Abstract
Enzymatic transformations of macromolecular substrates such as DNA repair enzyme/DNA transformations are commonly interpreted primarily by active-site functional-group chemistry that ignores their extensive interfaces. Yet human uracil–DNA glycosylase (UDG), an archetypical enzyme that initiates DNA base-excision repair, efficiently excises the damaged base uracil resulting from cytosine deamination even when active-site functional groups are deleted by mutagenesis. The 1.8-Å resolution substrate analogue and 2.0-Å resolution cleaved product cocrystal structures of UDG bound to double-stranded DNA suggest enzyme–DNA substrate-binding energy from the macromolecular interface is funneled into catalytic power at the active site. The architecturally stabilized closing of UDG enforces distortions of the uracil and deoxyribose in the flipped-out nucleotide substrate that are relieved by glycosylic bond cleavage in the product complex. This experimentally defined substrate stereochemistry implies the enzyme alters the orientation of three orthogonal electron orbitals to favor electron transpositions for glycosylic bond cleavage. By revealing the coupling of this anomeric effect to a delocalization of the glycosylic bond electrons into the uracil aromatic system, this structurally implicated mechanism resolves apparent paradoxes concerning the transpositions of electrons among orthogonal orbitals and the retention of catalytic efficiency despite mutational removal of active-site functional groups. These UDG/DNA structures and their implied dissociative excision chemistry suggest biology favors a chemistry for base-excision repair initiation that optimizes pathway coordination by product binding to avoid the release of cytotoxic and mutagenic intermediates. Similar excision chemistry may apply to other biological reaction pathways requiring the coordination of complex multistep chemical transformations.
One of the great promises of x-ray crystallography is that visualization of enzyme structures will reveal the basis for enzyme mechanisms. However, this promise is rarely realized (1–3), and most “structure-based” enzyme mechanisms are presented without key information from both enzyme–substrate and enzyme–product complexes or with insufficient resolution for detailed analysis. Enzymatic catalysis is traditionally explained by substrate recognition, preferential transition-state binding and stabilization, and chemical catalysis by active-site functional groups (4). More controversial interpretations of enzymatic mechanisms include enzyme-enforced positioning and conformational changes in the substrate promoting stereoelectronic, proximity, and strain effects, which can control the relationship between conformation and reactivity of simple organic molecules in solution. In organic chemistry, a difference between an experimentally determined fact and an expectation is termed an effect. The anomeric effect and the σ-πArom effect are two stereoelectronic processes not often included in explanations for enzyme mechanisms. The anomeric effect refers to the observation that electronegative substituents in the anomeric position (C1′ in sugars) prefer to be axial even though they are expected to be equatorial from steric considerations (5). The σ-πArom effect relates to an explanation for features of pyridoxal phosphate-dependent enzyme catalysis, where breaking σ-bonds are hypothesized to be perpendicular to the aromatic pyridinium ring to optimize σ-π interactions (6, 7). Stereoelectronic processes likely also dominate enzymatic processes in squalene cyclase (8) and chorismate mutase (9), but the absence of structural data for relevant enzyme–substrate complexes limits conclusive analysis. The binding energies generated by large macromolecular interfaces have the potential to affect catalysis in ways typically difficult for small molecules, but the catalytic implications of these interfaces have not been explored thoroughly. The role of stereoelectronic effects in the catalytic activity of intermacromolecular complexes, such as those that catalyze the critical reactions of multienzyme DNA repair pathways, is thus incompletely understood.
Human uracil–DNA glycosylase (UDG) is an archetypical DNA glycosylase that initiates the multienzymatic DNA base-excision repair pathway (10, 11). Both misincorporation and cytosine deamination put the RNA base uracil into DNA. UDG excises uracil from both single- and double-stranded DNA (12) by cleaving the N-C1′ glycosylic bond between the base and deoxyribose. UDG undergoes a global conformational change from an “open” unbound state to a “closed” DNA-bound state in the UDG product complex, which evidently creates the catalytically competent active center (13, 14). This open-to-closed transition is a feature of the UDG architecture and is centered on a β-zipper (15). In the product UDG–DNA complex, UDG has flipped its target uridine nucleotide out of the DNA base stack and into this active center, where the glycosylic bond is cleaved. This leaves an unstable and cytotoxic abasic site in DNA, which is further processed by at least apurinic/apyrimidinic (AP) endonuclease (APE1), polymerase β, and DNA ligase III (10, 16). Biochemical data suggest UDG coordinates the transfer of its abasic site product to APE1 (14, 17), as this unstable intermediate can autolytically degrade into abnormal DNA strand breaks (18), retard DNA polymerases, cause base misincorporation (19, 20), engage in suicide reactions with topoisomerase I (21), and cause DNA double-strand breaks (22). Thus, the abasic site product from UDG is more toxic to the cell than the original uracil base damage (23–25).
