Abstract
Bacillus subtilis has a single set of flagellar rotor proteins that interact with two distinct stator-force generators, the H+-coupled MotAB complex and the Na+-coupled MotPS complex, that energize rotation. Here, motility on soft agar plates and in liquid was assayed in wild-type B. subtilis and strains expressing only one stator, either MotAB, MotPS or hybrid MotAS or MotPB. The strains expressing MotAB or MotAS had an average of 11 flagella/cell while those expressing MotPS or MotPB had an average of 7 flagella/cell, while a Mot-less double mutant had 3-4 flagella/cell. MotAB had a more dominant role in motility than MotPS under most conditions, but MotPS supported comparable motility to MotAB on malate-containing soft agar plating media at elevated pH and Na+. MotAB supported much faster swimming speeds in liquid than MotPS, MotAS or MotPB under all conditions, but a contribution of MotPS to wild-type swimming was discernible from differences in swimming speeds of wild-type and MotAB at elevated viscosity, pH and Na+. Swimming supported by MotPS and MotAS was stimulated by Na+ and elevated pH whereas the converse was true of MotAB and MotPB. This suggests that MotAS is Na+-coupled and MotPB is H+-coupled and that MotB and MotS are major determinants of ion-coupling. However, the swimming speed supported by MotPB, as well as MotPS and MotAS, was inhibited severely at Na+ concentrations above 300 mM whereas MotAB-dependent swimming was not. The presence of either the MotP or MotS component in the stator also conferred sensitivity to inhibition by an amiloride analogue. These observations suggest that MotP contributes to Na+-coupling and inhibition by Na+ channel inhibitors. Similarly, a role for MotA in H+-dependent stator properties is indicated by the larger effects of pH on the Na+-response of MotAS vs MotPS. Finally, optimal function at elevated viscosity was found only in MotPS and MotPB and is therefore conferred by MotP.
Keywords: Bacillus, flagella, MotAB, MotPS, sodium
Introduction
Bacterial flagella are helical structures that function as propellers whose rotation depends upon membrane-associated rotary motors1. The motors are powered by electrochemical ion gradients across the cytoplasmic membrane2. Energy coupling is mediated by inward ion flux through channels in membrane-embedded flagellar stator complexes. The cytoplasmic domains of these complexes interact in some incompletely understood manner with the FliG protein of the rotor to cause rotation during ion flux3; 4; 5; 6. The stator complexes contain 4-TMS MotA and 1-TMS MotB proteins or their homologues4; 7 and require additional proteins in some bacteria8; 9. Together, MotA and MotB constitute the ion channels that are required for motility10; 11; 12. The MotB protein contains a conserved acidic residue that is critical for the ion pathway in E. coli13. MotB is additionally thought to support the stator function of the complexes via contacts with the rigid peptidoglycan layer of the cell wall14; 15. Electron microscopic evidence indicates that multiple stator complexes surround each flagellar rotor10; 16; 17 and recent biochemical studies suggest that each stator complex contains four MotA proteins and two MotB proteins so that a stator complex has a total of eighteen TMS and two ion channel pathways12; 18; 19.
Rotation of most bacterial flagella is coupled to an electrochemical gradient of H+ across the cytoplasmic membrane (pmf, negative and alkaline inside the cells )20; 21; 22, but rotation of others is coupled to an electrochemical gradient of Na+ (smf, negative and lower [Na+] inside the cell)23; 24. Thus far only extremely alkaliphilic Bacillus strains and Vibrio cholerae are known to rely solely on Na+-coupled motility24; 25; 26; 27, whereas other alkaline-tolerant bacteria have been shown or inferred from genomic evidence to possess both H+- and Na+-coupled flagellar motors23; 28; 29; 30. For example, Na+-coupled motility of extensively studied Vibrio alginolyticus and Vibrio parahaemolyticus utilizes a constitutively expressed, single polar flagellum, whereas H+-coupled motility is mediated by multiple lateral flagella that are produced under specific conditions (involving viscous and/or surface environments)30. The polar and lateral flagella of these Vibrio are encoded by distinct sets of genes29. A growing number of other bacteria exhibit dual motility systems only some of which are encoded by entirely distinct sets of genes or use different coupling ions30. In each bacterial setting the dual motility systems appear to optimize motility under different conditions (reviewed in 29; 30). The dual motility systems of Bacillus subtilis that are studied here are a recently discovered variation on the theme of dual motility systems. B. subtilis possesses two different stators, H+-coupled MotAB21; 22; 31 and Na+-coupled MotPS26, but apparently possesses only one set of flagellar rotor genes so that both stators would have to interact with a single form of FliG32; 33. In B. subtilis, all motility is abolished in mutants of the large fla/che operon that encodes FliG and other proteins that participate in the rotor “switch complex” 34. This dual motility system offers the opportunity to examine whether two stators with different ion-coupling properties interact differently with a single FliG and to probe the roles of each Mot protein in the properties conferred by the stator. As with other Na+–coupled stators, Na+ is a useful chemical probe of the ion pathway as are specific inhibitors of Na+ channels7; 35; 36; 37; 38. In this study, motility on soft agar plates and swimming speed in liquid were assayed in wild-type B. subtilis and in strains that each express only one of four different stator types, MotAB, MotPS, hybrid MotAS and hybrid MotPB. The wild-type strain was included for comparative purposes. The contribution of MotPS to the wild-type motility pattern can be inferred from differences between wild-type and MotAB patterns. Data from the other single stator strains cannot be extrapolated to the wild-type setting for two reasons: (i) the presence of hybrids in the wild-type has not been assessed; and (ii) in order to achieve sufficient motility of each single stator strain on soft agar plates and in liquid it was necessary to use two different promoters (motAB promoter for MotAB and MotPS vs an inducible synthetic promoter for MotAS and MotPB) and two up-motile variants (for the two hybrid forms). The resulting panel of motile single stator strains made it possible to assess the roles of the two stator components in ion-coupling specificity (Fig. 1), inhibition patterns and the viscosity response of the hybrid forms, and to follow up initial data indicating that MotPS has a poor capacity to support swimming in liquid compared to its ability to support motility on soft agar plates26.
