil Chem Soc. 1984;61:916–920. [Google Scholar]
- 3.Elkin R G, Freed M B, Hamaker B R, Zhang Y, Parsons C M. J Agric Food Chem. 1996;44:848–853. [Google Scholar]
- 4.Axtell J D, Kirleis A W, Hassen M M, D'Croz Mason N, Mertz E T, Munck L. Proc Natl Acad Sci USA. 1981;78:1333–1335. doi: 10.1073/pnas.78.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamaker B R, Mertz E T, Kirleis A W, Axtell J D. J Agric Food Chem. 1986;34:647–649. [Google Scholar]
- 6.Graham G G, MacLean W C, Morales E, Hamaker B R, Kirleis A W, Mertz E T, Axtell J D. J Nutr. 1986;116:978–984. doi: 10.1093/jn/116.6.978. [DOI] [PubMed] [Google Scholar]
- 7.MacLean W C, Lopez de Romanna G, Gastanaduy A, Graham G G. J Nutr. 1983;113:2171–2177. doi: 10.1093/jn/113.10.2071. [DOI] [PubMed] [Google Scholar]
- 8.Mukuru S Z, Butler L G, Rogler J C, Kirleis A W, Ejeta G, Axtell J D, Mertz E T. J Agric Food Chem. 1992;40:1172–1175. [Google Scholar]
- 9.Eggum B O, Moniwar L, Knudsen K E B, Munck L, Axtell J D. J Cereal Sci. 1983;1:127–137. [Google Scholar]
- 10.Mitaru B N, Reichert R D, Blair R. Poultry Sci. 1985;64:101–106. doi: 10.3382/ps.0622065. [DOI] [PubMed] [Google Scholar]
- 11.Hamaker B R, Mohamed A A, Habben J E, Huang C P, Larkins B A. Cereal Chem. 1995;72:583–588. [Google Scholar]
- 12.Shull J M, Watterson J J, Kirleis A W. J Agric Food Sci. 1991;39:83–87. [Google Scholar]
- 13.Watterson J J, Shull J M, Kirleis A W. Cereal Chem. 1993;70:452–457. [Google Scholar]
- 14.Shull J M, Watterson J J, Kirleis A W. Protoplasma. 1992;171:64–74. [Google Scholar]
- 15.Hamaker B R, Mertz E T, Kirleis A W, Axtell J D. Proc Natl Acad Sci USA. 1987;84:626–628. doi: 10.1073/pnas.84.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oria M P, Hamaker B R, Shull J M. J Agric Food Chem. 1995;43:2148–2153. [Google Scholar]
- 17.Oria M P, Hamaker B R, Shull J M. J Cereal Sci. 1995;22:85–93. [Google Scholar]
- 18.Rom D L, Shull J M, Chandrashekar A, Kirleis A W. Cereal Chem. 1992;69:178–181. [Google Scholar]
- 19.Weaver C A, Hamaker B R, Axtell J D. Cereal Chem. 1998;75:665–670. [Google Scholar]
- 20.Mohan D. Ph.D. thesis. W. Lafayette, IN: Purdue University; 1975. pp. 1–110. [Google Scholar]
- 21.Slot J W, Geuze H J. Eur J Cell Biol. 1985;38:87–95. [PubMed] [Google Scholar]
- 22.Wallace J C, Galili G, Kawata E E, Cuellar R E, Shotwell M A, Larlins B A. Science. 1988;240:664–666. doi: 10.1126/science.2834822. [DOI] [PubMed] [Google Scholar]
- 23.Lending C R, Larkins B A. Plant Cell. 1989;1:1011–1023. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geli M I, Torrent M, Ludevid D. Plant Cell. 1994;6:1911–1922. doi: 10.1105/tpc.6.12.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes M A, Coleman C E, Kodrzycki R, Lending C R, Larkins B A. Mol Gen Genet. 1994;245:537–547. doi: 10.1007/BF00282216. [DOI] [PubMed] [Google Scholar]
- 26.Coleman C G, Lopes M A, Gillikin J W, Boston R S, Larkins B A. Proc Natl Acad Sci USA. 1995;92:6828–6831. doi: 10.1073/pnas.92.15.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Barros E G, Takasaki K, Kirleis A W, Larkins B A. Plant Physiol. 1991;97:1606–1607. doi: 10.1104/pp.97.4.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]