Abstract
Pax3, a member of the paired class homeodomain family of transcription factors, is essential for early skeletal muscle development. Previously, others and we have shown that the stability of Pax3 is regulated on a post-translational level. Evidence in the literature and from our laboratory suggests that phosphorylation, a common form of regulation, may play a role. However, at present, the sites of Pax3 phosphorylation are not known. We demonstrate here the first evidence that Pax3 exists as a phosphoprotein in proliferating mouse primary myoblasts. Using an in vitro kinase assay, deletion, and point mutant analysis, we conclusively identify Ser205 as a site of phosphorylation. The phosphorylation of Ser205 on endogenously expressed Pax3 was confirmed in vivo using antibodies specific for phosphorylation at Ser205. Finally, we demonstrate for the first time that the phosphorylation status of endogenous Pax3 changes rapidly upon the induction of myogenic differentiation. The presence of phosphorylation in a region of Pax3 important for mediating protein–protein interactions, and the fact that phosphorylation is lost upon induction of differentiation, allow for speculation on the biological relevance of phosphorylation.
Keywords: Pax3, phosphorylation, myogenic differentiation, phosphopeptide analysis, post-translational modifications
Pax3, a member of the paired class homeobox family of transcription factors, plays an essential role in early skeletal muscle development. Pax3 is required for the formation of muscles of the trunk and for the delamination and migration of myogenic progenitor cells to the limb buds (Williams and Ordahl 1994; Tajbakhsh and Buckingham 2000; Buckingham and Relaix 2007). Pax3-deficient Splotch mice are embryonic lethal due to defects in skeletal muscle (Xia and Barr 2005), and human patients with PAX3 haploinsufficiency display limb muscle hypoplasia (Epstein et al. 1996). It has been demonstrated that the ectopic expression of Pax3 is capable of activating the myogenic program in mesoderm tissue by activating the expression of the myogenic regulatory factors MyoD, Myf-5, and myogenin (Maroto et al. 1997).
Because of the importance of Pax3 in early muscle development and in the expression of early myogenic genes, it is critical that the expression and activity of Pax3 be tightly regulated throughout differentiation. Recently, it was determined that the stability of Pax3 is regulated on a post-translational level during myogenic differentiation. We have shown that Pax3 protein levels decrease significantly in the first 24 h of myogenic differentiation and that this change in protein levels are regulated post-translationally since changes in mRNA levels and protein translation for Pax3 do not correlate with the decrease in Pax3 protein levels (Miller and Hollenbach 2007). Furthermore, it has been shown that Pax3 stability is regulated, in part, via the ubiquitin-proteasome system (Boutet et al. 2007).
In addition to the ubiquitin-proteasome system, it has been speculated that phosphorylation may also be important in the regulation of Pax3 biological activities (Boutet et al. 2007; Miller and Hollenbach 2007). Phosphorylation has been widely studied due to its various roles in transcription factor regulation (Hunter and Karin 1992). However, at present it is not known whether Pax3 is phosphorylated in a physiologically relevant cell type, nor have the sites of phosphorylation been identified. Therefore, in the present study, we demonstrate that Pax3 is indeed phosphorylated in proliferating mouse primary myoblasts. Furthermore, we use both in vitro and in vivo mapping techniques along with a phosphospecific antibody to identify Ser205 as a site of phosphorylation on Pax3 in proliferating mouse primary myoblasts. Finally, we demonstrate that Pax3 is only phosphorylated at Ser205 in proliferating mouse primary myoblasts and that the phosphorylation status of Pax3 changes rapidly upon the induction of myogenic differentiation. The fact that a site of phosphorylation occurs in a region of Pax3 required for mediating protein–protein interactions and that phosphorylation of Pax3 changes upon induction of differentiation allows for speculation into the role of this phosphorylation event in the regulation of Pax3 activities.
