Abstract
The human kallikrein-related peptidases (KLKs) comprise 15 members (KLK1–15) and are the single largest family of serine proteases. The KLKs are utilized, or proposed, as clinically important biomarkers and therapeutic targets of interest in cancer and neurodegenerative disease. All KLKs appear to be secreted as inactive pro-forms (pro-KLKs) that are activated extracellularly by specific proteolytic release of their N-terminal pro-peptide. This processing is a key step in the regulation of KLK function. Much recent work has been devoted to elucidating the potential for activation cascades between members of the KLK family, with physiologically relevant KLK regulatory cascades now described in skin desquamation and semen liquefaction. Despite this expanding knowledge of KLK regulation, details regarding the potential for functional intersection of KLKs with other regulatory proteases are essentially unknown. To elucidate such interaction potential, we have characterized the ability of proteases associated with thrombostasis to hydrolyze the pro-peptide sequences of the KLK family using a previously described pro-KLK fusion protein system. A subset of positive hydrolysis results were subsequently quantified with proteolytic assays using intact recombinant pro-KLK proteins. Pro-KLK6 and 14 can be activated by both plasmin and uPA, with plasmin being the best activator of pro-KLK6 identified to date. Pro-KLK11 and 12 can be activated by a broad-spectrum of thrombostasis proteases, with thrombin exhibiting a high degree of selectivity for pro-KLK12. The results show that proteases of the thrombostasis family can efficiently activate specific pro-KLKs, demonstrating the potential for important regulatory interactions between these two major protease families.
Keywords: kallikrein-related peptidases, KLK, activation cascade, thrombostasis, plasmin, thrombin, inflammation
The 15 kallikrein-related peptidase (KLK) members (KLK1–15) represent the largest cluster of S1 (or chymotrypsin-like) serine proteases within the human genome (Yousef and Diamandis 2001). KLK3 (“prostate specific antigen” or PSA) is a widely used cancer biomarker for prostate cancer screening (Stamey et al. 1987; Luderer et al. 1995; Catalona et al. 1997), and KLK2 is emerging as another important prostate cancer diagnostic and prognostic tool (Darson et al. 1999; Recker et al. 2000; Fuessel et al. 2003). Evidence suggests several other KLKs are differentially regulated in specific types of cancer (Clements 1989; Diamandis et al. 2000; Pampalakis and Sotiropoulou 2007; Ramsay et al. 2008; Singh et al. 2008) and may also be potentially useful as cancer biomarkers (Diamandis and Yousef 2001; Clements et al. 2004). Additionally, KLK family members have been shown to be intimately associated with physiological processes of skin desquamation (Lundstrom and Egelrud 1991; Brattsand and Egelrud 1999; Brattsand et al. 2005), inflammatory demyelination (Blaber et al. 2004; Scarisbrick et al. 2006a), neurodegeneration (Scarisbrick et al. 2008), and semen liquefaction (Kumar et al. 1997; Vaisanen et al. 1999; Malm et al. 2000; Michael et al. 2006). These lines of evidence point to the increasing importance of KLKs in the diagnosis and treatment of serious human medical disorders.
KLKs are secreted as inactive pro-forms that are subsequently processed extracellularly to their active form via proteolytic removal of their N-terminal pro-peptide. This is a key regulatory step in controlling levels of active KLK relevant to both normal physiologic function and disease mechanisms. The regulatory cascades of other protease families (e.g., the coagulation cascade) have prompted various researchers to suggest that the KLK family might similarly participate in cascades of activation that regulate their function (Lovgren et al. 1997; Takayama et al. 1997; Sotiropoulou et al. 2003; Brattsand et al. 2005; Michael et al. 2006). One of the earliest such regulatory relationships was elucidated for KLK2 and 3 in the prostate. KLK2 exhibits trypsin-like activity and its pro-sequence requires cleavage after Arg; KLK2 was subsequently shown to autolytically activate (Denmeade et al. 2001). KLK3 exhibits chymotrypsin-like activity (Akiyama et al. 1987; Christensson et al. 1990; Robert et al. 1997; Coombs et al. 1998), but its activation pro-sequence requires cleavage after Arg; thus, KLK3 is unlikely to efficiently self-activate. Lilja and coworkers (Lovgren et al. 1997) showed that KLK2 is able to cleave the pro-form of KLK3 to yield mature active KLK3. These activation relationships describe the elements of an activation cascade in the prostate, involving initial activation of KLK2 followed by amplification of KLK2 activity by autolysis and subsequent cross-activation of KLK3 by KLK2. This cascade is relevant for the physiological process of semen liquefaction, and thus fertility, as KLK3 degrades semenogelins I and II as well as fibronectin (Malm et al. 2000).
A more extensive activation cascade involving KLKs 5, 7, and 14 has been elucidated in the physiological process of desquamation in the stratum corneum. KLK5 (stratum corneum tryptic enzyme) and KLK7 (stratum corneum chymotryptic enzyme) have been shown to be coexpressed as the dominant enzymes present in stratum corneum (Lundstrom and Egelrud 1991). Also present in stratum corneum is KLK14, and these proteases are postulated to degrade cornodesmosome proteins as part of the regulation of the desquamation of skin (Caubet et al. 2004). As its name implies, KLK5 exhibits a preference for hydrolysis after basic residues (both Arg and Lys), while KLK7 exhibits a preference for hydrolysis after aromatic residues (Yousef et al. 2003a; Brattsand et al. 2005; Michael et al. 2005). Since these proteases are coexpressed in stratum corneum, and the activation pro-sequences require hydrolysis after either Arg (KLK5) or Lys (KLK7 and KLK14), Brattsand et al. (2005) postulated that KLK5 may be an activating protease for pro-KLKs 7 and 14, forming the basis of a regulatory activation cascade in skin. Furthermore, the aberrant inhibition of these KLKs in skin (due to mutation in the lymphoepithelial-Kazal-type 5 serine protease inhibitor) appears responsible for the ichthyotic skin disorder Netherton syndrome (Hachem et al. 2006).
Recent studies have expanded considerably the number of relevant KLK pairwise activation relationships through proteolytic screening of pro-KLK peptide-fusion proteins (Yoon et al. 2007) or soluble pro-KLK peptides (Emami and Diamandis 2008). The results from peptide or fusion-protein hydrolyses identify, in the majority of cases, relevant activation relationships for the intact pro-KLK proteins. Such studies have thus produced a substantially complete characterization of the KLK “activome” (Yoon et al. 2007) that identifies the most efficient pairwise activation relationships of interest for subsequent detailed enzymatic characterization using recombinant pro-KLK proteins. This information, in combination with knowledge of tissue-specific expression of the KLKs, permits hypotheses to be developed and tested regarding more extensive tissue-specific KLK activation cascades (Yoon et al. 2007; Emami and Diamandis 2008).
With these strides made in elucidation of the KLK activome, the question now turns toward the possible intersection of the KLK axis with other protease families. In the present study, we characterize the hydrolytic profile of proteases of the thrombostasis system toward pro-KLK sequences, using a previously described pro-KLK fusion protein system. The data show that plasmin, tPA, uPA, thrombin, factor Xa, and plasma kallikrein are each capable of activating a uniquely different set of pro-KLKs. Several of the activation relationships identified using this system are subsequently characterized and confirmed using recombinant pro-KLK proteins. Of the set of pairwise activation relationships evaluated, the activation of pro-KLK6 by plasmin exhibits the greatest reaction rate. We postulate that under conditions of injury or inflammation, when proteases of the thrombostasis system can become activated and in contact with the extracellular environment, specific activation cascades involving the secreted pro-KLKs can be initiated.
Results
Purification of pro-KLK fusion proteins
The pro-KLK fusion proteins purified to apparent homogeneity, in each case, as determined by resolution on 16.5% Tricine SDS-PAGE and visualized by Coomassie Brilliant Blue staining (see Supplemental Figs. 1–23). Furthermore, in each case, 24-h incubations at 37°C, at either pH 6.0 or 7.4, showed no evidence of degradation due to the presence of contaminating Escherichia coli proteases (see Supplemental Figs. 1–23). Thus, the purified pro-KLK fusion proteins appeared suitable for use as substrates in proteolytic studies.