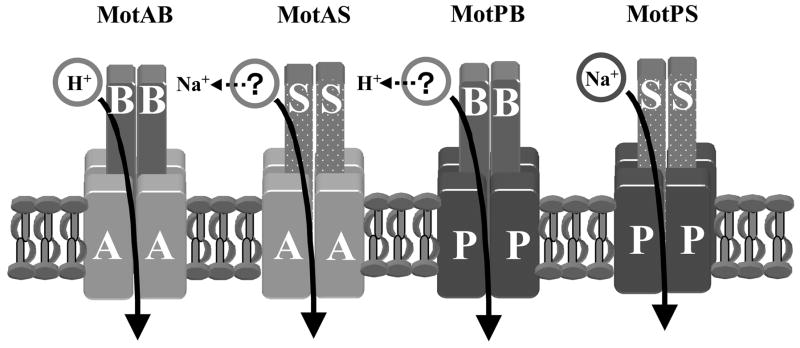
Figure 1.
Schematic diagram of the stators of MotAB, MotPS, hybrid MotAS and hybrid MotPB highlighting the question of ion-coupling of the hybrids and indicating, with dotted arrows, the results of this study.
Results
Expression of hybrid stators that support motility on soft agar plates
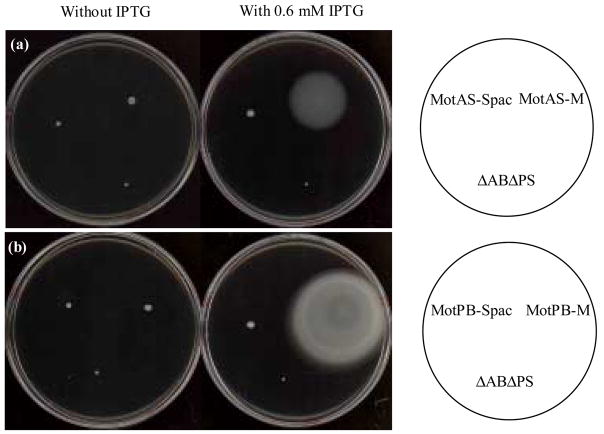
After construction of a mutant B. subtilis strain from which both the motAB and motPS genes were deleted (ΔABΔPS), motAB, motPS, motAS and motPB were returned to the chromosome in the amyE locus as described under Materials and Methods; the strains were designated by the flagellar pair that is expressed (MotAB, MotPS, MotAS and MotPB). In preliminary constructs, each mot locus was placed under control of an IPTG-inducible Pspac promoter. The strains with the hybrid Mot forms, MotAS and MotPB, exhibited very little spreading on soft agar plates relative to the ΔABΔPS whether or not IPTG was added (Fig. 2). Up-motile strains, MotAS-M and MotPS-M, were selected by several rounds of transfer from the motile edge of a colony on plates of the same composition. These strains were found to be free of mutations in the mot and fliG loci, including both the coding regions and the regions containing the promoters; the genetic change(s) in these strains are unknown. They were used for all the motility experiments, with the addition of 0.6 mM IPTG except where specifically indicated. The MotAB and MotPS strains with mot expression under Pspac exhibited motility in liquid but little or no motility on soft agar plates either in the presence or absence of inducer (data not shown). We have no explanation for this observation but an imbalance among proteins required for motility and chemotaxis had been suggested for a mutant strain with a similar phenotype in Pseudomonas aeruginosa39. New constructs were prepared with each of the two wild-type mot loci under control of the motAB promoter. These strains exhibited motility in liquid as well as on soft agar plates and were used in the experimental work. Strains in which the hybrid mot loci were expressed under control of PmotAB were not motile under any conditions (data not shown); the basis for this dependence of motility in MotAS and MotPB strains on an inducible, synthetic promoter was not examined.
Figure 2.
The motility behavior on soft agar plate of up-motile derivatives of MotAS and MotPB. A. Spreading of Δ ABΔ PS (negative control), MotAS-Spac (initial isolate), and MotAS-M (up-motile derivative) after 36 h of incubation at 37 C on TY medium plus 200mM NaCl, pH 8.5, without (left) or with (right) 0.6 mM IPTG, and solidified with 0.3% agar. B. Spreading of Δ ABΔ PS (negative control), MotPB-Spac (original isolate), and MotPB-M (up-motile derivative) after 24 h of incubation at 37 C on TY, pH 6.0, without (left) or with (right) 0.6 mM IPTG and solidified with 0.3% agar.
The number of flagella/cell was assessed using stained cells of the four single Mot stator strains used in the motility assays (MotAB, MotPS, MotAB-M and MotPB-M) as well as in the original (not up-motile) MotAS and MotPB hybrid strains. The MotAB and IPTG-induced MotAS-M strains had an average of 11 flagella/cell whereas the MotPS and IPTG-induced MotPB-M strains had an average of 7 flagella/cell (Table 1). The double mutant and the original MotAS and MotPB strains had significantly fewer flagella/cell (an average between 3-4); these strains also had shorter flagella than the strains that were used for the subsequent motility studies (Table 1). The number of flagella/cell was not increased by inclusion of IPTG in the growth medium of the original MotAS or MotPB strains whereas in their up-motile derivatives the basal number of flagella/cell was higher and further IPTG-induced increases were found; up-motile B. subtilis mutants had earlier been found to have increased numbers of flagella/cell40; 41. Although not shown, electron microscopic studies using negatively stained cells confirmed the patterns shown by staining.
