Abstract
Flavonoids are the major functional components of many herbal and insect preparations and demonstrate varied pharmacological functions including antibacterial activity. Here by enzymatic assay and crystal structure analysis, we studied the inhibition of three flavonoids (quercetin, apigenin, and (S)-sakuranetin) against the β-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori (HpFabZ). These three flavonoids are all competitive inhibitors against HpFabZ by either binding to the entrance of substrate tunnel B (binding model A) or plugging into the tunnel C near the catalytic residues (binding model B) mainly by hydrophobic interaction and hydrogen-bond pattern. Surrounded by hydrophobic residues of HpFabZ at both positions of models A and B, the methoxy group at C-7 of (S)-sakuranetin seems to play an important role for the inhibitor's binding to HpFabZ, partly responsible for the higher inhibitory activity of (S)-sakuranetin than those of quercetin and apigenin against HpFabZ (IC50 in μM: (S)-sakuranetin, 2.0 ± 0.1; quercetin: 39.3 ± 2.7; apigenin, 11.0 ± 2.5). Our work is expected to supply useful information for understanding the potential antibacterial mechanism of flavonoids.
Keywords: HpFabZ, quercetin, apigenin, (S)-sakuranetin, complex structure, inhibitory mechanism
Flavonoids, with the largest and most important group of polyphenolic compounds in plants, are major functional components of many herbal and insect preparations for medical use (Havsteen 2002; Cushnie and Lamb 2005). For decades of medical research, flavonoids have been reported to possess varied pharmacological and biochemical properties, including anti-inflammatory, antioxidant, antiallergic, antimicrobial, antitumor, antiplatelet, and estrogenic activities, as well as the inhibition of various enzymes, such as hydrolases, oxidoreductases, DNA synthetases, RNA polymerases, phosphatases, protein phosphokinases, oxygenases, and amino acid oxidases (Harborne and Williams 2000; Havsteen 2002; Ross and Kasum 2002; Cushnie and Lamb 2005).
Quercetin (3,3′,4′,5,7-pentahydroxyflavone), apigenin (4′,5,7-trihydroxyflavone), and sakuranetin (5,4′-dihydroxy-7-methoxyflavanone) are three representative flavonoids. Quercetin belongs to the flavonol class, which exists in foods such as citrus fruit, buckwheat, and onions. It has been demonstrated that quercetin has significant anti-inflammatory activity, antitumor activity, antioxidant activity, and vitamin C-sparing action and has been recognized as the most active flavonoid in study. Apigenin belongs to the flavone class and is isolated from parsley and celery. It has antioxidant, anti-inflammatory, and antitumor properties just like most of the other flavonoids. Sakuranetin falls into the flavanone class and is isolated from many plants (Hurabielle et al. 1982; Liu et al. 1992; Abdel-Sattar et al. 2000). It was discovered to possess anti-inflammatory activity and proved to be a phytoalexin (Nakazato et al. 2000; Hernandez et al. 2007). In addition, all these three flavonoids were reported to exhibit antibacterial activity, probably by acting on multiple cellular targets (Konstantinopoulou et al. 2003; Cushnie and Lamb 2005), such as DNA gyrase (Ohemeng et al. 1993) and cytoplasmic membrane (Mirzoeva et al. 1997). Recently, quercetin and apigenin were found to inhibit some enzymes involved in the type II fatty acid biosynthesis (FAS II) pathway. In detail, quercetin could inhibit β-ketoacyl-acyl carrier protein reductase (FabG) and enoyl–acyl carrier protein reductase (FabI) of Plasmodium falciparum as well as β-hydroxyacyl–acyl carrier protein dehydratase (FabZ) from P. falciparum and Helicobacter pylori, while apigenin could only inhibit FabI from P. falciparum and FabZ from H. pylori (Tasdemir et al. 2006; Zhang et al. 2008).
In this work, the flavonoid (S)-sakuranetin was identified as a new HpFabZ inhibitor. More importantly, to characterize the inhibitory mechanism of HpFabZ by quercetin, apigenin, and (S)-sakuranetin, complex crystal structure analyses with kinetically enzymatic assays were carried out. These flavonoids were found to be competitive inhibitors against HpFabZ by binding to the substrate tunnel and preventing the substrate from accessing the active site. Considering the fact that the reports on the antibacterial activity of flavonoids often show wide discrepancies from each other (Cushnie and Lamb 2005), the anti-H. pylori activities of these three inhibitors were also tested by the standard agar dilution method. Our work is expected to provide useful information for understanding the potential antibacterial mechanism of flavonoids.
