Abstract
Glutathione (GSH) is a major source of reducing equivalents in mammalian cells. To examine the role of GSH synthesis in development and cell growth, we generated mice deficient in GSH by a targeted disruption of the heavy subunit of γ-glutamylcysteine synthetase (γGCS-HStm1), an essential enzyme in GSH synthesis. Embryos homozygous for γGCS-HStm1 fail to gastrulate, do not form mesoderm, develop distal apoptosis, and die before day 8.5. Lethality results from apoptotic cell death rather than reduced cell proliferation. We also isolated cell lines from homozygous mutant blastocysts in medium containing GSH. These cells also grow indefinitely in GSH-free medium supplemented with N-acetylcysteine and have undetectable levels of GSH; further, they show no changes in mitochondrial morphology as judged by electron microscopy. These data demonstrate that GSH is required for mammalian development but dispensable in cell culture and that the functions of GSH, not GSH itself, are essential for cell growth.
All cells require a source of reducing equivalents for survival. In mammalian cells and tissues, glutathione (GSH) is the most abundant nonprotein thiol and is usually present in millimolar concentrations (≈1–10 mM) (1, 2). Despite extensive study, it is unclear whether GSH plays an essential role in mammals because significant functional redundancy in the supply of reducing equivalents appears to exist. For example, GSH, GSH reductase, and glutaredoxin supply reducing equivalents to many cytosolic proteins including ribonucleotide reductase (3, 4), an enzyme necessary for deoxyribonucleotide formation for DNA synthesis. However, the thioredoxin pathway also can function as a reducing system for this and other cellular processes (5, 6). Superoxide anion radicals are detoxified by superoxide dismutase, which forms hydrogen peroxide, which in turn is converted to water via the action of GSH peroxidase; however, this conversion also can be performed by catalase and thioredoxin peroxidase (7). Oxidized GSH (GSSG) has been assumed to be the major source of oxidizing equivalents for disulfide-bond formation associated with protein folding within the endoplasmic reticulum (ER) (8); however, the Ero1 gene of yeast can sustain the oxidized status of the ER in the absence of GSH/GSSG (9), and similar genes may exist in mammals.
All eukaryotes contain GSH except protozoans like Entamoeba histolytica and Giardia duodenalis, which function predominantly via anaerobic metabolism (10–12). These organisms do not synthesize GSH and lack mitochondria, suggesting that GSH may be essential for mitochondrial function (10). Mammals can survive substantial decreases in intracellular GSH levels as evidenced by inherited diseases in human and pharmacological models in animals (13, 14). For example, in our studies of families with GSH synthetase deficiency, affected individuals had GSH levels in their white blood cells as low as 5–10% of the controls (15). However, newborn rats (but not adult rats) and guinea pigs, which lack the ability to synthesize ascorbate, die with widespread mitochondrial damage if GSH synthesis is severely curtailed by an inhibitor (buthionine sulfoximine, BSO) (16, 17). In vitro studies have suggested an important role for GSH in mammalian development (18, 19); however, administration of BSO to pregnant rodents often has been ineffective in lowering intraembryonic GSH so that it has been difficult to study the effects of low GSH in embryonic development in vivo (20, 21). Cultured mammalian cells can tolerate GSH depletion down to ≈10% of control values, but studies of more severe reductions have been inconclusive (22, 23). GSH depletion-induced cell death via apoptosis has been observed in some recent studies (24, 25), but not in others (23). Thus, the issue of whether or not GSH provides essential functions in mammalian development and cell growth remains unsolved.
A more definitive approach to this problem can be achieved through the use of genetic tools. GSH is synthesized via the action of the γ-glutamylcysteine synthetase (γGCS) and GSH synthetase (2). γGCS catalyzes formation of γ-GC, and GSH synthetase adds glycine to complete the tripeptide. γGCS is the rate-limiting enzyme in GSH synthesis and is composed of a catalytic heavy subunit and a regulatory light subunit, which are encoded by different genes (26). We describe here the cloning of the gene encoding the mouse heavy subunit of γGCS (γGCS-HS) and the generation of mice carrying a null mutation in this gene. Embryos homozygous for this mutation fail to gastrulate and die before embryonic day (E) 8.5, demonstrating that GSH synthesis is essential for early mammalian development. However, permanent cell lines can be rescued from homozygous mutant blastocysts by the addition of GSH to the medium. These lines can be grown indefinitely in GSH-free medium supplemented with N-acetylcysteine (NAC), indicating that in vitro under culture conditions the participation of GSH itself is not required for cell growth.
Materials and Methods
Generation of γGCS-HS Mutant Mice.
