Abstract
Adrenal cytochrome b561 (cyt b561), a transmembrane protein that shuttles reducing equivalents derived from ascorbate, has two heme centers with distinct spectroscopic signals and reactivity towards ascorbate. The His54/His122 and His88/His161 pairs furnish axial ligands for the hemes, but additional amino acid residues contributing to the heme centers have not been identified. A computational model of human cyt b561 (Bashtovyy, D., Berczi, A., Asard, H., and Pali, T. (2003) Protoplasma 221, 31–40) predicts that His92 is near the His88/His161 heme and that His110 abuts the His54/His122 heme. We tested these predictions by analyzing the effects of mutations at His92 or His110 on the spectroscopic and functional properties. Wild type cytochrome and mutants with substitutions in other histidine residues or in Asn78 were used for comparison. The largest lineshape changes in the optical absorbance spectrum of the high-potential (bH) peak were seen with mutation of His92; the largest changes in the low-potential (bL) peak lineshape were observed with mutation of His110. In the EPR spectra, mutation of His92 shifted the position of the g=3.1 signal (bH) but not the g=3.7 signal (bL). In reductive titrations with ascorbate, mutations in His92 produced the largest increase in the midpoint for the bH transition; mutations in His110 produced the largest decreases in ΔA561 for the bL transition. These results indicate that His92 can be considered part of the bH heme center, and His110 part of the bL heme center, in adrenal cyt b561.
1. Introduction
Adrenal cytochrome (cyt1) b561 was first observed in bovine adrenal medullae [1] and later found in several other tissues [2–6]. Cyt b561 spans the chromaffin granule (CG) membrane in adrenal medullae [7, 8] and the cytochrome is believed to reduce monodehydroascorbate in the CG lumen (matrix) at the expense of cytoplasmic ascorbate [9], thus providing reducing equivalents for lumenal monooxygenases such as dopamine β-hydroxylase [10]. Genetic analyses have revealed that the adrenal cyt b561 is the prototype of a large protein family distributed widely in vertebrate, invertebrate and plant tissues [11].
Native cyt b561 has two molecules of heme per polypeptide [12, 13], and two major forms of ferric heme are apparent in the EPR spectra of cyt b561 in intact CG membranes and in the purified, detergent solubilized cytochrome [12–16]. One is a conventional low spin species with a signal at g = 3.1 that has a redox potential of ~160 mV (the “high-potential” or bH heme center), the other is a highly-axial low spin (HALS) species with a signal at g = 3.7 that has a redox potential of ~60 mV (the “low-potential” or bL heme center) [14, 17, 18]. Spectroscopic and mutagenic studies [12–16, 19–21] have supported earlier models [19, 22, 23] for adrenal cyt b561 in which each heme has two histidines as axial ligands, with the His54/His122 pair (residue numbering follows the system of Okuyama et al. [22] ) furnishing ligands for one heme and the His88/His161 pair furnishing ligands for the other heme. These four histidines in the bovine adrenal cytochrome are completely conserved among cyt b561 family members [11, 24]. Our recent mutagenic study of histidines 54, 88, 122 and 161 in bovine cyt b561 [15] are consistent with their roles as axial ligand to cyt b561 hemes.
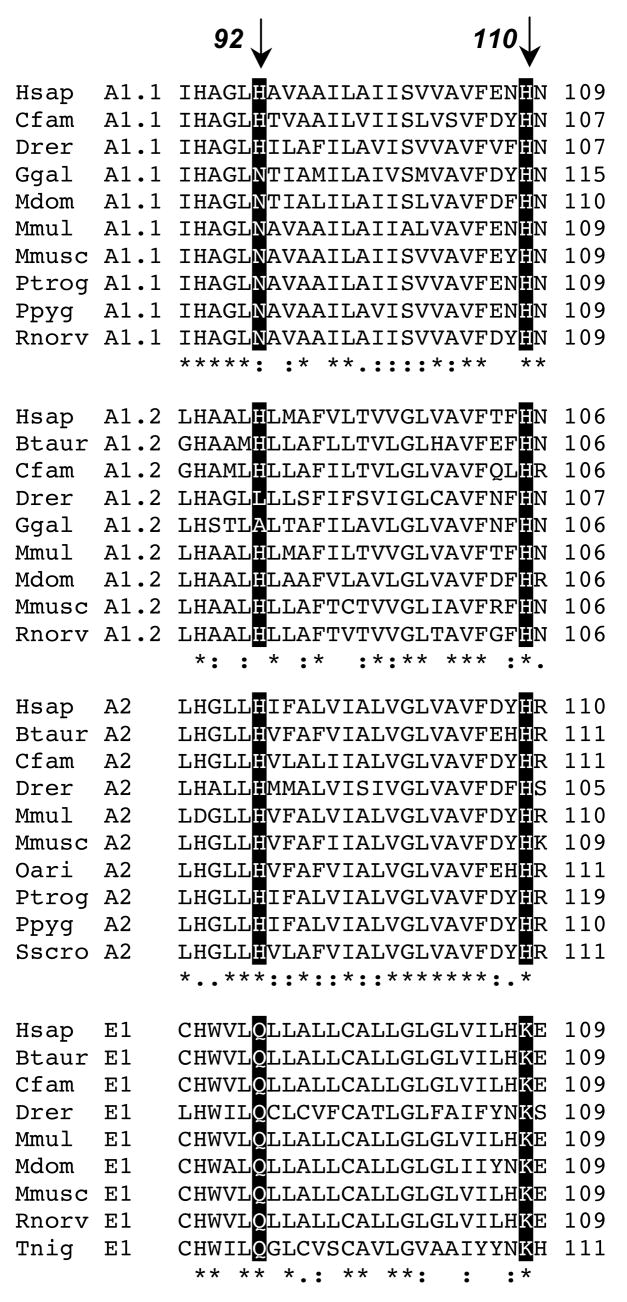
Besides the axial ligands to heme, bovine adrenal cyt b561 has three other histidine residues, at positions 92, 109, and 110. His109 is not conserved in the four vertebrate subfamily groups corresponding to the four major cyt b561 isoforms identified so far in humans (Fig. 1). Both His92 and His110 are fully conserved in the subset of vertebrate subfamily A members that we term group A2; this group includes adrenal cyt b561. Group A1.1 members, which include human duodenal cyt b561 [25], all have histidine at position 110 but most have asparagine at position 92. In group A1.2 members, His110 is again fully conserved but His92 is not. In group E1, which includes the putative human tumor suppressor protein (101F6) [26], position 92 is fully conserved as a glutamine and position 110 is fully conserved as a lysine. This pattern of generally strong conservation, whether as histidine or as another amino acid, within the subgroups at positions 92 and 110 suggests that the sidechain structures at these positions have some specialized functional importance.
Figure 1.
Sequence alignment for four human cyt b561 family members with analogous sequences from other vertebrate species. The classification scheme of Tsubaki et al. [11] recognized twelve cyt b561 subfamilies among eukaryotes. Sequence analysis by ClustalX [59] of currently known human cyt b561 family members indicates that three of the human proteins (termed A1.1, A1.2 and A2) are in subfamily A and one (E1) is in subfamily E described by Tsubaki et al. [11]; we have subdivided the original subfamiles A and E to reflect the divergent sequence patterns among their members. The shaded boxes and arrows indicate the residues corresponding to His92 and His110 in bovine adrenal cyt b561 (Btaur A2). Subfamily A1.1 includes duodenal cyt b561, subfamily E1 includes the putative tumor suppressor protein (101F6). Hsap, Homo sapiens; Btaur, Bos taurus; Cfam, Canis familiaris; Drer, Danio rerio; Ggal, Gallus gallus; Mdom, Monodelphia domesticus; Mmul, Macaca mulatta; Mmusc, Mus musculus; Oari, Ovis aries; Ptrog, Pan troglodytes; Ppyg, Pongo Pygmaeus; Rnorv, Rattus norvegicus; Sscro, Sus scrofa; Tnig, Tetraodon nigriviridis.
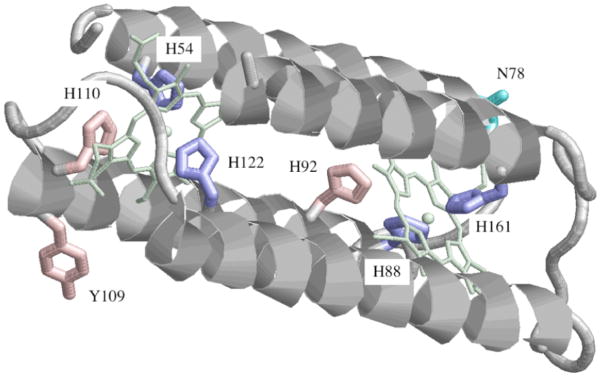
Crystallographic information is not available for cyt b561 but a computational model has been developed for the four transmembrane helices carrying the axial ligands for the two hemes [23]. This model was based on conservation, among the 26 members of the b561 family known at the time, of four histidine residues presumed to be ligands to the two hemes and of six predicted transmembrane segments. The modeling of the central four helices was constrained by the requirement for ligation of the hemes by the conserved histidines and for outward orientation of the helix faces predicted to be lipid-exposed. One remarkable prediction of this structural model is that three, and not two, histidine residues flank each heme (Fig. 2). Thus, His110 is predicted to be close to His54 near the face of one heme and His92 close to His88 near the face of the second heme. Such an arrangement suggests that His92 and/or His110 might be additional components of the heme centers in cyt b561. To evaluate this possibility, we have constructed cDNAs encoding substitutions of His92 or His110 in the bovine cytochrome. The corresponding mutant proteins, along with mutants in the five other histidines for comparison, were expressed in a newly developed bacterial system [27]. The recombinant proteins were examined to evaluate the effects of the mutations on expression levels, spectroscopic characteristics and functional interaction with the physiological reductant, ascorbate. The results indicate that His92 and His110 can indeed be considered components of the heme centers, namely the bH heme for His92 and the bL heme for His110. These data provide independent evidence for a revised topology of hemes in cyt b561 [15] in which the high-potential heme on the cytoplasmic surface of the chromaffin membrane serves as a donor of electrons for the low-potential heme on the lumenal side.