Results
Inhibition of quercetin, apigenin, and (S)-sakuranetin against HpFabZ
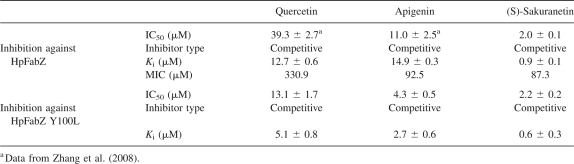
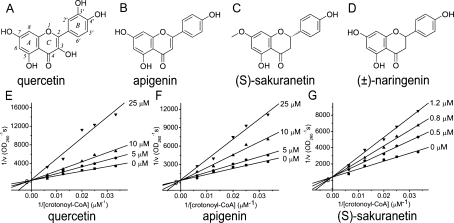
Quercetin and apigenin have been reported as the inhibitors of HpFabZ in our previous work (Zhang et al. 2008). Here we further discovered that another flavonoid, (S)-sakuranetin, could also inhibit the HpFabZ enzyme with IC50 of 2.0 ± 0.1 μM, much lower than those of quercetin and apigenin (IC50 in μM: quercetin: 39.3 ± 2.7; apigenin, 11.0 ± 2.5; Table 1). Inhibition mode investigation suggested that all three flavonoids functioned as competitive inhibitors against HpFabZ by competing with the substrate crotonoyl-CoA (Fig. 1). However, although (S)-sakuranetin displays a high structural similarity with quercetin and apigenin, it exhibits a distinct difference in binding affinity to HpFabZ. As indicated in Table 1, the K i value of (S)-sakuranetin (0.9 ± 0.1 μM) is ten times lower than that of quercetin (12.7 ± 0.6 μM) and apigenin (14.9 ± 0.3 μM). Such a stronger inhibition activity of (S)-sakuranetin against HpFabZ could be explained by the determination of complex crystal structures at the atomic level.
Table 1.
Enzymatic inhibition data of three flavonoids against HpFabZ
Figure 1.
(A–D) Chemical structures of quercetin, apigenin, (S)-sakuranetin, and (±)-naringenin. (E–G) The double-reciprocal plots of quercetin, apigenin, and (S)-sakuranetin for the mode of HpFabZ inhibition. Three rings are named, and positions are numbered according to the nomenclature.
Crystal structure of HpFabZ–flavonoid complex
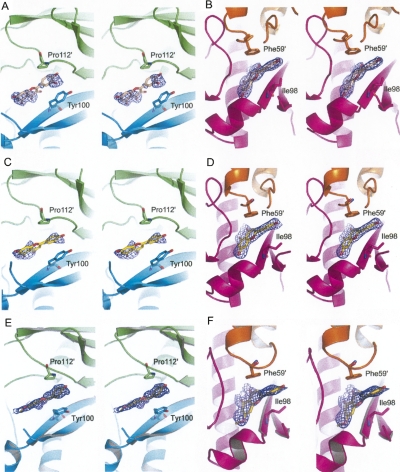
To investigate the potential inhibition mechanism of these three flavonoids against HpFabZ, the complex crystal structures of HpFabZ–quercetin, HpFabZ–apigenin, and HpFabZ–sakuranetin were determined by using the molecular replacement (MR) method. The coordinate of native HpFabZ (PDB code is 2GLL) was employed as the search model. Maximum-likelihood refinement was then carried out with CNS (Crystallography and NMR system; http://cns.csb.yale.edu/v1.1/) against 2.4 Å, 2.6 Å, and 2.4 Å level data, respectively. The following Procheck results showed that in these structures, 88.6%, 87.7%, and 88.0% of residue phi-psi angles were in the “core” area of the Ramachandran plot, and 11.4%, 12.3%, and 12.0% of residue phi-psi angles were in the “allow” area, respectively. These data indicated that the quality of the structures was good enough for further study. The stereoviews of the omit electron density maps contoured at 1.0 σ around the compounds are shown in Figure 2.
