Abstract
Growth factors such as insulin regulate phosphatidylinositol 3-kinase-dependent actin cytoskeleton rearrangement in many types of cells. However, the mechanism by which the insulin signal is transmitted to the actin cytoskeleton remains largely unknown. Yeast two-hybrid screening revealed that the phosphatidylinositol 3-kinase downstream effector phosphoinositide-dependent protein kinase-1 (PDK1) interacted with protein kinase N (PKN), a Rho-binding Ser/Thr protein kinase potentially implicated in a variety of cellular events, including phosphorylation of cytoskeletal components. PDK1 and PKN interacted in vitro and in intact cells, and this interaction was mediated by the kinase domain of PDK1 and the carboxyl terminus of PKN. In addition to a direct interaction, PDK1 also phosphorylated Thr774 in the activation loop and activated PKN. Insulin treatment or ectopic expression of the wild-type PDK1 or PKN, but not protein kinase Cζ, induced actin cytoskeleton reorganization and membrane ruffling in 3T3-L1 fibroblasts and Rat1 cells that stably express the insulin receptor (Rat1-IR). However, the insulin-stimulated actin cytoskeleton reorganization in Rat1-IR cells was prevented by expression of kinase-defective PDK1 or PDK1-phosphorylation site-mutated PKN. Thus, phosphorylation by PDK1 appears to be necessary for PKN to transduce signals from the insulin receptor to the actin cytoskeleton.
Actin cytoskeleton reorganization has been implicated in a variety of cellular functions, including motility, intracellular trafficking, cytokinesis, and cell survival (1–3). In many cell types, the growth factor-induced actin cytoskeleton rearrangement is known to be mediated by the activation of phosphatidylinositol 3-kinase (PI3-kinase) (4–6). However, the signaling components linking PI3-kinase to the actin cytoskeleton remain elusive. The stimulation of cells with insulin leads to the activation of PI3-kinase downstream effectors such as phosphoinositide-dependent protein kinase-1 (PDK1). PDK1 is a Ser/Thr protein kinase that phosphorylates and activates members of the AGC (protein kinase A, G, and C) protein kinase family, such as protein kinase B (PKB/Akt), protein kinase C (PKC), cAMP-dependent protein kinase (PKA), and p70 S6 kinase (7–13). Thus, PDK1 may play a key role in the diversification of upstream signals to regulate specific downstream biological activities in cells.
Sequence comparison of conserved PDK1 phosphorylation motifs in known substrates led us to speculate that there may be additional targets for PDK1 (11). To identify PDK1 interactive proteins, we screened a yeast two-hybrid cDNA library. In the present study, we report the identification of a substrate for PDK1, the Rho-activated Ser/Thr protein kinase N (PKN). Our results show that PKN interacts with PDK1 in vitro and in cells and is phosphorylated and activated by PDK1. We also found that overexpression of PKN or PDK1 mimics insulin action to induce actin cytoskeleton reorganization. Furthermore, we have shown that insulin-induced actin cytoskeleton reorganization can be prevented by ectopically expressing mutant forms of either PKN or PDK1. These findings suggest that phosphorylation of PKN by PDK1 may play a potential role in mediating insulin signaling to the actin cytoskeleton.
Materials and Methods
Yeast Two-Hybrid Studies.
Yeast two-hybrid plasmids encoding various regions of PDK1 were constructed by subcloning the PCR-amplified PDK1 cDNA fragments (Fig. 1B) into the EcoRI/XhoI sites of pGBT9 (CLONTECH), fused in-frame with the GAL4 DNA binding domain. The two-hybrid screen was done with a bait plasmid encoding full-length mouse PDK1 (amino acids 4–559) and a pretransformed cDNA library derived from HeLa cell mRNAs (CLONTECH) according to the protocol of the manufacturer. The interaction between PDK1 and the carboxyl terminus of PKN was determined by β-galactosidase filter assays as described (14).
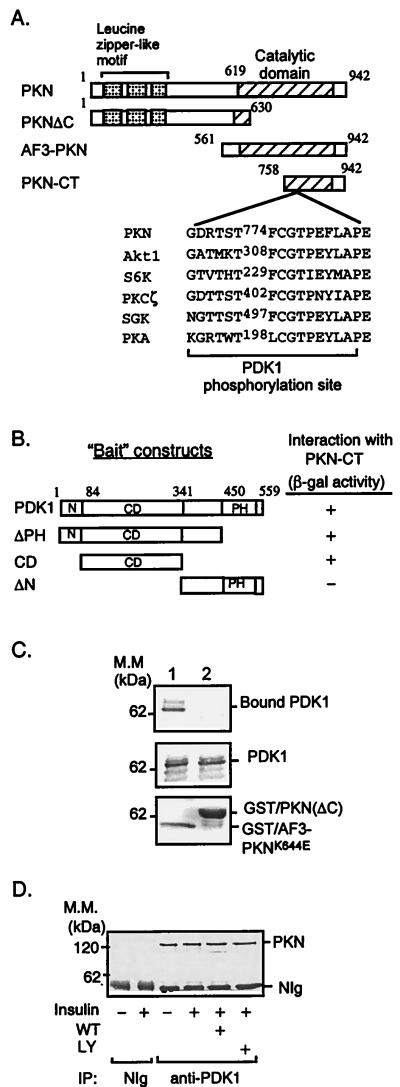
Figure 1.