Despite the importance of UDG and the many proposed mechanisms for its catalysis of glycosylic bond cleavage, the structural chemistry underlying efficient catalysis by UDG has remained elusive. Two groups, including our own, have proposed a traditional acid-base catalysis carried out by a cationic His-268 and an absolutely conserved Asp-145 (13, 26). Two proposals (27, 28) invoke only general base catalysis via the same catalytic aspartate and a neutral histidine. Another proposal (29), based on a computationally simulated uncleaved-substrate complex, predicts that Asp-145 is fully solvated in the substrate complex so that it cannot act as a general-base catalyst. These proposed mechanisms are inconsistent with each other and with the uncleaved-substrate crystal structure presented here. Moreover, they are problematic in that they rely entirely on functional group chemistry and thus do not adequately explain UDG's efficiency or the activity of mutants that alter evidently key functional groups (13, 27). UDG mutations that remove apparently key active site functional groups are actually faster enzymes than several wild-type DNA glycosylases (30). Thus, without knowledge of a substrate complex structure, the characterization of UDG mechanism is severely hindered. Here, we present the structures of UDG bound to both uncleaved-substrate analog at 1.8-Å resolution and cleaved-product DNA oligonucleotides at 2.0-Å resolution. These high-resolution cocrystal structures suggest how UDG applies part of the DNA-binding energy to glycosylic bond cleavage by conformationally altering its deoxyuridine substrate to take advantage of stereoelectronic and strain effects in what is then an apparently dissociative process.
Methods
DNA Synthesis.
2′-Deoxypseudouridine nucleotide was made as previously described (31), converted into its 5′-dimethoxytrityl (DMT) derivative, and phosphitylated to generate the 3′-phosphoramidite (G.W., unpublished results). Synthesis of 4′-thio-2′-deoxyuridine nucleotide was as previously described (32). A DMT group was added to the 5′ position and the molecule converted to the 3′-phosphoramidite. These modified phosphoramidites were incorporated into an oligonucleotide (5′-CTGTUATCTT-3′) by using an Applied Biosystems DNA synthesizer and phosphoramidites from Glen Research (Sterling, VA). Complementary strands were purchased from Midland Certified Reagent Company (Midland, TX). All DNA was HPLC purified, desalted with a Sep-Pak cartridge (Millipore) and the content verified by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry.
UDG Expression, Purification, and Activity Assay.
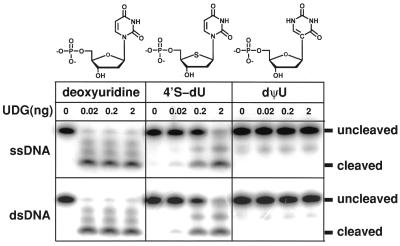
UDG is fully active recombinant human mitochondrial UDG consisting of residues 85–304 (numbered from UNG1) and an N-terminal methionine, glutamate, and phenylalanine coded by the expression vector. The deleted N-terminal peptide is involved in subcellular targeting and is not part of the catalytic domain. Expression and purification were as previously described (12). Labeling and annealing of oligonucleotide substrates for the activity assays were as previously described (33) by using the same DNA sequences as for the crystal structures. The two extra bands in the normal deoxyuridine lanes evidently result from minor 3′ piperidine cleavage of both the aldehyde and closed-ring forms of the abasic site created by UDG.
Crystallization and Data Collection.
Crystals were grown as previously described (14). Briefly, the synthesized DNA was mixed with UDG in a 3:1 ratio and incubated at room temperature for 30 min. Equal volumes of this UDG–DNA solution and reservoir solution [20% polyethylene glycol 4000 (Sigma)/100 mM Hepes buffer (pH 6.5)/10% dioxane/1 mM DTT] were mixed, with the crystals growing to full size within 2 mo. X-ray diffraction data for the 2′-deoxypseudouridine (dΨU) complex were collected from a single crystal flash cooled at −170°C at beamline 14-bmd of the Advanced Photon Source on a 2 × 2 Area Detector Systems Corporation (San Diego, CA) charge-coupled device detector. Diffraction data were processed with denzo and scalepack (34) to yield 116,160 observations of 26,594 unique reflections to 1.80 Å resolution (93% complete overall/76.9% last shell). The average I/σI was 14.9, with an overall Rsym between symmetry-related reflections of 0.052 (0.231 last shell). X-ray diffraction data for the 4′-thio-2′-deoxyuridine (4′S-dU) complex were collected under identical conditions at beamline 7–1 of the Stanford Synchrotron Radiation Laboratory on a 30-cm MAR image plate detector (MAR Research, Hamburg, Germany) and yielded 150,204 observations of 21,491 unique reflections to 2.0 Å resolution (99.9% complete/99.8% last shell). The average I/σI was 6.0 with an overall Rsym between symmetry-related reflections of 0.131 (0.464 last shell). Both the dΨU and 4′S-dU complexes are in space group P212121 (a = 48.7, b = 64.8, c = 95.0 Å), with one complex per asymmetric unit.
Structure Solution and Refinement.
The 4′S-dU complex structure was solved by molecular replacement with amore (35) with a UDG–DNA product complex (14) as a search model and all data from 4.0 to 15.0 Å resolution. The correct solution gave a correlation coefficient of 0.46 and an R value of 0.37, with the corresponding values for the next highest peak 0.25 and 0.51. This model was refined with cns (36). Cycles of stereochemically restrained simulated annealing and positional refinement were alternated with manual inspection and rebuilding of the structure performed with xfit (37). Individual atomic temperature values were assigned to all of the atoms and overall anisotropic temperature factor and bulk solvent corrections applied to the data set. Progress was monitored by the consistent decrease of the Rfree. The 4′S-dU structure, which has 1,809 nonhydrogen protein and 417 DNA atoms, was refined to an Rcryst of 0.228 and Rfree of 0.257 for 20,581 reflections from 20.0 to 2.0 Å resolution with rms deviations from ideality (38) of 0.010 Å for bond lengths and 1.48° for bond angles. The dΨU complex was solved with the 4′S-dU complex as a starting model and refined as with the 4′S-dU complex with 25,453 reflections from 20.0 to 1.8 Å resolution to an Rcryst of 0.216 and an Rfree of 0.233. The correctness of the current atomic structures was verified by agreement of electron density omit maps. Moreover, electron density difference maps of the active center are flat.