Results
Pax3 is phosphorylated in vivo
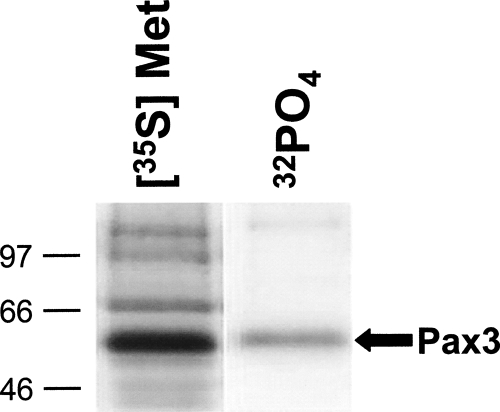
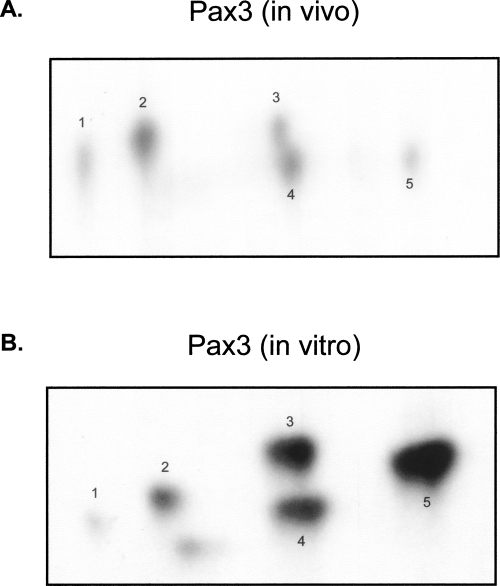
In order to demonstrate that Pax3 is phosphorylated in a physiologically relevant cell type, we stably transduced proliferating mouse primary myoblasts with a retroviral construct containing a FLAG epitope tagged Pax3 (FLAG-Pax3) construct, metabolically labeled the cells with [32P]-orthophosphate or [35S]-Methionine, immunoprecipitated FLAG-Pax3 with a FLAG-specific antibody, and examined incorporated radiolabel by SDS-PAGE analysis. We observed the specific incorporation of both radiolabels into Pax3 (Fig. 1), demonstrating that Pax3 is both expressed and phosphorylated in proliferating primary myoblasts. This result provides the first evidence that Pax3 exists as a phosphoprotein in a physiologically relevant cell type. In order to further characterize the in vivo phosphorylation of Pax3, we generated a two-dimensional phosphopeptide map of FLAG-Pax3 that had been metabolically labeled with [32P]-orthophosphate. The phosphopeptide analysis demonstrates the presence of five distinct radiolabeled peptides, suggesting multiple sites of phosphorylation may be present (Fig. 2A).
Figure 1.
Pax3 is phosphorylated in proliferating mouse primary myoblasts. Proliferating mouse primary myoblasts stably transduced with an amino-terminal FLAG epitope tagged Pax3 were metabolically labeled with either [35S]-Methionine or [32P]-orthophosphate, and FLAG-Pax3 was immunoprecipitated from total cell extracts using an anti-FLAG antibody, as described in Materials and Methods. The resulting immunoprecipitates were separated by 12% SDS-PAGE, and the radiolabeled species were detected by autoradiography.
Figure 2.
Development of an in vitro kinase assay to facilitate the identification of phosphorylation sites on Pax3. (A) Proliferating mouse primary myoblasts stably expressing FLAG-Pax3 were metabolically labeled with [32P]-orthophosphate, and FLAG-Pax3 was immunoprecipitated from total cell extracts using an anti-FLAG antibody. The resulting radiolabeled protein was isolated from the dried 12% SDS-PAGE gel and subjected to two-dimensional phosphopeptide analysis, as previously described (Boyle et al. 1991). (B) Bacterially expressed and purified GST-Pax3 was phosphorylated using our in vitro kinase assay. The radiolabeled protein was isolated from the dried 10% SDS-PAGE gel and subjected to two-dimensional phosphopeptide analysis. In both panels, the five distinctly migrating phosphopeptides are indicated by numbers one through five.
Development and validation of an in vitro kinase assay
Because the generation of an in vivo phosphopeptide map required a substantial amount of [32P]-orthophosphate and a minimum exposure period of at least 6 wk, using this technique was not an efficient method of identification of the sites of phosphorylation on Pax3. In addition, several attempts at using mass spectral analysis to identify the sites of phosphorylation both in vivo and in vitro did not yield usable results. Therefore, to facilitate the identification of the site(s) of phosphorylation on Pax3, we needed to develop an in vitro kinase assay that would use smaller amounts of radioactivity and require shorter exposure times expediting the analysis. We used bacterially expressed and purified GST-tagged Pax3 (GST-Pax3), phosphorylated the isolated protein in vitro using total cell extracts derived from proliferating mouse primary myoblasts, and analyzed the phosphorylated protein by two-dimensional phosphopeptide analysis in order to verify that the in vitro phosphopeptide map of Pax3 was similar to the in vivo map. The in vitro method required considerably less radioactivity and required an exposure period of only 12–24 h. The in vitro phosphopeptide map of Pax3 contained five distinct radiolabeled peptides with mobilities that are similar to that seen for the in vivo map (Fig. 2). Because the phosphopeptide maps of the in vitro and in vivo assays were essentially identical, the in vitro method is sufficient to perform an initial identification of the site(s) of phosphorylation on Pax3.
The phospho-amino acid is located on the region of Pax3 surrounding the octapeptide domain
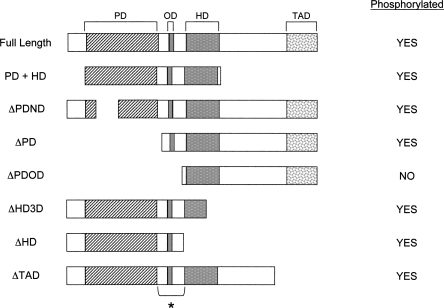
In an effort to identify the region of Pax3 that is phosphorylated, we created GST-Pax3 deletion mutants (Fig. 3) for use in the in vitro kinase assay. The deletion mutants targeted key regions of Pax3 that are required for its interaction with DNA (ΔPDND, ΔPD, ΔHD3D, and ΔHD), for transcriptional activity (ΔTAD and PD + HD), or for mediating protein–protein interactions (ΔPDOD). We observed no apparent change in the phosphorylation status in most of the deletion mutants when used in our in vitro kinase assay. Only when the region surrounding the octapeptide domain was deleted (ΔPDOD) did we observe a complete loss of phosphorylation (Fig. 3). This result demonstrates that the phosphorylation of Pax3 occurs primarily in the region surrounding the octapeptide domain.
