Abstract
The regulatory mechanism of Src tyrosine kinases includes conformational activation by a change in the catalytic domain tertiary structure and in domain–domain contacts between the catalytic domain and the SH2/SH3 regulatory domains. The kinase is activated when tyrosine phosphorylation occurs on the activation loop, but without phosphorylation of the C-terminal tail. Activation also occurs by allostery when contacts between the catalytic domain (CD) and the regulatory SH3 and SH2 domains are released as a result of exogenous protein binding. The aim of this work is to examine the proposed role of an electrostatic network in the conformational transition and to elucidate the molecular mechanism for long-range, allosteric conformational activation by using a combination of experimental enzyme kinetics and nonequilibrium molecular dynamics simulations. Salt dependence of the induction phase is observed in kinetic assays and supports the role of an electrostatic network in the transition. In addition, simulations provide evidence that allosteric activation involves a concerted motion coupling highly conserved residues, and spanning several nanometers from the catalytic site to the regulatory domain interface to communicate between the CD and the regulatory domains.
Keywords: activation, molecular dynamics, phosphorylation, enzyme kinetics, ionic strength, allostery
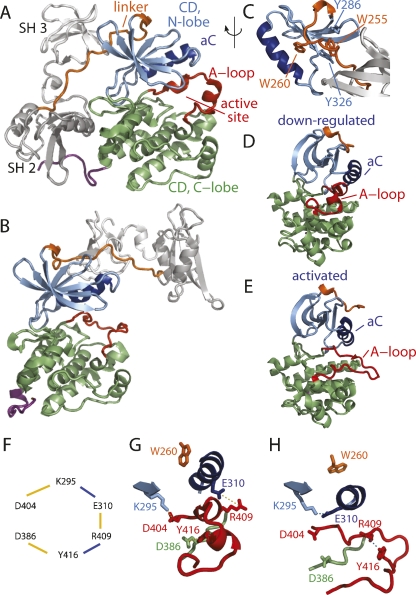
Enzymatic activity of Src tyrosine kinase is tightly regulated in the cell; a critical feature of regulation is the proper control of the relative populations of activated and down-regulated conformations (Hubbard 1999; Johnson and Lewis 2001; Huse and Kuriyan 2002; Nolen et al. 2004; Kannan and Neuwald 2005). A variety of activating signals, including tyrosine phosphorylation and intermolecular protein association, can lead to the conformation most competent for catalyzing phosphotransfer. The structures of Src kinases in either down-regulated (Fig. 1A; Schindler et al. 1999; Xu et al. 1999) or activated (Fig. 1B; Yamaguchi and Hendrickson 1996; Breitenlechner et al. 2005; Cowan-Jacob et al. 2005) forms determined from crystallization show that one component of conformational activation, which distinguishes active and down-regulated Src, is a change in the tertiary structure of the catalytic domain (CD). Noted differences include αC helix rotation, which alters an interaction between two fully conserved residues, Glu310 and Lys295 (chicken c-Src numbering is used throughout), rearrangement of the activation loop (A-loop), and a change in the relative orientation of the N- and C-lobes (Fig. 1D,E). An additional level of control is achieved through inhibitory intramolecular domain–domain contacts (Sicheri and Kuriyan 1997; Boggon and Eck 2004), also a central feature in signaling (Kuriyan and Cowburn 1997; Pawson 2004). The regulatory SH3 and SH2 domains in down-regulated Src contact the CD on the side away from the active site (Fig. 1A, gray). The autoinhibitory control was shown by enhanced Src activity in the presence of exogenous proteins that release the SH2/SH3 domains from the CD (Fig. 1B, gray) as a result of intermolecular association with the Src SH3 or SH2 domain (Cooper et al. 1986; Roussel et al. 1991; Pawson 1995; Moarefi et al. 1997). The numerous kinase structures reveal variations in interdomain orientations and alternative A-loop conformations (Nolen et al. 2004; Breitenlechner et al. 2005; Kornev et al. 2006), as well as structures that are a “mix” of these structural features typically associated with the active or down-regulated states (Cowan-Jacob et al. 2005; Levinson et al. 2006). Thus, as the crystallographic structural information on Src and other kinases continues to grow, the process of conformational activation appears to increase in complexity.
Figure 1.
Ribbon drawings of Src kinase in “assembled” down-regulated (A) and activated (B) forms. The linker (residues 242–263, orange) connects the regulatory domains (SH3, SH2, gray) to the CD. The CD comprises an N-lobe (residues 267–341, blue) with αC (dark blue), and the C-lobe (residues 342–520, green). The A-loop (residues 404–430, red) and the C-terminal tail (521–531, magenta) are highlighted. (C) Expanded view showing Trp260 along with Trp255, Tyr286, and Tyr326. Rotated views of the CD in down-regulated (D) and activated (E) forms. (F) Scheme for the switched electrostatic networks for either the down-regulated (gold lines) or activated (blue lines) states. Ribbon drawings (colored as in A) for the three-way network in the down-regulated state (G) (Asp404–Lys295, Glu310–Arg409, Tyr416–Asp386, dashed line) and the two-way network in the activated state (H) (Lys295–Glu310, Arg409–Tyr416, dashed line). Changes in Trp260 (orange) position are shown. The structure figures were generated using PyMOL (DeLano Scientific).
Analyses of sequence conservation and structural similarity provide the initial framework to understand the regulation of the conformational change between activated and down-regulated forms of Src (Johnson and Lewis 2001; Nolen et al. 2004; Kannan and Neuwald 2005; Kornev et al. 2006), while mutagenesis and kinetic studies afford tests to confirm or dispute the significance of the side chain interactions implicated by such analyses (Gonfloni et al. 1997, 1999, 2000; LaFevre-Bernt et al. 1998; Adams 2003; Masterson et al. 2008). In addition to evaluation of the activated and down-regulated end states, information on how the conformational change occurs and on the transition pathways connecting active and down-regulated states is required for a complete understanding of the forces that govern conformational activation. Theoretical and simulation methods can contribute by identifying probable transition pathways (Young et al. 2001; Banavali and Roux 2005; Levinson et al. 2006; Ozkirimli and Post 2006; Faraldo-Gómez and Roux 2007), and by providing atomic details about the underlying process and how motion of different regions is coupled. When these pathways can be probed experimentally, the combined exploration is a powerful one that can lead to new physical insights. The importance of better defining conformational transitions is further motivated by success in the treatment of chronic myelogenous leukemia of a small-molecule tyrosine kinase inhibitor, imatinib, discovered to act selectively by targeting a particular conformation of the kinase (Capdeville et al. 2002). An emerging theme is that differences in alternative conformational states, including intermediate metastable states, as well as down-regulated and active conformations, can be exploited in the design of inhibitors of ATP binding (Noble et al. 2004; McInnes et al. 2006; Okram et al. 2006).