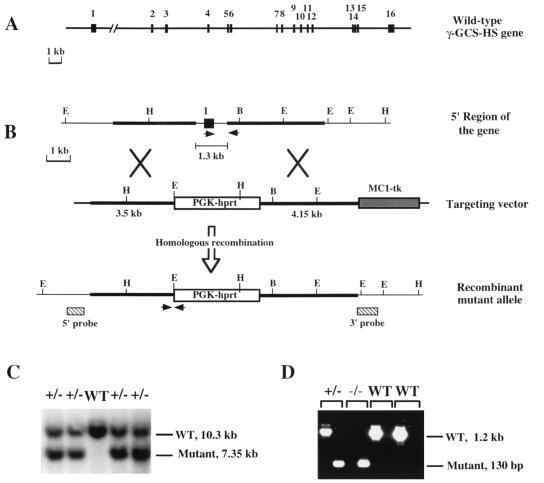
Genomic clones containing exons 2–16 of the γGCS-HS were obtained from a λ DNA library of mouse strain 129/Sv (Stratagene). A clone containing exon 1 was isolated from a 129/Sv bacterial artificial chromosome library (Genome Systems, St. Louis). A targeting vector was designed to replace a 1.3-kb genomic fragment containing TATA box and exon 1 (coding for the translation start site and the first 50 aa) with a PGK-hprt cassette (Fig. 1 A and B). The construct was linearized by digestion with SacII, and 25 μg of DNA was electroporated into 1 × 107 AB2.1 embryonic stem cells (27). Recombinant colonies were selected as described (27) and further confirmed by Southern blot analysis using 5′ and 3′ external probes (Fig. 1C). An hprt probe (not shown) also was used to demonstrate single-copy integration. Chimeric mice and mice heterozygous for the mutant allele were produced by standard techniques (27, 28).
Figure 1.
Structure of the γ-GCS-HS gene and targeted disruption of γ-GCS-HS. (A) Structure of the mouse γGCS-HS locus. Exons (solid boxes) and introns (solid line) are drawn to scale. The positions of the nucleotides at the 3′ limits of exons 1–15 are as follows: 150, 299, 482, 596, 655, 789, 864, 981, 1120, 233, 1326, 1431, 1504, 1617, and 1738 (see ref. 43 for cDNA sequence). (B) Targeting scheme for γGCS-HS disruption. The genomic structure of the 5′ portion of the gene is presented at the top. The targeting vector (middle) was designed such that the PGK-hprt cassette replaced exon 1. An MC1-tk cassette was used as a negative selectable marker. The predicted mutant allele is shown at the bottom. Two external probes used to determine targeting events are indicated as slashed boxes. They both were expected to hybridize with shortened “mutant” bands from the mutant allele. Restriction sites: B, BamHI; E, EcoRI; H, HindIII. (C) Southern blot analysis of genomic DNA isolated from mouse tails derived from heterozygote matings. Only the analysis using the 5′ probe is shown. (D) PCR genotype analysis of mouse embryos. Each DNA sample was subjected to two separate PCRs using the primer pairs designed for either the wild-type (WT) or the mutant band. Two primer pairs are indicated with arrows in B.
Manipulation of Embryos.
Mice heterozygous for γGCS-HStm1 were mated for timed pregnancies, with 1 p.m. of the day of the vaginal plug considered as E0.5. Blastocyst collection and dissection of E6.5–E8.5 embryos were carried out in the standard fashion (29). For histological analysis and in situ hybridization, intact decidua were isolated, fixed overnight with 4% paraformaldehyde in PBS at 4°C, dehydrated through a series of graded ethanols, and embedded in paraffin. Serial sections (6–7 μm) then were prepared.
Genotype Analysis.
Pups and embryos at E8.5 or older and cultured embryonic cells were genotyped by Southern blot analysis using the 5′ external probe (see Fig. 1 B and C). Embryos at E7.5 and E6.5 were genotyped by PCR using two sets of primers that were designed to detect the wild-type allele and the mutant allele, respectively (see Fig. 1 B and D). The primers used to detect the wild-type allele were as follows (sense, antisense): 5′-TGCTGTCCCAAGGCTCGCCACTG-3′ and 5′-ACCACTGTGCTTCGACTGCACTG-3′ for the primary PCR; 5′-ATCTACCACGCAGTCAAGGACCGG-3′ and 5′-CATCTGCGGATGAAGTTCAACTCC-3′ for the nested PCR. The primers for the mutant allele were: 5′-TCAGCCAGGCGTGGTAGTGCACA-3′ and 5′-TGGAATGTGTGCGAGGCCAGAGGC-3′ for the primary PCR; 5′-TCAGTCCTTATTCGCAGACAGCC-3′ and 5′-TGTGTAGCGCCAAGTGCCCAGCGG-3′ for the nested PCR. In histological analysis, in situ hybridization was used to distinguish homozygous mutant embryos from wild-type and heterozygous embryos. Digoxigenin-labeled RNA probes containing the sequences of γGCS-HS exon 1 [nucleotides −32 to 150 (GenBank accession no. AF184592)] were used to detect mRNA in the sectioned embryos. Hybridization was performed as described (30) except signals were visualized with an anti-digoxigenin antibody conjugated with alkaline phosphatase (Boehringer Mannheim).
BrdUrd Labeling of Cells.