Table 1.
Number and length of flagella of B. subtilis mot strains at pH 7.0.
| Number of flagella / cella |
Length of flagella ( μm)b |
|||
|---|---|---|---|---|
| Strains | Range of values | Average | Range of values | Average |
| wild type | 9-16 | 12.3 | 6.1-9.3 | 7.5 |
| Δ ABΔ PS, -IPTG | 1-6 | 3.6 | 1.8-5.7 | 4.0 |
| Δ ABΔ PS, +IPTG | 1-6 | 3.3 | 1.8-5.4 | 3.9 |
| MotAB | 9-15 | 10.8 | 5.0-8.9 | 7.0 |
| MotPS | 5-10 | 7.2 | 5.4-9.3 | 7.1 |
| MotAS, -IPTG | 2-5 | 3.1 | 1.8-7.5 | 4.3 |
| MotAS, +IPTG | 2-5 | 3.1 | 1.8-5.4 | 3.9 |
| MotAS-M, -IPTG | 5-9 | 6.9 | 3.2-7.9 | 5.6 |
| MotAS-M, +IPTG | 9-15 | 11.2 | 5.0-8.9 | 7.2 |
| MotPB, -IPTG | 2-5 | 3.3 | 1.8-6.4 | 4.5 |
| MotPB, +IPTG | 2-5 | 3.5 | 1.8-5.7 | 4.3 |
| MotPB-M, -IPTG | 3-7 | 5.2 | 2.9-8.2 | 5.2 |
| MotPB-M, +IPTG | 5-9 | 7.3 | 5.7-8.9 | 7.2 |
Flagella were counted in 50 cells. The standard deviations of the average values shown were less than 2 for all values.
Measurements were made on 50 cells. The standard deviations of the average values shown were between 0.8 and 1.3.
Motility of the strains-bearing four different mot loci was first assayed in comparison with wild-type on soft agar plates identical to those used in the original study of wild-type B. subtilis, single and double motAB and motPS mutant strains26. The ΔABΔPS strain was the negative control (Table 2). The dominance of MotAB in the motility profile of the wild-type laboratory strain was evident in the profile of spreading on soft agar plates as a function of pH, carbon source and Na+. Also evident, was the absence of motility in strains with hybrid stators in the absence of IPTG. In the presence of IPTG, MotAS-M exhibited significantly more motility than MotPB-M under all conditions except on glucose plates without added Na+. As highlighted by the colored bars in Table 2, the patterns of MotAB and MotPB-M were similar with respect to their better motility on glucose than on malate and with respect to the absence of a positive effect of elevated Na+ and pH. By contrast the MotPS and MotAS-M exhibited the opposite pattern of better motility on malate than glucose, better motility at elevated Na+ and somewhat better motility at elevated pH. On malate media with added Na+ at pH 8.3, MotPS exhibited comparable spreading to MotAB and wild-type.
Table 2.
Motility of B. subtilis mot strains on soft agar plates after 24h.a
| pH7.0 | pH8.3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Malate | Glucose | Malate | Glucose | ||||||
| train | 5mM Na+ | − | + | − | + | − | + | − | + |
| wild type | 33.1 | 31.5 | 55.4 | 51.9 | 31.4 | 29.3 | 51.5 | 48.7 | |
| MotAB | 32.1 | 30.1 | 52.2 | 47.3 | 32.9 | 28.3 | 52.7 | 49.3 | |
| MotPS | 13.8 | 21.7 | 13.1 | 18.4 | 18.5 | 24.9 | 15.9 | 22.1 | |
| MotAS-M (+IPTG) | 9.3 | 16.3 | 6.8 | 11.5 | 11.1 | 19.3 | 8.3 | 15.2 | |
| MotAS-M (no IPTG) | 2.3 | 2.4 | 2.2 | 2.3 | 2.5 | 2.7 | 2.4 | 2.6 | |
| MotPB-M (+IPTG) | 7.3 | 6.4 | 9.2 | 8.9 | 5.2 | 4.3 | 8.2 | 7.6 | |
| MotPB-M (no IPTG) | 2.4 | 2.3 | 2.5 | 2.3 | 2.2 | 2.1 | 2.2 | 2.1 | |
| ΔABΔPS | 2.1 | 2.2 | 2.2 | 2.2 | 2.1 | 2.2 | 2.3 | 2.1 | |
All values are the diameter of the indicated strain, in mm, after 24 h on media containing either glucose or malate ± 5 mM added Na+ at pH 7.0 or 8.3. The results shown are the mean values for five independent experiments with the wild-type,MotAB, MotPS, MotAS-M, MotPB-M and Δ ABΔ PS strains. The standard deviations of duplicate or triplicate measurements within experiments were well below 5%.