From a study using a nonequilibrium method of molecular dynamics (MD) to investigate conformational activation of the isolated CD, we recently identified a group of six residues that switched during the transition process between alternative networks of electrostatic interactions in the two end states, and we proposed that this switched electrostatic network was important in reducing the free-energy barriers along the transition pathway (Ozkirimli and Post 2006). The importance of electrostatic interactions has long been recognized for physical processes such as DNA-ligand binding (Sharp and Honig 1995), protein–protein association (Wade et al. 1998; Sheinerman et al. 2000; Nag and Dinner 2006), transport in ion channels (Gouaux and Mackinnon 2005), and rearrangement of enzyme active sites (Davydov et al. 2004) and regulatory units (Vigil et al. 2006). An electrostatic network to promote conformational activation has not been previously proposed and would add to this list. The residues comprising this proposed switched electrostatic network are highly conserved residues in the CD (Fig. 1F) so that the network spans across the C-lobe catalytic loop (catalytic Asp386), the A-loop (Tyr416, Arg409), the N-lobe αC (Glu310) and β-sheet (Lys295), and the metal binding loop (Asp404 in the DFG sequence) (Ozkirimli and Post 2006). The network couples all of the motions in the CD associated with activation: αC rotation, A-loop extension, and lobe–lobe opening. The switch is an exchange of partners from a three-way network of interactions (Lys295–D404, Glu310–Arg409, Tyr416–Asp386) (Fig. 1G) to a two-way network (Lys295–Glu310 and Tyr416–Arg409) (Fig. 1H), and is predicted to be important in guiding the multifaceted transition. In the two-way network, which corresponds to the activated form, the catalytically important residues are left available to interact with ATP-Mg2+ (Asp404) and the substrate (Asp386).
We report here kinetic experiments of enzyme activity to test the proposal of an electrostatic component to the activation process and the existence of the switched electrostatic network suggested from the simulation studies (Ozkirimli and Post 2006). Src kinase activity proceeds with an induction phase attributed to conformational activation and phosphorylation on the A-loop (Porter et al. 2000). We tested whether this induction phase exhibits dependence on ionic strength, as predicted if a significant electrostatic component is present in conformational activation.
We also build upon the theoretical work on the isolated CD by investigating the switched electrostatic network in the context of the nearly full-length kinase, comprising SH3, SH2, and the catalytic domains, using nonequilibrium, biased molecular dynamics (BMD) (Marchi and Ballone 1999; Paci and Karplus 1999). With BMD the conformational transition for Src activation occurs on a computationally feasible timescale by use of an external potential term to penalize thermal fluctuations that move the protein away from the target structure. BMD identifies possible activation pathways between two known end states, without prior knowledge of the transition.
Autoinhibition was also investigated using BMD methods. A steric hindrance mechanism, whereby the active site is blocked by the regulatory domain, as in the example of PKA (Kim et al. 2005) or CaMKII (Rosenberg et al. 2005), can be explained from the structure. In the case of Src kinases, however, the regulatory domains in the down-regulated state interact on the “backside” of the enzyme relative to the active site and do not occlude access to the active site located between the N- and C-lobes near the A-loop (Fig. 1A). Instead, allosteric activation of Src through release of the SH2 or SH3 contact with the CD by intermolecular binding is a long-distance effect of a more subtle nature (Sicheri and Kuriyan 1997; Boggon and Eck 2004). Biochemical studies have shown that the regulatory domains communicate bidirectionally with the catalytic site. Phosphorylation of the A-loop tyrosine enhances the accessibility of the regulatory domains for binding exogenous ligands (Gonfloni et al. 2000; Porter et al. 2000), while intramolecular binding of the SH3/2 domains down-regulates the kinase (Pawson 1995). Further evidence of long-range conformational coupling comes from the observation that αC mutations at the αC/A-loop contact interface prevent regulation by the C-terminal Tyr527 phosphorylation (Gonfloni et al. 1997), and H-D exchange studies have suggested that αC is implicated in the transmission of information from the nucleotide binding site to the SH2 domain (Hamuro et al. 2002). Of specific interest here, mutagenesis studies have identified Trp260 (LaFevre-Bernt et al. 1998) and Trp255 (Gonfloni et al. 1999) in the linker (Fig. 1C, orange) between the regulatory domains and the CD to be important in coupling between the regulatory domains and the catalytic site. Previous theoretical studies have contributed to understanding the equilibrium basis of regulation (Young et al. 2001; Banavali and Roux 2005) by defining the energetics for conformational compatibilities of the linker region and CD. Nonetheless, the question of how these residues work to transmit the long-range communication between the SH3/2-CD interfaces and the catalytic site remains unanswered, and the analysis reported here suggests molecular details underlying this coupled motion.
In what follows, we first present the experimental results to test for the presence of electrostatic contributions for activation from the salt dependence of the initial lag phase of substrate phosphorylation. The computational results are then described for the characterization of the switched electrostatic network for nearly full-length Src, and for the process leading to long-range regulation of Src activity. The role of the functionally important residue Trp260 in mediating domain interactions and allosteric activation is discussed.
Results and Discussion
Enzyme kinetics and MD simulations were employed to investigate a switched electrostatic network in the conformational activation of assembled Src kinase and its involvement along with Trp260 in long-range communication between the active site region and the regulatory domains. The Src family members from our previous studies, Hck for kinetic analysis and Lyn for molecular dynamics calculations, were used. Because the sequence and structural similarities among Src kinases imply a common activation mechanism for the Src kinase family, we use Src hereafter to refer to the family in general. Trp260, which abuts αC, and another conserved hydrophobic residue at position 255 (Trp255 in the case of Lyn, Lck, and Hck), which stacks between Tyr286 and Tyr326 of the N-lobe (Fig. 1C), were identified by mutagenesis studies to be functionally important for autoinhibition (LaFevre-Bernt et al. 1998; Gonfloni et al. 1999). These residues are at the C terminus of the SH2–CD linker and lie in the SH3/CD interface.