Interaction of PKN with PDK1. (A) Schematic representation of PKN constructs used to study PKN and PDK1 interaction and phosphorylation. The consensus PDK1 phosphorylation sequences of several PDK1 substrates are shown. (B) Interaction of PDK1 with PKN in the yeast two-hybrid system. cDNAs containing different regions of PDK1 were amplified by PCR and subcloned into the yeast two-hybrid plasmid pGBT9. The interaction between PDK1 fragments and PKN-CT in SFY526 yeast cells was detected by β-galactosidase filter assays as described (14). +, Positive interaction (blue color) was visualized within 30 min; −, no interaction was detected for 24 h. N, Amino terminus; CD, catalytic domain; PH, pleckstrin homology domain. (C) Interaction between PDK1 and PKN in vitro. Lysates of CHO/IR/PDK1 cells (11) were incubated with GST/AF3-PKNK644E (lane 1) or GST/PKN(ΔC) (lane 2) bound to glutathione-Sepharose beads (Sigma). The bound proteins were resolved by SDS/PAGE, transferred onto a nitrocellulose membrane, and examined by immunoblotting with antibody to the hemagglutinin tag (Top). Five percent of cell lysates used in the immunoprecipitation were loaded as a control (Middle). The GST fusion proteins used in the experiments were separated by SDS/PAGE and visualized by Coomassie blue staining (Bottom). Results are representative of three independent experiments. (D) Interaction of PDK1 and PKN in intact cells. CHO/IR cells were serum-starved overnight and treated with or without wortmannin (WT, 200 nM) or LY294002 (LY, 200 μM) for 1 h. Cells then were treated with (+) or without (−) 10 nM insulin for 10 min. Cell lysates were incubated for 8 h at 4°C with affinity-purified polyclonal antibody to PDK1 (11) or NIg bound to protein A beads. PKN coimmunoprecipitated with PDK1 was detected with a mAb to PKN (Transduction Laboratories).
Site-Directed Mutagenesis.
Mutants of PKN or AF3-PKN were generated by single-stranded site-directed mutagenesis, using pRc/CMV/PKN or pRc/CMV/AF3-PKN (15) as templates. Mutations were confirmed by DNA sequencing.
Cell Culture.
Chinese hamster ovary (CHO) cells stably expressing the insulin receptor (IR) (CHO/IR) have been described (11). CHO/IR cells stably expressing PKN, AF3-PKN, or AF3-PKNT774A were generated by a standard protocol (11) and selected with 10 μg/ml puromycin. Cells expressing recombinant proteins were grown in 60-mm plates, serum-starved for 4 h, and lysed in lysis buffer consisting of 50 mM Hepes (pH 7.6), 150 mM NaCl, 1% NP-40, 5 mM EDTA, 1 mM NaF, 1 mM sodium pyrophosphate, 1 mM orthovanadate, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μM microcystin-LR, and 1 mM PMSF. The homogenate was centrifuged (10,000 × g, 4°C, 10 min), and supernatants were used in binding or immunoprecipitation experiments.
In Vitro Binding Studies.
Glutathione S-transferase (GST)-PKN fusion proteins were generated by subcloning the PCR-amplified cDNA fragments encoding different regions of PKN (Fig. 1A) into pGEX-4T-3 vector (Amersham Pharmacia) in-frame with the sequence encoding GST. Fusion proteins expressed in Escherichia coli DH5α cells were purified by adsorption to glutathione agarose beads (Sigma). The beads were washed three times with buffer A consisting of 50 mM Hepes (pH 7.6), 150 mM NaCl, and 0.1% NP-40. Lysates of CHO/IR/PDK1 cells (11) were added to the beads and incubated for 4 h at 4°C with gentle shaking. The beads were washed three times with buffer A, and the bound proteins were eluted with sample loading buffer and boiled at 95°C for 3 min. Proteins were resolved by SDS/PAGE and transferred onto a nitrocellulose membrane, and the bound PDK1 was visualized by Western blot with anti-PDK1 antibody (11).
Phosphorylation of PKN by PDK1.
FLAG-tagged PKN, AF3-PKN, or AF3-PKNT774A was immunoprecipitated from cells transiently or stably expressing these proteins with anti-FLAG M2 antibody (IBI-Kodak) adsorbed onto protein G agarose beads. After incubation at 4°C for 4–6 h, the beads were collected and washed twice with ice-cold buffer A and once with ice-cold PDK1 kinase buffer (50 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/0.1 mM sodium orthovanadate/0.1 mM sodium pyrophosphate/1 mM NaF/1 mM PMSF). Unless otherwise indicated, phosphorylation of PKN was carried out at 30°C for 30 min in the presence of PDK1 kinase buffer (final volume 30 μl), 2 μCi [γ-32P]ATP, and immunoaffinity-purified PDK1 or PDK1K114G (11). The reaction was terminated by the addition of 5 μl of 0.5 M EDTA. After separation by SDS/PAGE, protein phosphorylation was detected by autoradiography.