Pregnant females at E6.5 were injected i.p. with BrdUrd (100 μg/g of body weight) and killed after 1 h. Decidua then were dissected out and prepared for tissue sections. In situ detection of BrdUrd-labeled cells was carried out on tissue sections with an in situ cell proliferation (FLUO) kit using a fluorescein-labeled anti-BrdUrd mAb (Boehringer Mannheim).
Whole-Mount in Situ Hybridization.
Dissected E7.5 embryos were processed for whole-mount in situ hybridization essentially as described by Conlon and Rossant (31). Digoxigenin-labeled Brachyury (T) probe (32) (antisense; nucleotide 1 − 1740) was used.
Culture of Blastocyst-Derived Cell Lines.
All studies used M15 complete medium [“knockout” DMEM (GIBCO/BRL) supplemented with 15% embryonic stem cell qualified FBS (GIBCO/BRL), 2 mM glutamine, 0.1 mM β-mercaptoethanol (BME), 100 units/ml of penicillin, and 100 μg/ml of streptomycin]. In GSH and NAC rescue experiments, various concentrations of GSH (Sigma) or NAC (Sigma) were added. All cultures were maintained at 37°C in humidified incubators containing 5% CO2 and 95% air. To establish embryonic cultures and cell lines, blastocysts were cultured individually in 24-well plates containing a preformed feeder layer of STO cells (SNL76/7 cells) in M15 medium supplemented with 5 mM GSH (fresh daily). After 6–7 days of culture, inner cell mass-derived clumps were disaggregated by treatment with trypsin-EDTA and transferred into wells of 96-well plates with feeder cells. Subsequent subcultures were performed, and cells were transferred into 48-well plates with feeder cells, 24-well plates with feeder cells, and finally gelatin-coated 6-well plates without feeder cells. Once established, cells were passed every 3–4 days by splitting 1:3 into gelatin-coated 6-well plates. Wild-type and heterozygous cells were maintained in medium without GSH, while homozygous cells were maintained in medium containing 2.5 mM GSH (changed daily).
Results
Disruption of γGCS-HS Results in Early Embryonic Lethality.
We cloned the gene for mouse γGCS-HS and analyzed its structure (Fig. 1A). It consists of 16 exons and encompasses more than 60 kb. A targeting vector was constructed to disrupt the gene in embryonic stem cells by homologous recombination (Fig. 1 B–D). Mice heterozygous for the mutant γGCS-HStm1 allele developed normally and were fertile. However, among ≈600 progeny from heterozygote intercrosses, no viable homozygous pups was found, indicating that homozygosity for γGCS-HStm1 results in embryonic lethality. By genotyping embryos on different days, we found that homozygous embryos died between E7.5 and E8.5. At E6.5, homozygous embryos could not be distinguished morphologically from their wild-type or heterozygous littermates (not shown). At E7.5, about one-fourth of all embryos were morphologically abnormal; genotype analysis revealed that they were homozygous mutants. E7.5 mutant embryos were less than half the size of their wild-type or heterozygous littermates, failed to differentiate beyond the egg cylinder stage, and showed indefinite boundaries between embryonic and extraembryonic regions (Fig. 2 A and B). All homozygous mutant embryos were in resorption at E8.5, and most disappeared in their degenerating decidua by E10.5. We unsuccessfully attempted to rescue mutant embryos by administration of NAC, a cysteine derivative and antioxidant, to pregnant dams.
Figure 2.
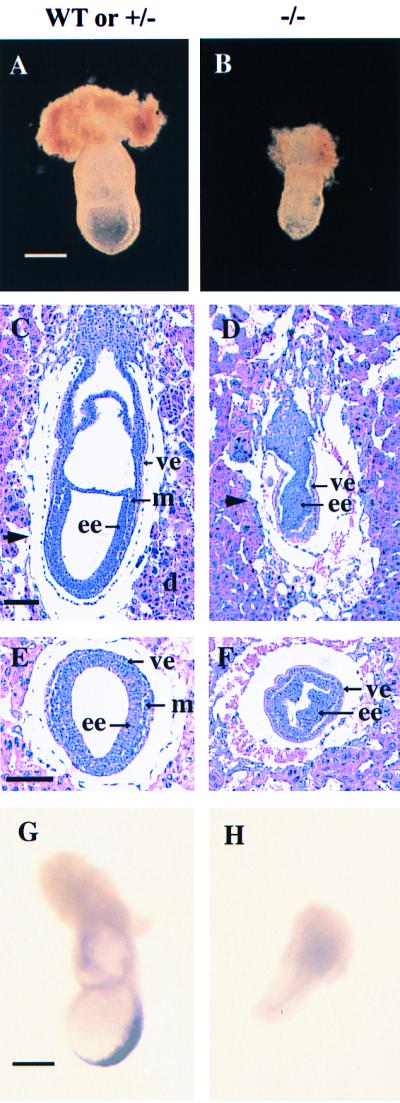
Developmental abnormalities in E7.5 γGCS-HS mutant embryos. (A and B) Whole-mount preparations of E7.5 normal and mutant embryos. The mutant embryos (B) are smaller than normal (A) and show lack of organization. (C–F) Histological comparison of normal (C and E) and mutant (D and F) embryos. The arrowheads in the sagittal sections (C and D) indicate the approximate position of the transverse sections (E and F). Note lack of mesoderm and ectoplacental, exocoelomic, and amniotic cavities in the mutant embryos. d, Decidua; ee, embryonic ectoderm; m, mesoderm; ve, visceral endoderm. (G and H) Whole-mount in situ hybridization analysis with a mesoderm marker, Brachyury (T). A normal expression pattern of T gene is shown in the wild-type embryo (G). No signal was detected in a mutant littermate (H). [Bar: 300 μm (A, B, G, and H); 100 μm (C–F).]