Results and Discussion
Generating a UDG–DNA Substrate Complex and Cocrystal Structure.
To understand glycosylic bond cleavage by UDG, we focused on solving the high-resolution structure of a UDG-uncleaved “substrate” DNA complex. The cocrystal structure of a catalytically impaired UDG Asp-145→Asn mutant with deoxyuridine-containing DNA (13) failed to capture the uncleaved substrate as even the diminished enzyme maintained significant residual activity, indicating that functional group chemistry is not the primary basis for UDG catalysis. From viral UDG studies with oligomers containing 2′,4′-dideoxy-4′-methylene-deoxyuridine and 4′S-dU (G.M.B., unpublished results), we deduced that the furanose oxygen is an essential component of any deoxyuridine analog for effective ligand studies with UDG. Moreover, UDG cleaves the glycosylic bond of 2′-fluoro-2′-deoxyuridine (2′F-dU) (39). Therefore, 2′-deoxypseudouridine (dΨU) was selected as a stable substrate isomer achieved solely by interchange of N1 and C5 in the uracil ring and having a C–C glycosylic bond (Fig. 1).
Figure 1.
UDG activity assays for substrate and product DNA constructs. Human UDG cleaves the glycosylic bonds of deoxyuridine and 4′S-dU but not the glycosylic bond of dΨU (see Methods). This is true even at high concentrations of UDG relative to DNA and over periods of weeks.
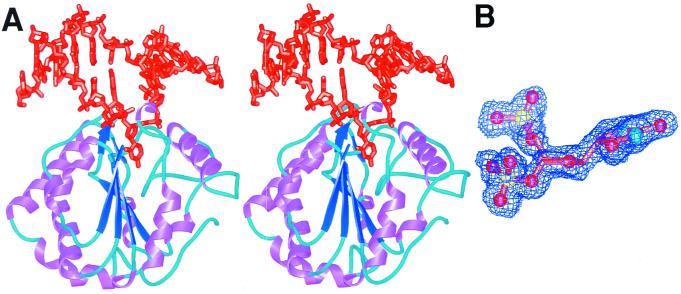
We solved high-resolution cocrystal structures of wild-type UDG in complex with DNA (Fig. 2A) containing the two substrate analogs, dΨU at 1.8 Å resolution (Fig. 2B) and 4′S-dU at 2.0 Å resolution. Human UDG cleaves uracil out of 4′S-dU at reduced but measurable rates relative to uracil from deoxyuridine to give a cleaved product complex. In contrast, and consistent with data from solution studies, UDG does not cleave the C–C glycosylic bond in dΨU (Figs. 1 and 2B) but provides an uncleaved transition-state-like UDG–DNA complex. This allows us to deduce an accurate chemical and structural mechanism for glycosylic bond cleavage by UDG, as the interchange of N1 and C5 between deoxyuridine and dΨU is isosteric. Moreover, this interchange does not affect the deoxyribose pucker, aromaticity, or hydrogen-bonding functionalities of the substrate to the enzyme [apart from N (5)H, which is passively directed at the face of a tyrosine in the complex].
Figure 2.
Cocrystal structures of UDG bound to uncleaved substrate and cleaved product DNA. (A) dΨU-containing DNA (orange) binds UDG near the C-terminal end of its central β-sheet (dark blue arrows), which is surrounded by eight α-helices (purple). (B) Experimental electron density defines the stereochemical deformation and intact bond for the substrate dΨU complex. The glycosylic bond of the dΨU (orange carbon tubes, red oxygens, blue nitrogens, yellow phosphorus) is not cleaved, as demonstrated by the simulated-annealed omit map (blue) contoured at 2σ. The normally trigonal planar 1-position is clearly distorted out of the plane of the uracil ring toward a tetrahedral geometry. Difference maps of the active-site center are flat, indicating that the distortion is accurately depicted by the crystal structure.
Coupling Enzyme Architecture and Active Center to Conformational Destabilization of Deoxyuridine.