Figure 3.
Pax3 is phosphorylated in the region surrounding the octapeptide domain. The in vitro kinase assay was performed on bacterially expressed and purified GST-Pax3 deletion mutants that had key Pax3 structural domains removed. The overall phosphorylation status of each of the mutants is indicated. The domains of Pax3 are as follows: The paired DNA-binding domain (PD) is depicted by the diagonal stripes, the octapeptide domain (OD) by the gray box, the homeodomain DNA-binding domain (HD) by the gray stippled box, and the transcriptional activation domain (TAD) by the white stippled box. The bracket and asterisk indicate the region of Pax3 that is phosphorylated.
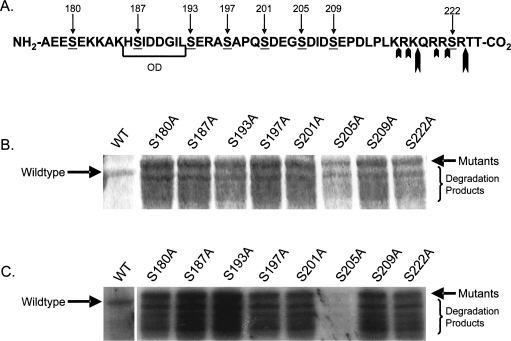
Ser205 is a site of Pax3 phosphorylation in vitro
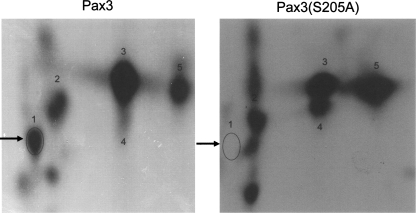
A close examination of the amino acids deleted in the ΔPDOD mutant revealed the presence of eight serines and two threonines that could act as sites of phosphorylation (Fig. 4A). An independent phospho-amino acid analysis demonstrated that only serines are phosphorylated on Pax3 (data not shown). Therefore, we focused our attention on the eight serines located in the region surrounding the octapeptide domain. We created phospho-incompetent GST-Pax3 point mutants by independently converting each serine to an alanine and then used each of these mutants in our in vitro kinase assay. As seen with the deletion mutants, a majority of the point mutants showed no apparent change in phosphorylation despite similar levels of total protein. Only when serine 205 is mutated to alanine (S205A) do we observe a significant decrease in phosphorylation compared with wild-type Pax3 (Fig. 4B,C), indicating that serine 205 is the primary site of phosphorylation on Pax3. A subsequent two-dimensional phosphopeptide analysis demonstrated that, consistent with the observed decrease in overall phosphorylation, Pax3(S205A) has a complete loss of a single spot relative to wild-type Pax3 (Fig. 5). Taken together, these results therefore confirm serine 205 as a site of phosphorylation on Pax3 in vitro.
Figure 4.
Pax3 is phosphorylated at serine 205 in vitro. (A) Schematic of the primary amino acid sequence of the region deleted in the ΔPDOD mutant. The octapeptide domain (OD) is indicated by the bracket, and the eight serines present in this region are underlined. The smaller bold arrows indicate the predicted sites of trypsin cleavage, and the larger bold arrows indicate the predicted major sites of cleavage. (B) Bacterially expressed and purified GST-Pax3 and the individual GST-Pax3 phospho-incompetent mutants were separated by 8% SDS-PAGE and visualized by Coomassie staining. (C) The same GST proteins were used in parallel in vitro kinase assays, separated by 8% SDS-PAGE, and visualized by autoradiography. The mobilities of the wild-type Pax3 and the Pax3 point mutants are indicated by the arrows. Wild-type Pax3 lacks the carboxyl terminus, which was demonstrated to not affect phosphorylation, and therefore migrates with a slightly faster mobility.
Figure 5.
Two-dimensional phosphopeptide analysis of wild-type Pax3 (left) and Pax3(S205A) (right). Bacterially expressed GST-Pax3 and GST-Pax3 S205A were phosphorylated in vitro, trypsinized using TPCK-trypsin, and analyzed by two-dimensional phosphopeptide analysis, as described in Materials and Methods. The arrow indicates the phosphopeptide that is no longer phosphorylated upon the mutation of serine 205.