Proteolysis of pro-KLK fusion proteins by thrombostasis proteases
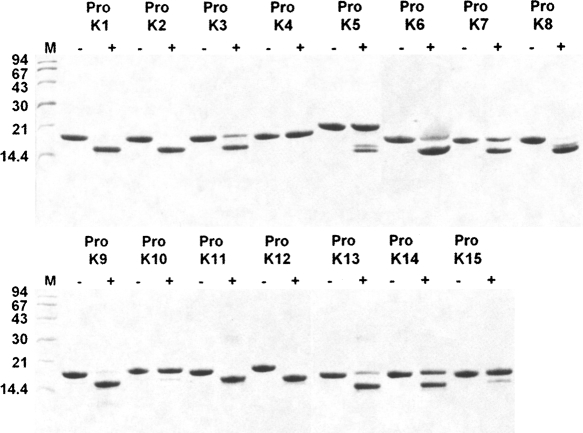
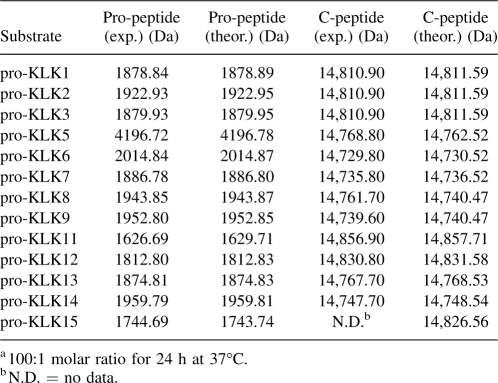
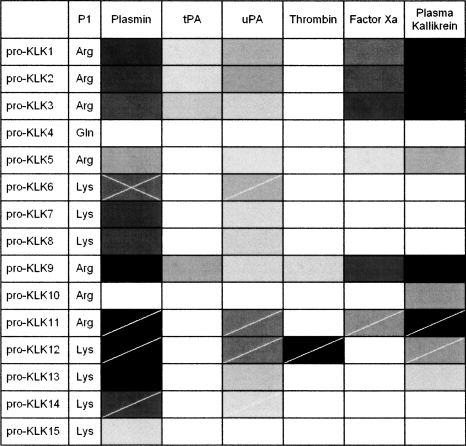
The 15 pro-KLK fusion proteins were subjected to hydrolysis by six different proteases of the thrombostasis system (plasmin, tPA, uPA, thrombin, factor Xa, and plasma kallikrein) at two different pH values (7.4 and 6.0), and for two different incubation periods (1 h and 24 h), resulting in the analysis of 360 samples. Scanned images of the SDS-PAGE gels (96 total) are provided as Supplemental figures; however, Figure 1 involving incubation of the pro-KLK1–15 fusion proteins with plasmin, at pH 7.4, and for the 24-h incubation period, is included here as an example. Under these conditions, incubation with plasmin resulted in complete hydrolysis of pro-KLK1, 2, 9, 11, and 12 fusion proteins; substantially complete hydrolysis of pro-KLK3, 6, 8, and 13 fusion proteins; partial hydrolysis of pro-KLK5, 7, and 14 fusion proteins; and minimal or no hydrolysis for pro-KLK4, 10, and 15 fusion proteins. While hydrolysis of the pro-KLK fusion protein results in release of the pro-KLK peptide and associated C-terminal fusion protein fragment, only the C-terminal fragment is typically visualized by staining with Coomassie Brilliant Blue. The pro-KLK fusion proteins exhibiting hydrolysis by plasmin were subjected to mass spectrometry analysis (Table 1). In this case, fragment masses corresponding to the released C-terminal fusion protein, as well as the correctly processed pro-peptide, were identified (with the exception of the C-terminal fusion protein for the pro-KLK15 fusion protein) and yielded excellent agreement with theoretical values. The hydrolysis of pro-KLK5 and 8 fusion proteins yielded evidence of minor secondary sites of hydrolysis (i.e., minor doublets); however, in neither of these cases, we were able to identify the minor sites of hydrolysis using mass spectrometry data.
Figure 1.
Coomassie Brilliant Blue stained 16.5% Tricine SDS-PAGE analysis of 100:1 molar ratio incubation of pro-KLK1–15 fusion proteins (abbreviated as “Pro K”) by plasmin for 24 h at pH 7.4.
Table 1.
Mass spectrometry data for the hydrolysis of pro-KLK1–15 fusion proteins by plasmina
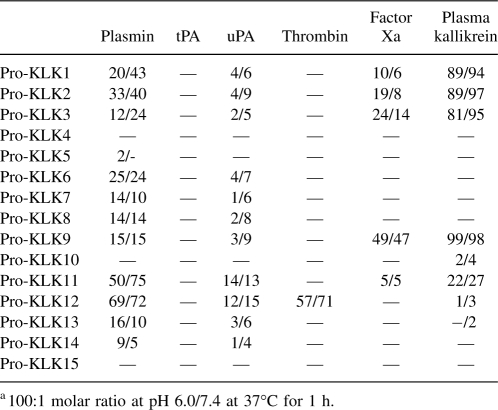
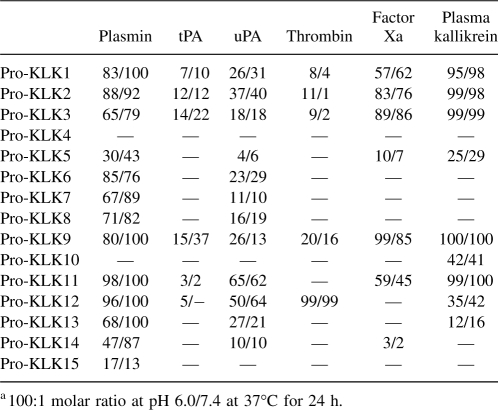
The mass spectrometry data for all reactions are provided as supplemental data (Supplemental Tables 1–14). In no case was the observed hydrolysis determined to be outside of the expected P1/P1′ junction of the pro-KLK fusion protein sequence, and in no case was an atypical mass observed for hydrolysis. For those protease/substrate combinations exhibiting hydrolysis, scanning densitometry was performed to quantify the extent of hydrolysis. Tables 2 and 3 list the percent hydrolysis for the pro-KLK1–15 fusion proteins by the set of thrombostasis proteases utilized, at both pH 6.0 and 7.4, and for both 1- and 24-h incubations.
Table 2.
Percentage of hydrolysis of pro-KLK1–15 fusion proteins by thrombostasis proteasesa
Table 3.
Percentage of hydrolysis of pro-KLK1–15 fusion proteins by thrombostasis proteasesa
Activation studies of recombinant pro-KLKs
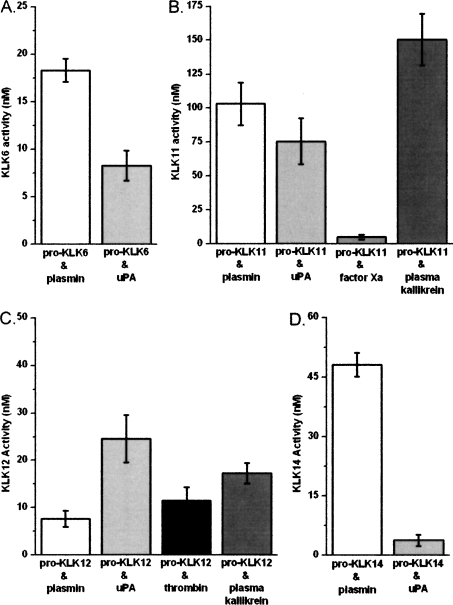
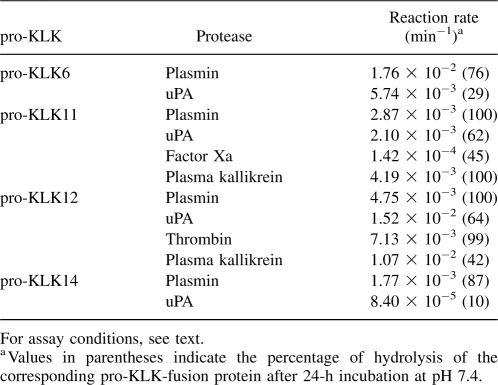
A representative subset of the most significant hydrolysis reactions observed using the pro-KLK fusion proteins was evaluated using available recombinant forms of pro-KLK6, 11, 12, and 14. In this case, reaction rates were quantified at substrate (i.e., pro-KLK protein) concentrations of 0.4 μM pro-KLK6 (with plasmin and uPA), 2.5 μM pro-KLK11 (with plasmin, uPA, factor Xa, and plasma kallikrein), 4.0 μM pro-KLK12 (with plasmin, uPA, thrombin, and plasma kallikrein), and 4.0 μM pro-KLK14 (with plasmin and uPA). The assay conditions utilized pH 7.4 for digestion of pro-KLK6, and pH 7.5–8.0 for the other pro-KLKs, based upon the manufacturer's recommended buffer conditions (see Materials and Methods). The raw activity data for these assays with substrate and enzyme controls subtracted are shown in Figure 2, and the reaction rates normalized to enzyme concentration are given in Table 4.