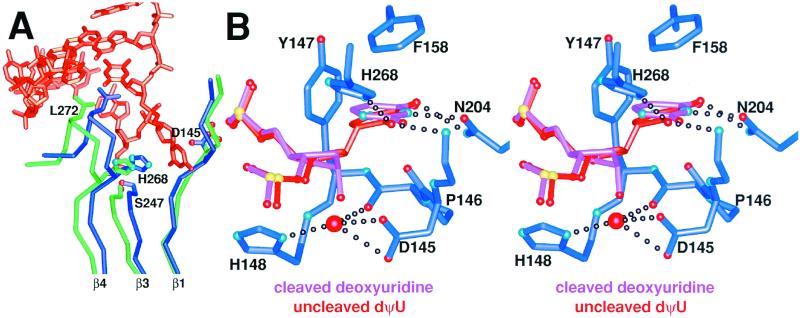
Our uncleaved “substrate” complex is markedly different from that derived by computer simulation (29), and the enzyme closely resembles the enzyme in the product complex, with a rms deviation of 0.26 Å for all Cα atoms. Thus UDG undergoes an architecturally determined conformational closing even in the uncleaved substrate analog complex. Shifts of the minor groove intercalation loop, insertion of the Leu-272 sidechain into the DNA base stack, and the β-zipper closing of β strands occur without bond cleavage in the UDG–DNA dΨU complex (Fig. 3A). Comparison of the substrate analog and product complexes shows that UDG's conformation is essentially unchanged throughout the reaction after closing on binding target DNA. Importantly, the UDG closing and coupled nucleotide flipping evidently funnel interaction energy into significant destabilizations of the substrate deoxyuridine via energetically demanding conformational distortions required by UDG active center binding.
Figure 3.
A UDG global conformational change on binding substrate DNA creates the enzyme active center, which thereafter remains unchanged during the glycosylic bond cleavage reaction. (A) Superposition of apo-UDG (green) and DNA-bound UDG (dark blue) with the uncleaved substrate DNA (orange) shows that UDG undergoes an architecturally determined conformational closing on binding substrate DNA. β1 and β3 increase the number of interstrand hydrogen bonds between them and thereby zip up the UDG β-zipper. This creates the catalytically competent active site by bringing L272 into the DNA base stack and H268 and D145 into the active center. (B) Superposition of the uncleaved-substrate (orange) and product (pink) UDG–DNA complexes reveals the basis of glycosylic bond cleavage by UDG. As the conformation of UDG (dark blue carbon tubes) remains unchanged throughout the reaction (see text), only the protein conformation from the dΨU structure is shown.
Structural features discernible in the 1.8-Å resolution UDG uncleaved-substrate complex suggest the enzyme uses both steric and stereoelectronic effects to achieve catalysis. DNA binds to UDG at the C-terminal end of its central four-stranded all-parallel β-sheet, which is surrounded by eight α-helices (Fig. 2A). The deoxyuridine analogs are flipped out of the DNA helix and into the enzyme active center, which is set in a cleft between β1 and β3 and partly formed by an Ω loop (13–15) (Fig. 2A). The enzyme–DNA macromolecular interface allows UDG to constrain and orient the uracil ring to stretch and weaken the N-C1′ glycosylic bond while simultaneously aligning orbitals for overlap from O4′ through the uracil O2. Three sets of interactions define a strikingly specific pocket for binding uracil that focuses the binding energy of the complex onto the deoxyuridine. First, direct hydrogen bond partners are available to every polar atom of the uracil ring (Fig. 3B). Second, a favorable face-to-face π-π stacking interaction is made with Phe-158. Finally, Tyr-147 prevents productive binding of thymine. However, none of these interactions evident in the product complex is optimized until after the glycosylic bond is cleaved (Fig. 3B).
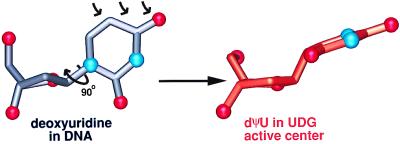
In the substrate analog complex, the UDG active center contorts the uracil and deoxyribose while the phosphates 5′ and 3′ of the uridine are anchored by interactions with a proline-rich loop and a glycine–serine loop. UDG rotates the uracil ring ≈90° on its N1–C4 axis to a position almost halfway between anti and syn (Fig. 4) with a χ angle of 177°. Importantly, this enzyme-induced deoxyuridine conformation causes steric clashes between the uracil C6 hydrogen and the deoxyribose O4′ and between uracil O2 and the deoxyribose C2′ hydrogen that together significantly stretch the glycosylic bond (40). As the uracil is still attached to the deoxyribose C1′ in the uncleaved dΨU UDG–DNA complex, the dΨU sugar is pulled ≈0.4 Å deeper into the enzyme pocket than in the product complex. Meanwhile, the dΨU uracil ring is sterically prevented from inserting as deeply into the active center as the cleaved uracil in the product complex (Fig. 3B). Therefore, in the dΨU–UDG uncleaved substrate complex, the C1–C6 (N1–C6 in normal uracil) edge is tilted ≈20° more toward the sugar, making the π-π interaction with Phe-158 less ideal and forming longer hydrogen bonds than in the cleaved-product complex (Fig. 3B). Thus, the cleaved uracil and DNA product complex achieves greater complementarity with the enzyme than does the uncleaved dΨU DNA substrate complex.
Figure 4.
Deoxyuridine (gray carbon tubes, red oxygens, blue nitrogens) in DNA is severely distorted by the UDG active center to achieve the observed conformation of the dΨU (orange carbon tubes, red oxygens, blue nitrogens). The left side of the large arrow is deoxyuridine in the conformation normally found in DNA. The arrow implies the observed flipping of the substrate nucleotide out of the DNA helix, which results in the altered position of the 5′P. When flipped into the UDG active center (right side of large arrow), the uracil ring is rotated ≈90° on its N1–C4 axis to a χ angle of 177°. Furthermore, the deoxyribose sugar of the enzyme bound substrate is flattened to a mild C3′-exo, which raises the uracil to a semiaxial position. The normally trigonal planar 1-position of uracil is strained to an almost tetrahedral geometry. The small arrows indicate the steric hindrance, which causes the deformation at the uracil 1-position. The conformation of deoxyuridine in DNA (gray) is derived from the conformation of deoxythymidine in a G/T mismatch (Protein Data Bank accession code 113D).