Figure 2.
Computational model of helices 2–5 of human cyt b561 [23] showing the residues targeted for mutagenesis. Y109 in human cyt b561 is a histidine in the bovine cytochrome. The heme center coordinated by His 54 and His122 is oriented toward the lumenal side of the CG membrane, and the heme coordinated by His 88 and His161 is oriented toward the cytoplasmic side [39, 40].
2. Materials and methods
Materials
Hemin, sodium ascorbate, δ-aminolevulinic acid, isopropyl-1-thio-β-D-galactopyranoside, guanidine, ampicillin, chloramphenicol, Sephacryl 300 HR beads, bovine pancreatic trypsin, soybean trypsin inhibitor type I-S, egg lysozyme and the redox dyes were from Sigma (St. Louis, MO). n-Dodecyl-β-D-maltoside was from Anatrace (Maumee, OH). Restriction enzymes and other DNA modifying enzymes were purchased from New England BioLabs (Beverly, MA). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Reagents for DNA extraction and purification were from Qiagen (Valencia, CA). Immunoblotting reagents were from Bio-Rad (Hercules, CA). Plasmid vector pET43.1a, Benzonase nuclease, rLysozyme Solution, and Protease Inhibitor Cocktail Set III (without EDTA) were purchased from Novagen (Madison, WI). Escherichia coli strains DH5α and BL21Star(DE3), and anti-His(C-term) monoclonal antibody were from Invitrogen (Carlsbad, CA). The chaperone plasmid pT-groE was generously provided by Dr. Lee-Ho Wang (University of Texas Health Science Center at Houston). TALON metal affinity resin was purchased from BD Biosciences Clontech (Palo Alto, CA). DNA sequencing was performed at Microbiology and Molecular Genetics Core Facility, University of Texas Health Science Center at Houston (Houston, TX).
Constructs for recombinant, His-tagged wild type and mutant cyt b561
Construction of a pET43.1a plasmid containing sequence coding for bovine adrenal cyt b561, designated pET43.1a-b561C6H, are described elsewhere [27]. Point mutations were introduced using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA) and the pET43.1a-b561C6H expression plasmid as the template. The mutagenic primers, together with their complementary (antisense) strands, were used in a PfuTurbo polymerase-initiated reaction. The original, methylated DNA strand was digested with DpnI, and the remaining DNA was transformed into E. coli XL-10 competent cells. The mutated clones were verified by DNA sequencing.
Expression and purification of His-tagged wild type and mutant cyt b561
Procedural details are described elsewhere [27]. Briefly, the BL21Star(DE3)/pT-groE E. coli strain was transformed with the desired expression plasmid and grown in the presence of heme, δ-aminolevulinic acid, and isopropyl-1-thio-β-D-galactopyranoside for 24 hr at 20 °C. The recombinant cytochrome was solubilized from the membrane fraction by stirring overnight at 0–4°C in 0.1 M potassium phosphate (pH 7.2) containing 5% glycerol and ~1.5 g DM/g total protein. Unextracted material was pelleted by centrifugation and the pH of the crude detergent extract was adjusted to 7.3–7.5. Purification of recombinant protein used chromatography on TALON Resin (BD Biosciences Clontech) and Sephacryl 300 HR [27]. The procedure resulted in ~90% electrophoretically pure recombinant cytochrome except for mutants at residues 54, 88, 122, and 161. To control for potential contributions of endogenous bacterial proteins in these less-purified mutants, detergent extracts of membranes from control bacteria were subjected to the same chromatographic steps and brought to a similar protein concentration as the recombinant cyt b561 samples for spectroscopic evaluation.
Assay of protein, cyt b561, and heme content
Total protein was assayed with a modified Lowry method using bovine albumin as the standard [28]. Recombinant wild type cyt b561 content was calculated using the reduced (dithionite-treated) minus oxidized (ferricyanide-treated) difference absorption coefficients (561–575 nm) determined for the individual purified wild type and mutant proteins. Dot blot assay [29] of recombinant cyt b561 used antibody against the His tag, with purified wild type protein as the standard. Heme content was determined by the pyridine hemochrome method [30], using a difference absorption coefficient (556–538 nm) of 24.5 mM−1 cm−1 [31].
Electrophoretic and immunoblot analyses
The general procedures have been described [13, 27]. Levels of recombinant cyt b561 mutant protein in partially purified samples were determined by dot blot assay [29] using a monoclonal antibody against a C-terminal His tag (Invitrogen) and a standard solution of homogeneous N78K mutant. The densitometry value for each dot was corrected for the adjacent background. The dot blot assay typically has a useful range of 0.5–200 ng of target protein.
Electronic absorption spectroscopy
Recombinant cytochrome samples purified by metal ion chromatography (in most cases followed by gel filtration chromatography) were oxidized by incubation with 0.1 mM potassium ferricyanide and then chromatographed on a 10DG desalting column (BioRad) in 0.1 M potassium phosphate, pH 7.2, containing 0.075% DM and 20% glycerol. Absorbance spectra were recorded at room temperature in 0.1 M potassium phosphate, pH 7.2, containing 0.075% DM and 10% glycerol in 1 cm pathlength quartz cuvettes in a Shimadzu Model 2101PC spectrophotometer. Spectra were scanned at 20 nm/min, with 20 data points/nm and a spectral band width of 0.5 nm. The approximation of the difference spectra of high- and low-potential heme centers by three Gaussian components was done with Origin 6.1 (OriginLab, Northampton, MA).
Ascorbate titrations of cyt b561 and mutants
For ascorbate titrations, the pre-oxidized cytochrome b561 sample (~ 4 μM) in 100 mM potassium phosphate buffer (pH 7.2) containing 0.075% (w/v) n-dodecyl-β-D-maltoside was placed in a 1-cm pathlength quartz cuvette and titrated with ascorbate. The standard titration uses 17 levels of ascorbate: 0, 0.13, 0.40, 0.80, 2.13, 4.78, 8.70, 21.9, 48.3, 87.7, 219, 481, 875, 2180, 4780, 9960, and 20200 μM. The system was allowed to equilibrate completely after each addition of ascorbate before the spectrum was recorded; ~5 min was found to be sufficient. All ascorbate stock solutions were freshly made, the pH adjusted to 7.2, and kept on ice and protected from light. The extent of reduction was calculated from the increase in A561, corrected for dilution. Some ascorbate titrations were performed under anaerobic conditions in a sealed titrator [32].
Potentiometric titrations
The procedure for potentiometric titrations of the ferric/ferrous heme couples in recombinant cyt b561 has been described previously [27]. Data for the fraction of maximal change in A561 (F) as a function of the redox potential (E) was fitted to the equation: F = (1 − FH)/(1 + 10^((E − Em(bL))/59)) + FH/(1 + 10^((E − Em(bH))/59)), where FH and (1 − FH) are the fractions of the maximal absorbance change associated with the high- and low-potential hemes, respectively, Em(bL) and Em(bH) are the midpoint potentials for the low- and high-potential hemes, respectively.
EPR spectroscopy
Each sample was concentrated by ultrafiltration, oxidized with ferricyanide and the buffer was exchanged to 0.1 M potassium phosphate, pH 7.2, containing 18% glycerol and 0.08% DM by chromatography on a 10-DG column before transfer to a quartz EPR tube and freezing in a dry ice/acetone bath. The samples were stored in liquid nitrogen until the EPR spectra were recorded with a Bruker EMX EPR spectrometer. The liquid helium system included an Oxford GFS 600 transfer line, an ITC 503 temperature controller, and an ESR 900 cryostat.
3. Results
Expression efficiency of wild type and mutant cyt b561
Expression vectors were constructed for wild type cyt b561 and for mutants with substitutions at His92 and His110. For comparison, mutants were also made in each of the other five histidine residues in the bovine cytochrome (residues 54, 88, 109, 122, and 161). As a control, an Asn ---> Lys substitution was made at residue 78, which is oriented away from both hemes in the 3D model (Fig. 2). Glutamine and tyrosine substitutions were made for His92 and His110, and a tyrosine substitution was made for His109. Four substitutions were designed for the histidines identified as axial ligands (residues 54, 88, 122 and 161): Gln, Leu, Tyr and Met.
The initial results indicated that the level of expression in the bacterial system was not sensitive to which amino acid replaced histidine (data not shown), and the glutamine mutants were chosen for a quantitative comparison of expression efficiency. The molecular mass of each recombinant cyt b561 protein was examined by immunoblot and the level of recombinant protein in whole cell lysates and in detergent extracts was quantified by a dot blot assay. The amount of ascorbate-reducible b-type cytochrome in the extracts was also determined as a measure of expression of functional protein.
An immunoreactive band with the same electrophoretic mobility as the wild type cytochrome, indicating expression of the full-size recombinant protein, was seen for each of the mutants except H54Q and H122Q (data not shown); the latter two recombinant proteins were, however, detected by the more sensitive dot blot assay (Table 1). The data in Table 1 also show that the wild type cytochrome and the negative control mutant (N78K) were expressed at high levels, fully extractable with detergent, and almost entirely functional (i.e., reduced by ascorbate). The H92Q, H110Q and H109Y mutants were also well expressed, and detergent extractable in functional form. The higher value for functional cytochrome than for recombinant protein with the H110Q mutant probably reflects slightly different responses for the immunological and spectroscopic assays. Recombinant protein expression was much lower (and a lower fraction was extractable) for the mutants at His 54, 88, 122 and 161 (Table 1), indicating that these four histidine residues are important for efficient expression of cyt b561. Almost identical, and very low, levels of expression were found for mutants at His 54 and 122, suggesting that these two residues function as a pair, consistent with their roles as axial ligands to one of the hemes (Fig. 2) [15]. Almost identical and somewhat higher levels of expression were found for the mutants at His 88 and 161, suggesting that these two residues function as a second pair, consistent with their identification as axial ligands for the second heme (Fig. 2) [15].