Swimming speed of the wild-type and hybrid B. subtilis stators as a function of viscosity, pH and [Na+]
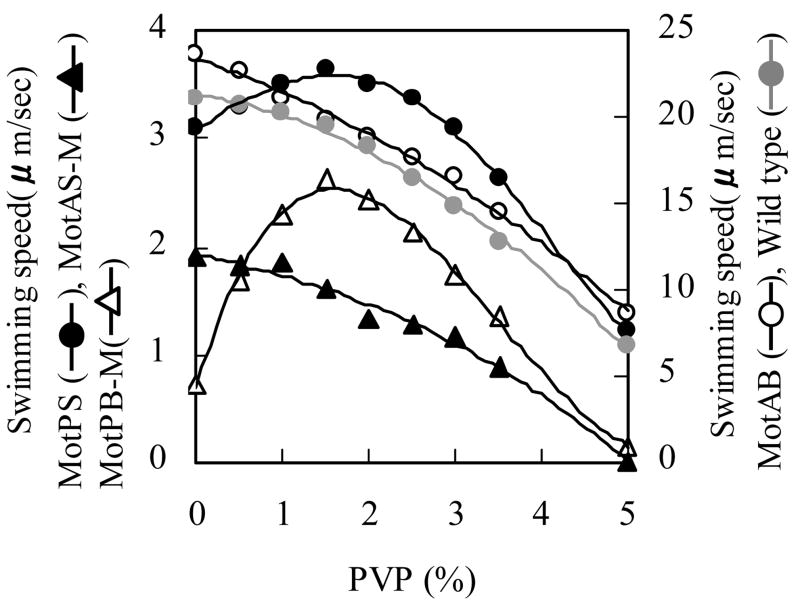
Preliminary assays of swimming in liquid showed that a richer medium than used for the soft agar assays is required to support optimal swimming in liquid, so TY medium (LB medium without added NaCl) was used; this medium contained 14-17 mM contaminating Na+. The assays also showed extremely slow swimming speeds for the MotPB-M strain. Since substantial motility was observed for the MotPS and MotPB-M on soft agar plates (Table 2), we explored the possibility that increasing the viscosity of the liquid medium would increase the swimming speed of some of the strains and of MotPB-M in particular. As shown in Fig. 3, MotAB exhibited a much faster swimming speed than any of the other strains at pH 7 and the swimming speed of MotPB-M was the slowest. Addition of PVP up to about 2% wt/vol increased the swimming speed of MotPB-M and MotPS and decreased the swimming speed of MotAB and MotAS-M. The wild-type strain exhibited slightly lower swimming speeds and its swimming was stimulated modestly by PVP at the same concentration at which swimming of MotPS and MotPB-M was stimulated. In subsequent experiments MotPB-M swimming was assayed both in the absence and in the presence of 1.5% PVP. When used, PVP was added to the assay medium but not to the pre-growth medium and motility was assayed immediately upon dilution of the cells into the motility assay medium.
Figure 3.
Effect of PVP on the swimming speed of wild-type and single Mot-bearing strains. Each strain was grown overnight on TY medium, pH 7.0, at 37 C. A 20 μl sample was used to inoculate 1ml of a fresh TY medium, and growth was continued at 37 C for 6 hours. For growth of MotAS-M and MotPB-M, 0.6mM IPTG was added during this period. After 6 hours, each cell culture (at an A600 of about 0.45-0.50) was diluted by 50-fold with TY medium, pH 7.0, with a range of PVP contents. The swimming speed was assayed by dark field microscopy, as described under Materials and Methods. The results represent the averages of three independent experiments in each of which the swimming speed of twenty independent cells was calculated.
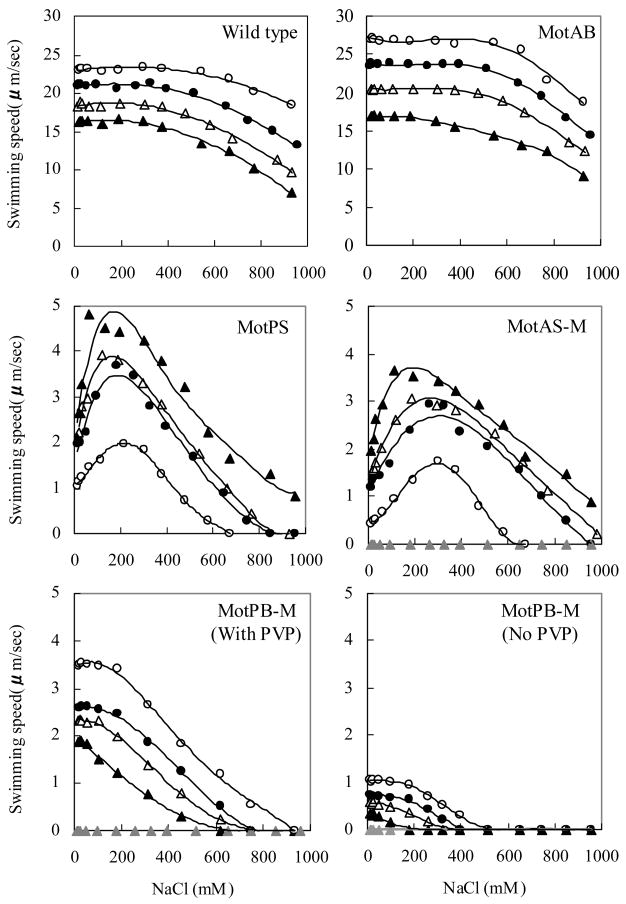
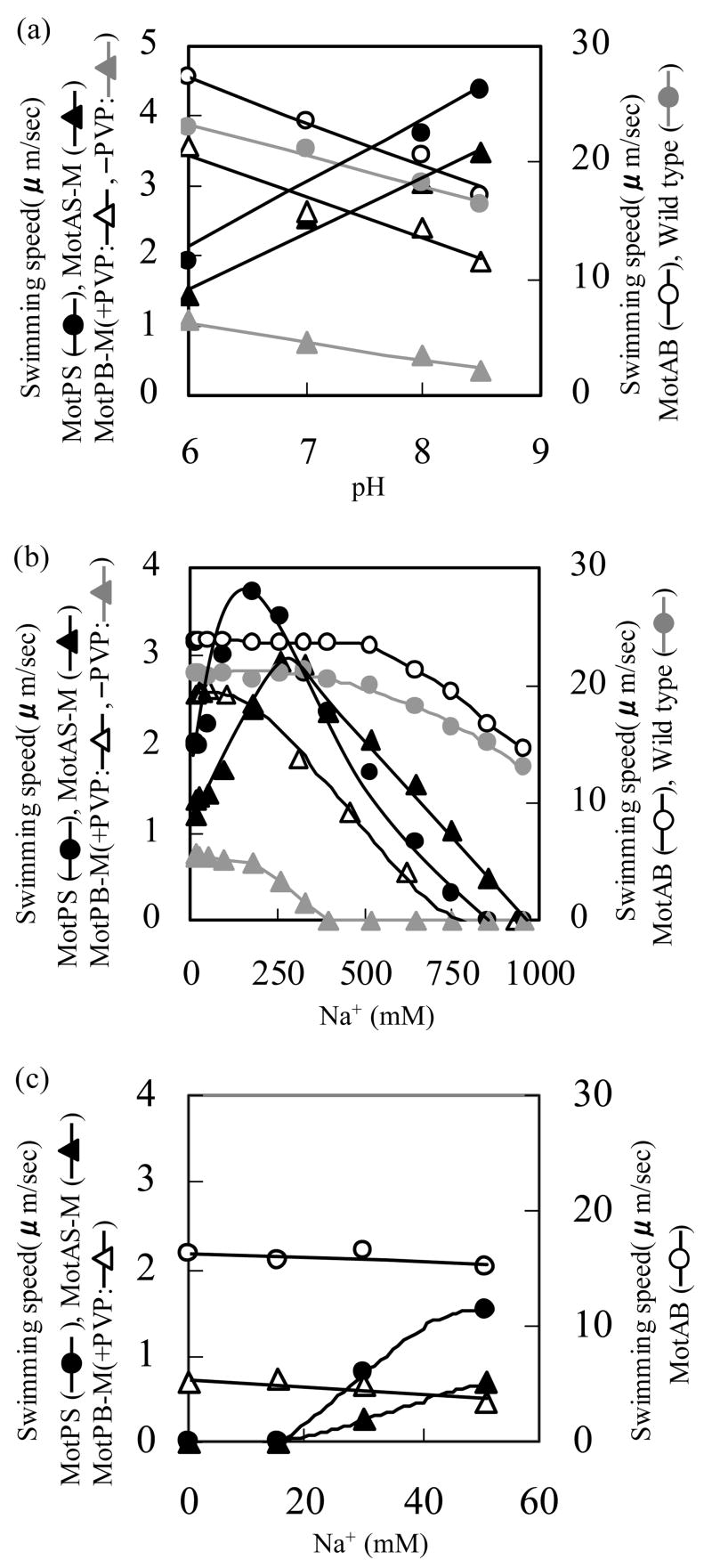
The swimming speed of the wild-type and four single Mot-bearing strains was next measured as a function of the Na+ concentration at four different pH values from 6 and 8.5. The entire data set is shown in Fig. 4. Fig. 5a and 5b respectively focus on the dependence of swimming speed on the pH, at the optimal Na+ concentration for each strain (Fig. 5a), and the dependence of swimming speed on Na+ concentration at pH 7.0 (Fig. 5b). As in the soft agar assays, no swimming was observed by MotAS-M and MotPB-M in the absence of IPTG. In contrast to the observations on soft agar, the swimming speed of wild-type and MotAB were much faster than that of any of the other strains under all