Figure 2.
Raw data for activation assays of pro-KLK6 (0.4 μM, 24 h; panel A), pro-KLK11 (2.5 μM, 24 h; panel B), pro-KLK12 (4.0 μM, 1 h; panel C), and pro-KLK14 (4.0 μM, 24 h; panel D) by plasmin (white), uPA (light gray), factor Xa (gray), plasma kallikrein (dark gray), and thrombin (black). Measured KLK activity has been normalized to account for enzyme and substrate controls.
Table 4.
Reaction rates for the activation of recombinant pro-KLK proteins by proteases of the thrombostasis system
A comparison of the reaction rates observed using recombinant pro-KLK proteins with the percentage of hydrolysis for the pro-KLK fusion proteins for individual pro-KLK proteins yields generally good agreement (i.e., greater percentage of hydrolysis for the pro-peptide fusion protein correlates with faster reaction rates using the actual pro-KLK recombinant proteins). The notable exception to this is seen with pro-KLK12; in this case, while hydrolysis by uPA and plasma kallikrein are slower than plasmin and thrombin in the pro-KLK12 fusion protein assay, they are several-fold faster than plasmin and thrombin when using actual pro-KLK12 protein (Table 4).
Discussion
The KLKs appear to reside primarily as inactive pro-forms in the extracellular environment, and their functional activation is dependent upon specific proteolytic release of their N terminus pro-peptide. Characterization of the potential autolytic and cross-activation potential among the members of the KLK family, and the elucidation of activation cascades regulating function, has been a subject of intense investigation (Lovgren et al. 1997; Denmeade et al. 2001; Takayama et al. 2001a; Brattsand et al. 2005; Debela et al. 2006; Michael et al. 2006; Memari et al. 2007; Yoon et al. 2007; Emami and Diamandis 2008). These studies have demonstrated the potential for complex regulatory activation cascades between members of the KLK family; however, almost nothing is known regarding the intersection of the KLK axis with other protease families. In this regard, of particular physiologic and clinical importance is the potential intersection of the KLK and thrombostasis family of proteases. The present study utilizes a pro-KLK fusion protein system to rapidly characterize the hydrolytic profile of potential KLK-activating proteases within the thrombostasis family. Positive results are used to identify those protease/pro-KLK combinations that merit further study using the actual pro-KLK protein. This system has previously been used to evaluate members of the KLK family as potential activating proteases, and a subset of hydrolyses were validated using recombinant pro-KLK protein (Yoon et al. 2007). The results of the present study indicate that plasmin and plasma kallikrein have the greatest potential as activators of the KLK axis; however, each protease tested exhibits activation potential for a specific subset of pro-KLKs.
The thrombostasis proteases are abundant in serum, and the majority of the KLKs have also been detected in this fluid (Catalona et al. 1991; Diamandis et al. 2002, 2003; Borgono et al. 2003; Dong et al. 2003; Kishi et al. 2003; Luo et al. 2003; Yousef et al. 2003b; Kapadia et al. 2004). Both families colocalize in a wide range of tissues, including liver, kidney, pancreas, and brain. Notably the brain is a rich source of several kallikreins (Clements et al. 2001; Scarisbrick et al. 2006a) and is also shown to express plasminogen, plasminogen activators, and prothrombin (Dihanich et al. 1991; Basham and Seeds 2001; Zhang et al. 2002). In addition to physiologic coexistence, inflammation and tissue damage can cause leakage of vascular proteins into the tissue extracellular matrix. This presents a particular threat to the central nervous system (CNS) since the blood–brain barrier, which normally prevents entry of serum proteins, breaks down with traumatic injury, stroke, and in CNS inflammatory conditions such as multiple sclerosis (MS). In such cases, proteases of the thrombostasis axis can easily come into contact with pro-KLKs.
While roles for the newly identified KLKs in health and disease are just beginning to emerge, there is already a long history of research related to the activities of the thrombostasis enzymes. In CNS, tPA has been demonstrated to be involved in hippocampal synaptic plasticity and long-term potentiation (Nicole et al. 2001; Pang et al. 2004); however, when levels become deregulated with respect to plasminogen activator inhibitors (α2-macroglobulin and α1-antitrypsin), as occurs in MS and in animal models of this disease, both tPA and uPA are thought to contribute to tissue injury and inflammation, with a net accumulation of fibrin (Akenami et al. 1999; Cuzner and Opdenakker 1999; Gveric et al. 2001; Lo et al. 2002; Lu et al. 2002; East et al. 2005). There is also abundant evidence that excess tPA contributes to hippocampal degeneration (Tsirka et al. 1995). Similarly, at low concentrations thrombin promotes neurite outgrowth but in excess is associated with neurotoxicity (Citron et al. 2000; Festoff et al. 2004). Furthermore, the KLK1-kinin system, which influences the permeability of the blood–brain barrier, is activated in stroke (Wagner et al. 2002). Given the growing evidence supporting coincident alterations in KLKs and thrombostasis enzymes in a range of CNS disorders, including Alzheimer's (Zarghooni et al. 2002; Paul et al. 2007), stroke (Chao and Chao 2006; Gravanis and Tsirka 2008), trauma (Festoff et al. 2004; Scarisbrick et al. 2006b), and MS (Gveric et al. 2001; Scarisbrick et al. 2002, 2008; Terayama et al. 2005), there appears to be tremendous potential for the intersection of these two important protease families.
KLK6 is up-regulated in response to CNS damage and in concert with the demyelination/remyelination processes that take place after such damage (Scarisbrick et al. 1997; He et al. 2001; Terayama et al. 2004). Subsequently, KLK6 has been shown to be robustly expressed at sites of active demyelination in human MS lesions (Scarisbrick et al. 2002). Most important, inhibition of KLK6 activity in animal models of experimental autoimmune encephalomyelitis (EAE) results in a delay of onset and a reduction in severity of inflammatory demyelination (Blaber et al. 2004). Together, these data indicate that KLK6 is involved in the normal turnover of myelin as well as in the progression of inflammatory demyelinating disease. These observations suggest stringent control of the activation status of KLK6 is critical to myelin homeostasis and normal CNS function. KLK6 exhibits broad specificity for hydrolysis after Arg, but not Lys residues (Bernett et al. 2002); however, its pro-sequence requires hydrolysis after Lys for release, and KLK6 is therefore unable to efficiently self-activate (Blaber et al. 2007). Of the thrombostasis proteases tested, plasmin exhibits the greatest efficiency for activation of pro-KLK6 (Fig. 3; Table 4) and is more efficient at activating pro-KLK6 than any mature KLK (Blaber et al. 2007). Therefore plasmin may be one of the most important activators of pro-KLK6. Given the expression of plasminogen and plasminogen activators in CNS and leakage of these proteins into CNS with inflammation (Gveric et al. 2001), these data further suggest that plasmin may be a critical activator of KLK6 in cases of CNS inflammatory disease. It is of considerable interest with regard to the potential interaction between KLK6 and plasmin that plasmin inhibitors, like those for KLK6, have been shown to slow the progress of EAE (Brosnan et al. 1980).
Figure 3.
Summary of pro-KLK fusion protein digest results (100:1 molar ratio pro-KLK fusion protein:enzyme at pH 7.4, 24 h at 37°C) represented in grayscale (white = 0%; black = 100% hydrolysis). The cross-hatches indicate those activation relationships reported in the literature for pro-KLKs (Blaber et al. 2007), and the single hatched marks indicate those substrate:enzyme combinations subsequently quantified in this study using recombinant intact pro-KLK protein.
Tissue kallikrein (KLK1) is known to be activated within CNS in stroke (Wagner et al. 2002) and elevated in the serum of MS patients (Scarisbrick et al. 2008). Additionally, activation of both plasma and tissue kallikreins occurs during activation of the contact system after cardiopulmonary bypass (Campbell et al. 2001). The hydrolysis results herein indicate that plasma kallikrein is an efficient activator of pro-KLK1 (Tables 2, 3; Fig. 3); thus, functional amplification of the kallikrein system may occur as a consequence of the activation of pro-KLK1 (“tissue kallikrein”) by plasma kallikrein. Both KLK1 and plasma kallikrein are capable of releasing kinins from kininogens. As kinins play an important role in inflammation, this observation suggests an important role for the activation of pro-KLK1 by plasma kallikrein in the inflammatory cascade. Plasma kallikrein appears capable of activating a significant subset of KLKs, notably including pro-KLK10. No mature KLK has yet been shown capable of activating pro-KLK10, and so the ability of plasma kallikrein to activate this KLK is of substantial interest. The function of KLK10 is not known; however, it has the second-highest mRNA expression level (after KLK6) in the CNS (Scarisbrick et al. 2006a).