Identification of Ser205 as a site of Pax3 phosphorylation in vivo
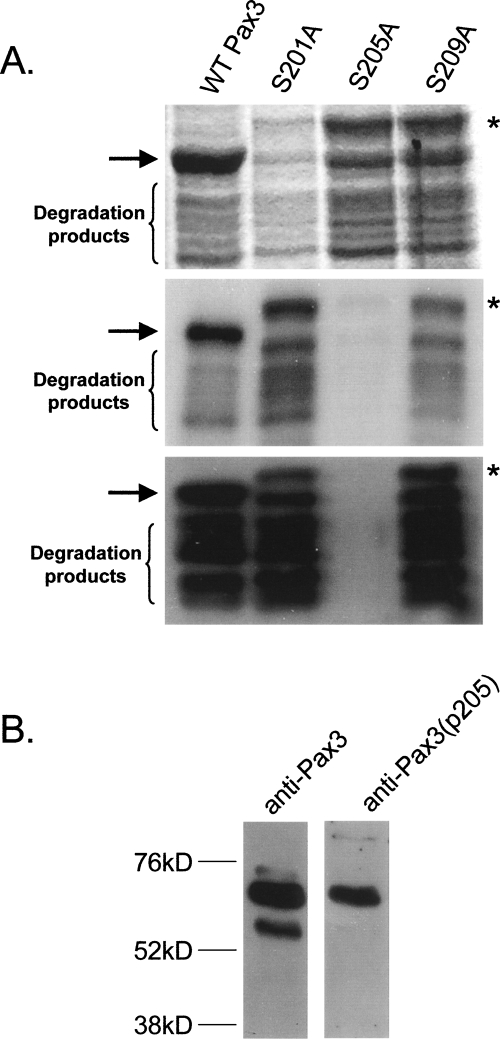
To facilitate the in vivo confirmation and biological analysis of phosphorylation at serine 205, we raised an antibody specific for Pax3 only when Pax3 is phosphorylated at serine 205. In order to confirm the specificity of this antibody, we performed a Western blot analysis with the anti-Pax3(p205) antibody on bacterially expressed and purified GST-Pax3 and GST-Pax3(S205A) that had been phosphorylated using our in vitro kinase assay. For negative controls, we also mutated the two serines immediately adjacent to serine 205 (S201A and S209A) and used them in parallel in the Western blot analysis. An independent radiolabeling experiment confirmed that the efficient phosphorylation of Pax3 is dependent on the presence of serine 205 (Fig. 6A, middle panel). Consistent with the phosphorylation of Pax3 at serine 205, the antibody showed a strong reactivity with wild-type Pax3 and the two negative controls, but not when serine 205 was mutated to alanine (Fig. 6A, bottom panel). This result therefore confirms the specificity of the antibody for Pax3 when phosphorylated at serine 205.
Figure 6.
Confirmation of the phosphorylation of serine 205 in vivo. (A) An antibody was raised against a synthetic peptide specifically phosphorylated at serine 205, as described in Materials and Methods. Equal amounts of bacterially expressed and purified GST-Pax3 and three GST-Pax3 phospho-incompetent point mutants were used in independent in vitro kinase assays using [γ-32P]-ATP to confirm phosphorylation or with cold ATP. The proteins that were nonradioactively labeled were subsequently used for Western blot analysis using the anti-Pax3(p205). (Top panel) Coomassie staining to demonstrate similar amounts of protein; (middle panel) independent radiolabeling to confirm the phosphorylation status; (bottom panel) Western blot analysis using the anti-Pax3(p205) antibody. The arrow indicates the mobility of wild-type Pax3, while the asterisk indicates the mobility of the phospho-incompetent point mutants. (B) Pax3 is phosphorylated at Ser205 in proliferating mouse primary myoblasts. Total cell extract was isolated from proliferating mouse primary myoblasts, as described in Materials and Methods; and a Western blot analysis was performed on 50 μg of total cell extract using either the general anti-Pax3 antibody (left panel) or anti-Pax3(p205) antibody (right panel).
To confirm that serine 205 is a site of phosphorylation on Pax3 in vivo, we performed a Western blot analysis on total cell extracts from proliferating mouse primary myoblasts, which have been demonstrated to endogenously express Pax3 (Miller and Hollenbach 2007). The analysis was performed using the previously described mono-specific Pax3 antibody (Lam et al. 1999) and our anti-Pax3(p205) antibody. Consistent with previous reports (Miller and Hollenbach 2007), the general Pax3 antibody identified two distinctly migrating species with apparent molecular weights of 56 kDa and 66 kDa (Fig. 6B, left panel). However, a Western blot analysis using the anti-Pax3(p205) antibody on the identical membrane reacted solely with the apparent 66-kDa species (Fig. 6B, right panel). This result conclusively demonstrates not only that Pax3 is phosphorylated at serine 205 in proliferating mouse primary myoblasts but that this phosphorylation event changes the electrophoretic mobility of Pax3.