We could not detect GSH in E7.5 embryos homozygous for γGCS-HStm1 by using an HPLC/electrochemical detection (ECD) system (33) with a limit of detection of 0.08 nmol/mg protein. In contrast wild-type embryos had GSH levels of 61.5 ± 10.97 nmol/mg protein (n = 4).
Histological Analysis of the Mutant Embryos.
We serial-sectioned 40 E6.5 embryos and were unable to distinguish homozygous mutant embryos from littermate controls. However, at E7.5, ≈25% of all (both 129Sv × C57BL/6, n = 103; and 129Sv inbred, n = 19) embryos showed developmental abnormalities. Mutant embryos, identified by in situ hybridization (see Materials and Methods), were developmentally retarded and misshapen and had egg cylinders composed of discrete ectodermal and endodermal layers with no evidence of mesoderm (Fig. 2 C–F). We confirmed lack of mesoderm induction by examining Brachyury (T) expression and were unable to detect it in mutant embryos (ref. 32; Fig. 2 G and H).
Mutant Embryos Have High Rates of Apoptosis.
Arrest of gastrulation, failure of mesoderm induction, and the reduced size of mutant embryos might be accounted for by a decline in cell number via increased cell death, decreased cell proliferation, or both. Thus we examined E6.5 embryos (the last time point at which mutant embryos appeared normal) for programmed cell death (apoptosis) by terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay and cell proliferation by analyzing BrdUrd incorporation.
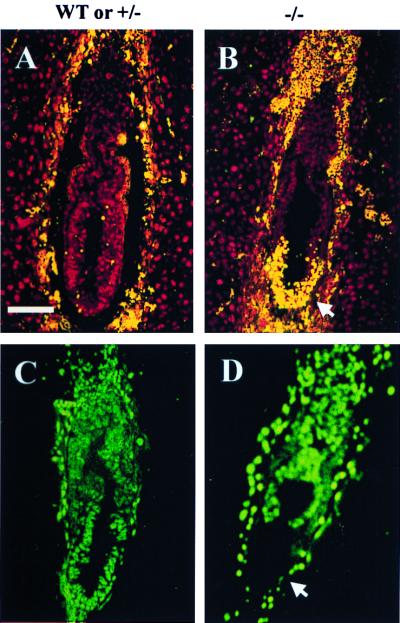
We examined 16 homozygous mutant embryos (from 10 different litters) and 20 wild-type and heterozygous littermates by the TUNEL assay and found that all mutant embryos had high concentrations of TUNEL-positive nuclei in their distal portions (Fig. 3 A and B). This U-shaped region covered approximately one-fourth of the egg cylinder and involved both embryonic ectoderm and visceral endoderm. In the apoptotic region, the rate of TUNEL-positive nuclei ranged from 10% to ≈100% (Fig. 3B). In the remainder (proximal region) of the mutant embryos, only scattered TUNEL-positive nuclei were observed; the number of apoptotic cells in this region was the same as that of wild-type or heterozygous embryos.
Figure 3.
In vivo apoptosis and proliferation in E6.5 γGCS-HS mutant embryos. (A and B) Sagittal sections from two embryos [wild-type (WT) or heterozygous (+/−) and homozygous mutant (−/−)] were assayed by the TUNEL reaction (ApopTaq fluorescein kit, Oncor/Intergen). Fluorescein-labeled nuclei (orange) indicate apoptotic cells. Unlabeled nuclei appear red as a result of counterstaining with propidium iodide. The normal embryo (A) shows few apoptotic nuclei, whereas the mutant embryo (B) shows severe distal apoptosis (arrow). (C and D) Sagittal sections from wild-type or +/− and −/− littermate embryos were analyzed by BrdUrd incorporation. Positive nuclei are visualized by green fluorescence. The mutant embryo (D) shows total absence of BrdUrd incorporation at its distal end (arrow), but the incorporation in other regions is comparable to the embryo (C). Genotypes of embryos were determined in adjacent sections by in situ hybridization with a γGCS-HS exon 1 probe (not shown). (Bar: 100 μM.)