In Vivo Labeling.
CHO/IR cells were cotransfected with cDNAs encoding PKN and the control vector pBEX, pBEX-PDK1, or pBEX-PDK1K114G (11). Thirty-six hours after transfection, cells were in vivo-labeled with 32P-orthophosphate according to a protocol described previously (16). Cells then were stimulated with (+) or without (−) 10 nM insulin for 10 min and lysed. PKN was immunoprecipitated with antibody to the FLAG tag, resolved by SDS/PAGE, and transferred to a nitrocellulose membrane. The phosphorylation of the protein was visualized by autoradiography and quantified by using a PhosphorImager. Equal expression of PKN in these cells was confirmed by Western blot analysis using the anti-FLAG antibody.
PKN Kinase Assays.
FLAG-tagged PKN was immunoprecipitated from serum-starved CHO/IR cells transiently expressing the protein and phosphorylated by PDK1 as described above, except that 2 μM ATP was included in the kinase buffer instead of [γ-32P]ATP. The phosphorylated PKN absorbed to the protein G beads was washed twice with buffer A and once with the PKN kinase buffer containing 20 mM Tris⋅HCl (pH 7.5), 20 μM ATP, and 8 mM MgCl2. Kinase assays were carried out at 30°C for 5 min in the presence of PKN kinase buffer (final volume 30 μl), 2 μCi [γ-32P]ATP, phosphorylated or nonphosphorylated PKN, and the ribosomal S6 peptide (amino acids 229–239). This peptide previously has been shown to be a preferred substrate for PKN (17), but not for PDK1 (data not shown).
Immunofluorescence Studies.
3T3-L1 or Rat1-IR cells were seeded in 24-well plates containing coverslips and transfected with plasmids encoding the wild-type or mutant forms of PDK1 or PKN by Lipofectamine (GIBCO/BRL). Twenty-four hours after transfection, cells were either fixed directly or were first serum-starved for 4 h, treated with 10 nM insulin for 10 min, and then fixed in PBS containing 4% paraformaldehyde for 1 h at 4°C. Fixed cells were blocked and permeabilized with PBS containing 0.05% Triton X-100 and 5% normal goat serum. Actin cytoskeleton was visualized by rhodamine-phalloidin staining (Molecular Probes), and images were captured by a Spot II digital camera attached to an Olympus IX70 fluorescence microscope. The expression of FLAG-PKN, myc-PDK1, or myc-PKCζ was detected by staining with M2 (IBI-Kodak) or 9E10 (Santa Cruz Biotechnology) antibodies, respectively, followed by Alexa Fluor 350 or 488-conjugated goat anti-mouse or rabbit antibodies (Molecular Probes).
Results and Discussion
To identify potential targets for PDK1, we screened a yeast two-hybrid cDNA library using full-length PDK1 as bait. This screen yielded positive clones encoding the carboxyl terminus of human PKN, a Ser/Thr protein kinase potentially implicated in various cellular events, including the phosphorylation of cytoskeletal components (18, 19). [Recently we identified an isoform of PKN (PKNβ), therefore the originally identified PKN also is called PKNα or PRK1 (20, 21).] The region in PKN that interacted with PDK1 contained amino acid residues 758–942 (PKN-CT, Fig. 1A) including a PDK1 consensus phosphorylation sequence and part of the catalytic domain homologous to that of the PKC family (22). However, unlike PKCs, PKN is activated by the small GTPase Rho but not by PKC activators such as Ca2+, phorbol esters, or phosphatidylserine (18, 19).
To map the sequence in PDK1 that interacted with PKN, we constructed yeast two-hybrid “bait” vectors that encoded different regions of PDK1 (Fig. 1B) and tested their interactions with PKN-CT by using the yeast two-hybrid system. Data showed that full-length, pleckstrin-homology (PH) domain deleted mutant (ΔPH), and a fragment containing the catalytic domain of PDK1 bound to PKN-CT (Fig. 1B). On the other hand, the carboxyl terminus of PDK1 (ΔN) showed no interaction. These findings indicated that the catalytic domain of PDK1 is sufficient to bind PKN in the yeast two-hybrid system.
To test whether PKN and PDK1 interact directly, we carried out pull-down studies. Initially, we tried to express PKN-CT as a GST-fusion protein. However, we found that this fusion protein was toxic to bacterial cells. A similar finding was obtained with the expression of the apoptotic fragment of PKN (AF3-PKN; ref. 15, Fig. 1A). To circumvent this problem, we produced a mutated GST-AF3-PKN fusion protein in which the critical lysine residue in the ATP-binding site of PKN was replaced with glutamate (GST/AF3-PKNK644E). As a control, we also generated a GST-PKN fusion protein in which the carboxyl terminus (residues 631–942) was deleted [GST/PKN(ΔC); Fig. 1 A and C, Lower]. We examined the ability of these fusion proteins to interact with proteins in lysates derived from CHO/IR/PDK1 cells. Our results showed that PDK1 interacted with the GST-AF3-PKNK644E fusion protein but not with GST/PKN(ΔC) (Fig. 1C, Top), further confirming that the region that interacted with PDK1 was located at the carboxyl terminus of PKN.