Before glycosylic bond cleavage, the normally trigonal planar C1 position (N1 in uracil) is distorted toward a tetrahedral geometry by the UDG active center. Unbiased 1.8-Å omit maps of the dΨU establish this high-energy distortion (Fig. 2B). Furthermore, electron density difference maps of the active center are flat, indicating that the structure accurately reflects the experimental x-ray diffraction data. Moreover, this observed tetrahedral distortion results from steric constraints imposed by the UDG active center and DNA binding. Phe-158 and Tyr-147 form the rigid walls of the extraordinarily specific UDG active center. These walls together with the tight anchoring of the DNA phosphates both 5′ and 3′ of the uridine nucleotide by the UDG–DNA interface interactions impose the tetrahedral geometry observed at the uracil 1-position (Fig. 4), and these UDG residues are unlikely to yield as they are buttressed by the bulk of the protein. Thus, the distortion seen in the enzyme–DNA complex dΨU structure is almost certainly effected in the natural DNA deoxyuridine substrate.
Although unlikely, the dΨU nucleotide in the UDG active center could be in a tautomeric form with a proton translocated from N5 to C1, thereby making C1 sp3 hybridized. This imino tautomer is calculated to be ≈7.5 kcal/mol higher in energy than the N (5)H tautomer (largely as a result of loss of aromaticity), but it is noteworthy that this energy lies within the range of energy generated through DNA binding to UDG (≈9 kcal/mol). If this tautomer were present, the C–C glycosylic bond of the dΨU might be cleaved by UDG, but no cleavage is observed (Fig. 1). Crucially, the strain energy required to generate this dΨU tautomer would equally distort the trigonal N1 of the normal deoxyuridine substrate toward a tetrahedral geometry. Thus, the presence of this high-energy tautomer would not affect the mechanistic arguments developed here for the biologically relevant deoxyuridine substrate.
Structural Implications for Two Coupled Stereoelectronic Effects in UDG Catalysis.
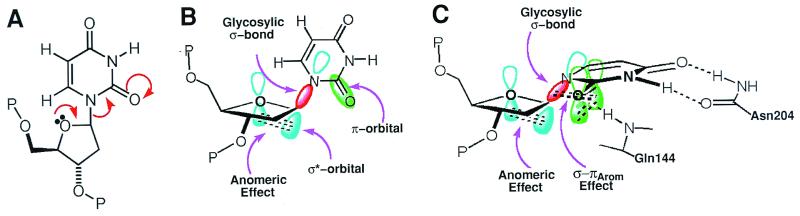
Importantly, the uncleaved dΨU complex shows that the UDG active center flattens the pucker of the uridine deoxyribose to a mild C3′-exo, thereby raising the glycosylic bond to a semiaxial position (Fig. 4). This deoxyribose flattening enables p-σ* overlap (41) with O4′ (Fig. 5), which is termed the anomeric effect. This electron orbital overlap is substantial because the angle between the glycosylic σ* orbital and the oxygen p orbital is ≈35°. Moreover, and consistent with the stronger anomeric stereoelectronic effect expected in the transition state (42), this angle between orbitals needing to transpose electrons is expected to decrease as the substrate is distorted further toward the transition state. In addition, a second stereoelectronic effect is achieved via the glycosylic bond rotation and pyramidalization of N1 (the glycosylic bond is bent ≈50° out of the plane of the uracil ring), which together create electron orbital overlap between the glycosylic bond and the C2=O and C4=O carbonyl π-systems (Fig. 5). This second effect is likely related to the σ-πArom stereoelectronic control, which is hypothesized to orient the breaking σ-bond to be perpendicular to the aromatic pyridinium ring in pyridoxal phosphate-dependent enzymes (6). Most importantly, the UDG–DNA structures show how an enzyme active center can implement and couple these two distinct stereoelectronic effects, the anomeric and σ-πArom effects, by distorting the substrate structure to increase sequential linear orbital overlap from O4′ to O2 (Fig. 5). In contrast, small molecule crystal structures showing how the N–C bonds in substituted uridines respond to electron withdrawal in the uracil show no departure from trigonal planar geometry at N1 (43). Therefore, UDG funnels macromolecular DNA substrate-binding energy into catalytic power by conformationally closing to enforce substrate distortions, which would be energetically difficult in small molecule systems that lack large interfaces and transitions between two distinct rigid conformational states. The architecturally stabilized closed UDG conformation and active center structure thus couple two distinct yet complementary stereoelectronic effects that promote efficient catalysis by altering three orthogonal nonoverlapping electron orbitals into stereochemically defined conformations, favoring the electron transpositions for glycosylic bond cleavage.
Figure 5.