Phosphorylation at serine 205 is rapidly lost upon myogenic differentiation
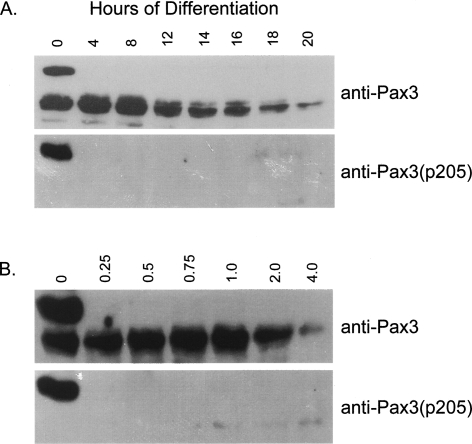
Finally, in order to determine if Pax3 is phosphorylated throughout myogenic differentiation, we induced differentiation, as described in the Materials and Methods section, and performed a Western blot analysis on equal amounts of total cell extracts from each time point of differentiation using the anti-Pax3 or the anti-Pax3(p205) antibodies. Consistent with the results just described, we observed two distinctly migrating species of Pax3 in proliferating primary myoblasts, of which only the apparent 66-kDa species was phosphorylated at serine 205 (Fig. 7). Consistent with previous results, we observed the complete loss of expression of Pax3 within 24 h of differentiation (Miller and Hollenbach 2007). More importantly, within 4 h of the induction of myogenic differentiation, we observed the complete loss of the apparent 66-kDa species and the corresponding reactivity of the anti-Pax3(p205) antibody (Fig. 7A). To further characterize this change in phosphorylation, we repeated the differentiation experiment using earlier time points. Surprisingly, we observed a complete loss of the apparent 66-kDa species and reactivity with the anti-Pax3(p205) antibody by 15 min of differentiation. Therefore, these results are the first to demonstrate that phosphorylation of Pax3 at serine 205 is rapidly abolished upon the induction of myogenic differentiation.
Figure 7.
Phosphorylation of Pax3 at serine 205 is rapidly abolished upon the induction of differentiation. Proliferating mouse primary myoblasts were induced to differentiate as described in Materials and Methods for 0–48 h (A) or for 0–4 h (B). Total cell extracts were created from the differentiated myoblasts at the indicated time points, and a standard Western blot analysis was performed on 50 μg of total cell extract using the anti-Pax3 antibody (top panels) or the anti-Pax3(p205) antibody (bottom panels).
Discussion
Pax3 is a member of the paired class homeodomain family of transcription factors that plays an essential role in early skeletal muscle development. Previous reports have shown that Pax3 protein levels are regulated on a post-translational level (Boutet et al. 2007; Miller and Hollenbach 2007). Post-translational modifications are common mechanisms for the regulation of the biological activities of transcription factors. In particular, the phosphorylation status of transcription factors has been widely studied due to its important role in cellular regulation. Evidence from our laboratory and others suggests that phosphorylation may contribute to the biological regulation of Pax3 (Boutet et al. 2007; Miller and Hollenbach 2007); however to date, it is not known if Pax3 is phosphorylated in a physiologically relevant system. Therefore, the focus of the present study was to examine the phosphorylation status of Pax3 in mouse primary myoblasts. In this report, we demonstrate that Pax3 is phosphorylated at serine 205 in the physiologically relevant proliferating mouse primary myoblasts. Furthermore, we demonstrate for the first time that phosphorylation of Pax3 at serine 205 is rapidly abolished upon the induction of myogenic differentiation.
Our data conclusively identifies serine 205 as a site of phosphorylation on Pax3 in proliferating primary myoblasts. Interestingly, our report is the first to demonstrate that, upon the induction of differentiation, Pax3 undergoes a rapid loss of phosphorylation at serine 205 (Fig. 7). This observation is reminiscent of how phosphorylation regulates the activity of other myogenic transcription factors as myoblasts shift from the proliferative to the differentiating state. It was shown that both MyoD and myogenin, two early myogenic transcription factors, are phosphorylated in proliferating myoblasts (Li et al. 1992; Kitzmann et al. 1999). Phosphorylation of these two factors inhibits their transcriptional activity, either through inhibition of DNA binding (Li et al. 1992) or by promoting their degradation (Kitzmann et al. 1999). The phosphorylation of MyoD and myogenin is subsequently decreased upon the induction of myogenic differentiation, resulting in activation of their transcriptional activity (Li et al. 1992; Kitzmann et al. 1999). Because the loss of phosphorylation of Pax3 at serine 205 resembles what is known for MyoD and myogenin, it is conceivable that the observed change in Pax3 phosphorylation alters the biological activity of Pax3 as the myoblasts begin to differentiate.
Although the exact mechanism by which phosphorylation regulates Pax3 is not known, the position of this phosphorylated amino acid within the primary amino acid structure indicates a possible effect on the biological activities of Pax3. Serine 205 is located immediately adjacent to the octapeptide domain (Fig. 4A) and is present in the region of Pax3 demonstrated to mediate protein–protein interactions with the transcriptional regulators hDaxx (Hollenbach et al. 1999), calmyrin (Hollenbach et al. 2002), and HIRA (Magnaghi et al. 1998). Phosphorylation of transcription factors is a common mechanism used to regulate protein–protein interactions. Therefore, the phosphorylation status of serine 205 may control the interaction of Pax3 with these and other cofactors, thereby regulating such biological activities as transcriptional activation (Magnaghi et al. 1998; Hollenbach et al. 1999) and DNA binding (Hollenbach et al. 2002).