To show that PDK1 and PKN interacted in mammalian cells, we examined the association of endogenous PDK1 and PKN in CHO/IR cells. Lysates from insulin-treated or nontreated cells were incubated with an affinity-purified antibody to PDK1 (11) or normal Ig (NIg) adsorbed onto protein A-Sepharose beads, and the bound proteins were examined by immunoblot analysis with a mAb to PKN (Transduction Laboratories, Lexington, KY). PKN was coimmunoprecipitated with the antibody to PDK1, but not with the NIg control (Fig. 1D). Neither insulin nor PI3-kinase inhibitors had a significant effect on the binding of PKN to PDK1 (Fig. 1D). These findings indicate that PDK1 and PKN are able to form a complex in mammalian cells. However, we detected little interaction between PDK1 and PKN in reciprocal experiments involving anti-PDK1 immunoblotting of anti-PKN immunoprecipitates. One possible explanation may be that the anti-PKN antibody blocked the interaction between these enzymes. Consistent with this, significant interaction between PDK1 and PKN could be observed in cells expressing hemagglutinin-tagged PDK1 and FLAG-tagged PKN when immunoprecipitations were carried out with antibodies to either tag (data not shown).
Several mechanisms, such as proteolytic digestion (15) and binding to Rho (18, 19), have been shown to activate PKN. However, whether this enzyme is activated by growth factor-stimulated phosphorylation remains unclear. PKN is phosphorylated in intact cells (17), and its activity is inhibited by dephosphorylation (23). PKN also contains a putative phosphorylation motif in the activation loop highly homologous to that of Akt and other downstream substrates for PDK1 (Fig. 1A). These findings suggested that PKN not only interacts directly with PDK1 but also may be a substrate for the enzyme. To test this idea, FLAG-tagged PKN was affinity-purified from CHO/IR/PKN cells and phosphorylated in vitro by affinity-purified PDK1 (11). In agreement with the findings of others (17), we found that PKN underwent significant autophosphorylation in vitro (Fig. 2A, Upper). Incubation of the enzyme with PDK1 led to a 2-fold increase in PKN phosphorylation (Fig. 2A, Upper). The increase of PKN phosphorylation depended on PDK1 kinase activity because the kinase-defective mutant PDK1K114G (11), which bound PKN as well as the wild-type enzyme, did not stimulate the phosphorylation of PKN in vitro (data not shown).
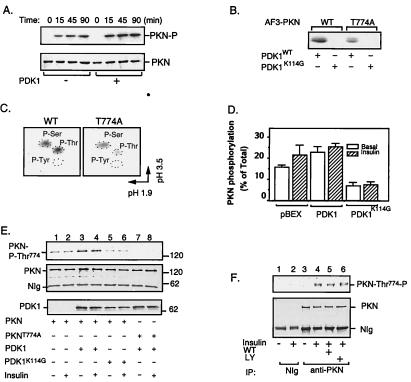
Figure 2.
Phosphorylation and activation of PKN by PDK1. (A) Time course study of PKN phosphorylation. The FLAG-tagged full-length PKN was immunoprecipitated from CHO/IR/PKN cells with the M2 anti-FLAG antibody. In vitro phosphorylation was carried out in the presence (+) or absence (−) of immunoaffinity-purified PDK1 (11). Reactions were stopped at indicated times. The phosphorylated proteins were eluted from beads by boiling in SDS-sample buffer, separated by SDS/PAGE, transferred to a nitrocellulose membrane, and visualized by autoradiography (Upper). The same membrane was reblotted with anti-FLAG antibody to ensure equal loading of proteins in each lane (Lower). (B) Phosphorylation of AF3-PKN by PDK1. AF3-PKN (15) was transiently expressed in CHO/IR cells, immunoprecipitated by anti-FLAG antibody, and phosphorylated in vitro in the presence of immunoaffinity-purified PDK1 or PDK1K114G, according to a protocol similar to that described previously (11). The phosphorylation of AF3-PKN was visualized by autoradiography. (C) Phosphoamino acid analysis of AF3-PKN. The 32P-labeled wild-type (WT) or T774A mutant of AF3-PKN bands were excised from the membrane, hydrolyzed, and analyzed by two-dimensional thin-layer electrophoresis, according to a protocol described previously (16). (D) The effect of PDK1 on PKN phosphorylation in cells. CHO/IR cells were transfected with PKN and PDK1, in vivo-labeled with 32P-orthophosphate, and treated with (+) or without (−) insulin as described in Materials and Methods. Data are mean ± SEM from four independent experiments. Background values obtained from NIg precipitates were subtracted from all values. (E) Overexpression of PDK1 stimulates PKN phosphorylation at Thr774 in intact cells. CHO/IR cells were cotransfected with FLAG-tagged PKN or PKNT774A and either myc-tagged PDK1 or PDK1K114G as indicated. Cell lysates were immunoprecipitated with antibody to the FLAG tag. One-eighth of the immunoprecipitates were separated by SDS/PAGE and analyzed by Western blot with antibody specific to phospho-Thr816 of PRK2 (Top). Two-thirds of the immunoprecipitates were used for Western blot with antibody to the FLAG tag (Middle). Expression of PDK1 in these cells was detected by Western blot of cell lysate (1/20 of total) using antibody to the myc tag (Bottom). (F) Insulin stimulates the phosphorylation of endogenous PKN at Thr774 in CHO/IR cells. CHO/IR cells were serum-starved overnight and treated with or without wortmannin (WT, 200 nM) or LY294002 (LY, 200 μM) for 1 h. Cells then were treated with (+) or without (−) 10 nM insulin for 10 min. Cell lysates were incubated for 8 h at 4°C with mAb to PKN (Transduction Laboratories) or NIg bound to protein G beads. One-eighth of the immunoprecipitates were separated by SDS/PAGE and analyzed by Western blot with antibody specific to phospho-Thr816 of PRK2 (Upper). Two-thirds of the immunoprecipitates were used for Western blot with a mAb to PKN (Transduction Laboratories) (Lower).