Structure-based reaction mechanism that resolves the apparent orthogonal paradox for electron transpositions by altering the substrate stereochemistry. (A) A simplified valence-bond representation of the glycosylic bond dissociation hides the paradox that the three electron pairs to be transposed are involved in orthogonal orbitals. (B) In the normal anti-conformation of deoxyuridine, the σ*-orbital involved in the anomeric effect and the π-orbital of the C2⩵O bond are orthogonal to one another, thus preventing orbital overlap. (C) Severe distortions of the deoxyribose and the glycosylic bond in the strained conformation of deoxyuridine enforced by the UDG active center align the pairs of atomic orbitals participating in each electron transposition, thereby electronically coupling the anomeric and σ-πArom effects to promote bond cleavage.
Unlike the computationally simulated uncleaved-substrate complex (29), our experimental 1.8-Å cocrystal structure shows that in the uncleaved substrate complex, the active center Asp-145 is in the closed conformation seen in the product complexes. A water is tightly positioned 3.54 Å below the deoxyribose C1′ by four hydrogen bonds (Fig. 3B) but is likely a poor nucleophile. Instead, the UDG–DNA substrate structure suggests this water is well positioned to become the 1′-α-OH after dissociation of the incipient deoxyribose oxocarbenium ion from the uracil ring. On cleavage, the UDG–DNA product structure shows that the uracil tilts to move deeper into the active site, shortens hydrogen bonds, and improves its stacking interaction with Phe-158. Furthermore, in the cleaved product complex, the abasic nucleotide relaxes to a more puckered C2′-endo form and withdraws from the enzyme (≈0.4 Å), thereby reducing sterically induced strain (Fig. 3B). Thus, the experimentally defined product complex is significantly less strained than the experimentally defined uncleaved-substrate complex. These structural results are consistent with biochemical results showing that UDG binds preferentially to its cleaved product (14). These experimental DNA-binding measurements indicate that human UDG binds with 11.2 kcal/mol of binding energy to product but with only 8.8 kcal/mol binding energy to its uncleaved substrate.
Control of Substrate and Product Stereochemistry by the UDG Active Center Structure.
The open-to-closed UDG conformational switch, which is stabilized by UDG architectural features (Fig. 3A), occurs in the uncleaved substrate complex so that the resulting tightly closed UDG active center controls and strains the stereochemistry for the DNA substrate. We propose that the UDG nucleophilic substitution reaction is substantially dissociative in nature and involves substrate strain induced by the macromolecular interface (Figs. 4 and 5) coupled to the formation of a stable uracil enolate anion. However, a simplified valence-bond representation of this dissociation reaction (Fig. 5A) hides the paradox that the three transposing electron pairs are in orthogonal orbitals that evidently preclude the overlap needed for the proposed electron transpositions (Fig. 5B). We term this lack of overlap between normally perpendicular orbitals involved in the electron transposition the “orthogonal paradox.” The structures of the UDG–DNA substrate analog and product complexes show that the strained deoxyuridine conformation effected by the UDG active center overcomes this orthogonal paradox by enforcing stereoelectronic coupling of the anomeric and σ-πArom effects (Fig. 5C) to create a concerted path for the electrons. As far as we are aware, this enzyme-enforced coupling of stereoelectronic effects, to ensure optimized molecular orbital overlap and thereby generate the concerted mechanism we have described, is unprecedented. These enzyme–DNA complex structures and this mechanism provide the basis for detailed computational, mechanistic, and structural analyses, which will ultimately quantitate where along the SN1 to SN2 continuum the UDG reaction mechanism lies. Although kinetic isotope measurements of the UDG reaction could quantitate the relative contributions of associative and dissociative processes to glycosylic bond cleavage, these will be challenging, as both nucleotide flipping and product dissociation are slower than bond cleavage.
We discern five features from the experimental UDG substrate and product complexes that suggest the UDG active center provides an environment for the uracil and deoxyribose that is characteristic of a substantially dissociative (SN1-like) process. (i) The substrate O4′ is effectively sp2 hybridized (C-O-C angle 120°), unsolvated, and oriented for p-overlap with σ* of the glycosylic bond targeted for cleavage (Figs. 4 and 5). (ii) The uracil ring is rotated ≈90° to a χ angle of 177°, and the glycosylic bond is deformed with respect to the pyrimidine ring, thereby making concerted p-σ* and σ-πArom interactions favorable in the transition state (Figs. 4 and 5). (iii) The leaving group (uracil) is more effectively activated by hydrogen bonding (four hydrogen bonds) than the ultimate nucleophile (water) and moves away ≈0.8 Å into the pyrimidine pocket after cleavage (Fig. 3B). Consequently, the uracil N1 ends up ≈3 Å away from the deoxyribose C1′. (iv) The ultimate water nucleophile is tightly held by hydrogen bonds to three amino acids and is 3.6 Å away from C1′. In the product structure, the anomeric carbon (C1′) has moved 0.8 Å toward the water nucleophile, which itself moves ≈2 Å toward the C1′ to become the C1′-α hydroxyl group (Fig. 3B). (v) The water, the anomeric carbon (C1′), and the leaving group (N1) are not “in line,” as is optimally required for an associative process (wat-C1′-N1 angle 137°).