Kinase activation salt dependence
The prediction that a switched electrostatic network enhances the conformational activation of Src-family kinase was tested experimentally by determining the dependence of activation on salt concentration using Hck. If the switched electrostatic network contributes to kinase activation by lowering the free-energy barrier, then an increase in ionic strength would weaken the electrostatic interactions, and the process of kinase activation is predicted to be slower at a higher salt concentration.
The salt dependence of the enzymatic activation was experimentally determined using kinetic assays of phosphorylation. The enzyme progress curves for Src-family kinase are known to be biphasic when unphosphorylated enzyme is incubated with a peptide substrate and ATP (Porter et al. 2000). The initial slow phase is attributed to activation of the enzyme by intermolecular autophosphorylation at Tyr416 on the A-loop, while the second phase reports on the more rapid phosphorylation of the peptide substrate by activated enzyme. Evidence that the slow induction phase results from activation of the enzyme by intermolecular autophosphorylation of the A-loop is that pre-incubation of the kinase with ATP results in the loss of the lag phase, and that the lag time depends on enzyme concentration (Porter et al. 2000).
In vitro kinase assays were conducted at different salt concentrations using purified Hck and a synthetic peptide substrate. The biphasic progress curves were measured by a continuous spectrophotometric assay (Porter et al. 2000). The induction time includes processes associated with catalytic autophosphorylation as well as the conformational transition for activation. To account for the salt dependence of the bimolecular diffusion encounter of Hck at low concentrations, assays were conducted as a function of Hck concentration, and the results were extrapolated to zero enzyme concentration to obtain the salt dependence for the unimolecular process. The salt dependence of the chemical steps associated with tyrosine phosphorylation and product dissociation was assessed from the change with ionic strength observed for the second phase of substrate phosphorylation.
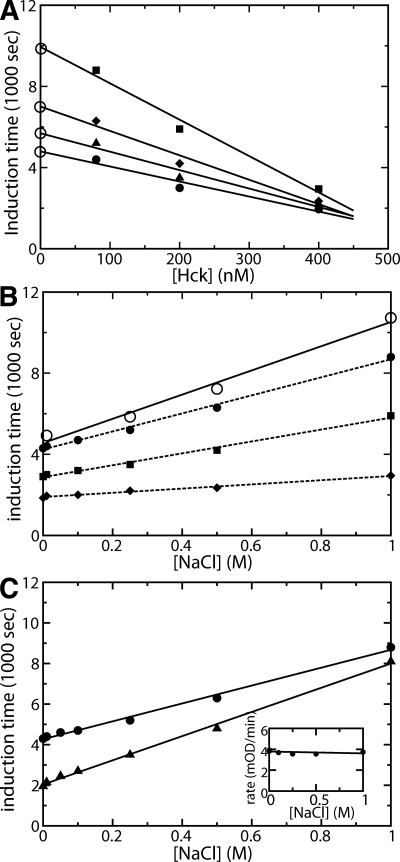
The results for the induction times as a function of increasing Hck concentration at different salt concentrations are shown in Figure 2A. The induction times decrease at higher Hck concentrations, indicating contributions from the bimolecular encounter at the nanomolar enzyme concentrations required by the experiment, as expected (Porter et al. 2000). In regard to the effect of added salt, the dependence on Hck concentration is diminished at lower ionic strength; as the salt concentration decreases from 1 M NaCl (Fig. 2A, squares) to 10 μM NaCl (Fig. 2A, circles), the magnitude of the slope decreases from −18.8 to −7.8 sec/nM. The reduced Hck-concentration dependence of the induction time at lower salt concentration indicates that electrostatics work against binding and serve as a repulsive component in the diffusion encounter. This behavior is expected for the association of two Hck molecules with similar (negative) charge.
Figure 2.
Hck induction (lag) time depends on ionic concentration. (A) Induction time as a function of down-regulated full-length Hck (SH3/2–CD) for different concentrations of NaCl: 1 M NaCl (▪), 0.5 M NaCl (♦), 0.25 M NaCl (▴), 0.01 M NaCl (●). (B) Induction time as a function of [NaCl] for values extrapolated to zero concentration of full-length Hck (○, solid lines) and at finite concentrations of full-length Hck (dotted lines): 80 nM Hck (●), 200 nM Hck (▪), 400 nM Hck (♦). (C) Induction time as a function of [NaCl] for down-regulated Hck (SH3–SH2–CD) (●) and for the isolated catalytic domain of Hck (▴). (Inset) Substrate tyrosine phosphorylation as a function of [NaCl]. Induction times were calculated from kinase progress curves as described in Materials and Methods.
Induction times, obtained by extrapolation to zero Hck concentration to remove the effects of collision encounter, are plotted in Figure 2B (solid line) as a function of NaCl concentration. A positive salt dependence consistent with electrostatic screening is observed; the [Hck]-independent induction time is 4700 sec at 0.01 M [NaCl] but increases with added salt to 10,100 sec at 1 M [NaCl]. The results at finite Hck concentrations are also shown (Fig. 2B, dotted lines). These lines demonstrate the diminished salt dependence at higher Hck concentration, and that the salt dependence on Hck concentration is of opposite sign, consistent with the repulsive electrostatic contribution of bimolecular association noted above. Thus, the Hck concentration-independent unimolecular process is slowed at high salt concentration, indicative of a process in which electrostatic contributions favor the reaction progress.
The isolated catalytic domain of Hck also displays biphasic activity, although the time period for activation is not as long. The induction time for the isolated kinase domain (Fig. 2C, triangles) shows a similar salt dependence to that of full-length down-regulated Hck (Fig. 2C, circles). For the CD, the induction time increased from 1950 sec to 8100 sec as the NaCl concentration was increased from 10 μM to 1 M. The slopes in Figure 2C are 4500 sec M−1 (circles) and 6150 sec M−1 (triangles). That the salt dependence is similar for the isolated CD and full-length down-regulated Hck indicates that the salt effect largely reflects activation of the CD, consistent with an electrostatic switch, and not release of the regulatory domain contacts.