Thrombin is notably unique in its activation profile for being able to activate pro-KLK12 (Fig. 3). Not much is known regarding the function of KLK12; however, KLK12 may be involved in the pathogenesis and/or progression of breast and prostate cancers (Yousef et al. 2000). KLK12 is known to be able to activate pro-KLK11 (Memari et al. 2007; Yoon et al. 2007), and subsequently, KLK11 has been shown capable of activating pro-KLK12 (Yoon et al. 2007). Thus, thrombin may intersect with the KLK axis via activation of pro-KLK11, and this can result in a more extensive KLK activation cascade.
KLK8 has been reported capable of activating two-chain tPA (tc-tPA) to single-chain tPA (sc-tPA). Additionally, KLK4 (Takayama et al. 2001b) and KLK2 (Frenette et al. 1997) have been reported to be capable of activating single-chain uPA (sc-uPA) to two-chain uPA. Thus, the potential exists for activated KLKs to feed back into, and amplify, the thrombostasis axis activity. In particular, the ability of plasmin to activate KLK2 and the ability of KLK2 to activate uPA create a potential feedback amplification circuit between the two proteolytic axes. Similarly, the ability of plasmin to activate KLK8 and the ability of KLK8 to activate tc-tPA provide an additional feedback amplification circuit between these axes.
Although the turnover numbers in Table 4 may be considered slow in comparison to other proteolytic systems, the temporal resolution of regulatory cascades involving the KLKs might be significantly slower compared with other proteolytic cascades such as thrombogenesis or thrombolysis. In these other proteolytic pathways, the relevant signals can resolve over a time frame of seconds (e.g., partial thromboplastin time) and are essential for the physiological function of the signaling cascade. However, functionally relevant regulatory cascades involving the KLKs may extend over days or weeks. An activation cascade has been proposed in human skin (stratum corneum) and involves KLK5, 7, and 14 in the regulation of desquamation (Lundstrom and Egelrud 1991; Brattsand and Egelrud 1999). This process resolves over a timescale of hours to days, regulated by pH changes in the skin, and is quite slow in comparison to more familiar proteolytic cascades. There is evidence to suggest that KLK6 activity associated with spinal cord trauma, including dynamic modification of the ECM affecting the capacity for axon outgrowth, occurs over a temporal period of days (Oka et al. 2005; Scarisbrick et al. 2006b). KLK6 has also been reported to be involved in the process of skin carcinogenesis via the alteration of ECM proteins to support tumor cell growth and invasion (Klucky et al. 2007). In this case, KLK6 appears functionally up-regulated throughout the entire precancerous and invasive cancerous stages (i.e., spanning a period of days and weeks). Indeed, one of the most important potential uses of the KLKs is diagnostic and prognostic cancer biomarkers (Diamandis and Yousef 2001; Sotiropoulou et al. 2003), suggesting functionally relevant activities that are temporally extended. It appears unlikely, therefore, that regulatory cascades involving the KLKs resolve over a timeframe of seconds, and comparatively slow reaction rates are of physiological relevance.
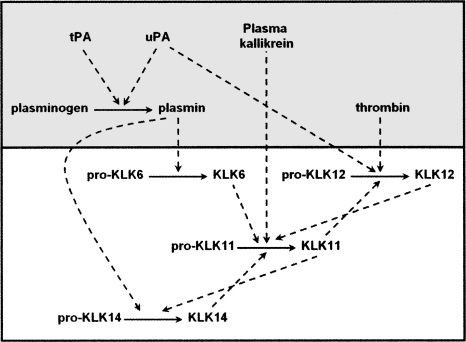
We note that the activation profile information reported herein, in combination with details of the thrombostasis cascade and the previously characterized KLK activome (Yoon et al. 2007), permits the construction of hypothetical activation cascades involving the intersection of the thrombostasis and KLK axes. Based upon our previous results with KLK activation profiles, we proposed a CNS-relevant cascade (Memari et al. 2007; Yoon et al. 2007). We propose a modification that shows how such a cascade can be initiated by intersection with the thrombostasis axis, as might occur in inflammatory demyelination (Fig. 4). Together, these data point to the need for additional characterization of the activation relationships between select KLKs and thrombostasis enzymes, not only to determine their role in disease processes but also to uncover novel therapeutically relevant regulatory targets.
Figure 4.
Proposed intersection of KLK and thrombostasis axes (shaded background) within the CNS relevant to inflammatory demyelination and nerve injury disease states.
Materials and Methods
Production of pro-KLK fusion proteins
The cloning, expression, and purification of pro-KLK fusion proteins have previously been described in detail (Yoon et al. 2007). Briefly, the N terminus pro-peptide region of KLK1–15, comprising the entire set of pro-peptide “P” positions and continuing through the P6′ position of the corresponding KLK mature N terminus, in each case, was inserted into the His-tagged N terminus of a mutant form of human fibroblast growth factor-1 (FGF*). The FGF* protein is highly soluble (thereby ensuring solubility of the pro-KLK peptide); readily expressed using an E. coli host; contains an unstructured, highly solvent-exposed N terminus (as with the KLKs) (Blaber et al. 1996; Gomis-Ruth et al. 2002); is engineered to be resistant to internal proteolysis; and has a mass appropriate for rapid SDS-PAGE quantitation of the hydrolytically released KLK pro-peptide.
All expression and purification steps were performed as previously described (Yoon et al. 2007). Purified pro-KLK fusion protein (1.0 mg/mL) was exchanged into 20 mM sodium phosphate, 0.15 M NaCl (PBS), pH 7.4, filtered through a 0.2 μ filter (Whatman Inc.), snap-frozen in dry ice/ethanol, and stored at −80°C prior to use. Samples of all pro-KLK fusion proteins were subjected to incubation, at a concentration of 50 μM, for 24 h at 37°C, followed by SDS-PAGE analysis, to confirm the absence of contaminating expression host proteases.
Production of pro-KLK proteins
Pro-KLK11, 12, and 14 were purchased from R&D Systems Inc. The purity of the pro-KLK11, pro-KLK12, and pro-KLK14 proteins was confirmed to be >95% as determined by silver-stained SDS-PAGE analysis. Recombinant pro-KLK6 was expressed from HEK293 human embryonic kidney epithelial cells as the expression host, as previously described (Blaber et al. 2007). Briefly, pro-KLK6 was expressed with the inclusion of a C-terminal Strep-tag and His-tag, respectively. X-ray structure data shows that the KLK6 C terminus is essentially antipodal to the N terminus, and as such, short C-terminal tags do not interfere with the N-terminal pro-region (Bernett et al. 2002; Gomis-Ruth et al. 2002). The cDNA encoding human pre-pro-KLK6 was cloned into the pSecTag2/HygroB expression vector (Invitrogen). In this construct, the native KLK6 secretion signal was utilized to direct secretion into the culture media. The HEK293 culture media was harvested 2 d after transfection and the pro-KLK6 purified by sequential affinity chromatography utilizing nickel-affinity resin and Strep-Tactin Superflow media (QIAGEN), respectively. Purity and homogeneity of recombinant pro-KLK6 was evaluated by 16.5% Tricine SDS-PAGE and N-terminal sequencing. To confirm the absence of contaminating host peptidase activity, purified pro-KLK6 was concentrated to 50 μM and incubated at 37°C for 6 h at pH 8.0, and analyzed using 16.5% Tricine SDS-PAGE.
Pro-KLK fusion protein hydrolysis assay
Plasmin (Roche, Applied Science), uPA, tPA (American Diagnostica Inc.), thrombin, factor Xa, and human plasma kallikrein (EMD Biosciences Inc.) were confirmed to be essentially homogenous by silver-stained SDS-PAGE. Pro-KLK fusion proteins and the above thrombostasis proteases were diluted into PBS (pH 6.0 or 7.4) and combined in a 100:1 molar ratio, respectively, with a final pro-KLK fusion protein concentration of 50 μM. Samples were incubated at 37°C for either 1 or 24 h, after which time they were immediately added to SDS-sample buffer and boiled. The digestion samples (5.0 μg) were subsequently resolved using 16.5% Tricine SDS-PAGE and visualized by staining with Coomassie Brilliant Blue. The stained gels were scanned and the extent of hydrolysis quantified against pro-KLK fusion protein standards using UN-SCAN-IT densitometry software (Silk Scientific). The normalized percentage of hydrolysis is for the intact pro-KLK fusion protein; thus, for samples with extensive fragmentation, the percentage of hydrolysis is given in reference to the residual intact pro-KLK fusion protein.