Table 1.
Expression of wild type and mutant cyt b561 in the bacterial system
| Construct | Total recombinant cyt b561 proteina (nmol/L culture) | Extractable recombinant cyt b561 proteinb (nmol/L culture) | Functional cyt b561c (nmol/L culture) |
|---|---|---|---|
| Wild type | 162 (100%) | 169 (100%) | 134 (100%) |
| N78K | 254 (157%) | 247 (146%) | 227 (169%) |
| H54Q | 1.4 (0.9%) | NDd | NDd |
| H88Q | 41 (25%) | 9.3 (5.5%) | NDd |
| H92Q | 78 (48%) | 69 (41%) | 52 (39%) |
| H109Y | 128 (79%) | 116 (69%) | 117 (87%) |
| H110Q | 120 (74%) | 115 (68%) | 152 (113%) |
| H122Q | 1.7 (1.0%) | NDd | NDd |
| H161Q | 40 (25%) | 13 (7.7%) | NDd |
Immunoreactive protein in cell lysate determined by dot blot assay; the percentage of the wild type value is given in parentheses.
Immunoreactive protein in detergent extract determined by dot blot assay; the percentage of the wild type value is given in parentheses.
Ascorbate-reducible cytochrome in detergent extract calculated from A561(red)-A575(ox) and the difference extinction coefficients determined for the individual purified proteins; the percentage of the wild type value is given in parentheses.
Not detected.
Effects of His92 and His110 mutations on heme content, absorbance spectra and ascorbate reactivity
Each recombinant protein, except the mutants at His54 and His122, was purified by metal ion affinity chromatography followed by size exclusion chromatography before characterization of their heme content, absorbance spectra and ascorbate reactivity. The heme contents of wild type cyt b561 and the negative control N78K mutant were 1.94 and 1.91 heme/monomer, respectively (Table 2). This is in good agreement with values obtained for the endogenous cytochrome purified from CG [12] and for the recombinant cytochrome expressed in insect and yeast cell systems [13]. The heme content of the glutamine and tyrosine mutants at His92 averaged 1.88 heme/monomer and that of the glutamine and tyrosine mutants at His110 averaged 1.92 heme/monomer (Table 2). The tyrosine substitution at His109 had 1.87 heme/monomer. Each of these recombinant proteins thus carried a full complement of the prosthetic group.
Table 2.
Optical spectral parameters and heme stoichiometry of oxidized (ox) and dithionite-reduced (red) states of wild type and mutant cyt b561a.
| WT | N78K | H88Q | H88Y | H92Q | H92Y | H109Y | H110Q | H110Y | H161Q | H161Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASoret/hemeb(ox) | 122 (415) | 113 (415) | 110 (411) | 120 (408) | 120 (416) | 110 (416) | 127 (415) | 115 (415) | 110 (415) | 110 (411) | 130 (414) |
| ASoret/hemeb(red) | 187 (427) | 174 (427) | 120 (427) | 130 (425) | 196 (428) | 183 (428) | 197 (427) | 171 (427) | 159 (427) | 130 (427) | 110 (427) |
| Abeta/hemeb(red) | 14 (530) | 14 (530) | 14 (530) | 14 (536) | 15 (530) | 14 (530) | 16 (529) | 14 (529) | 14 (530) | 14 (531) | 16 (530) |
| Aalpha/hemeb(red) | 30 (561) | 30 (561) | 16 (561) | 16 (561) | 31 (562) | 31 (561) | 33 (560) | 29 (561) | 26 (561) | 17 (560) | 17 (560) |
| ASoret/A280(ox) | 3.2 | 3.3 | nde | Nd | 2.9 | 3.0 | 3.1 | 3.3 | 2.8 | nd | Nd |
| ASoret/A280(red) | 4.9 | 5.2 | nd | Nd | 4.7 | 4.8 | 4.7 | 4.9 | 4.4 | nd | Nd |
| A561(red)-A575(ox)c | 54.4 | 48.9 | nd | Nd | 48.1 | 54.1 | 53.3 | 49.7 | 40.5 | nd | Nd |
| Heme/monomerd | 1.94 | 1.91 | 1.3 | 0.9 | 1.78 | 1.98 | 1.87 | 1.98 | 1.85 | 1.3 | 1.6 |
Values represent averages from at least two measurements. Purified protein was used except for the His88 and His161 mutants.
Extinction coefficient ((mM heme)−1cm−1); heme determined by pyridine hemochrome assay; peak position (nm) in parentheses.
Difference extinction coefficient ((mM monomer)−1cm−1)
Heme determined by pyridine hemochrome; recombinant protein determined by assay of total protein and densitometry of electrophoretic gels except for His88 and His161 mutants, where recombinant protein was determined by immunoblot.
Not determined.
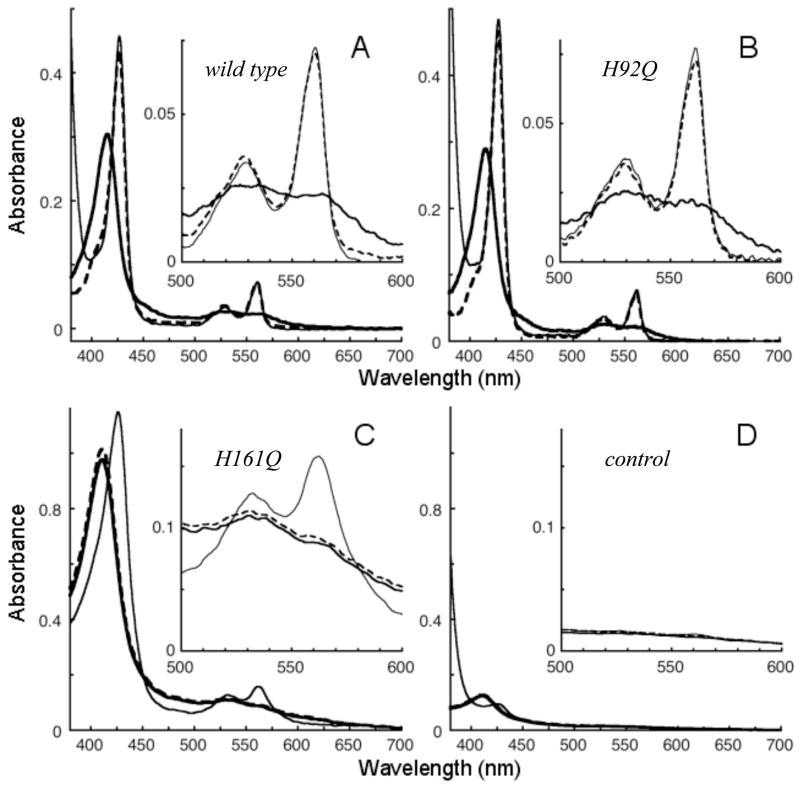
The absorbance spectrum of recombinant wild type cyt b561 is shown in Fig. 3A. The ferric cytochrome has a prominent Soret peak at 415 nm with an absorbance coefficient of 122 (mM heme)−1cm−1 (Table 2). Note that heme was used as the basis for the absorbance coefficients to facilitate comparison among mutants with differing heme stoichiometries (see below). Reduction with ascorbate shifted the Soret peak to 427 nm and produced alpha and beta bands at 561 and 530 nm. Further addition of dithionite increased the intensity of the Soret band by only a few percent, confirming full reduction by ascorbate. In this initial screening, recombinant cyt b561 with mutations at His92, His110, or His109 displayed optical properties similar to those of the wild type cytochrome (Table 2 and Fig. 3B). Each of these mutants was essentially completely reduced by addition of 17–20 mM ascorbate. The retention of grossly native optical properties and functional interaction with ascorbate confirm that His92, His110, and His109 do not furnish axial ligands to either of the hemes in cyt b561. The oxidized and reduced absorbance spectra for N78K, the negative control, were essentially the same as those of the wild type cytochrome (data not shown).
Figure 3.
Absorbance spectra of wild type and mutant cyt b561. Oxidized (thick line), ascorbate (17–21 mM) treated (dotted line), and dithionite treated (thin line) spectra are shown for wild type (A; 2.4 μM heme), H92Q (B; 2.5 μM heme), and H161Q (C; 8.9 μM heme, 1.55 mg total protein/mL). The control spectra (D) are from a detergent extract of control membranes processed through the same purification procedures as the other samples, with a total protein concentration of 1.64 mg/mL.
The heme content for two mutants at His88 and two mutants at His161 ranged from 0.9 to 1.6 heme/monomer (Table 2). This indicates a loss of some heme from one or both sites. In the oxidized state, each of the solubilized and partially purified mutants at His88 or His161 had a prominent Soret peak (spectra for H161Q are shown as examples in Fig. 3C), but the peak was shifted to shorter wavelengths than with the wild type (Table 2). This blue-shift could reflect heterogeneity in the low-spin species as well as a low-spin to high-spin conversion for a portion of the heme. In this connection, the EPR spectra of the His88 and His161 mutants exhibited prominent high spin signals near g = 6 (data not shown). Although not quantitated rigorously, comparison of the EPR spectra with those of high-spin and low-spin myoglobin derivatives suggest that the amount of high-spin heme in the His88 and His161 mutants is substantial, conceivably representing one-half of the endogenous heme. However, as both the g=3.7 and g=3.1 signals are modified in these mutant proteins (see below and Fig. 4B) the high-spin heme signal could arise from either heme center or a mixture of both.
Figure 4.