The possibility that the observed salt dependence arises from the chemical steps of phosphorylation or product release was addressed by considering the salt dependence of the second phase of the kinetics, corresponding to phosphorylation of the substrate tyrosine. We find no salt dependence (Fig. 2C, inset), and conclude that the lack of salt dependence for peptide phosphorylation argues against tyrosine phosphorylation as a possible source of salt dependence in the first phase. In addition, we tested for specific salt effects by measuring the kinetics in the presence of varying concentrations of RbCl (data not shown). The results show a similar salt dependence as that observed with NaCl, and therefore the salt effects are neither the consequence of a specific ion binding, nor an effect on overall protein stability.
Together, these data show that electrostatic interactions promote conformational activation and provide support for the hypothesis that an electrostatic switch regulates the activation of Src-family kinases. The shorter induction time at lower salt concentration is consistent with a network of electrostatic interactions being significant in conformational activation by restricting conformational space, while the interactions are screened at higher ionic strength to produce a longer induction time.
Assembled kinase molecular dynamics simulations
Molecular dynamics simulations were calculated to compare against the dynamic behavior of the switched electrostatic network observed with the isolated CD, and to elucidate the set of interactions between SH2/SH3 and the CD that collectively contribute to long-distance coupling in the conformational activation of Src-family kinase.
The simulation system included Lyn residues 85–531, ATP-Mg2+, and solvation by explicit solvent. Equilibrium MD of the down-regulated state established that the simulation system was stable based on constant time-averaged values for the total energy, temperature, and RMSD over the 300- to 400-psec equilibrium run period. Backbone coordinates from the structure at the end of the production run deviated from the initial structure by a RMSD of ∼1.3 Å for the CD4 and ∼1.8 Å for all kinase backbone atoms. The C-tail Tyr527 was not phosphorylated, in order to allow possible long-range effects of domain–domain contacts communicated in the absence of Tyr527 phosphorylation for activated Src. Tyr527 and the rest of the C-tail maintain contact with the SH2 domain during equilibrium MD. All interdomain interactions were also stable throughout the equilibration simulations.
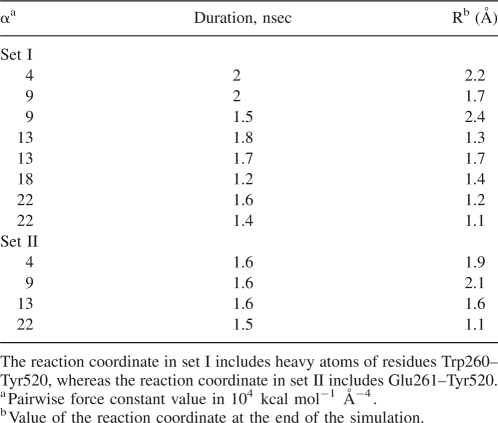
To promote a transition from the down-regulated form to the activated form, an external biasing potential (Paci and Karplus 1999; Ozkirimli and Post 2006) to favor fluctuations toward the open, activated conformation was applied to the CD heavy atoms following equilibrium MD of down-regulated Lyn. The target coordinates for the biasing potential corresponded to the equilibrium MD structure of the active form of CD (Ozkirimli and Post 2006). The sum in the RMS of the biasing potential was over the residues of either Trp260–Tyr520 (simulation set A, eight trajectories, 1.5–2.0 nsec each) or Glu261–Tyr520 (simulation set B, four trajectories, 1.5–1.6 nsec each) (Table 1). The SH2, SH3 domains, the SH2–CD linker, and the C-tail were not included in the biased set and were allowed to move freely in response to the CD motion. The progress of the BMD simulations was monitored from the value of the coordinate, R(t), the difference in the internal distances in the simulation and target structures (see Materials and Methods). Conclusions are based on the results from multiple trajectories with varying values for the external force constant. The calculations provide information on the CD conformational transition as it occurs in the context of the associated regulatory domains.
Table 1.
Summary of BMD simulations
Switched electrostatic network in assembled Lyn kinase
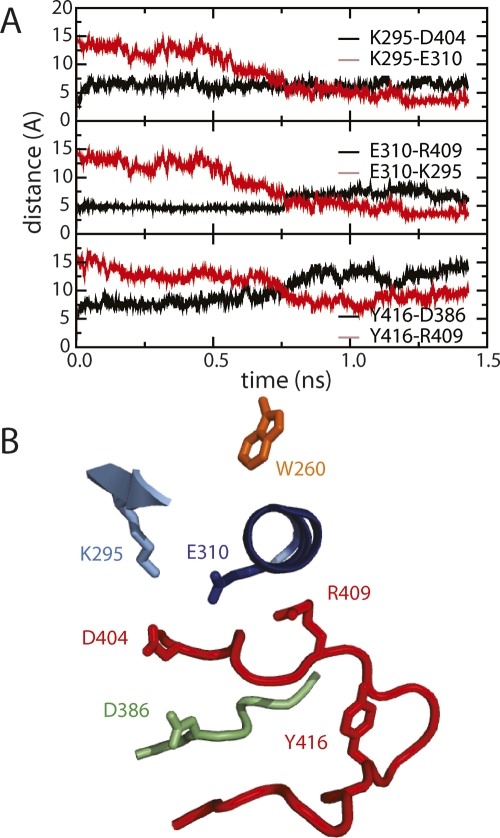
The original observation of the switched electrostatic network was made from BMD studies on the isolated CD. We, therefore, investigated the potential effect of contact with the regulatory domains in the assembled kinase on the switched electrostatic network. Overall, similar behavior is observed; alternative interactions of residues Asp404, Lys295, Glu310, Arg409, Tyr416, and Asp386 are switched in a temporally short time in the assembled kinase (Fig. 3A), as was previously observed for the isolated CD. Lys295 of the N-lobe switches from Asp404 of the metal binding loop to Glu310 of αC (Fig. 3A, top panel), Glu310 switches from Arg409 of the A-loop to Lys295 (Fig. 3A, middle panel), and Tyr416 of the A-loop switches from Asp386 of the catalytic loop to Arg409. Figure 3B is the snapshot at 1200 psec from the trajectory showing the transition where the E310–R409 interaction is broken but the switch to E310–K295 is not yet complete. The switched electrostatic network is therefore predicted to promote activation of both the isolated CD and assembled kinase by restricting the sampling of random interactions during the transition and anchoring intermediates to conformations with end-state contacts in the A-loop, αC, and active site region.