Figure 2.
Stereoview of the omit electron density maps contoured at 1.0 σ around quercetin (A,B), apigenin (C,D), and (S)-sakuranetin (E,F). Quercetin, apigenin, and (S)-sakuranetin are shown as sticks and colored in wheat, yellow, and olive, respectively. Monomers A, B, C, and D are colored in green, cyan, magenta, and orange, respectively. The pictures were generated using the program PyMOL (DeLano Scientific).
In these complex structures, the HpFabZ hexamer molecule displays the classical “trimer of dimers” conformation, similar to the native HpFabZ structure (PDB code is 2GLL). Two L-shaped active tunnels whose entrances are covered by door residue Tyr100 are located in the interface of dimer. Tyr100 has two different conformations (open and closed conformation). In the open conformation, the side chain of Tyr100 points toward Ile64′ and allows the chains of substrates entering the tunnel. In the closed conformation, the side chain of Tyr100 flops ∼120° around the Cα–Cβ bond pointing toward residue Pro112′, blocks the entrance of the tunnel, and stops the substrate from reaching the catalytic site. Each of the three flavonoids inhibits the activity of HpFabZ either by interacting with residue Tyr100 to occupy the entrance of the tunnel, or embedding into the tunnel near the catalytic site (residues Glu72 and His58′) to prevent the substrate from entering.
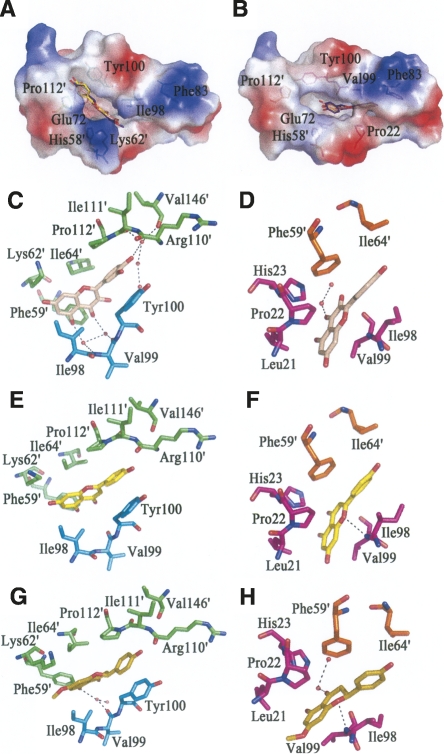
The structural analysis suggests that the three flavonoids bind to the HpFabZ hexamer in a similar way. They specifically bind to tunnels B and C rather than the other four active tunnels of HpFabZ hexamer with two distinct interaction models. In model A, each molecule of quercetin, apigenin, and (S)-sakuranetin binds to the entrance of the HpFabZ active tunnel B and interacts with door residue Tyr100 (Fig. 3A,C,E,G). The phenol ring of Tyr100 flops ∼120° from its original conformations (open and closed conformations) and sandwiches the phenol ring B of flavonoids with the pyrrolidine ring of Pro112′ (prime indicates the residue from the other subunit in the dimer). Meanwhile, rings A and C at the other end of each flavonoid also bind into a pocket formed by Phe59′, Lys62′, Ile64′, and Ile98 through hydrophobic interactions. Besides the hydrophobic interaction, the binding of quercetin or (S)-sakuranetin is also facilitated by some hydrogen bonds formed indirectly via water molecules (Fig. 3C,G). In model B, another molecule of quercetin, apigenin, or (S)-sakuranetin embeds into tunnel C near the catalytic site and plugs the tunnel to prevent the substrate from accessing the active site (Fig. 3B,D,F,H). The flavonoids stay in the right place via the hydrophobic interactions between their phenol rings and residues Leu21, Pro22, His23, Ile98, Val99, and Phe59′. Furthermore, either apigenin or (S)-sakuranetin also interacts with HpFabZ via the hydrogen bond between the oxygen atom O-1 and the backbone nitrogen of residue Val99 (Fig. 3F,H).
Figure 3.