Phosphopeptide mapping of rat PKN revealed at least seven autophosphorylation sites (17), five of which (Thr64, Ser372, Thr534, Ser576, and Ser608) are conserved in the human homolog. To avoid the high background encountered with autophosphorylation in studies of in vitro phosphorylation, we examined the PDK1-catalyzed phosphorylation of an amino-terminal truncated mutant of PKN (AF3-PKN; ref. 15 and Fig. 1A), which did not undergo autophosphorylation after immunoprecipitation (Fig. 2B). As shown in Fig. 2B, AF3-PKN was readily phosphorylated by the wild-type PDK1 but not by kinase-defective PDK1K114G. These findings suggest that AF3-PKN, which contains the conserved threonine residue in the activation loop of the enzyme, is a direct substrate for PDK1. Significant phosphorylation by PDK1 also was observed for the full-length kinase-defective mutant PKNK644D (data not shown). Phosphoamino acid analysis of AF3-PKN showed that the phosphorylation occurred equally at both serine and threonine residues (Fig. 2C, Left). Replacing Thr774 with alanine resulted in a significant decrease in AF3-PKN threonine phosphorylation by PDK1 (Fig. 2 B and C), suggesting that Thr774 was a potential site of phosphorylation. However, notable serine phosphorylation was still observed in AF3-PKNT774A, suggesting that in addition to Thr774, PDK1 may phosphorylate AF3-PKN at an additional site. Consistent with this idea, a double mutant of AF3-PKN in which both Thr774 and Ser773 were replaced with alanine was not phosphorylated by PDK1 in vitro (data not shown). However, although our findings suggest that PDK1 may be a direct upstream kinase for PKN, we cannot completely exclude the possibility that a heterologous kinase coimmunoprecipitated with the wild-type PDK1 might phosphorylate AF3-PKN in vitro.
To test whether insulin or PDK1 stimulates PKN phosphorylation at Thr774 in intact cells, in vivo phosphorylation studies were carried out. Fig. 2D shows the mean values from PhosphorImager analysis of four independent in vivo labeling experiments. PKN showed substantial constitutive phosphorylation in serum-starved CHO/IR cells. Also, the overexpression of PDK1 or the treatment of cells with insulin led to an approximate 40% increase in PKN phosphorylation. The modest increase in insulin and PDK1-stimulated phosphorylation of PKN was probably because of an elevated basal phosphorylation at Thr774 when the enzyme was overexpressed in cells (see below). On the other hand, overexpression of the kinase-defective mutant PDK1K114G resulted in an approximate 50% inhibition of PKN phosphorylation.
To test whether PKN was phosphorylated at Thr774 by PDK1 in intact cells, we carried out Western blot studies using a phospho-antibody against Thr816 of PRK2 (24). This antibody recognized wild-type PKN but not the mutant enzyme in which Thr774 was changed to alanine (Fig. 2E, lanes 3–8). The endogenous PKN in CHO/IR cells was not significantly phosphorylated at Thr774 under basal conditions (Fig. 2F, lane 3). Treatment of the cells with insulin led to notable increase in the phosphorylation of endogenous enzyme (Fig. 2F, lane 4). Preincubation of cells with either wortmannin (Fig. 2F, lane 5) or LY294002 (Fig. 2F, lane 6) had no significant effect on PKN phosphorylation at Thr774, suggesting that the insulin-stimulated phosphorylation of endogenous PKN in the activation loop may be PI3-kinase-independent.
To test whether phosphorylation of PKN at Thr774 was mediated by PDK1 in cells, we transfected FLAG-tagged PKN into CHO/IR cells together with myc-tagged wild-type or kinase-dead PDK1. In overexpressed cells, PKN showed a substantial constitutive phosphorylation at Thr774 under serum-starved conditions (Fig. 2E, lane 1). The high basal phosphorylation probably was caused by overexpression of PKN because the endogenous enzyme was not significantly phosphorylated under similar conditions (Fig. 2F, lane 3). Coexpression of the wild-type but not the kinase-dead mutant PDK1 led to a significant increase in PKN phosphorylation at Thr774 (Fig. 2E, lanes 3–6). The PDK1-induced phosphorylation of PKN at Thr774 also was confirmed by using a phospho-specific antibody to Thr500 in the activation loop of PKCβII (7), which recognizes only wild-type PKN but not PKNT774A (data not shown). Together, these findings indicate that PDK1 is an in vivo upstream kinase of PKN and that Thr774 in the activation loop is a PDK1-mediated phosphorylation site.