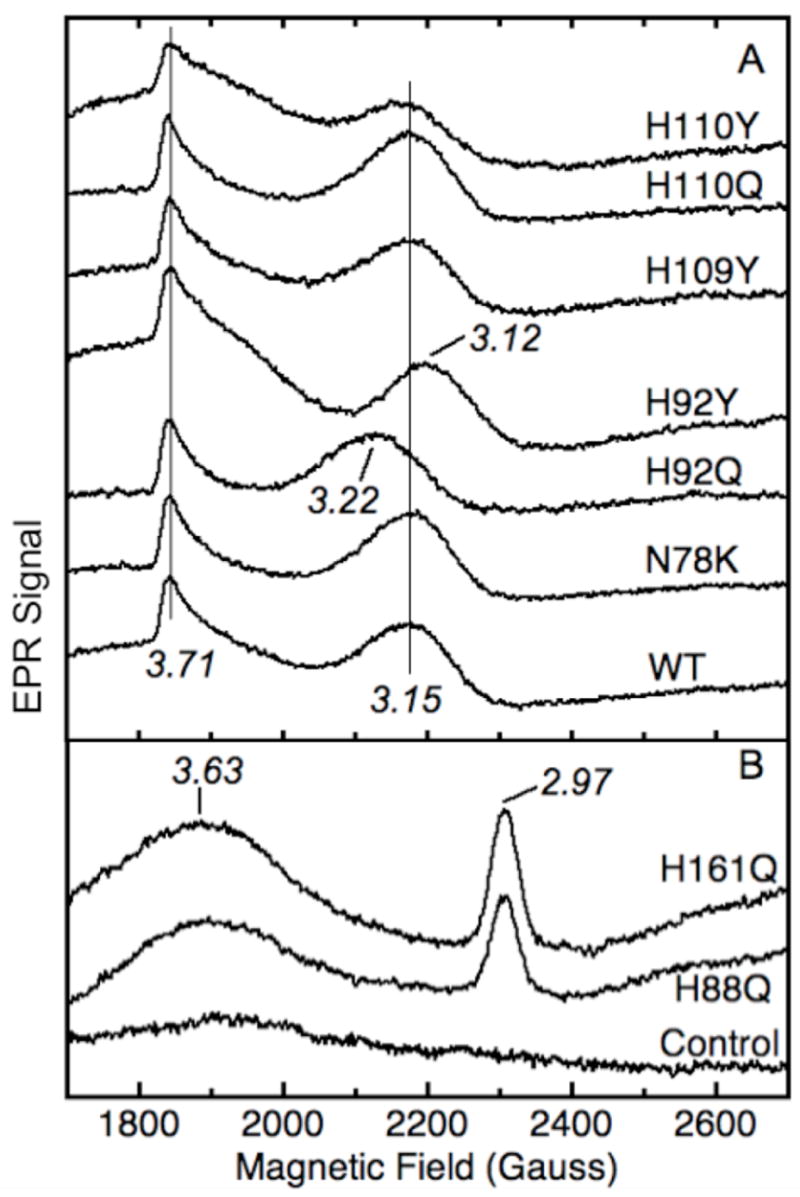
EPR spectra of low spin signals from purified wild type and mutant cyt b561. Panel A: homogeneous samples of wild type (WT, 143 μM heme), N78K (163 μM heme), H92Q (118 μM heme), H92Y (132 μM heme), H109Y (112 μM heme), H110Q (155 μM heme) and H110Y (106 μM heme). The signal amplitudes are normalized to the heme concentration and the g values are indicated. Panel B: partially purified samples of H88Q (69 μM heme, 8.7 mg total protein/mL), H161Q (61 μM heme, 9.3 mg total protein/mL), and a detergent extract from control membranes subjected to the same purification protocol (8.2 mg total protein/mL). The spectrometer conditions were: frequency, 9.60 GHz; modulation amplitude, 10.9 G; power, 4.0 mW; and temperature, 4 K.
None of the mutants at His88 or His161 was reduced significantly by ascorbate. Each of the His88 and His161 mutants was reduced by dithionite, but the absorption coefficients of the Soret and alpha bands were considerably less than those of the wild type (Table 2). The alpha band reveals the presence of low-spin species, while the decrease in absorption coefficient and the increased width of the Soret band suggests that these mutants comprise a mixture of low-spin species with different positions of the absorbance maximum, although the presence of some high-spin species is apparent in the EPR spectra. The optical background in host cell membrane extract put through the same purification steps was negligible (Fig. 3D), confirming that the absorbance spectra of the His88 and His161 mutants originate from the heme in the recombinant cytochromes and not from any co-purifying endogenous bacterial proteins.
Effects of His92 and His110 mutations on low spin EPR signals of cyt b561
The EPR spectrum of wild type cyt b561 purified from bacterial cells (Fig. 4A) shows prominent low spin ferric heme signals at g = 3.71 from the low-potential center (bL) and at g = 3.15 from the high-potential heme center (bH) [13–16, 33]. The high spin ferric heme signal at g = 6 and that of “adventitious” ferric iron at g = 4.3 are quite small (data not shown). Mutation of His92 perturbed the high-potential (bH) signal, with a shift to g = 3.12 in H92Y and a shift to g = 3.22 in H92Q (Fig. 4A). This indicates that the sidechain of residue 92 influences the geometry of the bH heme center. The proximity of His92 to the heme coordinated by the His88/His161 pair in the amino acid sequence and in the structural model (Fig. 2) suggests that this heme is the bH center. In contrast, the EPR spectra of both His110 mutants and the N78K and H109Y mutants are similar to that of the wild type (Fig. 4A). An important point here is that the retention of EPR signals for the bH and bL heme centers in all mutants at His92, His109, and His110 is further reason to rule out these histidines as axial ligands to the cyt b561 hemes.
EPR spectra were also recorded for partially purified recombinant cyt b561 mutants at His88 or His161 (Fig. 4B). In each of the four mutants, both the bH signal at g = 3.15 and the bL signal at g = 3.71 disappeared and were replaced by a broad, diffuse signal centered at g = 3.6 that is unlikely to represent a single species, and a signal at g = 2.97 that is probably a relaxed form of bis-histidine heme [15]. A control membrane extract subjected to the same purification procedure as the mutant extracts had negligible EPR signals in the range examined, demonstrating that these EPR signals originated from the recombinant cytochrome and not from endogenous bacterial host protein. These results support the assignment of His88 and His161 as axial ligands to one of the heme centers [15, 19, 22, 34] and further indicate that there is a structural linkage between the two heme centers so that disruption of either axial ligand at the His88/His161 bH heme center perturbs the bL heme center as well [15].
Effects of mutations of His92 and His110 mutations on cyt b561 reduction by ascorbate
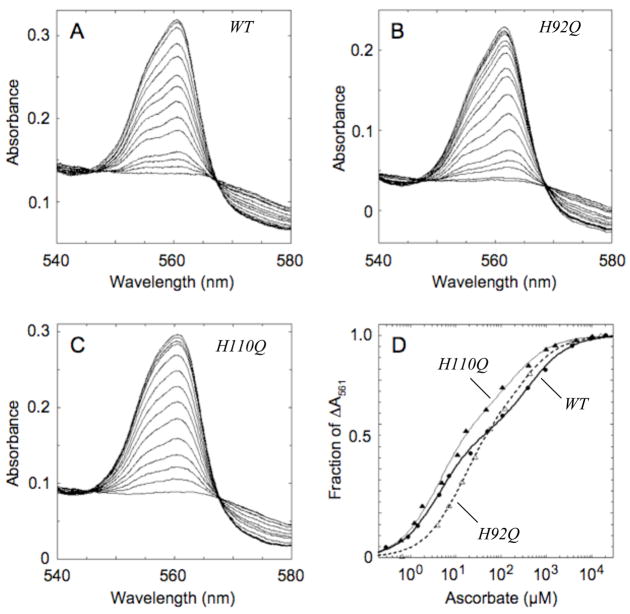
The reaction of ascorbate with cyt b561 mutants at His92, His110 and His109 was examined in more detail by following the increases in alpha band intensity during titrations with ascorbate (examples are shown in Fig. 5A–C). In each case, the increase in A561, expressed as a fraction of the overall absorbance increase at the end of the titration, appeared to go through two transitions (Fig. 5D). The first transition occurs at a low ascorbate level, reflecting reduction of the high-potential (bH) heme center; this midpoint ascorbate concentration is denoted as CH. The second transition occurs at higher ascorbate levels, reflecting reduction of the low-potential (bL) heme center; this midpoint ascorbate concentration is denoted CL. Fitting each titration as the sum of two hyperbolae yields estimates of the two apparent equilibrium constants (CH and CL), each of them a yet–to-be resolved composite of the redox and ascorbate binding properties of the two heme centers, and the fraction of the overall absorbance change associated with the high-potential transition (FH) and that associated with the low-potential transition (1−FH) (Table 3). The wild type cytochrome had average CH and CL values of 5.4 and 369 μM ascorbate, respectively, with 59% of the overall A561 associated with the first transition.
Figure 5.
Ascorbate titrations of recombinant cyt b561. Optical spectra were recorded during individual ascorbate titrations of wild type cyt b561 (A; 7.8 μM heme) and the H92Q (B; 7.8 μM heme) and H110Q (C; 9.4 μM heme) mutants. The fractional increases in A561 are plotted as a function of the ascorbate concentration in panel D for wild type (WT; filled circles), H92Q (open triangles) and H110Q (filled triangles). The lines are the non-linear regression fits to the equation given in Table 3.
Table 3.
Ascorbate titration parameters for wild type and mutant cyt b561 proteins
| Cyt b561a | FHb | CHb(μM) | CLb(μM) |
|---|---|---|---|
| Wild type (n=7) | 0.59 ± 0.03 | 5.4 ± 1.5 | 369 ± 119 |
| Wild type (n=3) (anaerobic) | 0.62 ± 0.02 | 5.2 ± 0.4 | 318 ± 50 |
| N78K (n=2) | 0.56 ± 0.04 | 5.2 ± 1.9 | 380 ± 190 |
| H92Q (n=2) | 0.59 ± 0.05 | 10.3 ± 3.7c | 270 ± 70 |
| H92Y (n=2) | 0.49 ± 0.02 c | 11.6 ± 1.7c | 1340 ± 170c |
| H109Y (n=2) | 0.64 ± 0.03 | 5.0 ± 0.1 | 460 ± 60 |
| H110Q (n=2) | 0.70 ± 0.03 c | 7.0 ± 2.8 | 250 ± 50 |
| H110Y (n=4) | 0.74 ± 0.04 c | 7.5 ± 1.3c | 620 ± 90c |
The number of titrations is given in parentheses. All titrations were performed aerobically except where indicated.