The most likely net result of the above-noted experimentally defined stereochemical relationships is that the leaving group is an enolate anion of uracil [pKa = 9.5 (44)]. The structures show that no definitive proton donors are within 4 Å of the uracil O2, which forms hydrogen bonds to the backbone amide of Gln-144 and to the sidechain of His-268. Deprotonation of the Gln-144 backbone amide would form an unstable enolate on the protein, which is less favorable than a uracil enolate. Moreover, His-268 is neutral (27) as measured by NMR (28). Yet, the hydrogen bond partners (Fig. 3B) likely stabilize the proposed uracil enolate anion leaving group without explicit protonation. Subsequent to bond cleavage, protonation on N1 gives the more stable amide and completes the reaction.
The chemical mechanism implicated by these two high-resolution UDG–DNA cocrystal structures explains how the substitution of the isomeric substrate dΨU for deoxyuridine arrests UDG activity, whereas the nonisomeric 4′S-dU modification is cleaved (Fig. 1). The 4′S-dU both has a normal N-C1′ glycosylic bond and also supports movement of electrons from S4′ to uracil O2 and therefore retards but does not prevent dissociation of the attached uracil. 2′F-dU is a similarly poor substrate for UDG (107-fold slower with t1/2 of ≈20 h) (39), and this result also biochemically supports our structure-based mechanistic description of glycosylic bond cleavage as a substantially dissociative process. The β-anomer of 2′F-dU is similar to deoxyuridine with respect to radius and sugar pucker but differs in electronic properties with an increased positive charge on C1′ (45). However, this electronic difference does not completely abrogate glycosylic bond cleavage for 2′F-dU substrates, because electrons can still flow from the sugar O4′ to the uracil. The resistance of dΨU to bond cleavage likely derives from the higher energy of a C–C bond (82.6 kcal/mol) relative to an N–C bond (72.8 kcal/mol) and to the high energy of formation of an unstable arene carbanion in the absence of simultaneous proton donation to C1 of dΨU. Indeed, the nearest proton donor to N1 (His-268) is held ≈4 Å away and is known to be neutral (28). In effect, the C–C glycosylic bond in the dΨU analog likely acts as an insulator to inhibit the flow of electrons from the sugar O4′ to the uracil ring, thus blocking the key coupling of the anomeric and σ-πArom stereoelectronic effects predicted here by the experimentally defined substrate and product stereochemistry.
Implications for Chemical Biology.
The conservation of UDG from bacteria to humans suggests that over 1.5 billion years of evolution, the appropriate polar functional groups for optimal stabilization of the transition state of glycosylic bond cleavage ought to have been achieved. These structural and biochemical results therefore provide a unique insight into the fundamental structural chemistry of catalysis for a dissociative substitution reaction. They show how an enzyme active center makes use of macromolecular substrate recognition and binding energy to distort the ground state to enforce stereoelectronic cooperativity between the anomeric and σ-πArom effects. This mechanism of promoted reactivity is likely rare in small molecule systems, where binding energy is normally insufficient to enforce the needed substantial substrate distortions. Many of the features revealed by juxtaposition of these DNA substrate and product complexes for UDG are recognizable as the basis of general electronic theory of reaction mechanisms (5). Thus, these high-resolution substrate and product cocrystal structures show how these electronic reaction mechanisms can be implemented cooperatively in an enzyme active center. Moreover, these results for the human BER enzyme UDG reveal a conservation of the tightly closed enzyme conformation throughout the reaction that is consistent with dominant roles for uridine nucleotide destabilization and product binding in initiating DNA repair excision. It is intriguing to consider how UDG's biological function may be served better by this type of strain-induced stereochemical cooperativity, which may be broadly applicable in reaction mechanisms for enzymatic transformations of macromolecules. The structurally and biochemically observed preference for tighter product than substrate binding suggests that the structurally defined reaction chemistry outlined here is optimized to couple the excision chemistry step initiating base-excision repair to DNA repair pathway progression by delaying the release of cytotoxic and mutagenic DNA abasic site products.
Acknowledgments
We thank C. Putnam, D. Daniels, C. Mol, M. Stroupe, K. Morgan, and the late R. T. Walker for helpful discussions, as well as the Stanford Synchrotron Radiation Laboratory and BioCARS Advanced Photon Source staff scientists for data collection facilities. We thank the Biotechnology and Biological Sciences Research Council (G.M.B.), the National Institutes of Health (J.A.T.), the Skaggs Institute for Chemical Biology (S.S.P.), the Norwegian Cancer Society (G.S.), and a Graduate Research Fellowship from the National Science Foundation (S.S.P.) for support.
Abbreviations
- UDG
uracil–DNA glycosylase
- dΨU
2′-deoxypseudouridine
- 4′S-dU
4′-thio-2′-deoxyuridine
- Arom
aromatic
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1emh and 1emj).