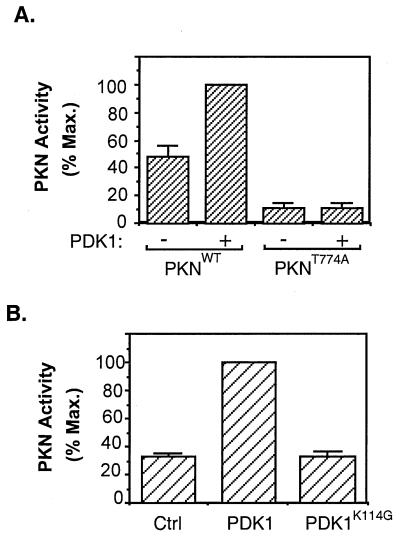
To determine whether phosphorylation by PDK1 could activate PKN, we carried out in vitro kinase assays. PKN isolated from serum-starved cells was constitutively active in vitro, and phosphorylation by PDK1 led to a 2-fold increase in the kinase activity of the enzyme (Fig. 3A). The increase in PKN activity was caused by phosphorylation rather than by binding to PDK1, because control experiments revealed that preincubation with PDK1K114G did not stimulate PKN activity (data not shown). Mutation of Thr774 to alanine resulted in a significant decrease in PKN activity. In addition, preincubation with PDK1 did not stimulate the activity of this mutant (Fig. 3A).
Figure 3.
Activation of PKN by PDK1. (A) Activation of PKN by PDK1 in vitro. The FLAG-tagged wild-type PKN or PKNT774A was immunoprecipitated from serum-starved CHO/IR cells transiently expressing these proteins, phosphorylated in vitro in the presence (+) or absence (−) of affinity-purified PDK1, and assayed for phosphorylation of the S6 peptide. Data are representative of three independent experiments, with bars representing means of duplicate determinations. (B) Activation of PKN by PDK1 in cells. CHO/IR cells were cotransfected with cDNAs encoding PKN, pcDNA/PDK1, or pcDNA/PDK1K114G and the control vector pcDNA. Twenty-four hours after transfection, cells were lysed and activity of the immunoprecipitated PKN was assayed as described in Materials and Methods. Data are mean ± SD of two independent experiments with duplicate determinations.
To test whether PDK1 activates PKN in intact cells, we cotransfected CHO/IR cells with FLAG-PKN and either myc-PDK1 or myc-PDK1K114G. Results showed that there was constitutive basal PKN activity in the absence of ectopically expressed PDK1. Overexpression of the wild-type but not the kinase-defective mutant of PDK1 led to a 3-fold increase in PKN activity (Fig. 3B). These findings provide further evidence that PKN is a downstream effector of PDK1. However, we were unable to test whether insulin and PDK1 can synergistically activate PKN in CHO/IR cells because overnight serum starvation of the cells overexpressing PDK1 and PKN led to significant cell death. Reduction of the serum-starvation time led to a higher basal PKN activity (data not shown).
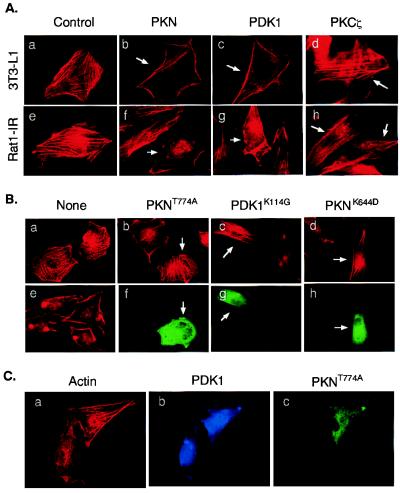
Because insulin activates PKN (25) and stimulates actin stress fiber depolymerization (6, 26), we sought to establish whether insulin signaling to the actin cytoskeleton could be mediated by PDK1/PKN interaction. As a control, rhodamine-phalloidin staining of 3T3-L1 cells revealed abundant actin stress fibers (Fig. 4Aa). Ectopic expression of PKN or PDK1 stimulated actin stress fiber depolymerization and membrane ruffling in approximately 65% of the cells examined (Fig. 4A b and c). On the other hand, only approximately 10% of cells overexpressing PKCζ showed significant actin stress fiber depolymerization (Fig. 4Ad). Similar results were obtained in Rat1-IR fibroblasts transiently expressing these proteins (Fig. 4A e–h).
Figure 4.