We studied BrdUrd incorporation as an index of proliferation in 16 pairs of E6.5 homozygous mutant embryos and their wild-type or heterozygous littermates (Fig. 3 C and D). In wild-type and heterozygous embryos, the percentage of BrdUrd-positive nuclei varied from litter to litter and ranged from 50% to 80% of all epiblast cells. In mutant embryos, the percentage of labeled nuclei was less than in their littermates and appeared to be inversely correlated with the severity of distal apoptosis (see above). In some mutant embryos, BrdUrd incorporation was almost absent in the distal region (Fig. 3D). However, in other regions, proliferation was comparable to that seen in wild-type embryos. This result indicates that the γGCS-HS null mutation is associated with a high rate of programmed cell death in a specific region of the embryo and has little effect on the overall cell proliferation.
Derivation of γGCS-HS-Deficient Cell Lines from Blastocysts.
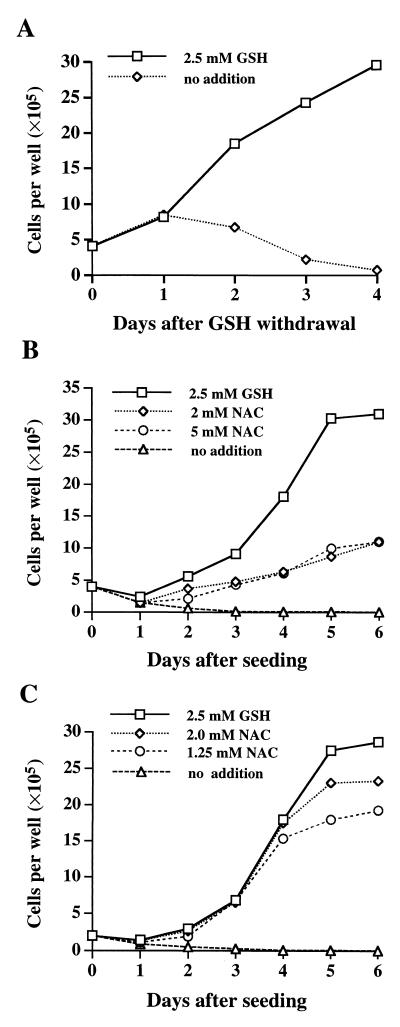
We were able to culture E3.5 blastocysts in medium containing 5 mM GSH. Repetitive passages gave rise to 17 embryonic cultures with the following genotypes: five wild type, eight heterozygous for γGCS-HStm1, and four homozygous for γGCS-HStm1. We derived cell lines from six of these and selected four for further study: a wild-type line (BDC-1) and three mutant lines (GCS-1, GCS-2, and GCS-3). We have passed these lines 50–100 times, and they are morphologically stable. GCS-1 grew as compact foci of small cells surrounded by epithelial cells. GCS-2 was epithelial whereas GCS-3 resembled GCS-1. BDC-1 was also epithelial. We confirmed the presence of epithelial cells in these lines by immunofluorescent staining for pan-cytokeratins (data not shown). Both wild-type and mutant embryonic lines required 15% FBS and 100 μM of BME. GCS-1, GCS-2, and GCS-3 also required exogenous GSH (optimal concentration 2.5 mM). BME alone (up to 1 mM) could not substitute for GSH in GCS cell lines (not shown). When GSH was withdrawn from the medium (containing 100 μM BME), cells proliferated at a normal rate for at least 24 h, the usual population doubling time; however, after this time the cell number dropped steadily (Fig. 4A).
Figure 4.
Growth of γGCS-HS-deficient cells. (A) Dependence of GCS-1 cells on exogenous GSH for growth. Cells were seeded in 6-well plates in duplicate with M15 medium containing 2.5 mM GSH. On day 0 they were refed with M15 medium with or without GSH. Trypan blue-excluding cells were counted at 24-h intervals. (B) Support of GCS-1 cell growth by NAC. On day 0, cells were seeded in M15 medium containing 2.5 mM GSH or 2 or 5 mM of NAC in place of GSH. (C) Growth of GCS-1nac subline cells in the presence of NAC. Cells were seeded in M15 medium containing the additions shown. In all experiments, medium was changed daily.
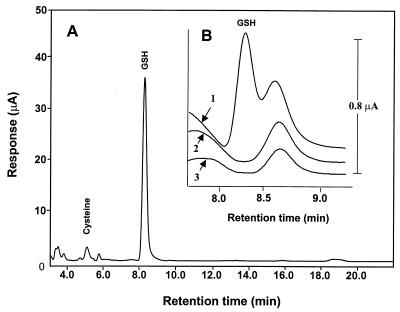
We measured GSH levels associated with wild-type (BDC-1) cells and γGCS-deficient (GCS-1) cells cultured with GSH and after GSH withdrawal (Table 1). GCS-1 cells cultured with 2.5 mM GSH had GSH values that were ≈1.8% of wild-type cells, and 24 h after GSH withdrawal, GSH was not detectable.
Table 1.