Figure 3.
The electrostatic switch mechanism. (A) Distances (Å) between K295NZ–D404CG (black) and K295NZ–E310CD (red) (left, top), between E310CD–R409CZ (black) and E310CD–K295NZ (red) (middle), and between Y416OH–D386CG (black) and Y416OH–R409CZ (red) (bottom) are plotted. (B) Ribbon drawing for an intermediate from the simulation near the transition from the three-way, down-regulated switch form to the two-way, activated switch form. K295, E310, R409, and Y416 are more equidistant to their electrostatic partners than in either Figure 1G or H.
Role of Trp260 in long-range allosteric coupling and the switched electrostatic network
Analysis of sequence conservation and mutagenesis studies (Gonfloni et al. 1997; LaFevre-Bernt et al. 1998) has identified Trp260 to be essential for the allosteric regulation mediated by SH3 and SH2. The crystallographic structures of active and down-regulated forms of Src kinase show a structural difference in the region surrounding Trp260 at the C terminus of the linker: In the down-regulated form with SH3 and SH2 in close contact with the CD, Trp 260 is packed on the surface of CD (Fig. 1C,G), while in the activated form with SH3 and SH2 released from the CD, Trp 260 has greater exposure to solvent and alternative interactions with the CD (Fig. 1H). Conformational free-energy calculations for the activated and down-regulated forms of the region around Trp 260 suggest that the structures are stable only in the context of the corresponding active or down-regulated forms of the CD (Banavali and Roux 2005). We examine here the allosteric role of Trp260 in long-distance communication during the conformational transition of the down-regulated to active forms of Src kinase.
Based on mutagenesis experiments indicating that Trp260 participates in the mechanism of regulation, it was suggested that interaction of αC with Trp260 stabilizes the down-regulated conformation of αC (LaFevre-Bernt et al. 1998). In the down-regulated conformation, Trp260 occupies a hydrophobic cavity next to αC (Fig. 4, top left), whereas in the activated state the contact of Trp260 with αC is reduced and the indole ring is solvated by a change in χ1 of Trp260 and reorientation of αC within the N-lobe (Fig. 4, top right). This outward displacement of Trp260 is correlated with the inward rotation of αC observed in active structures of both the isolated CD (Yamaguchi and Hendrickson 1996) and full-length Src kinase (Cowan-Jacob et al. 2005). A similar correlation between Trp260 and αC is observed for Abl kinase (Levinson et al. 2006) and Irk kinase (Hubbard 1997).
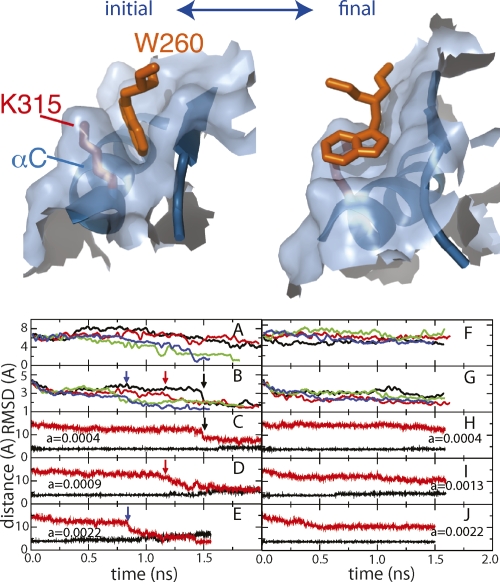
Figure 4.
Role of Trp260 in αC transition. (Top) The surface within 7 Å of Trp260 is shown (light blue) for the down-regulated (left) and activated (right) state. αC and one of the N-lobe β-strands are shown in dark blue. N-lobe β-sheets are overlaid to generate the figures. (Bottom) BMD results from set I, where the progress coordinate includes heavy atoms of Trp260–Tyr521 (A–E), and from set II including Glu261–Tyr521 without a biasing potential on Trp260 atoms (F–J). RMSD time profiles (A,B;F,G) for the simulations calculated with the pairwise force constant, α = 0.0004 (black), 0.0009 (red), 0.0013 (green), and 0.0022 (blue) are smoothed over a 25-psec running window. (A,F) Trp260 heavy-atom RMSD to target (active) after superimposition of αC heavy atoms. (B,G) αC heavy-atom RMSD to target after superimposition of C-lobe α-helices. (C–E,H–J) Distance between Glu310CD–Lys295NZ from multiple simulations (α is indicated in each panel). Arrows indicate the drop in αC RMSD values and the simultaneous formation of a Glu310–Lys295 interaction.
The role of Trp260 in the activation process was explored using two progress coordinates for BMD simulations (Table 1). In the first set of simulations (set I), Trp260 was part of the progress coordinate (residues Trp260–Tyr520), whereas in the second set (set II) the progress coordinate excluded Trp260 (residues Glu261–Tyr520). By including Trp260 in the progress coordinate in simulation set I, Trp260 fluctuations in the direction of the activated structure are energetically favored, while no bias is exerted on Trp260 in set II. The two progress coordinates therefore differ only in the inclusion, or not, of Trp260 atoms, so that all CD residues are included in the biasing potential for both sets I and II. Trp260 displacement from the hydrophobic cavity near αC was monitored by the RMSD from the active structure for Trp260 coordinates after superposition of αC coordinates. For set I, the RMSD decreases to values <2 Å in some trajectories within the simulation time period (Fig. 4A). As expected, less progress toward the active state is made in set II (Fig. 4F).
Comparison of the structural changes observed in the two sets of BMD simulations (Fig. 4, cf. B–E and G–J) finds that Trp260 displacement is coupled to the rotation of C. The activation transition requires translation of the αC N terminus and rotation that brings Glu310 into the catalytic site. Figure 4B illustrates for varying values of the BMD force constant in set I the progress of αC by the RMSD value to the target coordinates in the active structure. The initial decay in the RMSD profile in the first 250 psec is due to translation and is independent of the biasing force constant. A second, force constant-dependent component indicated by the arrows corresponds to helix rotation and is linked to Trp260 displacement. As Trp260 is displaced from the hydrophobic cavity in the simulation set I, αC rotates toward the active conformation. When Trp260 is not displaced from contact with αC (Fig. 4F, set II), the dependence on the force constant is lost and there is no rotation (Fig. 4G), despite the fact that αC is included in the biasing potential. Moreover, a drop in the time profile for αC RMSD to target, from ∼4 Å to 2 Å (Fig. 4B, arrows) coincides with the switch in the Glu310 interactions of the electrostatic network indicated by a decrease in the Glu310CD–Lys295NZ distance from 12 Å to 7 Å (Fig. 4C–E) in most set I simulations, but was not observed in set II simulations.