conditions. The patterns of response to pH, concentrations of Na+ up to about 100-250 mM and concentrations of Na+ from 300 to almost 1000 mM fell into distinct groups. The swimming of both MotAB and MotPB-M was highest at the most acid pH tested (pH 6.0) and declined with increasing pH, and the converse was true of MotPS and MotAS-M (Fig. 4 and 5a). As shown most clearly in Fig. 5a, a contribution of MotPS to wild-type motility was discernible both in the slower swimming speed in liquid of wild-type compared to MotAB and in the retention of higher swimming speed by wild-type than MotAB as the pH was raised. The swimming speeds of MotPS and MotAS-M were both significantly stimulated by Na+ (Fig. 4 and 5b). The positive response of MotAS-M was more pH-sensitive than that of MotPS. Both strains required the lowest Na+ concentrations, below 200 mM, for optimal swimming when the pH was 8.5 (Fig. 4), but at pH 7.0, the optimal Na+ for MotPS was 180 mM while that for MotAS-M was 260 mM (Fig. 5b). The swimming speed of wild-type, MotAB and MotPB-M was not enhanced by Na+ and was affected very little by concentrations of Na+ up to 100 mM. The pattern of response of the four single-stator strains to Na+ and pH suggested that the hybrid MotPB-M and MotAB use the same coupling ion, i.e. H+, and that the hybrid MotAS-M and MotPS use the same coupling ion, i.e. Na+. In order to further test this hypothesis, the strains expressing single stators were assayed in TY medium diluted 1:20 with water so that the medium contained < 1 mM contaminating Na+. The pH was adjusted to the optimal for each strain, i.e. pH 6.0 for MotAB and MotPB-M and pH 8.5 for MotPS and MotAS-M. Mot PB-M did not exhibit motility in the absence of added PVP under these conditions. As shown in Fig. 5c, although the swimming speeds were lower in the diluted than in the undiluted medium, a clear dependence of MotPS and MotAS-M motility on added Na+ was observed.
Figure 4.
The swimming speed of the wild-type and four single Mot-bearing strains in liquid media as a function of the Na+ concentration and pH. Each strain was pre-grown overnight on TY medium, pH 7.0, and then inoculated into fresh TY medium and grown for 6 more hours as described in the legend to Figure 3, except that the TY medium for the 6 hour growth period contained several Na+ concentrations at four different pH values, pH 6.0 (open circles), 7.0 (closed circles), 8.0 (open triangles), and 8.5 (closed triangles). For the growth of MotAS-M and MotPB-M, 0.6mM IPTG was added in the each culture medium during this period; data for assays at pH 8.0 without added IPTG are shown by the gray triangles. After 6 hours, each cell culture (at A600 of 0.45-0.50) was diluted by 50 times by same growth medium. When PVP was added to the MotPB-M medium, it was added to 1.5% wt/vol. The swimming speed was analyzed as described under Materials and Methods. Note that the swimming speed scale of the bottom four panels is different from that of the top two panels.