References
- 1.Davies G J, Mackenzie L, Varrot A, Dauter M, Brzozowski A M, Schulein M, Withers S G. Biochemistry. 1998;37:11707–11713. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 2.Sidhu G, Withers S G, Nguyen N T, McIntosh L P, Ziser L, Brayer G D. Biochemistry. 1999;38:5346–5354. doi: 10.1021/bi982946f. [DOI] [PubMed] [Google Scholar]
- 3.Uitdehaag J C, Mosi R, Kalk K H, vanderVeen B A, Dijkhuizen L, Withers S G, Dijkstra B W. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 4.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. [Google Scholar]
- 5.Kirby A J. The Anomeric Effect and Related Stereoelectronic Effects at Oxygen. Berlin: Springer; 1983. [Google Scholar]
- 6.Dunathan H C. Proc Natl Acad Sci USA. 1966;55:712–716. doi: 10.1073/pnas.55.4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weyand M, Schlichting I. Biochemistry. 1999;38:16469–16480. doi: 10.1021/bi9920533. [DOI] [PubMed] [Google Scholar]
- 8.Abe I, Zheng Y F, Prestwich G D. J Enzym Inhib. 1998;13:385–398. doi: 10.3109/14756369809020544. [DOI] [PubMed] [Google Scholar]
- 9.Lee A Y, Stewart J D, Clardy J, Ganem B. Chem Biol. 1995;2:195–203. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 11.Parikh S S, Mol C D, Tainer J A. Structure (London) 1997;5:1543–1550. doi: 10.1016/s0969-2126(97)00303-1. [DOI] [PubMed] [Google Scholar]
- 12.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle N M, Haug T, Levine D W, Krokan H E. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 13.Slupphaug G, Mol C D, Kavli B, Arvai A S, Krokan H E, Tainer J A. Nature (London) 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 14.Parikh S S, Mol C D, Slupphaug G, Bharati S, Krokan H E, Tainer J A. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Putnam C D, Shroyer M J, Lundquist A J, Mol C D, Arvai A S, Mosbaugh D W, Tainer J A. J Mol Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 16.Parikh S S, Mol C D, Hosfield D J, Tainer J A. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 17.Mol C D, Izumi T, Mitra S, Tainer J A. Nature (London) 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T. Mutat Res. 1990;238:305–311. doi: 10.1016/0165-1110(90)90022-4. [DOI] [PubMed] [Google Scholar]
- 19.Xiao W, Samson L. Proc Natl Acad Sci USA. 1993;90:2117–2121. doi: 10.1073/pnas.90.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinedinst D K, Drinkwater N R. Mol Carcinog. 1992;6:32–42. doi: 10.1002/mc.2940060107. [DOI] [PubMed] [Google Scholar]
- 21.Pourquier P, Ueng L-M, Kohlhagen G, Mazumder A, Gupta M, Kohn K W, Pommier Y. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 22.Kingma P S, Osheroff N. J Biol Chem. 1997;272:1148–1155. doi: 10.1074/jbc.272.2.1148. [DOI] [PubMed] [Google Scholar]
- 23.Demple B, Johnson A, Fung D. Proc Natl Acad Sci USA. 1986;83:7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb L A. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 25.Kingma P S, Corbett A H, Burcham P C, Marnett L J, Osheroff N. J Biol Chem. 1995;270:21441–21444. doi: 10.1074/jbc.270.37.21441. [DOI] [PubMed] [Google Scholar]
- 26.Savva R, McAuley-Hecht K, Brown T, Pearl L. Nature (London) 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 27.Shroyer M J, Bennett S E, Putnam C D, Tainer J A, Mosbaugh D W. Biochemistry. 1999;38:4834–4845. doi: 10.1021/bi982986j. [DOI] [PubMed] [Google Scholar]
- 28.Drohat A C, Xiao G, Tordova M, Jagadeesh J, Pankiewicz K W, Watanabe K A, Gilliland G L, Stivers J T. Biochemistry. 1999;38:11876–11886. doi: 10.1021/bi9910880. [DOI] [PubMed] [Google Scholar]
- 29.Luo N, Mehler E, Osman R. Biochemistry. 1999;38:9209–9220. doi: 10.1021/bi990262h. [DOI] [PubMed] [Google Scholar]
- 30.Krokan H E, Standal R, Slupphaug G. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H C, Daves G D. J Org Chem. 1992;57:4690–4696. [Google Scholar]
- 32.Otter G P, Elzagheid M I, Jones G D, MacCulloch A C, Walker R T, Oivanen M, Klika K D. J. Chem. Soc. Perkin Trans. 2. 1998. , 2343–2349. [Google Scholar]
- 33.Nilsen H, Yazdankhah S P, Eftedal I, Krokan H E. FEBS Lett. 1995;362:205–209. doi: 10.1016/0014-5793(95)00244-4. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 36.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.McRee D E. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 38.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 39.Stivers J T, Pankiewicz K W, Watanabe K A. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 40.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 41.Andrews C W, Fraser-Reid B, Bowen J P. The Anomeric Effect and Associated Stereoelectronic Effects. 1992. , in ACS Symposium Series No. 204, ed. Thatcher, G. R. J. (Am. Chem. Soc.), pp. 114–125. [Google Scholar]
- 42.Benner S A. In: Mechanistic Principles of Enzyme Activity. Liebman J F, Greenberg A, editors. New York: VCH; 1988. pp. 27–74. [Google Scholar]
- 43.Egert E, Linder H J, Hillen W, Böhm M C. J Am Chem Soc. 1980;102:3707–3713. [Google Scholar]
- 44.Dawson R M C, Elliott D C, Elliott W H, Jones K M. Data for Biochemical Research. Oxford: Clarendon; 1959. [Google Scholar]
- 45.Schärer O D, Kawate T, Gallinari P, Jiricny J, Verdine G L. Proc Natl Acad Sci USA. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]