The interaction between HpFabZ and inhibitors. (A,B) Binding positions of quercetin (salmon), apigenin (yellow), and (S)-sakuranetin (purple-blue) around the tunnel entrance (model A) or near the catalytic residues (model B) are shown. In model A, the inhibitors bind to the entrance of tunnel B linearly through hydrophobic interactions and are stacked between residues Tyr100 and Pro112′. In model B, inhibitors embed into tunnel C near the catalytic residues and are located in the hydrophobic pocket. The electrostatic surface of the active tunnel is rendered by a color ramp from red to blue. (C–H) Quercetin colored in wheat (C,D), apigenin colored in yellow (E,F), and (S)-sakuranetin colored in olive (G,H) interact with surrounding residues and water molecules in HpFabZ. Hydrogen bonds are shown as black dashes. Residues are labeled and colored in green, cyan, magenta, and orange for monomers A, B, C, and D, respectively.
HpFabZ Y100L site-directed mutagenesis
The above-mentioned structure analysis against HpFabZ–flavonoid complex has revealed that the door residue Tyr100 contributed a lot to the flavonoids' binding as a key tyrosine-binding residue. To test whether or not our flavonoids are specific to the Tyr100 of HpFabZ, HpFabZ Y100L site-directed mutagenesis and additional inhibition assay were performed. The inhibition activities as evaluated by IC50 of these flavonoids against HpFabZ Y100L were 13.1 ± 1.7 (quercetin), 4.3 ± 0.5 (apigenin), and 2.2 ± 0.2 μM ((S)-sakuranetin), respectively, indicating our flavonoids were not specific to the Tyr100 of HpFabZ.
Anti-H. pylori activities of the flavonoids
Besides assaying the inhibition of the three flavonoids against HpFabZ in vitro, we also tested their antibacterial activities against H. pylori strain ATCC 43504 with the standard agar dilution method (Osato 2000). The results showed that quercetin, apigenin, and (S)-sakuranetin inhibited the growth of H. pylori with minimum inhibitory concentration (MIC) values of 330.9, 92.5, and 87.3 μM, respectively. However, it was reported that no obvious antibacterial activity of apigenin was observed by using liquid culturing method while quercetin showed a lower MIC (165.4 μM) against H. pylori (Konstantinopoulou et al. 2003). This contradictory result might come from the different solubility of the inhibitors in liquid and solid culture media and the different variation of MIC judgment in these two methods (Cushnie and Lamb 2005).
Discussion
As subsequent research following our preliminary inhibitory effect assay of the flavonoids against HpFabZ in vitro (Zhang et al. 2008), the inhibition mode analysis and crystal structure characterization of HpFabZ–inhibitor complex in this work might help shed light on the potential inhibition mechanisms for flavonoids against FabZ. These three flavonoids were identified as competitive inhibitors against HpFabZ (Fig. 1), suggesting that they may interfere with the binding of substrate crotonoyl-CoA as further proved by the inhibitor binding model in the complex structures. Since the chemical structures of quercetin, apigenin, and sakuranetin are quite similar to each other (Fig. 1), it is not surprising that they locate in HpFabZ at similar positions in the substrate tunnel (Fig. 3), which is in good agreement with their competitive inhibitory properties against HpFabZ. Complex structure analysis indicated that the enzymatic activity of HpFabZ could be inhibited either by occupying the entrance of the tunnel (model A) or plugging the tunnel to prevent the substrate from accessing the active site (model B). These two binding models are nearly identical to those of the published HpFabZ inhibitors compounds 1 and 2 (PDB codes 2GLP and 2GLM) (Zhang et al. 2008). In model A, each of the five inhibitors (quercetin, apigenin, (S)-sakuranetin, compound 1, and compound 2) has a ring (phenyl ring or phenol ring) sandwiched between Tyr100 and Pro112′ at the entrance of the tunnel mainly by hydrophobic interactions, preventing the substrate chain from accessing the active site. In model B, the four inhibitors (quercetin, apigenin, (S)-sakuranetin, and compound 1) enter into the middle of the tunnel to locate near the active site of HpFabZ (His58 and Glu72′) and stay in the right place via hydrophobic interactions. Since compound 2 is larger than the other four inhibitors, it cannot enter the tunnel to form the binding model B successfully (Zhang et al. 2008).