The normalized A561 increases (F) as a function of the ascorbate concentration ([Asc]) for each titration were fitted to the equation: F = FH/(1+ CH/[Asc]) + (1 − FH)/(1+ CL/[Asc]), where FH and (1 − FH) are the fractions of overall absorbance change associated with the high- and low-potential hemes, respectively, and CH and CL are the midpoints ascorbate levels for the high- and low-potential hemes, respectively. The values shown represent the mean ± standard deviation.
p < 0.05 by Student’s t test (compared to aerobic wild type value)
Ascorbate solutions can decompose in air if trace metals are present [35], so parallel ascorbate titrations of wild type cytochrome were carried out under anaerobic conditions for comparison. The resulting average parameter values from the anaerobic titrations were statistically indistinguishable from those for the aerobic titrations (Table 3), confirming that ascorbate decomposition was not a significant problem under the conditions used for the present titrations. It is also worth noting that the standardized ascorbate titration protocol has given consistent results for several operators working on many batches of wild type cytochrome over the course of more than two years. The ascorbate titration thus seems to be a reliable, quantitative and convenient method to assess the functionality of the two hemes in cyt b561.
The negative control mutant, N78K, had ascorbate titration parameters essentially the same as the wild type (Table 3). Substitution with glutamine or tyrosine at His92 led to more than doubling of the CH value, but only the tyrosine substitution perturbed the CL value significantly; neither mutation significantly affected the relative proportions of the two transitions. Substitution with glutamine or tyrosine at His110 significantly decreased the absorbance change associated with the bL transition, seen as an increase in the value of FH (Table 3). The alterations in CH and CL values seen with the H110Y mutant may be consequences of the decreased contribution of the bL absorbance change. The titration parameters of the H109Y mutant were indistinguishable from the wild type, suggesting that His109 does not contribute to ascorbate interactions or to the redox behavior at either heme center. Overall, the N78K and H109Y mutations had little effect on the ascorbate titration parameters, whereas both mutations at His92 affected the ascorbate midpoint for the bH transition and both the His110 mutations affected the magnitude of the bL absorbance change. Tyrosine substitutions at either His92 or His110 affected both the bH and bL transitions, suggesting that these particular substitutions at one heme center affected the behavior of the other heme center.
Effects of His92 and His110 mutations on visible absorbance bands
The ascorbate concentrations needed to reduce the high- and low-potential heme centers differ significantly for wild type and mutant cytochromes (Table 3). Accordingly, the differences between successive spectra at the beginning of the present titrations were used to obtain the spectra of bH and the differences between successive spectra toward the end of the titrations were used to obtain the spectra of bL. The resulting spectra for the wild type cytochrome and six mutants, normalized to the same alpha band intensity, are shown in Fig. 6. For the bH spectra (Fig. 6A), only the H92Q and H92Y mutants are shifted and have a different shape from the wild type cytochrome. This difference in shape is also evident in the peak width values, which are narrower than those of other mutants (Table 4). The differences among the bL spectra (Fig. 6B) were more subtle but changes in shape were evident in the width of the alpha band (Table 4). The peak widths for the H110Q and H110Y mutants were notably wider than those of the wild type and the other mutants. Clearly, substitutions at His92 had a selective effect on the alpha band characteristics of bH, whereas substitutions at His110 selectively affected the bL alpha band.
Figure 6.
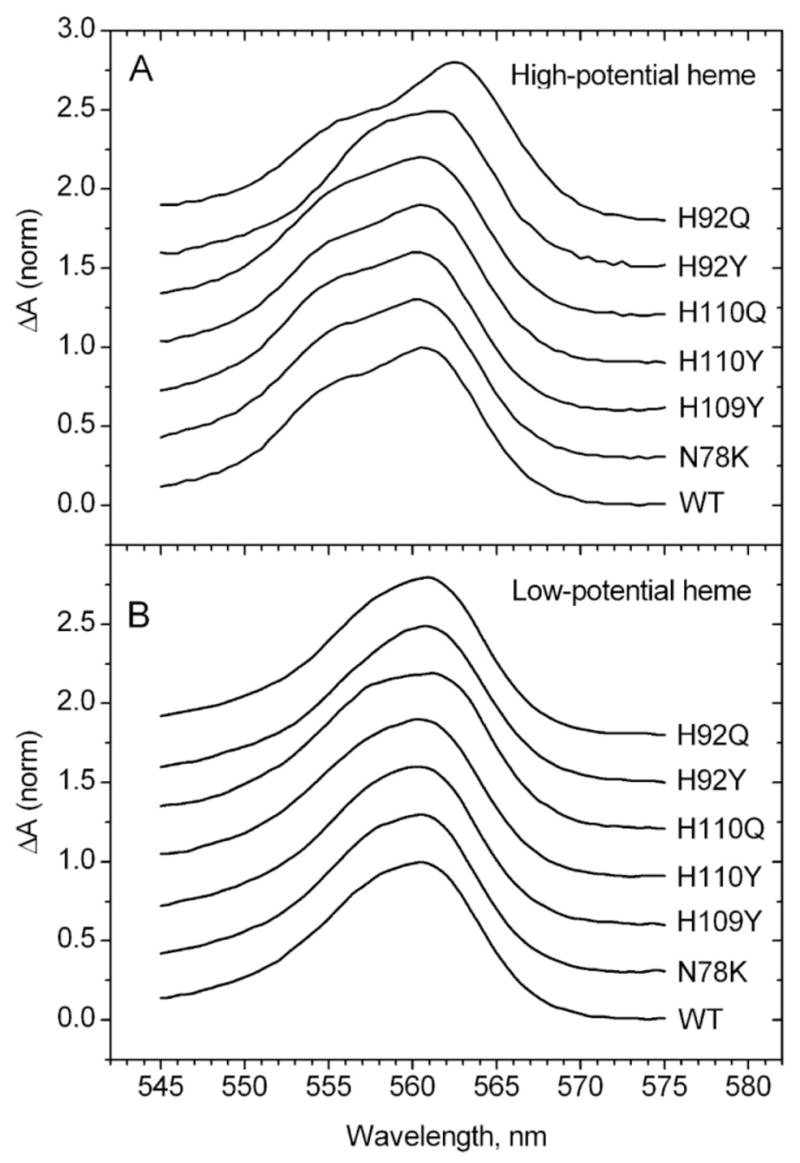
Alpha band difference spectra of high-potential (A) and low-potential (B) heme centers of wild type and mutant cyt b561 proteins obtained from ascorbate titrations. Each spectrum for the high potential heme was obtained by averaging the differences in three pairs of spectra from early points in the titrations (e.g., for wild type: Fig. 5A #5 - #1, #6 - #2, #7 - #3). Each spectrum for the low potential heme was obtained by averaging the differences in three pairs of spectra from later points in the titrations (e.g., for wild type: Fig. 5A #15 - #10, #14 - #9, #13 - #8).
Table 4.
Effects of mutations on widths of bH and bL alpha bands and the positions of their resolved components.
| Cyt b561 mutant | Width at 75% Height (nm)a | Shift in peak position, relative to wild typeb(nm) | ||||
|---|---|---|---|---|---|---|
| bH | bL | bH-1 | bH-2 | bL-1 | bL-2 | |
| Wild type | 8.3 | 7.1 | 0 | 0 | 0 | 0 |
| N78K | 8.6 | 7.0 | −0.2 | +0.0 | −0.1 | +0.4 |
| H92Q | 6.1 | 7.0 | +1.7 | +0.8 | +0.3 | +0.4 |
| H92Y | 7.6 | 6.6 | +0.7 | +1.7 | +0.0 | +0.0 |
| H109Y | 8.8 | 7.1 | −0.1 | +0.1 | −0.2 | +0.0 |
| H110Q | 8.8 | 8.2 | +0.1 | +0.1 | +0.3 | +0.6 |
| H110Y | 8.4 | 7.5 | +0.6 | +0.9 | +1.0 | +1.0 |
From spectra in Fig. 6.
The positions of the major resolved bands in wild type cyt b561 were: bH-1, 561.0 nm; bH-2, 554.6 nm; bL-1, 560.8; bL-2, 555.3 nm. Shifts were rounded to the nearest integer.
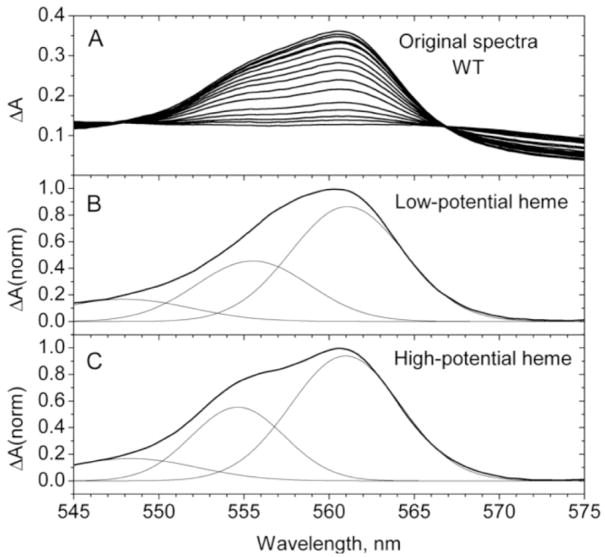
The alpha band components of bH and bL for wild type cyt b561 and each of the mutants obtained from ascorbate titration spectra as described above were further analyzed by resolution into three Gaussian components, two from transitions in the alpha band itself and one extending from a transition of the beta band, as shown for the recombinant wild type cytochrome in Fig. 7. The positions of the two main maxima in these resolved spectra, relative to the wild type maxima near 561 and 555 nm, are shown in Table 4. Both mutations of His110 led to shifts in the two major bL alpha band components, but did not affect the major bH components. In contrast, both mutations of His92 showed shifts in the positions of the two major bH alpha band components. The H92Q mutant did produce a shift in the bL-2 component similar to that seen with the N78K control. The H109Y mutant did not affect the position of any of the resolved spectral components, and the N78K mutant led to a small shift in only one component of bL. As for the comparison of the overall alpha band characteristics described above, the results of the resolution show that substitutions at His92 had a selective effect on the alpha band characteristics of bH, whereas substitutions at His110 selectively affected the bL alpha band.