That rotation of αC and Glu310 switching of partners are coupled to the displacement of Trp260 is supported by the contrasting results observed by exclusion of Trp260 from the biasing potential (set II vs. set I), so that any displacement toward the target position occurs in response to the changes in the rest of the CD. Without displacement of Trp260 from contact with αC, different transition progress curves are observed for αC and the Glu310 switch, even though the external biasing potential encompasses αC residues in both sets of simulations. That is, the translation of αC occurs, but αC rotation and the concomitant Glu310 switch do not (Fig. 4F–J). Therefore, the rotation of αC within the N-lobe as well as the Glu310 switch are linked to the displacement of Trp260 from the hydrophobic cavity.
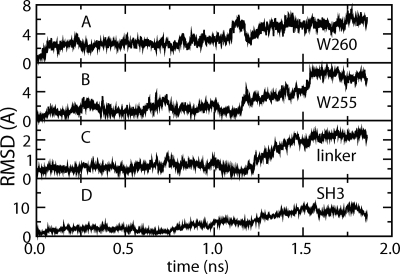
The coupling of Trp260 displacement with reorientation of αC provides a mechanism to link the CD active site to the regulatory SH3 domain. We find that the coupled movement of Trp260 and αC (with the Glu310 switch) affects the motion of the SH2–CD linker, mediated by Trp255 and thus SH3. Figure 5 illustrates this long-range communication between these nonsequential but spatially close segments for one BMD simulation. Following αC rotation and Glu310 switch to the inside, Trp260 displacement from αC (starting at ∼1.2 nsec) is communicated, through changes in the position of Trp255 and the linker region, to SH3. The change in SH3 is a movement away from the CD, consistent with the release of the regulatory domains seen in recent activated crystallographic structures (Fig. 1; Cowan-Jacob et al. 2005). The simultaneous increase suggests that the allosteric mechanism for regulation by SH3 occurs by a path in which Trp260 couples αC and the Glu310 switch with the SH2–CD linker and SH3. We propose that the important role of Trp260 is to communicate the CD activation state, specified through the switched electrostatic network, to the regulatory domains. These transitions are not observed in simulation set II where the displacement of Trp260 does not occur.
Figure 5.
Trp260 couples the catalytic site to SH3 through the SH2–CD linker. (A) RMSD to initial structure for Trp260 heavy atoms. The RMSD is calculated after superposition based on αC. (B) RMSD to initial structure for Trp255 heavy atoms. The RMSD is calculated after superposition based on the N-lobe. (C) RMSD to initial structure for the polyPro helix of the SH2–CD linker (residues 250–254). (D) RMSD to initial structure for SH3 heavy atoms. RMSD is calculated after superposition of structures based on the N-lobe.
Conclusions
Kinetics experiments and nonequilibrium molecular dynamics simulations reported here examined conformational activation of Src and the potential role of a switched electrostatic network of residues Asp386, Arg409, Tyr416, Glu310, Lys295, and Asp404 to guide the conformational transition of the CD. The possible role of electrostatic interactions in the rate-limiting step for activation is supported by the observation that the activation step is salt dependent; the lag time, independent of enzyme concentration, increases at higher salt concentration as predicted for a significant electrostatic contribution to conformational activation, while the faster substrate phosphorylation phase is independent of salt. These kinetic results support the proposal that the electrostatic switch residues are a central element of the transition between activated and down-regulated states of Src kinase, in addition to their recognized roles in catalysis and binding. It is worth noting that the importance of these residues in catalysis and binding complicates the use of site-directed mutagenesis as a means to test their role in a switched electrostatic network and that other experimental approaches may be more effective for comparison.
The interface of the linker C terminus with the N-lobe of the CD has long been suggested to be part of the mechanism of allosteric regulation. Mutational analysis established the inhibitory role of specific residues in the linker region (Gonfloni et al. 1997, 1999, 2000; LaFevre-Bernt et al. 1998), and the contact between Trp260 and αC observed in the crystallographic structures (Schindler et al. 1999; Xu et al. 1999), suggested how the SH2 and SH3 regulatory domains might communicate with the active site and the A-loop. Computational studies to calculate the free-energy surface of the C terminus linker region conformation confirm the incompatibility of the local geometry of the linker conformation with Trp260 packed against αC with the activated structure of the CD (Banavali and Roux 2005). We report here the first direct demonstration of the coupling of Trp260 and the linker region with αC rotation. A concerted motion of Trp260 and Trp255 in the linker was found to function in cooperation with αC and the electrostatic network to link SH3/2 to the catalytic loop. SH3/2 are released upon transition of Trp260, αC rotation, and therefore the switch of Glu310 along with the rest of the electrostatic network, which incorporates the A-loop and catalytic site residues. When the progress of Trp260 toward the activated state is slowed, the transition of αC is also retarded and the electrostatic switch centered on Glu310 does not occur. These results illustrate a molecular mechanism to explain the essential role of Trp260 in the allosteric regulation by SH3/2, and demonstrate the potential of simulations to elucidate allosteric communication motifs that have been inferred from evolutionary patterns of sequence conservation (Süel et al. 2003; Zheng et al. 2005).
Materials and Methods
Simulation systems
Initial coordinates of the down-regulated assembled Lyn kinase, residues 85–531, were built using the homology modeling program MODELLER 4.0 (Sali and Blundell 1993) and the down-regulated Hck kinase.
The AMPPNP position in the down-regulated Src kinase structure (2SRC.pdb) (Xu et al. 1999) was used to place the ATP. The protein, with or without the ATP and Mg2+ bound, was then solvated in an equilibrated rhombic dodecahedron water box carved from a cubic box of 134 Å on one side with at least 14 Å of water between the protein and the edge of the box. The final system included about 7000 protein atoms and 20,000 water molecules.
The target structure for BMD is the Lyn CD in the activated state. The active conformation of the Lyn CD was built based on the active Lck CD structure (3LCK.pdb) (Yamaguchi and Hendrickson 1996). Details of the equilibration of active Lyn can be found in our previous report (Ozkirimli and Post 2006).