Levels of GSH and cysteine in blastocyst-derived cell lines
| Cell Type | Genotype | Additions to medium | GSH | Cysteine |
|---|---|---|---|---|
| BDC-1 | Wild type | None | 18.07 ± 1.83 | 1.30 ± 0.18 |
| GCS-1 | −/− | GSH, 2.5 mM* | 0.32 ± 0.02 | 0.34 ± 0.05 |
| GSH withdrawn for 24 h* | ND | 0.50 ± 0.08 | ||
| GSH withdrawn for 48 h† | ND | 0.40 ± 0.02 | ||
| GSH withdrawn for 72 h† | ND | 0.29 ± 0.03 | ||
| GSH replaced with 2 mM NAC for 72 h* | ND | 0.63 ± 0.09 | ||
| GCS-1nac | −/− | NAC, 2 mM* | ND | 0.36 ± 0.02 |
| NAC, 1.25 mM* | ND | 0.15 ± 0.02 | ||
| NAC withdrawn for 72 h† | ND | 0.13 ± 0.01 |
Rescue of GSH-Deficient Cells by NAC.
To determine whether mutant cells have an absolute requirement for GSH, we attempted to rescue GCS-1 cells deprived of GSH. We found that 2–5 mM NAC could substitute for GSH (Fig. 4B). GCS-1 cells in medium containing NAC grew more slowly than in medium with GSH. However, by culturing cells continuously in NAC, we were able to develop a subline of GCS-1 (termed GCS-1nac) in which morphology was unchanged and which grew almost as rapidly in 1.25–2 mM NAC as in GSH (Fig. 4C). In other experiments, we used GCS-2 and GCS-3 and found that these lines were also able to grow in NAC in the absence of GSH. Withdrawal of NAC from all three sublines resulted in cell death after 2 days. We confirmed that these γGCS-deficient cells grown in NAC completely lacked GSH (Table 1 and Fig. 5). We also tried to rescue GCS-1 cells with DTT, ascorbic acid, α-tocopherol, and butylated hydroxytoluene and found that these agents failed to rescue cells (data not shown).
Figure 5.
Determination of cellular GSH by HPLC/ECD. (A) Chromatographic profile of acid soluble extract of BDC-1 (wild-type) cells. (B) Portions of chromatograms expanded between 7.5 and 9.5 min to show the presence and absence of GSH in γGCS-deficient cells: 1, GCS-1 cells grown in the presence of 2.5 mM GSH; 2, GCS-1 cells after withdrawal of GSH from the medium for 24 h; 3, GCS-1nac cells grown in the presence of 2 mM NAC. Note absence of GSH peak in 2 or 3. The small peak to the right of the GSH peak is an unidentified compound found in all cells. Note the difference in the μA scales for A and B.
Cysteine levels were determined in mutant and wild-type cells (Table 1). We found that γGCS-deficient cells maintained in GSH had about one-fourth the cysteine levels of wild-type cells. Withdrawal of GSH did not result in a further lowering of cysteine levels, and addition of NAC in place of GSH did not markedly raise cysteine levels. NAC itself was not detectable in cells cultured with 1.25–2.0 mM NAC (data not shown).
Cell Death Associated with GSH Deficiency.
We analyzed cell death 2 days after GSH withdrawal in GCS-1 cells by using visual inspection, the TUNEL assay, and DNA laddering and failed to find evidence of apoptosis (not shown, however, see Fig. 3B). In a control experiment, apoptotic cells were found at a rate of >50% in GSH-deprived GCS-1 cells 16 h after the addition of 1 μM sodium arsenite (not shown). These data support the interpretation that cell death after GSH withdrawal under the conditions studied is not primarily the result of apoptosis.
To determine whether mitochondrial damage occurred in the complete absence of GSH, we performed electron microscopy on mutant cells collected 2 or 3 days after GSH withdrawal and found no evidence of mitochondrial damage (e.g., swelling and degeneration) (not shown). Similarly, examination of cells grown in the presence of NAC with absent GSH showed no changes in mitochondrial structure.
Discussion
We describe here the generation of a null mutation of the γGCS-HS gene by homologous recombination in the mouse. The mutation results in complete GSH deficiency by E7.5, 4 days after the expected initiation of GSH synthesis in normal embryos (34), and embryonic lethality before E8.5. In contrast, γGCS-deficient cell lines rescued from mutant embryos grow indefinitely in vitro if supplemented with GSH or NAC.
Our data suggest that the lethality observed in γGCS-deficient embryos may result predominantly from apoptotic cell death rather than reduced cell proliferation (Fig. 3). The cause of this highly localized (distal) apoptosis is unclear. It may be that cells in the distal region are especially sensitive to GSH depletion or that this pattern is a common feature of “programmed developmental failure” stemming from insult or mutation (35). Our results raise the possibility that GSH and redox status may be involved in signaling events like embryonic remodeling and mesodermal induction (6).
Here, we demonstrate that γGCS-deficient mouse embryo cells can grow in culture with only minimal levels of cellular GSH and that NAC can support cell growth in the absence of GSH. GSH concentrations detected in mutant cells supplemented with 2.5 mM GSH were ≈2% of that seen in wild-type cells (Table 1). This minimal level is presumably attained by uptake of GSH from the medium and is necessary for cell viability. The large excess of GSH present in most cells probably functions to protect them against oxidative stress and in detoxification reactions. We attempted to rescue γGCS-deficient cells with reductants other than NAC and found that only this agent was effective. This finding is in contrast to studies on a GSH-deficient Saccharomyces cerevisiae mutant in which other thiols (BME and DTT) as well as cysteine support growth (36). We note that the conditions we have used to rescue GCS cells involve extracellular thiol concentrations that are ≈1,000-fold higher than those found in vivo and thus our findings cannot be easily extrapolated to the in vivo situation.