Figure 7.
Alpha band difference spectra during ascorbate titration of wild type (WT) cyt b561 (panel A) and resolution of alpha band components of low-potential heme (bL, panel B) and high-potential heme (bH, panel C). The resolution of alpha band spectra (from Fig. 6) into three Gaussian components was accomplished with Origin 6.1 software using a Levenberg-Marquardt nonlinear regression algorith. The only constraint on the fitting was that the position of the maximum of the third Gaussian component remained below 552 nm.
Overall, three separate aspects of the ascorbate titration data were analyzed: the concentrations of ascorbate required for the bH and bL redox transitions, the bH and bL alpha band characteristics as a whole, and the characteristics of the bH and bL alpha band components. The consistent, general pattern observed was that substitutions at His92 affected the bH heme center, substitutions at His110 affected the bL heme center, and substitutions at His109 had only modest affects on either heme center. This pattern indicates that His92 is in the vicinity of the bH heme center and His110 is near the bL heme center. His92 is just four residues away from one of the axial ligands, His88, with the two residues on the same face of an alpha helix; His92 is positioned close to His88 in the 3D model shown in Fig. 2. Optical spectral changes during the functional reaction with ascorbate thus link the bH center to His88 and its partner, His161, and the bL center to the His54/His122 ligand pair. Supporting the latter assignment is the close proximity of His110 to His54 in the 3D model (Fig. 2) and in models of the arrangement of the transmembrane helices [19].
Effects of His92 and His110 mutations on heme redox potentials
Potentiometric titrations monitoring alpha-band absorbance changes were used to examine the effects of mutations on the ferric/ferrous heme couples in recombinant cyt b561 (Table 5). Wild type cyt b561 had average midpoint potentials at 60 and 151 mV, with approximately half of the A561 change in each transition. These redox potentials fall in the ranges of 30–70 mV and 145–170 mV reported for the cytochrome from CG [17, 18, 33]. None of the cyt b561 mutants had a large difference with the wild type cytochrome in terms of the relative proportions contributed by the two heme centers. All but one of the cyt b561 mutants had midpoint potentials for the two heme centers within 27 mV of the wild type values. The exception was H92Y, where the Em(bL) value was decreased to 13 mV. Still, none of the mutations appear to have a dramatic effect on the redox properties of the heme centers in cyt b561.
Table 5.
Redox parameters of wild type and mutant cyt b561.
| FHa | Em(bL)(mV)a | Em(bH) (mV)a | |
|---|---|---|---|
| Wild type (n=3) b | 0.45±0.01 | 60±4 | 151±17 |
| N78K (n=3) | 0.49±0.02c | 87±6c | 174±10 |
| H92Q (n=3) | 0.54±0.08 | 72±13 | 162±16 |
| H92Y (n=5) | 0.39±0.04 | 13±12c | 157±20 |
| H109Y (n=3) | 0.45±0.04 | 84±6c | 176±15 |
| H110Q (n=6) | 0.48±0.05 | 77±7c | 171±19 |
| H110Y (n=3) | 0.45±0.01 | 76±1c | 165±7 |
Values of the fraction of maximal change in A561 were analyzed as a function of the redox potential as described in Section 2. FH and (1− FH) are the fractions of the maximal absorbance change associated with the high- and low-potential hemes, respectively; Em(bL) and Em(bH) are the midpoint potentials for the low- and high-potential hemes, respectively. The values shown represent the mean ± standard deviation.
Number of reductive or oxidative titrations analyzed.
p < 0.05 by Student’s t test (compared to wild type value).
Comparison of the potentiometric titration results in Table 5 with those from the ascorbate titrations in Table 3 reveal that when the ratios of CL to CH are converted to millivolts, the values are comparable to the differences in redox potentials of the two hemes. This linkage between CL/CH and ΔEm indicates that the redox reactions with ascorbate approach a thermodynamic equilibrium. CH and CL thus reflect a combination of heme redox and ascorbate binding properties and are not simply affinity or kinetic constants for ascorbate binding as suggested by Berczi et al. [21]. The lack of major changes in redox potentials in the mutations of His92 and His110 that do markedly alter CH and CL (Tables 3 and 5) suggests that these mutations impact ascorbate binding and reactivity more than the intrinsic redox characteristics of the hemes.
4. Discussion
Heme content and protein structure in His92 and His110 mutants
Substitutions of His92 and His110 with glutamine or tyrosine produced distinct effects on the spectroscopic properties and the functioning of adrenal cyt b561. These effects are useful in examining the roles of His92 and His110 themselves, and also for characterizing the individual heme centers, where mutations in the axial ligands have proven generally disruptive for both heme centers and for the protein overall (Tables 1 and 2, Figs. 3 and 4, Refs. [15, 21]). All mutants tested at His92 or His110 contained two molecules of heme, both of which retained functional reaction with ascorbate (Table 2 and Fig. 3). Murine cyt b561 was reported to retain reactivity with ascorbate when the residue corresponding to His110 was replaced with an alanine; the residue corresponding to His92 was not tested [21].
His92 and His110 and characteristics of the heme centers
The bH and bL heme centers of cyt b561 can be distinguished by their characteristic signals in EPR and optical spectra [12, 14–16]. The EPR spectra of the mutants at His92 and His110 were quite informative, with the low spin signal of the bH center (g ~3.1) shifted in both H92Q and H92Y, but unchanged in both H110Q and H110Y (Fig. 4), unambiguously connecting His92 to the bH heme center. The low spin bL EPR signal at g ~3.7 was not affected by mutations at His92 or at His110. The optical absorption spectra of the His92 and His110 mutants also proved informative. The alpha bands for the two heme centers resolved from the ascorbate titration spectra (Fig. 6 and Table 4) revealed that the bH alpha band was narrower in H92Q and H92Y but not in H110Q or H110Y, whereas the bL alpha band was altered in the His110 mutants but not in the His92 mutants. Overall, these results make it clear that the sidechains of His92 and His110 can indeed affect the properties of the heme centers, supporting the structural model of Bashtovyy et al. (Fig. 2) [23]. Given the differing conservation of His92 and His110 between the four cyt b561 family subgroups in Fig. 1, we speculate that the structures of residues 92 and 110 have evolved for a specialized function peculiar to each group. Several other hemoproteins are known to have a histidine residue that is not an axial ligand located near a heme residue. These include cytochrome c554 [36], cyt c3 [37], and yeast cyt c peroxidase [38]. The functional impact of the “shadow” histidine residue has not been evaluated so far for any of these proteins.
His92 and His110 and reaction of heme centers with ascorbate
Ascorbic acid is the physiological reductant for the adrenal cytochrome [9] and the titrations with ascorbate provide a useful probe of functional changes in the mutants (Fig. 5 and Table 3). The exact nature of the constants CL and CH extracted from the titration of cyt b561 with ascorbate remains to be clarified. They appear to be pseudo-equilibrium constants that are determined by a combination of the hemes’ redox properties and the affinity of ascorbate for each of its two binding centers. Berczi et al. [21] recently analyzed the dependence of the reduction of murine cyt b561 and its mutants on ascorbate concentration. They interpreted the parameters extracted by fitting as apparent affinity constants for ascorbate. However, additional experiments are needed to establish the exact contributions of ascorbate binding and heme redox properties to the values of empirical constants such as CL and CH.
Topology of cyt b561 heme centers in chromaffin granules
The observed pattern of changes in the EPR and optical spectra and in the ascorbate titrations for the His92 and His110 mutants described above connects His92 to bH and His110 to bL. These assignments are important because they have implications for the topology of the two cyt b561 heme centers in the chromaffin granule membrane. The topology of the adrenal cyt b561 polypeptide has not been directly characterized, but the currently accepted model from sequence analysis has six transmembrane helices, with the N- and C-termini exposed on the cytoplasmic (C) side of the CG membrane [39]. The C-terminus is known to be accessible to proteases from the C-side of intact CG vesicles [40]. This topology positions His54 and His122 towards the matrix (M) side, and His88 and His161 toward the C-side, of the CG membrane. The primary structure puts His92 near His88, so the bH heme is that coordinated by His88 and His161, on the C-side of the CG membrane. By elimination, the bL heme must be coordinated by His54 and His122, on the M-side of the membrane. Further evidence for the assignment of bL to the His54/His122 heme is the observation that murine adrenal cyt b561 with alanine substitutions of the residues corresponding to His88 and His161 retained only the heme center (bL) that is less reactive with ascorbate in equilibrium titrations [21]. The disposition of the two heme centers relative to the membrane indicated by these results is the opposite of that assumed in some current models [41], but it is consistent with conclusions drawn from the observation of some g = 3.7 EPR signal (from the bL center) in H88Q and H161Q mutants of cyt b561 expressed in Sf9 cells [15].