Effects of PDK1 and PKN on actin cytoskeleton reorganization. (A) Ectopic expression of PDK1 and PKN induces actin cytoskeleton reorganization. 3T3-L1 (Upper) or Rat1-IR (Lower) cells were transfected with plasmids encoding FLAG-PKN, myc-PDK1, or myc-PKCζ, respectively. Arrows indicate cells expressing the recombinant proteins as determined by staining with antibodies to the tags. Actin cytoskeleton was detected by a rhodamine-phalloidin stain. (B) Ectopic expression of mutant PKN or PDK1 blocks insulin-induced actin cytoskeleton reorganization. Rat1-IR cells (a and e) or cells transiently expressing FLAG-PKNT774A (b and f), myc-PDK1K114G (c and g), or FLAG-PKNK644D (d and h) were serum starved for 4 h and treated with 10 nM insulin for 10 min (b–h). Actin cytoskeleton was detected by a rhodamine-phalloidin stain (a–e). The expression of the recombinant proteins in cells was determined by staining with the anti-FLAG or anti-myc antibodies, followed by Alexa Fluor 488-conjugated goat anti-mouse Ig (f–h). Images are representative of those in three independent experiments. (C) Overexpression of PKNT774A inhibits PDK1-induced actin stress fiber reorganization. Rat1-IR cells were cotransfected with FLAG-tagged PKNT774A and myc-tagged PDK1. The expression of the recombinant proteins in cells was determined by staining with the anti-FLAG or anti-myc antibodies, followed by Alexa Fluor 488-conjugated goat anti-mouse Ig (green, for PKNT774A) or Alexa Fluor 350-conjugated goat anti-rabbit Ig (blue, for PDK1), respectively. Actin cytoskeleton was detected by a rhodamine-phalloidin stain (red). Results are representative of two independent experiments in which the inhibition of PDK1-induced actin stress fiber reorganization was observed in approximately 65% of cells coexpressing PKNT774A.
To further test the potential roles of PDK1 and PKN in insulin-induced actin cytoskeleton reorganization, we transfected Rat1-IR cells with plasmids encoding kinase-defective PDK1 (PDK1K114G) or phosphorylation site-mutated PKN (PKNT774A) and tested their effects on insulin-induced cytoskeleton rearrangement. In agreement with the findings of others (4–6), we found that the treatment of Rat1-IR cells with insulin induces actin stress fiber breakdown and membrane ruffling (Fig. 4Be), which could be blocked by pretreatment of cells with PI3-kinase inhibitors wortmannin or LY294002 (data not shown). The insulin-induced actin stress fiber rearrangement was notably prevented in approximately 60–70% cells transiently expressing PKNT774A (Fig. 4Bb), PDK1K114G (Fig. 4Bc), or PKNK644D (Fig. 4Bd), suggesting that PDK1 and PKN normally are involved in insulin signal transduction to the actin cytoskeleton.
To test whether PDK1 is upstream of PKN in mediating actin stress fiber depolymerization, we coexpressed PDK1 and PKNT774A in Rat1-IR cells and examined the PDK1-mediated actin cytoskeleton change. We found that a significant amount of actin stress fibers remained in approximately 65% of Rat1-IR cells coexpressing both PDK1 and PKNT774A (Fig. 4C). These findings suggest that the PDK1-induced actin cytoskeleton change probably was mediated by PKN.
Together, our results indicate that PKN is an in vivo substrate for PDK1 and that the activation of the enzyme may play an important role in insulin-induced actin cytoskeleton reorganization. However, PKN may not be the only target for PDK1 in this signaling pathway. A recent study showed that PDK1 binds to the carboxyl terminus of PRK2 (27), a PKN-related protein kinase that is implicated in growth factor-stimulated actin cytoskeleton reorganization (28). Because PRK2 shares an extensive homology with PKN and contains a PDK1 consensus phosphorylation motif in the activation loop, it is likely that this enzyme also may be a substrate for PDK1. Consistent with this, we have found that PRK2 interacts with and is phosphorylated by PDK1 (unpublished observations). It is interesting to note that both PKN and PRK2 are activated by small GTPases, such as Rho and Rac (18, 19, 28, 29). Thus, PKN and PRK2 may lie at a critical juncture where the small GTPase and PI3-kinase signaling pathways converge. A question that remains to be answered is whether these two enzymes play similar or different roles in insulin-stimulated actin cytoskeleton reorganization. In addition, it remains to be established how upstream signals are further transmitted downstream from these kinases. The finding that ectopic expression of wild-type PKN, but not the phosphorylation site-mutated or kinase-defective PKN, resulted in actin cytoskeleton reorganization suggests that the kinase activity of the enzyme may be critical in the signaling process. Although the in vivo substrates for both PKN and PRK2 remain elusive, it is possible that, like other Rho-activated kinases, such as Rho-kinase/ROCK (30), activated PKN and PRK2 may initiate further downstream kinase cascades or directly phosphorylate the cytoskeletal components necessary for actin cytoskeleton reorganization. Similar approaches as described in this study may enable us to identify the missing links in the signaling process.
Acknowledgments
We thank Dr. D. R. Alessi for phospho-specific antibody to Thr816 of PRK2, and Dr. A. C. Newton for phospho-specific antibody to Thr500 of PKCβII. We thank Dr. Y. Huang and D. Hu for their technical assistance. This work was supported in part by National Institutes of Health Grant DK52933 and a research grant from the American Diabetes Association. L.R.L. was supported by a supplemental research grant for minority undergraduate students from the National Institutes of Health.