The pro-KLK fusion proteins that exhibited proteolytic cleavage were subjected to MALDI-TOF mass spectrometry analysis using a matrix of α-cyano-4-hydroxy-cinnamic acid and performed on an Axima CFR-plus mass spectrometer (Shimadzu and Biotech).
Activation assays of pro-KLKs
All assays were designed to hydrolyze less than 10% of the pro-KLK substrate over the assay period; in this way, reaction rates were determined under conditions of essentially constant substrate concentration, thus permitting determination of intrinsic turnover numbers for the substrate concentration evaluated. The ability of plasmin and uPA to activate pro-KLK6 was evaluated using a coupled assay procedure with the KLK6-sensitive internally-quenched fluorogenic peptide Abz-AFRFSQ-EDDnp (Angelo et al. 2006). In the first step (comprising the pro-KLK6 “activation step”) 400 nM of pro-KLK6 in 100 mM Tris, 0.1 mM EDTA (pH 7.4) was mixed with 4nM (i.e., 100:1 molar ratio) of plasmin or uPA, and the sample was incubated at 37°C for 24 h. Following this activation step was the “detection step,” where the KLK6-sensitive fluorogenic substrate Abz-AFRFSQ-EDDnp was added to a concentration of 2.0 μM, and the generation of released fluorescence due to hydrolysis was measured after 5-min incubation, using a Cary Eclipse fluorescence spectrophotometer with excitation wavelength of 320 nm and emission wavelength of 420 nm. The resulting concentration of mature KLK6 was quantified using a standard curve of mature KLK6 protein. Controls included pro-KLK6 in the absence of added plasmin or uPA, and plasmin and uPA in the absence of added pro-KLK6, as well as buffer control.
Pro-KLK11 (2.5 μM) was incubated with 25 nM of either plasmin, tPA, thrombin, or factor Xa in 50 mM Tris, 0.15 M NaCl, 0.05% Brij-35, pH 7.5 (TCNB buffer) at 37°C for 24 h. This buffer condition followed the manufacturer's recommendation for pro-KLK11 activation. The generated KLK11 enzyme activity was quantified using a coupled D-Val-Leu-Lys-thiobenzyl/5, 5′-dithio-bis(2-nitrobenzoic acid) (DTNB) assay. In this assay, 0.1 mM each of D-Val-Leu-Lys-thibenzyl substrate (MP Biomedicals) and DTNB (Ellman's reagent) chromogen were directly added to the reaction mixture. Hydrolysis of the Lys-thiobenzyl substrate by active KLK11 was quantified by monitoring generation of the DTNB thiolate ion at 405 nm with an extinction coefficient of ε = 13,600 M−1cm−1 and a standard curve of mature KLK11. Controls included pro-KLK11 in the absence of added plasmin, tPA, thrombin, and factor Xa; plasmin, tPA, thrombin, or factor Xa in absence of added pro-KLK11; as well as buffer control. Pro-KLK11 activated by thermolysin according to the manufacturer's instructions was confirmed to yield a specific activity within the range quoted by the manufacturer (>450 pmoles/min/μg).
Pro-KLK12 (4.0 μM) was incubated with 40 nM of plasmin, tPA, or thrombin in 0.1 M Tris, 0.15 M NaCl, 10 mM CaCl2, 0.05% Brij-35, pH 8.0, for 1 h at 37°C. This buffer condition followed the manufacturer's recommendation for pro-KLK12 activation. The sample digests were subsequently diluted 1:10 into 0.1 M Tris, 0.15 M NaCl, 10 mM CaCl2, 0.05% Brij-35, pH 7.5 containing 0.1 mM Boc-Val-Pro-Arg-7-amino-4-methyl coumarin (Boc-Val-Pro-Arg-AMC; R&D Systems Inc.) substrate. The AMC fluorophore generated was quantified on a Varian Cary Eclipse fluorescence spectrophotometer using a standard of the Boc-Val-Pro-Arg-AMC substrate subjected to complete hydrolysis by overnight incubation with bovine trypsin. Normalization of KLK12 activity was performed using standards of KLK12 activated according to the manufacturer's protocol. Controls included pro-KLK12 in the absence of added plasmin, tPA, or thrombin; plasmin, tPA, or thrombin in the absence of added pro-KLK12; as well as buffer control. Pro-KLK12 activated by overnight self-incubation according to the manufacturer's instructions was confirmed to yield a specific activity within the range quoted by the manufacturer (>4000 pmoles/min/μg).
Pro-KLK14 (4.0 μM) was incubated with 40 nM of either plasmin or factor Xa in TCNB buffer for 24 h at 37°C. This buffer condition followed the manufacturer's recommendation for pro-KLK14 activation. The sample digest was subsequently diluted 1:10 into 50 mM Tris, 0.15 M NaCl, 0.05% Brij-35, pH 8.0 containing 0.1 mM Boc-Val-Pro-Arg-AMC. The KLK14 activity generated was assayed as described above for KLK12. Controls included pro-KLK14 in the absence of added plasmin or factor Xa, plasmin or factor Xa in the absence of added pro-KLK14, as well as buffer control. Pro-KLK14 activated by thermolysin according to the manufacturer's instructions was confirmed to yield a specific activity within the range quoted by the manufacturer (>3000 pmoles/min/μg).
Acknowledgments
This work was supported by NIH grant 1R15NS057771-01 (M.B.), National Multiple Sclerosis Society grants PP1113 (M.B.) and RG3367 (I.A.S), and a grant from Fundação de Amparo a Pesquisa do estado de São Paulo (FAPESP) to M.A.J.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Michael Blaber, 3350G Biomedical Sciences Building, College of Medicine, Florida State University Tallahassee, FL 32306-4300, USA; e-mail: michael.blaber@med.fsu.edu; fax: (850) 644-5781.
Abbreviations: KLK, kallikrein-related peptidase; FGF, fibroblast growth factor; SDS PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; HEK293, human embryonic kidney epithelial cells; PBS, phosphate buffered saline; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; DTNB, dithionitrobenzoate; Tris, trishydroxyaminoethyl; CNS, central nervous system.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036715.108.