The equilibrated structures of the assembled Lyn or active Lyn CD were the initial and target conformations used in BMD in order to eliminate effects that could result from the force field. Equilibrium simulations were calculated over 0.3–0.4 nsec with starting structures for unphosphorylated down-regulated assembled Lyn or active isolated CD of Lyn in the presence or absence of cofactors. Details of the simulation protocol were previously reported (Ozkirimli and Post 2006). The structures used were without phosphorylated tyrosine in the A-loop (Tyr416) or the C-terminal tail (Tyr527) in order to have the same chemical structure for both starting and target states. The use of unphosphorylated end states provides the basis for deciphering the molecular details of the activation pathway without the chemistry step.
Biased molecular dynamics
The conformational transition pathway was simulated by biased molecular dynamics (Marchi and Ballone 1999; Paci and Karplus 1999) using an external potential to efficiently move the system toward a known target state. The external potential was a time-dependent function of R(t), the difference in the internal distances in the simulation and target structures as described (Ozkirimli and Post 2006). The force constant governing the rate at which the conformational transition takes place was chosen such that the pairwise force constant, α, was 4, 9, 13, 18, or 22 × 10−4 kcal/(mol Å4).
BMD was performed as described (Ozkirimli and Post 2006) starting with homology-modeled coordinates for Lyn kinase plus ATP and Mg+2 in the assembled, down-regulated state. The external potential was referenced to the active state of the CD, allowing observation of the response of the regulatory domains to the CD conformational change. The external potential reaction coordinate included internal distances between all heavy atoms in CD, or ∼2.2 × 106 pairs. The BMD simulations were carried out until the change in reaction coordinate with time was 0.0005 Å/psec or less for a 30-psec running average.
Protein expression and purification
The following His-tagged versions of Hck were used in this study: (1) down-regulated Hck (SH3–SH2 catalytic domains) and (2) the isolated Hck catalytic domain. The proteins were expressed in Sf9 cells and purified essentially as described (Porter et al. 2000). C-terminal phosphorylation of down-regulated Hck was carried out by coinfection of Sf9 cells with Csk baculovirus (Porter et al. 2000). Sf9 cells were lysed in a French pressure cell, and lysates were applied to a Source Q column. Hck was eluted using a linear NaCl gradient. Fractions showing kinase activity were pooled and purified using Ni-NTA affinity chromatography.
Kinase assay
A continuous spectrophotometric kinase assay was employed in which ADP production is coupled to NADH oxidation (Barker et al. 1995; Porter et al. 2000). The assay was performed in the presence of 500 μM ATP and 600 μM substrate peptide (Ala-Glu-Glu-Glu-Ile-Tyr-Gly-Glu-Phe-Glu-Ala-Lys-Lys-Lys-Lys-Gly) for 4 h at 30°C. Data points were recorded every 7 sec. Induction times were determined from the intersection between the lines describing the initial and maximum enzymatic rates, as described (Favelyukis et al. 2001). Induction times were determined at various concentrations of NaCl (10 μM–1 M), and experiments were performed in duplicate. The induction times were replotted versus NaCl concentration and were analyzed by linear regression.
Acknowledgments
We thank Professor Robert Geahlen for insightful discussions. This work was supported by National Institutes of Health (NIH) grants GM39478 (C.B.P.) CA58530 (W.T.M), a Purdue University reinvestment grant, and the Purdue Cancer Center (CA23568). E.O. was supported by a Purdue Research Foundation Fellowship.
Footnotes
Reprint requests to: Carol Beth Post, Medicinal Chemistry and Molecular Pharmacology Department, Markey Center for Structural Biology and Purdue Cancer Center, Purdue University, 575 Stadium Mall Drive, West Lafayette, IN 47907-2091, USA; e-mail: cbp@purdue.edu; fax: (765) 496-1189.
Abbreviations: CD, catalytic domain; SH2, Src homology domain 2; SH3, Src homology domain 3; N-lobe, N-terminal lobe; C-lobe, C-terminal lobe; MD, molecular dynamics; BMD, biased molecular dynamics; A-loop, activation loop; αC, α-helix C; RMSD, root mean squared deviation; α, pairwise force constant.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037457.108.
4RMS differences in atomic coordinates were determined after least-squares superposition based on the same set of atoms used in the RMS sum unless otherwise noted.
References
- Adams, J.A. Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry. 2003;42:601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- Banavali, N., Roux, B. The N-terminal end of the catalytic domain of Src kinase Hck is a conformational switch implicated in long-range allosteric regulation. Structure. 2005;13:1715–1723. doi: 10.1016/j.str.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Barker, S., Kassel, D., Weigl, D., Huang, X., Luther, M., Knight, W. Characterization of pp60c-Src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- Boggon, T., Eck, M. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- Breitenlechner, C., Kairies, N., Honold, K., Scheiblich, S., Koll, H., Greiter, E., Koch, S., Shafer, W., Huber, R., Engh, R. Crystal structures of active Src kinase domain complexes. J. Mol. Biol. 2005;353:222–231. doi: 10.1016/j.jmb.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Capdeville, R., Buchdunger, E., Zimmermann, J., Matter, A. Glivec (sti571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A., Gould, K., Cartwright, C., Hunter, T. Tyr527 is phosphorylated in pp60cSrc: Implications for regulation. Science. 1986;231:1431–1434. doi: 10.1126/science.2420005. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob, S., Fendrich, G., Manley, P., Jahnke, W., Fabbro, D., Liebetanz, J., Meyer, T. The crystal structure of a c–Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Davydov, D., Botchkareva, A., Kumar, S., He, Y., Halpert, J. An electrostatically driven conformational transition is involved in the mechanisms of substrate binding and cooperativity in cytochrome p450eryf. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gómez, J.D., Roux, B. On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases. Proc. Natl. Acad. Sci. 2007;104:13643–13648. doi: 10.1073/pnas.0704041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favelyukis, S., Till, J., Hubbard, S., Miller, W.T. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat. Struct. Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- Gonfloni, S., Williams, J., Hattula, K., Weijland, A., Wierenga, R., Superti-Furga, G. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J. 1997;16:7261–7271. doi: 10.1093/emboj/16.24.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonfloni, S., Frischknecht, F., Way, M., Superti-Furga, G. Leucine 255 of Src couples intramolecular interactions to inhibition of catalysis. Nat. Struct. Biol. 1999;6:760–764. doi: 10.1038/11537. [DOI] [PubMed] [Google Scholar]