Our analysis of mitochondria from GSH-deficient cells by electron microscopy did not reveal changes in morphology observed when animals were treated with BSO (14). Because mitochondria have no catalse, GSH and GSH peroxidase have been thought to be the only system they use to metabolize hydrogen peroxide. Our data suggest other detoxification mechanisms (e.g., thioredoxin peroxidase; refs. 7 and 37) exist and that GSH itself is not required to maintain the redox balance necessary for mitochondrial function. Some studies have shown that when the mitochondrial GSH pool is severely depleted in mammalian cells by BSO and electrophilic chemicals cells lose viability (22, 38); however, in other studies, some types of cells seem to be more resistant to BSO (23). These data suggest that the ability to induce alternative antioxidant enzyme(s) in mitochondria and the time available for induction may determine survival of cells subjected to GSH depletion.
GSH depletion has been implicated as a mediator of the apoptotic pathway (24, 25, 39, 40). In the embryo, disruption of GSH synthesis is correlated with distal apoptosis (Fig. 3); however, in cultured embryonic cells, complete removal of GSH from the medium and replacement with NAC allows long-term viability. Further, when GSH is removed without NAC replacement, cell death is not accompanied by apoptosis. These data demonstrate that depletion of GSH alone is insufficient to induce apoptosis in these cells and indicate that the role of the redox status in apoptosis is complex.
The fact that cultured γGCS-HStm1/tm1 embryonic cells grow indefinitely in medium in which GSH has been replaced by NAC demonstrates that GSH itself is not required by mammalian cells maintained under standard culture conditions. It follows that many housekeeping functions, including DNA synthesis, protein synthesis, metabolism and respiration, cell cycling and division, do not absolutely require GSH. Rather GSH appears to function by providing essential reducing equivalents for these processes. It also follows that the coenzyme functions of GSH, like its role in the glyoxalase pathway (41), can be replaced by other cofactors or that the pathways themselves are not essential. The availability of these mutant cells deficient in GSH synthesis will allow further exploration of the role of cellular redox status in signal transduction, posttranslational regulation of proteins (e.g., chaperone activity; ref. 42) cell growth, differentiation, and cell death.
Acknowledgments
We thank Drs. Hiram F. Gilbert, Kathleen A. Mahon, Mark W. Majesky, and Yuji Mishina for their thoughtful comments and Cathy Guo and Pei Wang for their technical assistance. This work was supported in part by National Institutes of Health Grant ES 08668 to M.W.L.
Abbreviations
- GSH
glutathione
- γGCS
γ-glutamylcysteine synthetase
- γGCS-HS
γ-GCS heavy subunit
- NAC
N-acetylcysteine
- BSO
buthionine sulfoximine
- E(n)
embryonic day
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling
- BME
β-mercaptoethanol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kosower N S. Int Rev Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 2.Meister A. J Biol Chem. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 3.Holmgren A. In: Glutathione: Metabolism and Physiological Function. Viña J, editor. Boca Raton, FL: CRC; 1990. pp. 145–154. [Google Scholar]
- 4.Luthman M, Eriksson S, Holmgren A, Thelander L. Proc Natl Acad Sci USA. 1979;76:2158–2162. doi: 10.1073/pnas.76.5.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prinz W A, Aslund F, Holmgren A, Beckwith J. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 6.Sen C K. Biochem Pharmacol. 1998;55:1747–1758. doi: 10.1016/s0006-2952(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 7.Chae H Z, Chung S J, Rhee S G. J Biol Chem. 1994;273:27670–27678. [PubMed] [Google Scholar]
- 8.Hwang C, Sinskey A J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 9.Frand A R, Kaiser C A. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 10.Fahey R C, Newton G L, Arrick B, Overdank-Bogart T, Aley S B. Science. 1984;224:70–72. doi: 10.1126/science.6322306. [DOI] [PubMed] [Google Scholar]
- 11.Fahey R C, Sundquist A R. Relat Areas Mol Biol. 1991;64:1–53. doi: 10.1002/9780470123102.ch1. [DOI] [PubMed] [Google Scholar]