References
- Akenami, F.O.T., Siren, V., Wessman, M., Koskiniemi, M., Vaheri, A. Tissue plasminogen activator gene expression in multiple sclerosis brain tissue. J. Neurol. Sci. 1999;165:71–76. doi: 10.1016/s0022-510x(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Akiyama, K., Nakamura, T., Iwanaga, S., Hara, M. The chymotrypsin-like activity of human prostate-specific antigen, γ-seminoprotein. FEBS Lett. 1987;225:168–172. doi: 10.1016/0014-5793(87)81151-1. [DOI] [PubMed] [Google Scholar]
- Angelo, P.F., Lima, A.R., Alves, F.M., Blaber, S.I., Juliano, L., Juliano, M.A. Substrate specificity of human kallikrein 6: Salt and glycosaminoglycan effects. J. Biol. Chem. 2006;281:3116–3126. doi: 10.1074/jbc.M510096200. [DOI] [PubMed] [Google Scholar]
- Basham, M.E., Seeds, N.W. Plasminogen expression in the neonatal and adult mouse brain. J. Neurochem. 2001;77:318–325. doi: 10.1046/j.1471-4159.2001.t01-1-00239.x. [DOI] [PubMed] [Google Scholar]
- Bernett, M.J., Blaber, S.I., Scarisbrick, I.A., Dhanarajan, P., Thompson, S.M., Blaber, M. Crystal structure and biochemical characterization of human kallikrein 6 reveals a trypsin-like kallikrein is expressed in the central nervous system. J. Biol. Chem. 2002;277:24562–24570. doi: 10.1074/jbc.M202392200. [DOI] [PubMed] [Google Scholar]
- Blaber, M., DiSalvo, J., Thomas, K.A. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry. 1996;35:2086–2094. doi: 10.1021/bi9521755. [DOI] [PubMed] [Google Scholar]
- Blaber, S.I., Ciric, B., Christophi, G.P., Bernett, M., Blaber, M., Rodriguez, M., Scarisbrick, I.A. Targeting kallikrein 6 proteolysis attenuates CNS inflammatory disease. FASEB J. 2004;18:920–922. doi: 10.1096/fj.03-1212fje. [DOI] [PubMed] [Google Scholar]
- Blaber, S.I., Yoon, H., Scarisbrick, I.A., Juliano, M.A., Blaber, M. The autolytic regulation of human kallikrein-related peptidase 6. Biochemistry. 2007;46:5209–5217. doi: 10.1021/bi6025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgono, C.A., Grass, L., Soosaipillai, A., Yousef, G.M., Petraki, C.D., Howarth, D.H., Fracchioli, S., Katsaros, D., Diamandis, E.P. Human kallikrein 14: A new potential biomarker for ovarian and breast cancer. Cancer Res. 2003;63:9032–9041. [PubMed] [Google Scholar]
- Brattsand, M., Egelrud, T. Purification, molecular cloning, and expression of a human stratum corneum trypsin-like serine protease with possible function in desquamation. J. Biol. Chem. 1999;274:30033–30040. doi: 10.1074/jbc.274.42.30033. [DOI] [PubMed] [Google Scholar]
- Brattsand, M., Stefansson, K., Lundh, C., Haasum, Y., Egelrud, T. A proteolytic cascade of kallikreins in the stratum corneum. J. Invest. Dermatol. 2005;124:198–203. doi: 10.1111/j.0022-202X.2004.23547.x. [DOI] [PubMed] [Google Scholar]
- Brosnan, C.F., Cammer, W., Norton, W.T., Bloom, B. Protease inhibitors suppress the development of experimental allergic encephalomyelitis. Nature. 1980;285:235–237. doi: 10.1038/285235a0. [DOI] [PubMed] [Google Scholar]
- Campbell, D.J., Dixon, B., Kladis, A., Kemme, M., Santamaria, J.D. Activation of the kallikrein-kinin system by cardiopulmonary bypass in humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R1059–R1070. doi: 10.1152/ajpregu.2001.281.4.R1059. [DOI] [PubMed] [Google Scholar]
- Catalona, W.J., Smith, D.S., Ratliff, T.L., Dodds, K.M., Coplen, D.E., Yuan, J.J., Petros, J.A., Andriole, G.L. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N. Engl. J. Med. 1991;324:1156–1161. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- Catalona, W.J., Besser, J.A., Smith, D.S. Serum free prostate specific antigen and prostate specific antigen measurements for predicting cancer in men with prior negative prostatic biopsies. J. Urol. 1997;158:2162–2167. doi: 10.1016/s0022-5347(01)68187-4. [DOI] [PubMed] [Google Scholar]
- Caubet, C., Jonca, N., Brattsand, M., Guerrin, M., Bernard, D., Schmidt, R., Egelrud, T., Simon, M., Serre, G. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J. Invest. Dermatol. 2004;122:1235–1244. doi: 10.1111/j.0022-202X.2004.22512.x. [DOI] [PubMed] [Google Scholar]
- Chao, J., Chao, L. Experimental therapy with tissue kallikrein against cerebral ischemia. Front. Biosci. 2006;11:1323–1327. doi: 10.2741/1886. [DOI] [PubMed] [Google Scholar]
- Christensson, A., Laurell, C.-B., Lilja, H. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine proteinase inhibitors. Eur. J. Biochem. 1990;194:755–763. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]
- Citron, B.A., Smirnova, I.V., Arnold, P.M., Festoff, B.W. Upregulation of neurotoxic serine proteases, prothrombin, and protease-activated receptor 1 early after spinal cord injury. J. Neurotrauma. 2000;17:1191–1203. doi: 10.1089/neu.2000.17.1191. [DOI] [PubMed] [Google Scholar]
- Clements, J.A. The glandular kallikrein family of enzymes: Tissue-specific expression and hormonal regulation. Endocr. Rev. 1989;10:393–419. doi: 10.1210/edrv-10-4-393. [DOI] [PubMed] [Google Scholar]
- Clements, J., Hooper, J., Dong, Y., Harvey, T. The expanded human kallikrein (KLK) gene family: Genomic organisation, tissue-specific expression and potential functions. Biol. Chem. 2001;382:5–14. doi: 10.1515/BC.2001.002. [DOI] [PubMed] [Google Scholar]
- Clements, J.A., Willemsen, N.M., Myers, S.A., Dong, Y. The tissue kallikrein family of serine proteases: Functional roles in human disease and potential as clinical biomarkers. Crit. Rev. Clin. Lab. Sci. 2004;41:265–312. doi: 10.1080/10408360490471931. [DOI] [PubMed] [Google Scholar]
- Coombs, G.S., Bergstrom, R.C., Pellequer, J.-L., Baker, S.I., Mavre, M., Smith, M.M., Tainer, J.A., Madison, E.L., Corey, D.R. Substrate specificity of prostate-specific antigen (PSA) Chem. Biol. 1998;5:475–488. doi: 10.1016/s1074-5521(98)90004-7. [DOI] [PubMed] [Google Scholar]
- Cuzner, M.L., Opdenakker, G. Plasminogen activators and matrix metalloproteases, mediators of extracellular proteolysis in inflammatory demyelination of the central nervous system. J. Neuroimmunol. 1999;94:1–14. doi: 10.1016/s0165-5728(98)00241-0. [DOI] [PubMed] [Google Scholar]
- Darson, M.F., Pacelli, A., Roche, P., Rittenhouse, H.G., Wolfert, R.L., Saeid, M.S., Young, C.Y., Klee, G.G., Tindall, D.J., Bostwick, D.G. Human glandular kallikrein 2 expression in prostate adenocarcinoma and lymph node metastases. Urology. 1999;53:939–944. doi: 10.1016/s0090-4295(98)00637-2. [DOI] [PubMed] [Google Scholar]
- Debela, M., Magdolen, V., Schechter, N., Valachova, M., Lottspeich, F., Craik, C.S., Choe, Y., Bode, W., Goettig, P. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 2006;281:25678–25688. doi: 10.1074/jbc.M602372200. [DOI] [PubMed] [Google Scholar]
- Denmeade, S.R., Lovgren, J., Khan, S.R., Lilja, H., Isaacs, J.T. Activation of latent protease function of pro-hK2, but not pro-PSA, involves autoprocessing. Prostate. 2001;48:122–126. doi: 10.1002/pros.1088. [DOI] [PubMed] [Google Scholar]
- Diamandis, E.P., Yousef, G.M. Human tissue kallikrein gene family: A rich source of novel disease biomarkers. Expert Rev. Mol. Diagn. 2001;1:182–190. doi: 10.1586/14737159.1.2.182. [DOI] [PubMed] [Google Scholar]
- Diamandis, E.P., Yousef, G.M., Luo, L.Y., Magklara, A., Obiezu, C.V. The new human kallikrein gene family: Implications in carcinogenesis. Trends Endocrinol. Metab. 2000;11:54–60. doi: 10.1016/s1043-2760(99)00225-8. [DOI] [PubMed] [Google Scholar]
- Diamandis, E.P., Okui, A., Mitsui, S., Luo, L.Y., Soosaipillai, A., Grass, L., Nakamura, T., Howarth, D.J., Yamaguchi, N. Human kallikrein 11: A new biomarker of prostate and ovarian carcinoma. Cancer Res. 2002;62:295–300. [PubMed] [Google Scholar]
- Diamandis, E.P., Scorilas, A., Fracchioli, S., Van Gramberen, M., De Bruijn, H., Henrik, A., Soosaipillai, A., Grass, L., Yousef, G.M., Stenman, U.H., et al. Human kallikrein 6 (hK6): A new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J. Clin. Oncol. 2003;21:1035–1043. doi: 10.1200/JCO.2003.02.022. [DOI] [PubMed] [Google Scholar]