- Gonfloni, S., Weijland, A., Kretzschmar, J., Superti-Furga, G. Crosstalk between the catalytic and regulatory domains allows bidirectional regulation of Src. Nat. Struct. Biol. 2000;7:281–286. doi: 10.1038/74041. [DOI] [PubMed] [Google Scholar]
- Gouaux, E., Mackinnon, R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- Hamuro, Y., Wong, L., Shaffer, J., Kim, J., Stranz, D., Jennings, P., Woods, V.J., Adams, J. Phosphorylation driven motions in the COOH-terminal Src kinase, CSK, revealed through enhanced hydrogen-deuterium exchange and mass spectrometry (DXMS) J. Mol. Biol. 2002;323:871–881. doi: 10.1016/s0022-2836(02)01003-3. [DOI] [PubMed] [Google Scholar]
- Hubbard, S. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, S. Src autoinhibition: Let us count the ways. Nat. Struct. Biol. 1999;6:711–714. doi: 10.1038/11468. [DOI] [PubMed] [Google Scholar]
- Huse, M., Kuriyan, J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N., Lewis, R.J. Structural basis for control by phosphorylation. Chem. Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- Kannan, N., Neuwald, A.F. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J. Mol. Biol. 2005;351:956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- Kim, C., Xuong, N.-H., Taylor, S.S. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- Kornev, A.P., Haste, N.M., Taylor, S.S., Eyck, L.F.T. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan, J., Cowburn, D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- LaFevre-Bernt, M., Sicheri, F., Pico, A., Porter, M., Kuriyan, J., Miller, W.T. Intramolecular regulatory interactions in the Src family kinase Hck probed by mutagenesis of a conserved Tryptophan residue. J. Biol. Chem. 1998;273:32129–32134. doi: 10.1074/jbc.273.48.32129. [DOI] [PubMed] [Google Scholar]
- Levinson, N.M., Kuchment, O., Shen, K., Young, M.A., Koldobskiy, M., Karplus, M., Cole, P., Kuriyan, J. A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi, M., Ballone, P. Adiabatic bias molecular dynamics: A method to navigate the conformational space of complex molecular systems. J. Chem. Phys. 1999;110:3697–3702. [Google Scholar]
- Masterson, L.R., Mascioni, A., Traaseth, N.J., Taylor, S.S., Vegli, G. Allosteric cooperativity in protein kinase A. Proc. Natl. Acad. Sci. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, C., Mezna, M., Kontopidis, G. Catch the kinase conformer. Chem. Biol. 2006;13:693–694. doi: 10.1016/j.chembiol.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Moarefi, I., LaFevre-Bernt, M., Sicheri, F., Huse, M., Lee, C., Kuriyan, J., Miller, W.T. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- Nag, A., Dinner, A. Enhancement of diffusion-controlled reaction rates by surface-induced orientational restriction. Biophys. J. 2006;90:896–902. doi: 10.1529/biophysj.105.072785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, M., Endicott, J., Johnson, L. Protein kinase inhibitors: Insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- Nolen, B., Taylor, S.S., Ghosh, G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Okram, B., Nagle, A., Adrian, F., Lee, C., Ren, P., Wang, X., Sim, T., Xie, Y., Wang, X., Xia, G., et al. A general strategy for creating “inactive-conformation” Abl inhibitors. Chem. Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Ozkirimli, E., Post, C.B. Src kinase activation: A switched electrostatic network. Protein Sci. 2006;15:1051–1062. doi: 10.1110/ps.051999206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paci, E., Karplus, M. Forced unfolding of fibronectin type 3 modules: An analysis by biased molecular dynamics simulations. J. Mol. Biol. 1999;288:441–459. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
- Pawson, T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pawson, T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- Porter, M., Schindler, T., Kuriyan, J., Miller, W.T. Reciprocal regulation of Hck activity by phosphorylation of Tyr(527) and Tyr(416)—effect of introducing a high affinity intramolecular SH2 ligand. J. Biol. Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- Rosenberg, O., Deindl, S., Sung, R.-J., Nairn, A., Kuriyan, J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Roussel, R., Brodeur, S., Shalloway, D., Laudano, A. Selective binding of activated pp60cSrc by an immobilized synthetic phosphopeptide modeled on the carboxyl terminus of pp60cSrc . Proc. Natl. Acad. Sci. 1991;88:10696–10700. doi: 10.1073/pnas.88.23.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A., Blundell, T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schindler, T., Sicheri, F., Pico, A., Gazit, A., Levitzki, A., Kuriyan, J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- Sharp, K.A., Honig, B. Salt effects on nucleic acids. Curr. Opin. Struct. Biol. 1995;5:323–328. doi: 10.1016/0959-440x(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Sheinerman, F.B., Norel, R., Honig, B. Electrostatic aspects of protein–protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Sicheri, F., Kuriyan, J. Structures of Src-family tyrosine kinases. Curr. Opin. Struct. Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- Süel, G., Lockless, S., Wall, M., Ranganathan, R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Vigil, D., Lin, J.-H., Sotrifier, C., Pennypacker, J., McCammon, J., Taylor, S.S. A simple electrostatic switch important in the activation of type I protein kinase A by cyclic AMP. Protein Sci. 2006;15:113–121. doi: 10.1110/ps.051723606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, R.C., Gabdoulline, R.R., Ludemann, S.K., Lounnas, V. Electrostatic steering and ionic tethering in enzyme-ligand binding: Insights from simulations. Proc. Natl. Acad. Sci. 1998;95:5942–5949. doi: 10.1073/pnas.95.11.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Doshi, A., Lei, M., Eck, M.J., Harrison, S.C. Crystal structure of c-Src reveals features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, H., Hendrickson, W.A. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- Young, M.A., Gonfloni, S., Superti-Furga, G., Roux, B., Kuriyan, J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- Zheng, W., Brooks, B.R., Doniach, S., Thirumalai, D. Network of dynamically important residues in the open/closed transition in polymerases is strongly conserved. Structure. 2005;13:565–577. doi: 10.1016/j.str.2005.01.017. [DOI] [PubMed] [Google Scholar]