The inhibition mode study indicates that the binding affinity of quercetin (K i = 12.7 ± 0.6 μM) is very close to that of apigenin (K i = 14.9 ± 0.3 μM). The minor difference might be caused by the fact that more residues are forming a hydrophobic interaction with quercetin in model A than those in the case of apigenin (in model B, the binding patterns of these two compounds are almost the same). Notably, the only structural difference between quercetin and apigenin is the two additional hydroxyl groups of quercetin at position C-3 and C-3′ (Fig. 1A). As revealed in the crystal structures, these two hydroxyls make no direct contact with HpFabZ, indicating that these groups might make little contribution in the inhibition of HpFabZ, which is consistent with the close binding affinities of the two flavonoids.
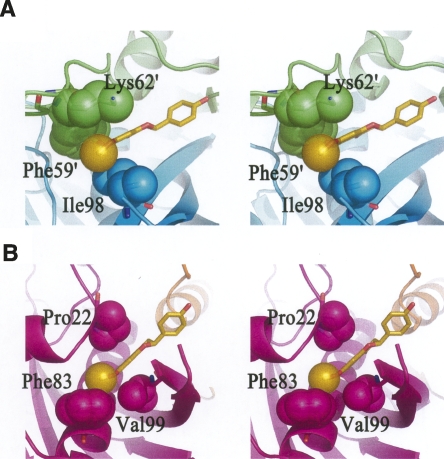
Although the three flavonoids share similar chemical structures and binding models against HpFabZ, (S)-sakuranetin displays a much better binding affinity (K i = 0.9 ± 0.1 μM) than the other two flavonoids based on the kinetic assays. The chemical structure comparison of (S)-sakuranetin with the other two flavonoids suggests that the main difference comes from the methoxy group at C-7 position (Fig. 1). Interestingly, it was also reported that the methoxy group at C-7 in the structure of (S)-sakuranetin is important for its high activity as a phytoalexin (Aida et al. 1996). As indicated in Figure 4, the methoxy group at C-7 position of (S)-sakuranetin is surrounded by hydrophobic residues in both models A and B. The strong hydrophobic interactions between the methoxy group and hydrophobic residues nearby would stabilize (S)-sakuranetin and make it more competitive. In model A, the methoxy group extends into the hydrophobic pocket composed of Ile98, Phe59′, and Lys62′ with a buried surface area of ∼136 Å2 [calculated by the software Ligplot (Wallace et al. 1995)] (Fig. 4A). Hence the illation might be reached that the hydrophobic interactions between the methoxy group and HpFabZ in this pocket may correlate with the suitable binding of (S)-sakuranetin in this site. In model B, the methoxy group embeds into the hydrophobic pocket composed of Pro22, Phe83, and Val99 with a buried surface area of ∼120 Å2 (Fig. 4B). The hydrophobic interactions between the methoxy group of (S)-sakuranetin and these residues might also be attributable to the stronger binding affinity. Accordingly, it could be hypothesized that inhibitors with a suitably sized hydrophobic group (such as the methoxy group of (S)-sakuranetin) located in these two hydrophobic pockets could increase their binding affinities and inhibitory effect. In addition, as shown in Figure 2, the omit electron density map of (S)-sakuranetin is better than those of quercetin and apigenin in the binding models of the complex structures, in accord with the higher binding affinity of (S)-sakuranetin in the kinetic assays.
Figure 4.
Stereoview of the hydrophobic interactions between the methoxy group of (S)-sakuranetin and HpFabZ. The color scheme is the same as in Figure 3. The protein is shown as cartoon, (S)-sakuranetin and the contacting residues are drawn as sticks. The methyl on the methoxy group of (S)-sakuranetin, the atoms involved in hydrophobic interactions such as side chains of Phe59′, Lys62′, and Ile98 in model A (A), and side chains of Pro22, Phe83, and Val99 in model B (B) are represented as transparent spheres.