Figure 5.
Dependence of the swimming speed on the pH at the optimal Na+ concentration and the dependence of swimming speed on the Na+ concentration at pH 7.0. (a) Relationship between the swimming speed ant the pH at the optimal Na+ concentration. The swimming speed of wild-type, MotAB and MotPB-M were measured at the indicated pH values in the absence of added Na+; MotPB-M was assayed in the presence and absence of 1.5% PVP. The swimming speed of MotPS and MotAS-M was measured in the presence of 180 mM Na+. The data are extracted from the data shown in Figure 4b. Relationship between the swimming speed and [Na+] at pH 7.0. The data are extracted from the data shown in Figure 4c. The swimming speed for the single stator strains was determined at pH 6.0, for MotAB and MotPB-M (in the presence of 1.5% PVP), and at pH 8.5 for MotPS and MotAS-M in assays conducted in 1/20TY at the indicated concentrations of Na+.
The response profiles of MotAB and MotPB-M, the two H+-coupled strains, diverged at Na+ concentrations above 100 mM. At higher concentrations of Na+, the swimming speed of wild-type and MotAB decreased, but not greatly. Even after a fall-off in speed at Na+ concentrations greater than 600 mM, wild-type and MotAB still exhibited robust swimming. By contrast, the swimming speed of all the other strains, MotPS, MotAS-M as well as MotPB-M, dramatically decreased as the Na+ concentration rose above the optimum. That is, motility by single-stator strains using either MotPS protein component was sensitive to inhibition by very high Na+ concentrations. We hypothesized that this inhibition by Na+ was mediated by elevated intracellular Na+. High cytoplasmic Na+ has been shown to reduce the swimming speed supported by Na+-coupled flagellar motors just as elevated intracellular H+ was found to reduce the speed supported by H+-coupled flagellar motors 42; 43. The similar inhibition pattern of MotPB-M, MotPS and MotAS-M to high extracellular Na+ suggested that use of either MotPS protein component allowed binding of Na+, most likely on the cytoplasmic side. Further, this Na+ binding apparently inhibited energizing influx of either coupling ion, i.e. Na+ influx mediated by MotPS and MotAS-M as well as H+ influx mediated by MotPB-M.
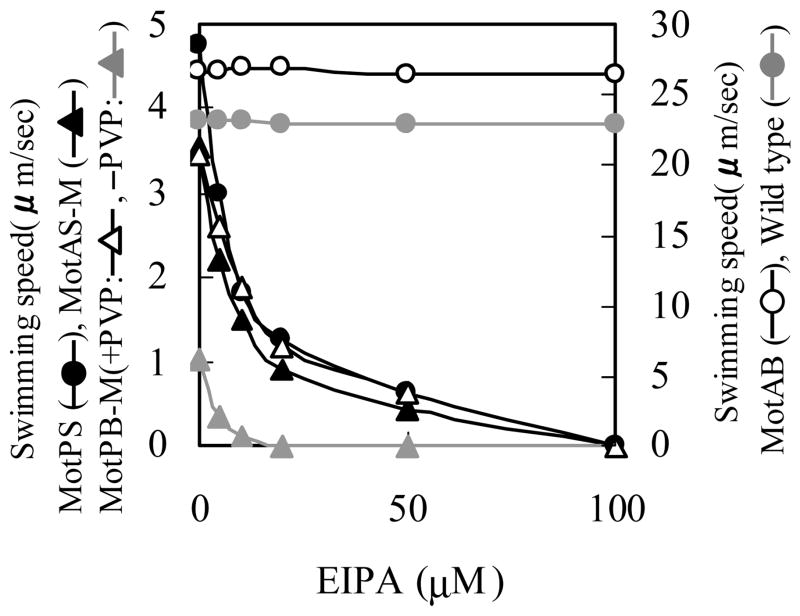
The effect of EIPA on the swimming speed
The Na+ channel blocker EIPA, an amiloride compound, had earlier been shown to inhibit MotPS-dependent motility in alkaliphilic Bacillus pseudofirmus OF4 and in a B. subtilis strain with a disrupted motAB locus26. We hypothesized that if the inhibitory effects of elevated Na+ on swimming by MotPS, MotAS-M and MotPB-M reflected Na+ binding sites on each MotPS protein, EIPA would inhibit those same three strains. We examined the effect of added EIPA on the swimming speed of the strains expressing the different single types of stators. As shown in Fig. 6, the swimming speeds of MotPS, MotAS-M and MotPB-M were all extremely sensitive to inhibition by EIPA whereas those of wild-type and MotAB were not.
Figure 6.
Effect of the Na+ channel inhibitor EIPA on motility. Wild-type, MotAB and MotPB-M strains were grown overnight on TY medium, pH 6.0, at 37 C and the MotPS and MotAS-M strains were grown overnight on TY medium, pH 8.5, at 37 C. Each strain was then inoculated into fresh TY medium at the same pH and grown for 6 hours at 37 C. During this growth period 0.6 mM IPTG was added to the culture medium for MotAS-M and MotPB-M. After 6 hours, each culture was dilute 50-fold into TY medium that was prepared as follows for particular strains: MotAB, TY medium, pH 6.0; MotPB-M, TY medium, pH 6.0, with/without 1.5% (w/v) PVP; MotPS, TY medium, pH 8.5, containing 50 mM NaCl; and MotAS-M, TY medium, pH 8.5, containing 100 mM NaCl. The swimming speed of each strain was measured in the presence of EIPA at concentrations over a range from 0-100 μM. The results represent the averages of three independent experiments in which the swimming speed of twenty independent cells was calculated by each experiment.