Although our data conclusively identifies serine 205 as a site of phosphorylation on Pax3, our results also suggest that serine 205 may not be the only site. If serine 205 were the sole site of phosphorylation, mutation of serine 205 would be expected to completely abrogate all of the radiolabeled peptides on the two-dimensional phosphopeptide map. Instead, the phosphopeptide maps of Pax3(S205A) from the in vitro kinase assay (Fig. 5) and in vivo metabolic labeling (data not shown) retain four of the five radiolabeled peptides seen with wild-type Pax3. This result suggests that in addition to serine 205, Pax3 contains additional sites of phosphorylation. An examination of the amino acids in the region of Pax3 that contains Ser205 and the eight potential phosphorylated serine residues indicates that trypsin cleavage would produce, at most, four individual peptides. We observed five individually phosphorylated peptides in our two-dimensional analysis. Taken together, this information indicates that minimally one peptide must contain multiple sites of phosphorylation.
A close examination of the biochemical characteristics of the tryptic peptide containing Ser205 provides a theoretical basis to explain how a single radioactively labeled peptide could potentially give rise to five distinctly migrating species in the two-dimensional analysis. The tryptic peptide contains three serines in addition to Ser205, each of which could serve as a site of phosphorylation (Fig. 4A). In addition to containing four potential sites of phosphorylation, this peptide contains six potential tryptic cleavage sites at its carboxyl terminus, of which two are predicted to be favored (Fig. 4A; Boyle et al. 1991). The alternative use of these two preferred sites, combined with the potential for multiple phosphorylation events on each of the possible tryptic products, would result in peptides with different charge and hydrophobicity characteristics. These differences in biochemical characteristics would ultimately result in alternative migrations for the same peptide fragment dependent on the exact tryptic site used and the number of phosphorylation events present (Boyle et al. 1991).
Finally, although serine 205 may not be the only site of phosphorylation on Pax3, our results suggest that this site may be the primary site of phosphorylation. We demonstrate that phosphorylation occurs only in the region of Pax3 surrounding the octapeptide domain (Figs. 3 and 4A). If all of the amino acids were to be phosphorylated independent of each other, then mutation of a single site, such as serine 205, should not alter subsequent phosphorylation events. Therefore, mutation of a single site would not be expected to significantly alter the global phosphate radiolabeling of Pax3 using our in vitro kinase assay. However, we observed that mutation of serine 205 to an alanine resulted in an approximately 80%–90% loss of global phosphate radiolabeling of Pax3 (Fig. 4C). Taken together, these results indicate that the inability of Pax3 to be phosphorylated at serine 205 greatly reduces the efficiency of phosphorylation at additional sites. In this manner, serine 205 would act as the primary site of phosphorylation on Pax3 that then regulates subsequent phosphorylation events. Studies are presently being performed to identify these additional sites of phosphorylation and to elucidate how phosphorylation regulates the biological activities of Pax3.
Materials and Methods
Cell culture conditions
Mouse primary myoblasts were isolated from 2- to 4-d-old C57/B16 mice as previously described (Rando and Blau 1997; Miller and Hollenbach 2007). Proliferation medium for the mouse primary myoblasts consisted of Ham's F-10 nutrient medium (Mediatech Cellgro) supplemented with 20% FBS (HyClone Laboratories, Inc.), 2.5 ng/mL bFGF (Promega Corp.), 15 mM HEPES, and penicillin-streptomycin. Differentiation medium consisted of Dulbecco's modification of Eagle's medium (DMEM, GIBCO BRL) supplemented with 2% horse serum (HyClone). All media contained penicillin G (200 U/mL) and streptomycin (200 μg/mL). DMEM was additionally supplemented with L-glutamine (2 mM, GIBCO BRL), and when prepared in this manner referred to as DMEM-complete. Cells were grown in a humidified incubator at 37°C in 5% CO2. All cells were grown on collagen-coated dishes (Becton Dickinson Labware), were passage-matched to prevent possible differences due to different passage conditions, were not used past passage 9 to prevent the cells entering crisis, and were not allowed to grow past ∼80% confluency to maintain the cells in an undifferentiated state. To induce the differentiation of primary myoblasts, the proliferation media was removed, the cells were washed twice with PBS, the media was replaced with 10 mL of differentiation media, and the cells were grown as described above until needed for further analysis.
Retroviral stocks and the stable transduction of mouse primary myoblasts
Retroviral stocks were generated by the transient transfection of the ecotropic Phoenix packaging cell line (Swift et al. 1999) with 8 μg of the MSCV-IRES-GFP retroviral construct containing either a FLAG-epitope tagged Pax3 (FLAG-Pax3) or FLAG-Pax3(S205A) in which serine 205 has been mutated to an alanine by the Fugene™6 method (Roche Applied Science) according to the manufacturer's specifications. Culture supernatants containing virus were collected between 36 and 72 h after transfection, filtered, and subsequently used for a single transduction of mouse primary myoblasts. Three to seven days post-transduction, primary myoblasts were harvested in F10 media supplemented with collagen (10 ng/mL; Sigma), and cells expressing GFP were selected by fluorescence activated cell sorting (FACS) analysis. Cells selected in this manner were cultured and expanded as described above.