Effects of mutations at Asn78 and His109
Residue 109 (a histidine in bovine cyt b561 and a tyrosine in the human cytochrome) is positioned on the periphery of the four-helix bundle in the computational model (Fig. 2). Thus, it was not surprising that substitutions of His109 had little effect on expression of the cytochrome or on the spectroscopic and functional properties (Figs. 4 and 6; Tables 1–4). Asn78 was selected as a negative control target for mutagenesis because it is predicted to lie on the periphery of the four-helix bundle (Fig. 2). Surprisingly, the N78K mutation was found to increase expression of the recombinant cytochrome (Table 1), which may make this protein useful for structural studies. Asn78 is predicted to be in the cytoplasmic loop connecting helices 2 and 3 and may well have important structural interactions with cytoplasmically-exposed residues in the N- or C-terminal segments or the loop connecting helices 4 and 5. The N-terminal segment preceding helix 1 is positioned next to the loop between helices 2 and 3 in one variant of the computational model [23].
Functional integration of the two heme centers
The effects of point mutations at His88 and His161 (Fig. 3) show that just a single mutation at either histidine is sufficient to prevent reduction of all heme in the cytochrome by ascorbate. The proportion of ascorbate-reducible cytochrome was also greatly diminished in murine cyt b561 mutants with alanine substitutions at the residues corresponding to His88 or His161 in the bovine cytochrome, with only 5% of the wild type response to ascorbate retained in the double mutant [21]. Overall, it is clear that the axial ligands to heme contributed by His88 and His161 are crucial for physiological reduction of the adrenal cyt b561 and that both heme centers are impacted by the loss of structural integrity of the bH heme. We take this coordinated behavior to reflect the integration of the two hemes as part of a structural unit that we have termed the “kernel” [15]. Additional evidence for integration of the two heme centers comes from the observation that the point mutants H92Y and H110Y affected some properties of both heme centers (Tables 3 and 4).
Heme centers and cyt b561 expression
Only a fraction of recombinant cyt b561 protein with substitutions in His88 or His161 could be extracted by nonionic detergent (Table 1), indicating that most of these mutant proteins were not properly folded and/or integrated into membrane. Similar low detergent extractability was observed when these mutants were expressed in Sf9 insect cells [15]. Any substitutions in any one of the histidines furnishing axial ligands to either of the cyt b561 hemes decreases expression of the bovine (Fig. 3 and Table 1) [15], murine [21] and thale cress [42] cytochromes, with comparable results in bacterial, yeast and insect cell systems. The integrity of the ligation of both hemes is thus linked to expression of functional cytochrome. Adrenal cyt b561 is an integral membrane protein and carries an uncleaved signal peptide for targeting the nascent polypeptide to the translocon machinery in the endoplasmic reticulum membrane [27]. In the translocon-mediated process, most polytopic proteins are integrated into the ER membrane co-translationally [43], and interactions between adjacent transmembrane segments are known to promote stable membrane integration of these segments [44]. Incorporation of heme by a myeloperoxidase precursor is a key step in that protein’s maturation in the ER lumen [45], and nascent globin polypeptides can bind heme [46]. A pair of hemes with bis-histidine axial ligation has been found or proposed for a number of membrane bound cytochromes, involving ligands on either two [47–52], three [53], or four [54, 55] transmembrane helices. In some cases, mutation in the histidines furnishing axial ligands to the hemes leads to decreased levels of functional protein, suggesting a role for properly coordinated heme in protein synthesis or stability [55, 56].
A striking observation, found with both bacterial and yeast expression systems, is that mutations in the His54/His122 pair decrease expression of recombinant cyt b561 much more than substitutions in the His88/His161 pair (Table 1) [21]. As ribosomal translation of the His54/His122 pair is completed before that of the His88/His161 pair, the differential effects on expression of mutations in the two pairs suggest that insertion of the two hemes may be important in cotranslational assembly of the transmembrane cytochrome.
There is some precedent for diheme ligation being required for proper expression of adrenal cyt b561, in that cofactor binding can accelerate protein folding [57]. Direct examples for comparison with cyt b561 are not plentiful. Other examples of diheme b cytochromes with a 4-helix structural unit are found as subunits of hetero-oligomeric complexes, making it difficult to examine the expression of these b cytochromes in isolation [54, 55]. Cyt b562 from E. coli is a monomeric 4-helix bundle protein with a single b type heme, and its folding behavior suggests that heme binds only after folding of the 4-helix bundle [58]. This bacterial cyt b562, however, is a globular protein and the constraints of folding in an aqueous environment are likely very different from those encountered by the adrenal cytochrome during membrane insertion. The small size, convenient expression, and sensitive spectroscopic characteristics of cyt b561 make it an attractive experimental model for more detailed characterization of cofactor influences on membrane protein biogenesis.
In summary, the present results indicate that the sidechains of His92 and His110 in adrenal cyt b561, although not axial ligands to either heme, nonetheless selectively influence the cytochrome’s spectroscopic properties and its interactions with ascorbate, supporting the identification of His92 and His110 as components of the bH and bL heme domains, respectively.
Acknowledgments
We are grateful to Dr. Han Asard (University of Antwerp, Belgium) and Dr. Tibor Pali (Biological Research Center, Hungarian Academy of Sciences) for sharing the coordinates from their cyt b561 computational model.
Footnotes
This work was supported by NIH grants GM44911 (ALT), GM53508 (VS) and GM080575 (GP), and American Heart Association Texas Affiliate grant 0455107Y (GP).
The abbreviations used are: cyt b561, adrenal cytochrome b561, CG, chromaffin granules; CD, circular dichroism; EPR, electron paramagnetic resonance; HALS, highly axial low spin EPR signal; DM, dodecyl maltoside.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spiro MJ, Ball EG. Studies on the respiratory enzymes of the adrenal gland. I. The medulla. J Biol Chem. 1961;236:225–30. [PubMed] [Google Scholar]
- 2.Duong LT, Fleming PJ, Russell JT. An identical cytochrome b561 is present in bovine adrenal chromaffin vesicles and posterior pituitary neurosecretory vesicles. J Biol Chem. 1984;259:4885–9. [PubMed] [Google Scholar]
- 3.Pruss RM, Shepard EA. Cytochrome b561 can be detected in many neuroendocrine tissues using a specific monoclonal antibody. Neuroscience. 1987;22:149–57. doi: 10.1016/0306-4522(87)90205-3. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava M, Gibson KR, Pollard HB, Fleming PJ. Human cytochrome b561: a revised hypothesis for conformation in membranes which reconciles sequence and functional information. Biochem J 303 (Pt. 1994;3):915–21. doi: 10.1042/bj3030915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flatmark T, Lagercrantz H, Terland O, Helle KB, Stjarne L. Electron carriers of the noradrenaline storage vesicles from bovine splenic nerves. Biochim Biophys Acta. 1971;245:249–52. doi: 10.1016/0005-2728(71)90031-4. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava M, Pollard HB, Fleming PJ. Mouse cytochrome b561: cDNA cloning and expression in rat brain, mouse embryos, and human glioma cell lines. DNA Cell Biol. 1998;17:771–7. doi: 10.1089/dna.1998.17.771. [DOI] [PubMed] [Google Scholar]
- 7.Abbs MT, Phillips JH. Organisation of the proteins of the chromaffin granule membrane. Biochim Biophys Acta. 1980;595:200–21. doi: 10.1016/0005-2736(80)90084-x. [DOI] [PubMed] [Google Scholar]
- 8.Duong LT, Fleming PJ. The asymmetric orientation of cytochrome b561 in bovine chromaffin granule membranes. Arch Biochem Biophys. 1984;228:332–41. doi: 10.1016/0003-9861(84)90074-2. [DOI] [PubMed] [Google Scholar]
- 9.Njus D, Kelley PM. The secretory-vesicle ascorbate-regenerating system: a chain of concerted H+/e(−)-transfer reactions. Biochim Biophys Acta. 1993;1144:235–48. doi: 10.1016/0005-2728(93)90108-r. [DOI] [PubMed] [Google Scholar]
- 10.Klinman JP. The copper-enzyme family of dopamine beta-monooxygenase and peptidylglycine alpha-hydroxylating monooxygenase: resolving the chemical pathway for substrate hydroxylation. J Biol Chem. 2006;281:3013–6. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 11.Tsubaki M, Takeuchi F, Nakanishi N. Cytochrome b561 protein family: expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753:174–90. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Tsubaki M, Nakayama M, Okuyama E, Ichikawa Y, Hori H. Existence of two heme B centers in cytochrome b561 from bovine adrenal chromaffin vesicles as revealed by a new purification procedure and EPR spectroscopy. J Biol Chem. 1997;272:23206–10. doi: 10.1074/jbc.272.37.23206. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Kamensky Y, Kakkar R, Foley E, Kulmacz RJ, Palmer G. Purification and characterization of bovine adrenal cytochrome b561 expressed in insect and yeast cell systems. Protein Expr Purif. 2005;40:429–39. doi: 10.1016/j.pep.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 14.Burbaev DS, Moroz IA, Kamenskiy YA, Konstantinov A. Several forms of chromaffin granule cytochrome b-561 revealed by EPR spectroscopy. FEBS Lett. 1991;283:97–9. doi: 10.1016/0014-5793(91)80562-h. [DOI] [PubMed] [Google Scholar]