Abbreviations
- PDK1
phosphoinositide-dependent protein kinase-1
- CHO
Chinese hamster ovary
- IR
insulin receptor
- GST
glutathione S-transferase
- PI3-kinase
phosphatidylinositol 3-kinase
- PKC
protein kinase C
- NIg
normal Ig
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090491897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090491897
References
- 1.Zigmond S H. Curr Opin Cell Biol. 1996;8:66–73. doi: 10.1016/s0955-0674(96)80050-0. [DOI] [PubMed] [Google Scholar]
- 2.Anand-Apte B, Zetter B. Stem Cells. 1997;15:259–267. doi: 10.1002/stem.150259. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt A, Hall M N. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 4.Kotani K, Yonezawa K, Hara K, Ueda H, Kitamura Y, Sakaue H, Ando A, Chavanieu B, Calas B, Grigorescu F, et al. EMBO J. 1994;13:2313–2321. doi: 10.1002/j.1460-2075.1994.tb06515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wennstrom S, Siegbahn A, Yokote K, Arvidsson A K, Heldin C H, Mori S, Claesson W L. Oncogene. 1994;9:651–660. [PubMed] [Google Scholar]
- 6.Martin S S, Haruta T, Morris A J, Klippel A, Williams L T, Olefsky J M. J Biol Chem. 1996;271:17605–17608. doi: 10.1074/jbc.271.30.17605. [DOI] [PubMed] [Google Scholar]
- 7.Chou M M, Hou W, Johnson J, Graham L K, Lee M H, Chen C-S, Newton A C, Schaffhausen B S, Toker A. Curr Biol. 1998;8:1069–1077. doi: 10.1016/s0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- 8.Le Good J A, Ziegler W H, Parekh D B, Alessi D R, Cohen P, Parker P J. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 9.Dutil E M, Toker A, Newton A C. Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X, Ma Y, Moore M, Hemmings B A, Taylor S S. Proc Natl Acad Sci USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L Q, Zhang L-B, Langlais P, He H, Clark M, Zhu L, Liu F. J Biol Chem. 1999;274:8117–8122. doi: 10.1074/jbc.274.12.8117. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T, Cohen P. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Leong M L, Buse P, Maiyar A C, Firestone G L, Hemmings B A. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L Q, Farris S, Christal J, Liu F. Mol Endocrinol. 1997;11:1757–1765. doi: 10.1210/mend.11.12.0014. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi M, Mukai H, Toshimori M, Miyamoto M, Ono Y. Proc Natl Acd Sci USA. 1998;95:11566–11571. doi: 10.1073/pnas.95.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong L Q, Du H-Y, Porter S, Kolakowski L F, Jr, Lee A V, Mandarino L J, Fan J B, Yee D, Liu F. J Biol Chem. 1997;272:29104–29112. doi: 10.1074/jbc.272.46.29104. [DOI] [PubMed] [Google Scholar]
- 17.Peng B, Morrice N A, Groenen L C, Wettenhall R E. J Biol Chem. 1996;271:32233–32240. doi: 10.1074/jbc.271.50.32233. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 19.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 20.Oishi K, Mukai H, Shibata H, Takahashi M, Ono Y. Biochem Biophys Res Commun. 1999;261:808–814. doi: 10.1006/bbrc.1999.1116. [DOI] [PubMed] [Google Scholar]
- 21.Palmer R H, Ridden J, Parker P J. Eur J Biochem. 1995;227:344–351. doi: 10.1111/j.1432-1033.1995.tb20395.x. [DOI] [PubMed] [Google Scholar]
- 22.Mukai H, Ono Y. Biochem Biophys Res Commun. 1994;199:897–904. doi: 10.1006/bbrc.1994.1313. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa M, Mukai H, Shibata H, Ono Y. Biochem J. 1995;310:657–664. doi: 10.1042/bj3100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balendran, A., Biondi, R. M., Cheung, P., Casamayor, A., Deak, M. & Alessi, D. R. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 25.Standaert M, Bandyopadhyay G, Galloway L, Ono Y, Mukai H, Farese R. J Biol Chem. 1998;273:7470–7477. doi: 10.1074/jbc.273.13.7470. [DOI] [PubMed] [Google Scholar]
- 26.Martin S S, Rose D W, Saltiel A R, Klippel A, Williams L T, Olefsky J M. Endocrinology. 1996;137:5045–5054. doi: 10.1210/endo.137.11.8895379. [DOI] [PubMed] [Google Scholar]
- 27.Balendran A, Casamayor A, Deak M, Paterson A, Gaffney P, Currie R, Downes C P, Alessi D R. Curr Biol. 1999;9:393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- 28.Vincent S, Settleman J. Mol Cell Biol. 1997;17:2247–2256. doi: 10.1128/mcb.17.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Settleman J. Genes Dev. 1999;13:1168–1180. doi: 10.1101/gad.13.9.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]