Discussion
In Vibrio species, the rotation rate and swimming speed supported by the Na+-coupled polar flagellum are very fast relative to other bacterial flagella 23; 44; 45; 46. By contrast the swimming speed supported by MotPS or either hybrid form that contained a MotPS component was well below that for MotAB-dependent swimming, let alone the much higher speeds reported for V. alginolyticus and V. paraheamolyticus 23; 44; 45; 46. Interestingly, the maximal swimming speed reported for Na+-coupled flagellar motility in alkaliphilic Bacillus YN-1 was in the same range as that shown in this study for MotAB-dependent swimming of B. subtilis 24 although its stator is probably MotPS. These observations on Na+-coupled swimming speed in Bacillus strains indicate that Na+-coupling per se is not a predictor of fast swimming rates; rotation rates have not yet been assayed for MotPS-supported systems.
The observations also raise the question of why the swimming speeds supported by B. subtilis MotPS and the hybrid versions are particularly slow in liquid relative to MotAB-dependent motility in B. subtilis even though MotPS can support comparable rates of spreading to MotAB on soft agar plates24; 26. We assessed the number and length of flagella in cells of the single stator strains under conditions of liquid growth in order to examine the possibility that the flagella of MotPS, MotAS-M and MotPB-M might simply have fewer and/or shorter flagella than MotAB when maintained in liquid. This was not the case. All the strains used for the motility assays had higher numbers of flagella/cell and greater flagellar length than found in the stator-less control strain or the original (non up-motile) hybrid strains whose stators did not support motility (Table 1).The lower number and length of flagella in strains lacking functional stators is consistent with an intriguing recent study suggesting that these same flagellar parameters in Salmonella typhimurium are modulated by an environmental sensing role of functional flagella themselves47. For the purposes of this study, the important finding was that there was no correlation between either flagellar number/cell or flagellar length and swimming speed of the assay strains. We hypothesize that the particularly poor motility of MotPS, MotAS-M and MotPB-M relative to MotAB in liquid does not reflect differences in the rotor or flagella of MotAB from the other strains but results from a difference in how MotAB interacts with the flagellar rotor. Perhaps the interactions between the MotPS stator and the two hybrid forms with B. subtilis FliG are qualitatively different from those of MotAB with the same FliG so that motility in liquid is selectively disadvantaged relative to motility on surfaces or in viscous environments. A comparable scenario appears to obtain in P. aeruginosa which has recently been shown to have two stators, MotAB and MotCD, that are apparently both H+-coupled and power a single polar flagellum39; 48. As with B. subtilis MotPS and MotAB, both hybrids of the P. aeruginosa MotAB and MotCD can support motility in liquid48, but MotCD appears to have a crucial role under conditions of high viscosity which suggests that there is something different in its interactions with the flagellar rotor that affects this function39; 48. We also note that E. coli MotAB has been reported to support weak H+-coupled motility of V. cholerae 49, reenforcing the capacity of one FliG species to interact productively with more than one stator but also suggesting that the interaction was sub-optimal relative to the native stator-rotor pair. Further clarification of the molecular interactions of FliG with alternate B. subtilis stators and of the biophysical impact of different interactions will be needed before it will be possible to advance a mechanistic hypothesis for differential outcomes with respect to surface and liquid motility.
This is the first time that both hybrids of differently coupled flagellar stators were functional so that their properties could be assessed. The activity profiles of motility as a function of pH and Na+ concentration, both on soft agar plates and in liquid, support the conclusion that MotPB-M is H+-coupled and MotAS-M is Na+-coupled. In the soft agar motility experiments, the single stator strains that are Na+-coupled, MotPS and MotAS-M, exhibited less motility on glucose-containing plates than on malate-containing plates in the presence of added Na+ (Table 2). This was also observed in the earlier comparison of single stator strains expressing either MotAB or MotPS under control of their native promoters. Since motPS is expressed in an apparent operon together with the glucose catabolite repressor gene ccpA50; 51, the inference was drawn that the poorer motility of MotPS in the presence of glucose resulted in part from a repressive effect of glucose26. However, a comparable effect of glucose on MotPS and MotAS-M in this study, when neither stator is expressed from its natural promoter, rules out a contribution of glucose repression. Rather, we hypothesize that the poorer motility supported by Na+-coupled stators in the presence of glucose as opposed to malate relates to changes in the medium pH, with acidification during glucose metabolism adversely affecting Na+-coupled motility and alkalinization during malate metabolism favoring it.
The finding that MotAB and MotPB-M are H+-coupled whereas MotPS and MotAS-M are Na+-coupled implicates the 1-TMS MotB and MotS components as dominant determinants of ion-selectivity. This is consistent with the conclusion from studies of a hybrid and several chimeric stator forms of Na+- and H+-coupled stators in V. alginolyticus52; 53; 54. For example, a functional hybrid was constructed in a laf (lateral flagella) mutant that also lacked PomA (the MotA homologue of the Na+-coupled stator) of Vibrio. This strain was non-motile but Na+-coupled motility was restored by expression of MotA from the H+-coupled Rhodobacter spheroides, implicating PomB as a major determinant of ion-specificity52.
The presence of MotA vs MotP influences the activity profile of B. subtilis stator hybrids as a function of pH and [Na+], albeit in a more nuanced manner than the presence of MotB vs. MotS. First, while the swimming speed of both MotAS-M and MotPS was enhanced by increasing Na+ up to about 250 mM, the optimal Na+ is more pH-dependent in MotAS-M than MotPS, i.e. MotAS-M required relatively higher concentrations of Na+ at low pH than MotPS (Fig. 4). This suggests that the MotA component confers H+-responsiveness that is absent in MotP-containing stators. Conversely, severe inhibition of swimming at Na+ concentrations above about 300 mM was observed for each stator that has either MotP, MotS or both; at these high Na+ concentrations, cytoplasmic levels of the cation are the presumed inhibitory agent43. Inhibition of MotPB-M as well as MotPS and MotAS-M suggests that both MotP and MotS participate in Na+ responsiveness. This was re-enforced by the finding that the amiloride compound EIPA inhibited motility of the same three stators, MotPS, MotAS and MotPB, even though one of them, MotPB, is H+-coupled (Fig. 6). We hypothesize that a functional site for Na+ and inhibitory amiloride compounds is retained by MotP in the MotPB hybrid except that in the hybrid it is the inward flux of H+ rather than Na+ that is now inhibited. This is consistent with reports of inhibition of the amiloride analogue phenamil at more than one site in the Na+-coupled Vibrio stator and demonstration of mutations conferring phenamil-resistant motility in both pomA and pomB55; 56; 57.