- 15.Kamensky Y, Liu W, Tsai AL, Kulmacz RJ, Palmer G. Axial Ligation and Stoichiometry of Heme Centers in Adrenal Cytochrome b(561) Biochemistry. 2007;46:8647–58. doi: 10.1021/bi700054g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamensky YA, Palmer G. Chromaffin granule membranes contain at least three heme centers: direct evidence from EPR and absorption spectroscopy. FEBS Lett. 2001;491:119–22. doi: 10.1016/s0014-5793(01)02173-1. [DOI] [PubMed] [Google Scholar]
- 17.Apps DK, Boisclair MD, Gavine FS, Pettigrew GW. Unusual redox behaviour of cytochrome b-561 from bovine chromaffin granule membranes. Biochim Biophys Acta. 1984;764:8–16. doi: 10.1016/0005-2728(84)90134-8. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi F, Kobayashi K, Tagawa S, Tsubaki M. Ascorbate inhibits the carbethoxylation of two histidyl and one tyrosyl residues indispensable for the transmembrane electron transfer reaction of cytochrome b561. Biochemistry. 2001;40:4067–76. doi: 10.1021/bi002240x. [DOI] [PubMed] [Google Scholar]
- 19.Degli Esposti M, Kamensky Yu A, Arutjunjan AM, Konstantinov AA. A model for the molecular organization of cytochrome beta-561 in chromaffin granule membranes. FEBS Lett. 1989;254:74–8. doi: 10.1016/0014-5793(89)81012-9. [DOI] [PubMed] [Google Scholar]
- 20.Kamensky Y, Arutjunian A, Ksenzenko M, Chertkova E, Konstantinov A. Axial ligands of haem iron in chromaffin granule cytochrome b561. Biological Membranes (USSR) 1990;4:667–671. [Google Scholar]
- 21.Berczi A, Su D, Lakshminarasimhan M, Vargas A, Asard H. Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Arch Biochem Biophys. 2005;443:82–92. doi: 10.1016/j.abb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Okuyama E, Yamamoto R, Ichikawa Y, Tsubaki M. Structural basis for the electron transfer across the chromaffin vesicle membranes catalyzed by cytochrome b561: analyses of cDNA nucleotide sequences and visible absorption spectra. Biochim Biophys Acta. 1998;1383:269–78. doi: 10.1016/s0167-4838(97)00216-1. [DOI] [PubMed] [Google Scholar]
- 23.Bashtovyy D, Berczi A, Asard H, Pali T. Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma. 2003;221:31–40. doi: 10.1007/s00709-002-0065-0. [DOI] [PubMed] [Google Scholar]
- 24.Verelst W, Asard H. A phylogenetic study of cytochrome b561 proteins. Genome Biol. 2003;4:R38. doi: 10.1186/gb-2003-4-6-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–9. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 26.Ji L, Nishizaki M, Gao B, Burbee D, Kondo M, Kamibayashi C, Xu K, Yen N, Atkinson EN, Fang B, Lerman MI, Roth JA, Minna JD. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62:2715–20. [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, Rogge CE, Kamensky Y, Tsai AL, Kulmacz RJ. Development of a bacterial system for high yield expression of fully functional adrenal cytochrome b(561) Protein Expr Purif. 2007;56:145–152. doi: 10.1016/j.pep.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Peterson GL. Review of the Folin phenol protein quantitation method of Lowry, Rosebrough. Farr and Randall, Anal Biochem. 1979;100:201–20. doi: 10.1016/0003-2697(79)90222-7. [DOI] [PubMed] [Google Scholar]
- 29.Bambai B, Rogge CE, Stec B, Kulmacz RJ. Role of Asn-382 and Thr-383 in activation and inactivation of human prostaglandin H synthase cyclooxygenase catalysis. J Biol Chem. 2004;279:4084–92. doi: 10.1074/jbc.M304762200. [DOI] [PubMed] [Google Scholar]
- 30.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 31.Kulmacz RJ, Tsai AL, Palmer G. Heme spin states and peroxide-induced radical species in prostaglandin H synthase. J Biol Chem. 1987;262:10524–31. [PubMed] [Google Scholar]
- 32.Williams CH, Jr, Arscott LD, Matthews RG, Thorpe C, Wilkinson KD. Methodology employed for anaerobic spectrophotometric titrations and for computer-assisted data analysis. Methods Enzymol. 1979;62:185–98. doi: 10.1016/0076-6879(79)62217-6. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi F, Hori H, Obayashi E, Shiro Y, Tsubaki M. Properties of two distinct heme centers of cytochrome b561 from bovine chromaffin vesicles studied by EPR, resonance Raman, and ascorbate reduction assay. J Biochem (Tokyo) 2004;135:53–64. doi: 10.1093/jb/mvh006. [DOI] [PubMed] [Google Scholar]
- 34.Tsubaki M, Kobayashi K, Ichise T, Takeuchi F, Tagawa S. Diethyl pyrocarbonate modification abolishes fast electron accepting ability of cytochrome b561 from ascorbate but does not influence electron donation to monodehydroascorbate radical: identification of the modification sites by mass spectrometric analysis. Biochemistry. 2000;39:3276–84. doi: 10.1021/bi991883d. [DOI] [PubMed] [Google Scholar]
- 35.Buettner GR. Ascorbate autoxidation in the presence of iron and copper chelates. Free Radic Res Commun. 1986;1:349–53. doi: 10.3109/10715768609051638. [DOI] [PubMed] [Google Scholar]
- 36.Iverson TM, Arciero DM, Hsu BT, Logan MS, Hooper AB, Rees DC. Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea. Nat Struct Biol. 1998;5:1005–12. doi: 10.1038/2975. [DOI] [PubMed] [Google Scholar]
- 37.Dolla A, Arnoux P, Protasevich I, Lobachov V, Brugna M, Giudici-Orticoni MT, Haser R, Czjzek M, Makarov A, Bruschi M. Key role of phenylalanine 20 in cytochrome c3: structure, stability, and function studies. Biochemistry. 1999;38:33–41. doi: 10.1021/bi981593h. [DOI] [PubMed] [Google Scholar]
- 38.Fulop V, Phizackerley RP, Soltis SM, Clifton IJ, Wakatsuki S, Erman J, Hajdu J, Edwards SL. Laue diffraction study on the structure of cytochrome c peroxidase compound I. Structure. 1994;2:201–8. doi: 10.1016/s0969-2126(00)00021-6. [DOI] [PubMed] [Google Scholar]
- 39.Perin MS, Fried VA, Slaughter CA, Sudhof TC. The structure of cytochrome b561, a secretory vesicle-specific electron transport protein. Embo J. 1988;7:2697–703. doi: 10.1002/j.1460-2075.1988.tb03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent UM, Fleming PJ. Cytochrome b561 is fatty acylated and oriented in the chromaffin granule membrane with its carboxyl terminus cytoplasmically exposed. J Biol Chem. 1990;265:16422–7. [PubMed] [Google Scholar]
- 41.Takeuchi F, Hori H, Tsubaki M. Selective Perturbation of the Intravesicular Heme Center of Cytochrome b561 by Cysteinyl Modification with 4,4′-Dithiodipyridine. J Biochem (Tokyo) 2005;138:751–62. doi: 10.1093/jb/mvi174. [DOI] [PubMed] [Google Scholar]
- 42.Berczi A, Asard H. Characterization of an ascorbate-reducible cytochrome b561 by site-directed mutagenesis. Acta Biol Szeged. 2006;50:55–59. [Google Scholar]
- 43.Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J Biol Chem. 2004;279:22787–90. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- 44.Mackenzie KR. Folding and stability of alpha-helical integral membrane proteins. Chem Rev. 2006;106:1931–77. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 45.Arnljots K, Olsson I. Myeloperoxidase precursors incorporate heme. J Biol Chem. 1987;262:10430–3. [PubMed] [Google Scholar]
- 46.Komar AA, Kommer A, Krasheninnikov IA, Spirin AS. Cotranslational folding of globin. J Biol Chem. 1997;272:10646–51. doi: 10.1074/jbc.272.16.10646. [DOI] [PubMed] [Google Scholar]
- 47.Buschlen S, Choquet Y, Kuras R, Wollman FA. Nucleotide sequences of the continuous and separated petA, petB and petD chloroplast genes in Chlamydomonas reinhardtii. FEBS Lett. 1991;284:257–62. doi: 10.1016/0014-5793(91)80698-3. [DOI] [PubMed] [Google Scholar]
- 48.Esposti MD, De Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A. Mitochondrial cytochrome b: evolution and structure of the protein. Biochim Biophys Acta. 1993;1143:243–71. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- 49.Biberstine-Kinkade KJ, DeLeo FR, Epstein RI, LeRoy BA, Nauseef WM, Dinauer MC. Heme-ligating histidines in flavocytochrome b(558): identification of specific histidines in gp91(phox) J Biol Chem. 2001;276:31105–12. doi: 10.1074/jbc.M103327200. [DOI] [PubMed] [Google Scholar]
- 50.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NC. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol. 2003;10:681–7. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 51.Finegold AA, Shatwell KP, Segal AW, Klausner RD, Dancis A. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J Biol Chem. 1996;271:31021–4. doi: 10.1074/jbc.271.49.31021. [DOI] [PubMed] [Google Scholar]
- 52.Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-Ray Structure of Rhodobacter Capsulatus Cytochrome bc (1): Comparison with its Mitochondrial and Chloroplast Counterparts. Photosynth Res. 2004;81:251–75. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 53.Jormakka M, Tornroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–8. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 54.Lancaster CR, Kroger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–85. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 55.Hederstedt L. Succinate:quinone oxidoreductase in the bacteria Paracoccus denitrificans and Bacillus subtilis. Biochim Biophys Acta. 2002;1553:74–83. doi: 10.1016/s0005-2728(01)00231-6. [DOI] [PubMed] [Google Scholar]
- 56.Kuras R, de Vitry C, Choquet Y, Girard-Bascou J, Culler D, Buschlen S, Merchant S, Wollman FA. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J Biol Chem. 1997;272:32427–35. doi: 10.1074/jbc.272.51.32427. [DOI] [PubMed] [Google Scholar]
- 57.Wittung-Stafshede P. Role of cofactors in protein folding. Acc Chem Res. 2002;35:201–8. doi: 10.1021/ar010106e. [DOI] [PubMed] [Google Scholar]
- 58.Garcia P, Bruix M, Rico M, Ciofi-Baffoni S, Banci L, Ramachandra Shastry MC, Roder H, de Lumley Woodyear T, Johnson CM, Fersht AR, Barker PD. Effects of heme on the structure of the denatured state and folding kinetics of cytochrome b562. J Mol Biol. 2005;346:331–44. doi: 10.1016/j.jmb.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 59.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]