To investigate the detail of the structure-activity relationship (SAR), the inhibition activity of another flavonoid, (±)-naringenin against HpFabZ was also tested. Surprisingly, although naringenin shares similar chemical structure with quercetin, apigenin, and (S)-sakuranetin, the assay result showed that (±)-naringenin had no inhibition against HpFabZ (IC50 > 500 μM). These experimental data are in very good agreement with recently published results (Tasdemir et al. 2006) that suggested that the loss of the double bond between C-2 and C-3 abolishes the activity of naringenin, indicating the importance of the planarity of flavonoids with respect to activity. However, the presence of the methoxy group at C-7 position improves the activity of (S)-sakuranetin greatly, revealing the importance of the methoxy group at C-7 position, as well as the double bond between C-2 and C-3. In addition, it was also pointed out that the stereochemistry at C-2 does not play an important role in the inhibition of FabZ (Tasdemir et al. 2006). Thus, it is likely that the stereochemistry of naringenin and sakuranetin contributes little to their inhibition activities.
Apart from the current reported inhibition of the three flavonoids against HpFabZ, a series of flavonoids were also published to be inhibitors against Plasmodium falciparum FabZ (PfFabZ) (Tasdemir et al. 2006). Interestingly, unlike its high inhibitory activity against HpFabZ, apigenin showed no inhibition against PfFabZ even at a concentration of 100 μM (Tasdemir et al. 2006). The structure analysis against HpFabZ–flavonoid complex revealed that the door residues Tyr100 contributed a lot to the flavonoid's binding as a key tyrosine-binding residue. By considering the result that the key residue Tyr100 in HpFabZ was replaced by a leucine in PfFabZ, as indicated from the structure-based amino acid sequence alignment of HpFabZ and PfFabZ (Zhang et al. 2008), we has tested whether or not our flavonoids have specific binding to the Tyr100 of HpFabZ by HpFabZ Y100L site-directed mutagenesis and relevant inhibition assays. The results suggested that the side chain of Leu100 could perform a similar function as the side chain of Tyr100 (Table 1) and interact with ring B of flavonoids via strong hydrophobic interactions. The data also proved that our flavonoids were not specific to the Tyr100 of HpFabZ.
In addition, the anti-H. pylori activities of quercetin, apigenin, and (S)-sakuranetin were confirmed with MIC values of 330.9, 92.5, and 87.3 μM, respectively. The disagreement between K i against HpFabZ and MIC values might be due to the fact that these flavonoids probably act on other bacterial targets, such as the DNA gyrase (Ohemeng et al. 1993) and D-alanine-D-alanine ligase (Wu et al. 2008). On the other hand, the permeation of these flavonoids through the cell wall and membrane is also crucial for their antibacterial activity, especially when the cytoplasmic membrane itself has also been found as one target of flavonoids (Mirzoeva et al. 1997).
In conclusion, three flavonoids, quercetin, apigenin, and (S)-sakuranetin, were identified as competitive inhibitors of HpFabZ. More importantly, the complex structures of HpFabZ with these flavonoids were determined, and two binding models were also discovered. On the basis of the complex structure information, the importance of the methoxy group at the C-7 position, with respect to activity, was pointed out and proved by the following inhibition assay of another flavonoid naringenin against HpFabZ. In addition, the HpFabZ Y100L site-directed mutagenesis assay indicated that our flavonoids are not specific to the Tyr100 of HpFabZ. Our work reveals the inhibitory mechanisms of three flavonoids, quercetin, apigenin, and (S)-sakuranetin against HpFabZ and is expected to provide some useful information for structure-based drug design in the flavonoid-research field.
Materials and Methods
Materials
Standard H. pylori strain ATCC 43504 was obtained from the Shanghai Institute of Digestive Disease. Escherichia coli strain BL21 (DE3) was purchased from Stratagene. All chemicals were of reagent grade or ultra-pure quality and commercially available.
HpFabZ enzymatic inhibition assay
The expression, purification, and enzymatic inhibition assay of HpFabZ enzyme were performed as reported previously (Liu et al. 2005; Zhang et al. 2008). The compounds dissolved in 1% DMSO were incubated with the enzyme for 1 h before the assay started. The IC50 value of (S)-sakuranetin was estimated by fitting the inhibition data to a dose-dependent curve using a logistic derivative equation. The inhibitor types of the three flavonoids against HpFabZ were determined in the presence of various inhibitor concentrations. After 2-h incubation, the reaction was started by the addition of crotonoyl-CoA. The K i values were obtained from Lineweaver-Burk double-reciprocal plots and subsequent secondary plots.