Creation of expression constructs
The retroviral constructs MSCV-FLAG-Pax3-IRES-GFP and FLAG-Pax3(S205A) and the GST-fusion constructs pGEX-5X-1-Pax3 and its corresponding domain deletion mutants were a kind gift from Dr. Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN). The MSCV-FLAG-IRES-GFP constructs contain either the cDNA for Pax3 or Pax3 in which serine 205 has been mutated to an alanine with a FLAG-epitope tag engineered onto its amino terminus. They also contain the cDNA for the green fluorescent protein (GFP). The presence of the IRES allows the dual production of GFP and FLAG-Pax3, both under control of the Murine Stem Cell Virus (MSCV) promoter (Laker et al. 1998).
Phospho-incompetent pGEX-5X-1-Pax3 point mutants, in which the indicated serine has been mutated to an alanine (S180A, S187A, S193A, S197A, S201A, S205A, S209A, S222A), were created using overlap extension PCR as described previously (Ho et al. 1989). The resulting PCR product was cloned into the pCRII vector using the TA Cloning Kit (Invitrogen) according to the manufacturer's protocol. DNA sequencing was used to confirm the presence of the desired mutation and to confirm that no additional mutations were introduced during PCR. Following a StuI-XhoI digestion of pCRII Pax3 point mutant constructs, the inserts containing the mutation were gel extracted and ligated into the pGEX-5X-1-Pax3 parent vector, which had previously been digested with the same enzymes. The resulting wild-type and the point mutant vectors were individually transformed into Rosetta(DE3)pLysS chemically competent cells (EMD Chemicals) and subsequently used for expression and purification, as previously described (Hollenbach et al. 1999, 2002). Bacterially expressed and purified GST-Pax3 or the GST-Pax3 deletion or point mutants were used without elution from the resin. Protein expression and purity were confirmed by SDS-PAGE analysis, and the relative protein concentrations on the resin were estimated by comparison to proteins of known concentration (data not shown).
[32P]-Orthophosphate metabolic labeling
Mouse primary myoblasts isolated as described above were grown to 70%–80% confluency, washed twice with filter-sterilized Tris-buffered saline (TBS), and starved of phosphates by incubating them for 30 min at 37°C in 5% CO2 with phosphate-free DMEM-complete supplemented with 2.5 ng/mL bFGF. [32P]-Orthophosphate (MP Biomedicals) was then added to the media (0.25 mCi/mL) and allowed to incubate for an additional 2 h under identical conditions. After metabolic labeling, the cells were washed three times with sterile TBS and lysed by the addition of 500 μL of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing the complete mini protease inhibitor cocktail (Roche Applied Science), phosphatase cocktail I specific for serine/threonine phosphatases (Sigma), and phosphatase cocktail II specific for tyrosine phosphatases (Sigma) followed by incubation at room temperature with shaking for 15–30 min. Following this incubation, the lysed cells were scraped from the dish using a cell lifter and transferred to a 1.5-mL microfuge tube, and the cellular debris was removed by centrifugation at maximum speed for 10 min at 4°C in an Eppendorf refrigerated microfuge. The resulting supernatant was transferred to a fresh 1.5-mL microcentrifuge tube. To immunoprecipitate the FLAG-Pax3 proteins, 40 μL anti-FLAG M2 affinity gel suspension (Sigma) was added to 1 mL of the cell lysate, and the mixture was incubated with rotation for at least 2 h at 4°C. Following this incubation, the tubes were centrifuged to pellet the resin, which was subsequently washed three times with 500 μL TBS. SDS-PAGE loading buffer was added, the samples were boiled for 5 min, and the eluted proteins were separated on a 10% SDS-PAGE gel. The gel was then dried and visualized by autoradiography.
In vitro kinase assay and two-dimensional phosphopeptide analysis
GST-Pax3 or the GST-Pax3 mutants present on the resin (8 μL of resin—approximately1 μg of protein), prepared as described above, were mixed with 26 μL of the kinase stock solution (2× kinase buffer [80 mM HEPES, 20 mM MgCl2, 100 mM KCl, 2 mM DTT], 2× phosphatase inhibitor cocktails described above, 84 μM ATP, 50 μCi [γ-32P]-ATP [MP Biomedicals]). The kinase reaction was initiated by the addition of 25 μL of proliferating mouse primary myoblast total cell extracts (2 μg/μL), prepared as previously described (Miller and Hollenbach 2007), and incubated for 1 h at 30°C. After incubation, the beads were washed 3× with 100 μL PBS, and the radiolabeled protein was eluted by boiling in 25 μL SDS-PAGE loading buffer and separated by 10% SDS-PAGE. The resulting gels were dried and exposed to film at −80°C overnight.
Following the in vitro kinase assay or metabolic labeling described above, the radiolabeled protein band corresponding to the phosphorylated Pax3 was extracted from the gel and submitted to two-dimensional phosphopeptide analysis as previously described (Boyle et al. 1991).