- Dihanich, M., Kaser, M., Reinhard, E., Cunningham, D., Monard, D. Prothrombin mRNA is expressed by cells of the nervous system. Neuron. 1991;6:575–581. doi: 10.1016/0896-6273(91)90060-d. [DOI] [PubMed] [Google Scholar]
- Dong, Y., Kaushal, A., Brattsand, M., Nicklin, J., Clements, J.A. Differential splicing of KLK5 and KLK7 in epithelial ovarian cancer produces novel variants with potential as cancer biomarkers. Clin. Cancer Res. 2003;9:1710–1720. [PubMed] [Google Scholar]
- East, E., Baker, D., Pryce, G., Lijnen, H.R., Cuzner, M.L., Gveric, D. A role for the plasminogen activator system in inflammation and neurodegeneration in the central nervous system during experimental allergic encephalomyelitis. Am. J. Pathol. 2005;167:545–554. doi: 10.1016/S0002-9440(10)62996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami, N., Diamandis, E.P. Human kallikrein-related peptidase 14 (KLK14) is a new activator component of the KLK proteolytic cascade. J. Biol. Chem. 2008;283:3031–3041. doi: 10.1074/jbc.M707253200. [DOI] [PubMed] [Google Scholar]
- Festoff, B.W., Ameenuddin, S., Santacruz, K., Morser, J., Suo, Z., Arnold, P.M., Stricker, K.E., Citron, B.A. Neuroprotective effects of recombinant thrombomodulin in controlled contusion spinal cord injury implicates thrombin signaling. J. Neurotrauma. 2004;21:907–922. doi: 10.1089/0897715041526168. [DOI] [PubMed] [Google Scholar]
- Frenette, G., Tremblay, R.R., Lazure, C., Dube, J.Y. Prostatic kallikrein hK2, but not prostate-specific antigen (hK3), activates single-chain urokinase-type plasminogen activator. Int. J. Cancer. 1997;71:897–899. doi: 10.1002/(sici)1097-0215(19970529)71:5<897::aid-ijc31>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Fuessel, S., Sickert, D., Meye, A., Klenk, U., Schmidt, U., Schmitz, M., Rost, A.K., Eweigle, B., Kiessling, A., Wirth, M.P. Multiple tumor marker analyses (PSA, HK2, PSCA, tryp-p8) in primary prostate cancers using quantitative RT-PCR. Int. J. Oncol. 2003;23:221–228. [PubMed] [Google Scholar]
- Gomis-Ruth, F.X., Bayes, A., Sotiropoulou, G., Pampalakis, G., Tsetsenis, T., Villegas, V., Aviles, F.X., Coll, M. The structure of human prokallikrein 6 reveals a novel activation mechanism for the kallikrein family. J. Biol. Chem. 2002;277:27273–27281. doi: 10.1074/jbc.M201534200. [DOI] [PubMed] [Google Scholar]
- Gravanis, I., Tsirka, S.E. Tissue-type plasminogen activator as a therapeutic target in stroke. Expert Opin. Ther. Targets. 2008;12:159–170. doi: 10.1517/14728222.12.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gveric, D., Hanemaaijer, R., Newcombe, J., van Lent, N.A., Sier, C.F., Cuzner, M.L. Plasminogen activators in multiple sclerosis lesions: Implications for the inflammatory response and axonal damage. Brain. 2001;124:1978–1988. doi: 10.1093/brain/124.10.1978. [DOI] [PubMed] [Google Scholar]
- Hachem, J.P., Wagberg, F., Schmuth, M., Crumrine, D., Lissens, W., Jayakumar, A., Houben, E., Mauro, T.M., Leonardsson, G., Brattsand, M., et al. Serine protease activity and residual LEKTI expression determine phenotype in Netherton syndrome. J. Invest. Dermatol. 2006;126:1609–1621. doi: 10.1038/sj.jid.5700288. [DOI] [PubMed] [Google Scholar]
- He, X.-P., Shiosaka, S., Yoshida, S. Expression of neuropsin in oligodendrocytes after injury to the CNS. Neurosci. Res. 2001;39:455–462. doi: 10.1016/s0168-0102(01)00200-0. [DOI] [PubMed] [Google Scholar]
- Kapadia, C., Yousef, G.M., Mellati, A.A., Magklara, A., Wasney, G.A., Diamandis, E.P. Complex formation between human kallikrein 13 and serum protease inhibitors. Clin. Chim. Acta. 2004;339:157–167. doi: 10.1016/j.cccn.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Kishi, T., Grass, L., Soosaipillai, A., Shimizu-Okabe, C., Diamandis, E.P. Human kallikrein 8: Immunoassay development and identification in tissue extracts and biological fluids. Clin. Chem. 2003;49:87–96. doi: 10.1373/49.1.87. [DOI] [PubMed] [Google Scholar]
- Klucky, B., Mueller, R., Vogt, I., Teurich, S., Hartenstein, B., Breuhahn, K., Flechtenmacher, C., Angel, P., Hells, J. Kallikrein 6 induces E-cadherin shedding and promotes cell proliferation, migration and invasion. Cancer Res. 2007;67:8198–8206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Mikolajczyk, S.D., Goel, A.S., Millar, L.S., Saedi, M.S. Expression of pro-form of prostate-specific antigen by mammalian cells and its conversion to mature, active form by human kallikrein 2. Cancer Res. 1997;57:3111–3114. [PubMed] [Google Scholar]
- Lo, E.H., Wang, X., Cuzner, M.L. Extracellular proteolysis in brain injury and inflammation: Role for plasminogen activators and matrix metalloproteinases. J. Neurosci. Res. 2002;69:1–9. doi: 10.1002/jnr.10270. [DOI] [PubMed] [Google Scholar]
- Lovgren, J., Rajakoski, K., Karp, M., Lundwall, A., Lilja, H. Activation of the zymogen form of prostate-specific antigen by human glandular kallikrein 2. Biochem. Biophys. Res. Commun. 1997;238:549–555. doi: 10.1006/bbrc.1997.7333. [DOI] [PubMed] [Google Scholar]
- Lu, W., Bhasin, M., Tsirka, S.E. Involvement of tissue plasminogen activator in onset and effector phases of experimental allergic encephalomyelitis. J. Neurosci. 2002;22:10781–10789. doi: 10.1523/JNEUROSCI.22-24-10781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer, A., Chen, Y.-T., Soraino, T.F., Kramp, W.J., Carlson, G., Cuny, C., Sharp, T., Smith, W., Petteway, J., Brawer, K.M., et al. Measurement of the proportion of free to total prostate-specific antigen improves diagnostic performance of prostate-specific antigen in the diagnostic gray zone of the total prostate-specific antigen. Urology. 1995;46:187–194. doi: 10.1016/s0090-4295(99)80192-7. [DOI] [PubMed] [Google Scholar]
- Lundstrom, A., Egelrud, T. Stratum corneum chymotryptic enzyme: A proteinase which may be generally present in the stratum corneum and with a possible involvement in desquamation. Acta Derm. Venereol. 1991;71:471–474. [PubMed] [Google Scholar]
- Luo, L.Y., Katsaros, D., Scorilas, A., Fracchioli, S., Bellino, R., van Gramberen, M., de Bruijn, H., Henrik, A., Stenman, U.H., Massobrio, M., et al. The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res. 2003;63:807–811. [PubMed] [Google Scholar]
- Malm, J., Hellman, J., Hogg, P., Lilja, H. Enzymatic action of prostate-specific antigen (PSA or hK3): Substrate specificity and regulation by Zn2+, a tight-binding inhibitor. Prostate. 2000;45:132–139. doi: 10.1002/1097-0045(20001001)45:2<132::aid-pros7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Memari, N., Jiang, W., Diamandis, E.P., Luo, L.Y. Enzymatic properties of human kallikrein-related peptidase 12 (KLK12) Biol. Chem. 2007;388:427–435. doi: 10.1515/BC.2007.049. [DOI] [PubMed] [Google Scholar]
- Michael, I.P., Sotiropoulou, G., Pampalakis, G., Magklara, A., Ghosh, M., Wasney, G.A., Diamandis, E.P. Biochemical and enzymatic characterization of human kallikrein 5 (hK5), a novel serine protease potentially involved in cancer progression. J. Biol. Chem. 2005;280:14628–14635. doi: 10.1074/jbc.M408132200. [DOI] [PubMed] [Google Scholar]
- Michael, I.P., Pampalakis, G., Mikolajczyk, S.D., Malm, J., Sotiropoulou, G., Diamandis, E.P. Human tissue kallikrein 5 (hK5) is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J. Biol. Chem. 2006;281:12743–12750. doi: 10.1074/jbc.M600326200. [DOI] [PubMed] [Google Scholar]
- Nicole, O., Docagne, F., Ali, C., Margaill, I., Carmeliet, P., Mackenzie, E.T., Vivien, D., Buisson, A. The proteolytic activity of tissue-plasminogen activator enhances NMDA receptor-mediated signaling. Nat. Med. 2001;7:59–64. doi: 10.1038/83358. [DOI] [PubMed] [Google Scholar]
- Oka, Y., Uchida, A., Aoyama, M., Fujita, M., Hotta, N., Tada, T., Katano, H., Mase, M., Asai, K., Yamada, K. Expression of myelencephalon-specific protease after cryogenic lesioning of the rat parietal cortex. J. Neurotrauma. 2005;22:501–510. doi: 10.1089/neu.2005.22.501. [DOI] [PubMed] [Google Scholar]