- 12.Brown D M, Upcroft J A, Upcroft P. Mol Biochem Parasitol. 1993;1:155–158. doi: 10.1016/0166-6851(93)90169-x. [DOI] [PubMed] [Google Scholar]
- 13.Meister A, Larsson A. In: The Metabolic Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1461–1477. [Google Scholar]
- 14.Meister A. Pharmacol Ther. 1991;51:155–194. doi: 10.1016/0163-7258(91)90076-x. [DOI] [PubMed] [Google Scholar]
- 15.Shi Z-Z, Habib G M, Rhead W K, Gahl W A, He X, Sazer S, Lieberman M W. Nat Genet. 1996;14:361–365. doi: 10.1038/ng1196-361. [DOI] [PubMed] [Google Scholar]
- 16.Martensson J, Meister A. Proc Natl Acad Sci USA. 1991;88:4656–4660. doi: 10.1073/pnas.88.11.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister A. Biochim Biophys Acta. 1995;1271:35–42. doi: 10.1016/0925-4439(95)00007-q. [DOI] [PubMed] [Google Scholar]
- 18.Legge M, Sellens M H. Hum Reprod. 1991;6:867–871. doi: 10.1093/oxfordjournals.humrep.a137442. [DOI] [PubMed] [Google Scholar]
- 19.Slott V L, Hales B F. Pharmacology. 1987;36:683–688. doi: 10.1016/0006-2952(87)90719-2. [DOI] [PubMed] [Google Scholar]
- 20.Reyes E, Ott S, Robinson B, Contreras R. Pharmacol Biochem Behav. 1995;50:491–497. doi: 10.1016/0091-3057(94)00320-3. [DOI] [PubMed] [Google Scholar]
- 21.Martensson J, Steinherz R, Jain A, Meister A. Proc Natl Acad Sci USA. 1989;86:8727–8731. doi: 10.1073/pnas.86.22.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dethmers J K, Meister A. Proc Natl Acad Sci USA. 1981;78:7492–7496. doi: 10.1073/pnas.78.12.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato N, Iwata S, Nakamura K, Hori T, Mori K, Yodoi J. J Immunol. 1995;154:3194–3203. [PubMed] [Google Scholar]
- 24.Merad-Boudia M, Nicole A, Santiard-Baron D, Saillé C, Ceballos-Picot I. Biochem Pharmacol. 1998;56:645–655. doi: 10.1016/s0006-2952(97)00647-3. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi Y, Matsukawa S. Arch Biochem Biophys. 1999;363:33–42. doi: 10.1006/abbi.1998.1067. [DOI] [PubMed] [Google Scholar]
- 26.Huang C-S, Chang L-S, Anderson M E, Meister A. J Biol Chem. 1993;268:19675–19680. [PubMed] [Google Scholar]
- 27.Matzuk M M, Finegold M J, Su J G, Hsueh A J, Bradley A. Nature (London) 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman M W, Wiseman A L, Shi Z-Z, Carter B Z, Barrios R, Ou C N, Chevez-Barrios P, Wang Y, Habib G M, Goodman J C, et al. Proc Natl Acad Sci USA. 1996;93:7923–7926. doi: 10.1073/pnas.93.15.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 30.Albrecht U, Eichele G, Helms J A, Lu H-C. In: Molecular and Cellular Methods in Developmental Toxicology. Daston G P, editor. Boca Raton, FL: CRC; 1997. pp. 23–48. [Google Scholar]
- 31.Conlon R A, Rossant J. Development (Cambridge, UK) 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann B G, Labeit S, Poustka A, King T R, Lehrach H. Nature (London) 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 33.Kleinman W A, Richie J P., Jr J Chromatogr B Biomed Appl. 1995;672:73–80. doi: 10.1016/0378-4347(94)00194-a. [DOI] [PubMed] [Google Scholar]
- 34.Gardiner C S, Reed D J. Arch Biochem Biophys. 1995;318:30–36. doi: 10.1006/abbi.1995.1200. [DOI] [PubMed] [Google Scholar]
- 35.Chen W S, Manova K, Weinstein D C, Duncan S A, Plump A S, Prezioso V R, Bachvarova R F, Darnell J E. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 36.Grant C M, MacIver F H, Dawes I W. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 37.Kowaltowski A J, Netto L E S, Vercesi A E. J Biol Chem. 1998;273:12766–12769. doi: 10.1074/jbc.273.21.12766. [DOI] [PubMed] [Google Scholar]
- 38.Wüllner U, Seyfried J, Groscurth P, Beinroth S, Winter S, Gleichman M, Heneka M, Loschmann P A, Schulz J B, Weller M, Klockgether T. Brain Res. 1999;826:53–62. doi: 10.1016/s0006-8993(99)01228-7. [DOI] [PubMed] [Google Scholar]
- 39.Wright S C, Wang H, Wei Q S, Kinder D H, Larrick J W. Cancer Res. 1998;58:5570–5576. [PubMed] [Google Scholar]
- 40.Celli A, Que F G, Gores G J, LaRusso N F. Am J Physiol. 1998;275:G749–G757. doi: 10.1152/ajpgi.1998.275.4.G749. [DOI] [PubMed] [Google Scholar]
- 41.Thornalley P J. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakob U, Muse W, Eser M, Bardwell J C A. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 43.Reid L L, Botta D, Lu Y, Gallagher E P, Kavanagh T J. Biochim Biophys Acta. 1997;1352:233–237. doi: 10.1016/s0167-4781(97)00058-4. [DOI] [PubMed] [Google Scholar]