HpFabZ Y100L site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. pQE30-HpFabZ plasmid was used as the template for construction of the HpFabZ (Y100L) mutant. On the basis of the sequence of HpFabZ, two polymerase chain reaction (PCR) primers (forward: 5′-GCCAAAACAAAAATCGTGTTATTCATGACGATTGACAAGG-3′ and reverse: 5′-CCTTGTCAATCGTCATGAATAACACGATTTTTGTTTTGGC-3′) were designed to amplify the corresponding region. The mutant was then sequenced to confirm the desired mutation site. The expression and purification of HpFabZ (Y100L) were similar to those of HpFabZ as described above. The enzymatic characterization of HpFabZ (Y100L) mutant was determined according to the previously published method (Zhang et al. 2008).
HpFabZ–inhibitor complex crystallization and data collection
The crystallization of HpFabZ was performed using the hanging-drop vapor diffusion method as previously described (Zhang et al. 2008). Inhibitors (quercetin, apigenin, and (S)-sakuranetin) were added into the original drops to a final concentration of about 10 mM and soaked for 24 h, respectively. Diffraction data were collected at −180°C on an in-house R-Axis IV++ image-plate detector equipped with a Rigaku rotating-anode generator. The data sets were integrated with MOSFLM (Leslie 1999) and scaled with programs from the CCP4 suite (Collaborative Computational Project Number 4 1994). A summary of the diffraction data statistics is given in Table 2.
Table 2.
Summary of diffraction data and structure refinement statistics
Structure determination and refinement
The structures of HpFabZ–inhibitor complex were solved by molecular replacement (MR) with the programs in CCP4 (Collaborative Computational Project Number 4 1994) using the coordinate of native HpFabZ (PDB code 2GLL) as the search model. Structure refinement was carried out using CNS (Brunger et al. 1998) standard protocols (energy minimization, simulated annealing, water picking, and B-factor refinement), and model building was facilitated using the program Coot (Emsley and Cowtan 2004). The stereochemical quality of the structure models during the course of refinement and model building was evaluated with the program PROCHECK (Wallace et al. 1995). The statistics of structure refinement are summarized in Table 2. The complex structures of HpFabZ–quercetin, HpFabZ–apigenin, and HpFabZ–sakuranetin have been deposited in the RCSB Protein Data Bank under accession codes 3CF8, 3CF9, and 3D04, respectively.
Anti-H. pylori activity assay
The MIC values were determined by the standard agar dilution method using Columbia agar supplemented with 10% sheep blood containing twofold serial dilutions of three inhibitors. The plates were inoculated with a bacterial suspension (108 cfu/mL) in Brain-Heart Infusion broth with a multipoint inoculator (Sakuma Seisakusho). Compound-free Columbia agar media were used as controls. Inoculated plates were incubated at 37°C under a microaerobic condition and examined after 3 d. The MIC value was defined as the lowest concentration of agents that completely inhibited visible bacterial growth.
Acknowledgments
This work was supported by the State Key Program of Basic Research of China (grants 2004CB518905, 2006AA09Z447, and 2007CB914304), the National Natural Science Foundation of China (grants 30525024, 90713046, and 20721003), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (grants 06JC14080 and 03DZ19228), and CAS Foundation (grant KSCX1-YW-R-18).
Footnotes
Reprint requests to: Xu Shen, Drug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; e-mail: xshen@mail.shcnc.ac.cn; fax: 86-21-50806918; or Lihong Hu, Drug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; e-mail: simmhulh@mail.shcnc.ac.cn; fax: 86-21-50806918.
Abbreviations: FabZ, β-hydroxyacyl-acyl carrier protein dehydratase; HpFabZ, Helicobacter pylori FabZ; FAS, fatty acid biosynthesis; PfFabZ, Plasmodium falciparum FabZ.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036186.108.