- Pampalakis, G., Sotiropoulou, G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim. Biophys. Acta. 2007;1776:22–31. doi: 10.1016/j.bbcan.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Pang, P.T., Teng, H.K., Zaitsev, E., Woo, N.T., Sakata, K., Zhen, S., Teng, K.K., Yung, W.-H., Hempstead, B.L., Lu, B. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Paul, J., Strickland, S., Melchor, J.P. Fibrin deposition accelerates neurovascular damage and neuroinflammation in mouse models of Alzheimer's disease. J. Exp. Med. 2007;204:1999–2008. doi: 10.1084/jem.20070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, A.J., Dong, Y., Hunt, M.L., Linn, M., Samaratunga, H., Clements, J.A., Hooper, J.D. Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J. Biol. Chem. 2008;283:12293–12304. doi: 10.1074/jbc.M709493200. [DOI] [PubMed] [Google Scholar]
- Recker, F., Kwiatkowski, M.K., Piironen, T., Pettersson, K., Huber, A., Lummen, G., Tscholl, R. Human glandular kallikrein as a tool to improve discrimination of poorly differentiated and non-organ-confined prostate cancer compared with prostate-specific antigen. Urology. 2000;55:481–485. doi: 10.1016/s0090-4295(99)00611-1. [DOI] [PubMed] [Google Scholar]
- Robert, M., Gibbs, B.F., Jacobson, E., Gagnon, C. Characterization of prostate-specific antigen proteolytic activity on its major physiological substrate, the sperm motility inhibitor precursor/semenogelin I. Biochemistry. 1997;36:3811–3819. doi: 10.1021/bi9626158. [DOI] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Towner, M.D., Isackson, P.J. Nervous system specific expression of a novel serine protease: Regulation in the adult rat spinal cord by excitotoxic injury. J. Neurosci. 1997;17:8156–8168. doi: 10.1523/JNEUROSCI.17-21-08156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Blaber, S.I., Lucchinetti, C.F., Genain, C.P., Blaber, M., Rodriguez, M. Activity of a newly identified serine protease in CNS demyelination. Brain. 2002;125:1283–1296. doi: 10.1093/brain/awf142. [DOI] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Blaber, S.I., Tingling, J.T., Rodriguez, M., Blaber, M., Christophi, G.P. Potential scope of action of tissue kallikreins in CNS immune-mediated disease. J. Neuroimmunol. 2006a;178:167–176. doi: 10.1016/j.jneuroim.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Sabharwal, P., Cruz, H., Larsen, N., Vandell, A., Blaber, S.I., Ameenuddin, S., Papke, L.M., Fehlings, M.G., Reeves, R.K., et al. Dynamic role of kallikrein 6 in traumatic spinal cord injury. Eur. J. Neurosci. 2006b;24:1457–1469. doi: 10.1111/j.1460-9568.2006.05021.x. [DOI] [PubMed] [Google Scholar]
- Scarisbrick, I.A., Linbo, R., Vandell, A.G., Keegan, M., Blaber, S.I., Blaber, M., Sneve, D., Lucchinetti, C.F., Rodriguez, M., Diamandis, E.P. Tissue kallikreins are associated with secondary progressive multiple sclerosis and promote neurodegeneration. Biol. Chem. 2008 doi: 10.1515/BC.2008.085. (in press). doi: abs/10.1515/BC.2008.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J., Naran, A., Misso, N.L., Rigby, P.J., Thompson, P.J., Bhoola, K.D. Expression of kallikrein-related peptidases (KRP/hK5, 7, 6, 8) in subtypes of human lung carcinoma. Int. Immunopharmacol. 2008;8:300–306. doi: 10.1016/j.intimp.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou, G., Rogakos, V., Tsetsenis, T., Pampalakis, G., Zafiropoulos, N., Simillides, G., Yiotakis, A., Diamandis, E.P. Emerging interest in the kallikrein gene family for understanding and diagnosing cancer. Oncol. Res. 2003;13:381–391. doi: 10.3727/096504003108748393. [DOI] [PubMed] [Google Scholar]
- Stamey, T.A., Yang, N., Hay, A.R., McNal, J.E., Freiha, F.S., Redwine, E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 1987;15:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- Takayama, T.K., Fujikawa, K., Davie, E.W. Characterization of the precursor of prostate-specific antigen. Activation by trypsin and by human glandular kallikrein. J. Biol. Chem. 1997;272:21582–21588. doi: 10.1074/jbc.272.34.21582. [DOI] [PubMed] [Google Scholar]
- Takayama, T.K., Carter, C.A., Deng, T. Activation of prostate-specific antigen precursor (pro-PSA) by prostin, a novel human prostatic serine protease identified by degenerate PCR. Biochemistry. 2001a;40:1679–1687. doi: 10.1021/bi002129r. [DOI] [PubMed] [Google Scholar]
- Takayama, T.K., McMullen, B.A., Nelson, P.S., Matsumura, M., Fujikawa, K. Characterization of hK4 (prostase), a prostate-specific serine protease: activation of the precursor of prostate specific antigen (pro-PSA) and single-chain urokinase-type plasminogen activator and degradation of prostatic acid phosphatase. Biochemistry. 2001b;40:15341–15348. doi: 10.1021/bi015775e. [DOI] [PubMed] [Google Scholar]
- Terayama, R., Bando, Y., Takahashi, T., Yoshida, S. Differential expression of neuropsin and protease M/neurosin in oligodendrocytes after injury to the spinal cord. Glia. 2004;48:91–101. doi: 10.1002/glia.20058. [DOI] [PubMed] [Google Scholar]
- Terayama, R., Bando, Y., Yamada, M., Yoshida, S. Involvement of Neuropsin in the pathogenesis of experimental autoimmune encephalomyelitis. Glia. 2005;52:108–118. doi: 10.1002/glia.20226. [DOI] [PubMed] [Google Scholar]
- Tsirka, S.E., Gualandris, A., Amaral, D.G., Strickland, S. Excitotoxin induced neuronal degeneration and seizure are mediated by tissue plasminogen activator. Nature. 1995;377:340–344. doi: 10.1038/377340a0. [DOI] [PubMed] [Google Scholar]
- Vaisanen, V., Lovgren, J., Hellman, J., Piironen, T., Lilja, H., Pettersson, K. Characterization and processing of prostate specific antigen (hK3) and human glandular kallikrein (hK2) secreted by LNCaP cells. Prostate Cancer Prostatic Dis. 1999;2:91–97. doi: 10.1038/sj.pcan.4500289. [DOI] [PubMed] [Google Scholar]
- Wagner, S., Kalb, P., Lukosava, M., Hilgenfeldt, U., Schwaninger, M. Activation of the tissue kallikrein-kinin system in stroke. J. Neurol. Sci. 2002;202:75–76. doi: 10.1016/s0022-510x(02)00208-3. [DOI] [PubMed] [Google Scholar]
- Yoon, H., Laxmikanthan, G., Lee, J., Blaber, S.I., Rodriguez, A., Kogot, J.M., Scarisbrick, I.A., Blaber, M. Activation profiles and regulatory cascades of the human kallikrein-related proteases. J. Biol. Chem. 2007;282:31852–31864. doi: 10.1074/jbc.M705190200. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Diamandis, E.P. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr. Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Magklara, A., Diamandis, E.P. KLK12 is a novel serine protease and a new member of the human kallikrein gene family-differential expression in breast cancer. Genomics. 2000;69:331–341. doi: 10.1006/geno.2000.6346. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Kapadia, C., Polymeris, M.E., Borgono, C., Hutchinson, S., Wasney, G.A., Soosaipillai, A., Diamandis, E.P. The human kallikrein protein 5 (hK5) is enzymatically active, glycosylated and forms complexes with two protease inhibitors in ovarian cancer fluids. Biochim. Biophys. Acta. 2003a;1628:88–96. doi: 10.1016/s0167-4781(03)00116-7. [DOI] [PubMed] [Google Scholar]
- Yousef, G.M., Polymeris, M.E., Grass, L., Soosaipillai, A., Chan, P.C., Scorilas, A., Borgono, C., Harbeck, N., Schmalfeldt, B., Dorn, J., et al. Human kallikrein 5: A potential novel serum biomarker for breast and ovarian cancer. Cancer Res. 2003b;63:3958–3965. [PubMed] [Google Scholar]
- Zarghooni, M., Soosaipillai, A., Grass, L., Scorilas, A., Mirazimi, N., Diamandis, E.P. Decreased concentration of human kallikrein 6 in brain extracts of Alzheimer's disease patients. Clin. Biochem. 2002;35:225–231. doi: 10.1016/s0009-9120(02)00292-8. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Seiffert, D., Fowler, B.J., Jenkins, G.R., Thinnes, T.C., Loskutoff, D.J., Parmer, R.J., Miles, L.A. Plasminogen has a broad extrahepatic distribution. Thromb. Haemost. 2002;87:493–501. [PubMed] [Google Scholar]