Antibodies and Western blot analysis
An antibody specific for phosphorylation of Pax3 at serine 205, anti-Pax3(p205), was produced by rabbit immunization using the following synthetic phosphopeptide: NH2-CAPQSDEG(pS)DIDSEP-CO2 (QCB Custom Immunology Group). The antibody was affinity purified by QCB Custom Immunology Group, and the specificity was confirmed by Western blot analysis, as described in the Results section. The Pax3-specific antibody was described previously (Lam et al. 1999) and was used without further purification.
Total cell extracts from proliferating primary myoblasts or myoblasts that were induced to differentiate for a specific period of time were prepared as described above. A constant amount of total cell extract (50 μg) was separated by 10% SDS-PAGE, proteins were transferred to Immobilon-P membrane (Millipore), and the presence of Pax3 or Pax3 phosphorylated at serine 205 was detected using the affinity purified, monospecific Pax3 antibody or the Pax3(p205) antibody, using previously described conditions (Lam et al. 1999).
Acknowledgments
We thank Dr. Gerard Grosveld, St. Jude Children's Research Hospital, for kindly providing us with laboratory and office space and allowing us access to reagents and supplies during the aftermath of Hurricane Katrina. This work was supported in part by grant no. 1-P20-RR020152-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH); grant no. LEQSF(2004-07) from the Louisiana Board of Regents (BoRSF); and the Louisiana Cancer Research Consortium (LCRC) Immediate Response Fund. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR, NIH, BoRSF, or the LCRC.
Footnotes
Reprint requests to: Andrew D. Hollenbach, Louisiana State University Health Sciences Center, Department of Genetics, 533 Bolivar Street, New Orleans, LA 70112, USA; e-mail: aholle@lsuhsc.edu; fax: (504) 568-8500.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035956.108.
References
- Boutet, S.C., Disatnik, M.H., Chan, L.S., Iori, K., Rando, T.A. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Boyle, W.J., van der Geer, P., Hunter, T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., Relaix, F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Epstein, J.A., Shapiro, D.N., Cheng, J., Lam, P.Y., Maas, R.L. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hollenbach, A.D., Sublett, J.E., McPherson, C.J., Grosveld, G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach, A.D., McPherson, C.J., Lagutina, I., Grosveld, G. The EF-hand calcium-binding protein calmyrin inhibits the transcriptional and DNA-binding activity of Pax3. Biochim. Biophys. Acta. 2002;1574:321–328. doi: 10.1016/s0167-4781(02)00230-0. [DOI] [PubMed] [Google Scholar]
- Hunter, T., Karin, M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Kitzmann, M., Vandromme, M., Schaeffer, V., Carnac, G., Labbe, J.C., Lamb, N., Fernandez, A. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: Role in modulating MyoD half-life and myogenic activity. Mol. Cell. Biol. 1999;19:3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laker, C., Meyer, J., Schopen, A., Friel, J., Heberlein, C., Ostertag, W., Stocking, C. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J. Virol. 1998;72:339–348. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, P.Y., Sublett, J.E., Hollenbach, A.D., Roussel, M.F. The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol. Cell. Biol. 1999;19:594–601. doi: 10.1128/mcb.19.1.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhou, J., James, G., Heller-Harrison, R., Czech, M.P., Olson, E.N. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- Magnaghi, P., Roberts, C., Lorain, S., Lipinski, M., Scambler, P.J. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 1998;20:74–77. doi: 10.1038/1739. [DOI] [PubMed] [Google Scholar]
- Maroto, M., Reshef, R., Munsterberg, A.E., Koester, S., Goulding, M., Lassar, A.B. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Miller, P.J., Hollenbach, A.D. The oncogenic fusion protein Pax3-FKHR has a greater post-translational stability relative to Pax3 during early myogenesis. Biochim. Biophys. Acta. 2007;1770:1450–1458. doi: 10.1016/j.bbagen.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T.A., Blau, H.M. Methods for myoblast transplantation. Methods Cell Biol. 1997;52:261–272. doi: 10.1016/s0091-679x(08)60382-9. [DOI] [PubMed] [Google Scholar]
- Swift, S., Lorens, J., Achacoso, P., Nolan, G.P. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. In: Coligan J.E., editor. Current protocols in immunology. Wiley; Boston, MA: 1999. pp. 10–17. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh, S., Buckingham, M. The birth of muscle progenitor cells in the mouse: Spatiotemporal considerations. Curr. Top. Dev. Biol. 2000;48:225–268. doi: 10.1016/s0070-2153(08)60758-9. [DOI] [PubMed] [Google Scholar]
- Williams, B.A., Ordahl, C.P. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development. 1994;120:785–796. doi: 10.1242/dev.120.4.785. [DOI] [PubMed] [Google Scholar]
- Xia, S.J., Barr, F.G. Chromosome translocations in sarcomas and the emergence of oncogenic transcription factors. Eur. J. Cancer. 2005;41:2513–2527. doi: 10.1016/j.ejca.2005.08.003. [DOI] [PubMed] [Google Scholar]