References
- Abdel-Sattar, E., Kohiel, M.A., Shihata, I.A., el-Askary, H. Phenolic compounds from Eucalyptus maculata . Pharmazie. 2000;55:623–624. doi: 10.1002/chin.200047190. [DOI] [PubMed] [Google Scholar]
- Aida, Y., Tamogami, S., Kodama, O., Tsukiboshi, T. Synthesis of 7-methoxyapigeninidin and its fungicidal activity against Gloeocercospora sorghi . Biosci. Biotechnol. Biochem. 1996;60:1495–1496. doi: 10.1271/bbb.60.1495. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cushnie, T.P., Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Harborne, J.B., Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002;96:67–202. doi: 10.1016/s0163-7258(02)00298-x. [DOI] [PubMed] [Google Scholar]
- Hernandez, V., Recio, M.C., Manez, S., Giner, R.M., Rios, J.L. Effects of naturally occurring dihydroflavonols from Inula viscosa on inflammation and enzymes involved in the arachidonic acid metabolism. Life Sci. 2007;81:480–488. doi: 10.1016/j.lfs.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Hurabielle, M., Eberle, J., Paris, M. Flavonoids of Artemisia campestris, ssp. glutinosa . Planta Med. 1982;46:124–125. doi: 10.1055/s-2007-970035. [DOI] [PubMed] [Google Scholar]
- Konstantinopoulou, M., Karioti, A., Skaltsas, S., Skaltsa, H. Sesquiterpene lactones from Anthemis altissima and their anti-Helicobacter pylori activity. J. Nat. Prod. 2003;66:699–702. doi: 10.1021/np020472m. [DOI] [PubMed] [Google Scholar]
- Leslie, A.G. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- Liu, Y.L., Ho, D.K., Cassady, J.M., Cook, V.M., Baird, W.M. Isolation of potential cancer chemopreventive agents from Eriodictyon californicum . J. Nat. Prod. 1992;55:357–363. doi: 10.1021/np50081a012. [DOI] [PubMed] [Google Scholar]
- Liu, W., Luo, C., Han, C., Peng, S., Yang, Y., Yue, J., Shen, X., Jiang, H. A new β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Helicobacter pylori: Molecular cloning, enzymatic characterization, and structural modeling. Biochem. Biophys. Res. Commun. 2005;333:1078–1086. doi: 10.1016/j.bbrc.2005.05.197. [DOI] [PubMed] [Google Scholar]
- Mirzoeva, O.K., Grishanin, R.N., Calder, P.C. Antimicrobial action of propolis and some of its components: The effects on growth, membrane potential and motility of bacteria. Microbiol. Res. 1997;152:239–246. doi: 10.1016/S0944-5013(97)80034-1. [DOI] [PubMed] [Google Scholar]
- Nakazato, Y., Tamogami, S., Kawai, H., Hasegawa, M., Kodama, O. Methionine-induced phytoalexin production in rice leaves. Biosci. Biotechnol. Biochem. 2000;64:577–583. doi: 10.1271/bbb.64.577. [DOI] [PubMed] [Google Scholar]
- Ohemeng, K.A., Schwender, C.F., Fu, K.P., Barrett, J.F. DNA gyrase inhibitory and antibacterial activity of some flavones (1) Bioorg. Med. Chem. Lett. 1993;3:225–230. [Google Scholar]
- Osato, M.S. Antimicrobial susceptibility testing for Helicobacter pylori: Sensitivity test results and their clinical relevance. Curr. Pharm. Des. 2000;6:1545–1555. doi: 10.2174/1381612003399059. [DOI] [PubMed] [Google Scholar]
- Ross, J.A., Kasum, C.M. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Annu. Rev. Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- Tasdemir, D., Lack, G., Brun, R., Ruedi, P., Scapozza, L., Perozzo, R. Inhibition of Plasmodium falciparum fatty acid biosynthesis: Evaluation of FabG, FabZ, and FabI as drug targets for flavonoids. J. Med. Chem. 2006;49:3345–3353. doi: 10.1021/jm0600545. [DOI] [PubMed] [Google Scholar]
- Wallace, A.C., Laskowski, R.A., Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wu, D., Kong, Y., Han, C., Chen, J., Hu, L., Jiang, H., Shen, X. D-alanine: D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents. 2008 doi: 10.1016/j.ijantimicag.2008.06.010. (in press). [DOI] [PubMed] [Google Scholar]
- Zhang, L., Liu, W., Hu, T., Du, L., Luo, C., Chen, K., Shen, X., Jiang, H. Structural basis for catalytic and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) J. Biol. Chem. 2008;283:5370–5379. doi: 10.1074/jbc.M705566200. [DOI] [PubMed] [Google Scholar]