A property of the stator to which the 4-TMS MotP or MotA made a dominant contribution was the response to increases in viscosity upon addition of PVP up to 1-2% wt/vol (Fig. 3). The effect of PVP did not depend upon exposure during growth before the motility assay; it was observed immediately under assay conditions, indicating that high viscosity is favorable for formation and/or function of bundles of flagella that are powered by MotP-containing stators. This correlates, in turn, with the larger contribution of MotPS to swimming behavior of B. subtilis on soft agar surfaces than in liquid, with MotPS having a dominant role at high pH and Na+ when expressed from its native promoter in an up-motile strain26. In Vibrio species, it is the lateral, H+-coupled flagella that support motility in viscous environments rather than the Na+-coupled polar flagellum that supports fast motility in liquid23; 29; 58. By contrast, as noted above, one of two polar, H+-coupled stators of P. aeruginosa supports swarming and swimming at high viscosity39; 48. The mechanism whereby particular Mot complexes support motility especially well on surfaces and at high viscosity is yet to be clarified but it apparently does not depend upon coupling to polar vs. lateral/peritrichous flagella or to H+ vs. Na+. The importance of surface motility and motility in viscous niches is an emerging theme in environmental bacteriology and bacterial pathogenesis59. Maintenance media for laboratory strains of B. subtilis do not create conditions of viscosity, non-fermentative carbon sources, elevated pH and Na+ that could readily select for a dominant MotPS role, but such conditions are found in its natural niches in association with plants 60. Perhaps MotPS will have a more prominent role in undomesticated B. subtilis strains than observed here, just as they display behaviors such as swarming motility and fruiting body production that are absent or less robust features of laboratory strains61; 62. Even in laboratory strains, a benefit of MotPS is indicated by its retention in the B. subtilis genome even though it reduces the swimming speed in liquid relative to a strain expressing only MotAB (Fig. 3-5).
Our studies of the dual stators of B. subtilis have not yet addressed two questions of interest: (i) whether MotPS and MotAB can be assembled together in a single stator complex or must assemble into their own discrete 4MotP-2MotS and 4MotA-2MotB complexes; and (ii) if only discrete MotAB- and MotPS-containing stators can be formed, whether individual flagella are powered by a mixture of MotAB and MotPS stator complexes as opposed to there being discrete MotAB- and MotPS-powered flagella. The swimming speed pattern of the wild-type largely paralleled the pattern of MotAB while showing some contribution of MotPS, i.e. evidence by PVP stimulation and by less inhibition at elevated pH relative to MotAB. We note, though, that wild-type swimming speed was not inhibited at all by EIPA (Fig. 6) and was less inhibited than any of the strains, including MotAB, by Na+ concentrations above 600 mM (Fig. 5B). These properties of wild-type motility diverge from those expected if wild-type motility in liquid represents the sum of a MotAB-dominated swimming pattern and a small contribution of MotPS. Perhaps this divergence indicates the effect of a modulatory interaction between the two stator types as they function together within single stator complexes and/or together contribute to the rotor motility of individual flagella.
In summary, this study demonstrated that the presence of Mot stators influences the number of flagella/cell and that the native and two hybrid forms of the Na+-coupled MotPS and H+-coupled MotAB stators of B. subtilis all support motility. The relative effectiveness of MotAB compared to MotPS in supporting motility was much greater in liquid media than on soft agar plating media. MotAB-dependent motility was favored by low viscosity, low Na+ and low pH whereas MotPS-dependent motility exhibited opposite optima. The Na+ and pH profiles suggested that the MotAS-M is Na+-coupled and MotPB-M is H+-coupled. However, features of the Na+ response curves as a function of pH indicated that MotA and MotP components contribute, respectively, to sensitivity to H+ and Na+ (as well as to EIPA). Motility supported by MotP-containing stators was stimulated by increases in viscosity even though their coupling ion is different, one of several pieces of evidence that swimming speed under particular conditions is not directly related to the use of H+ or Na+. The two stators of B. subtilis offer an opportunity to further probe the basis for the different properties, the interactions and the roles of two stators working with a common flagellar rotor.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 3. E. coli strains were routinely grown at 37 C in LB medium63. B. subtilis 168 strain BR151MA (wild-type) and its derivatives were routinely grown at 37 C in LB medium or TY medium (1% Bacto Tryptone and 0.5% Yeast Extract) for liquid cultures. TY medium contained 14-17 mM Na+. TY medium was also used for assays of swimming speed in liquid, with the pH, Na+ content, PVP content and EIPA addition indicated for specific experiments. When Na+ was added, the actual final Na+ concentration of each assay medium was determined with a flame photometer (Model AMA-175, Tokyo Koden, Tokyo, Japan) calibrated with standard Na+ solutions of known concentrations. Several plating media were used. Spore stocks were prepared on agar plates of Difco Nutrient Sporulation Medium and grown out on tryptose blood agar base (TBAB). Assays of motility on soft agar plates were conducted on Spizizen-salt based plating media64, except that sodium citrate was omitted. The salts were supplemented with 0.05% yeast extract and either glucose or potassium malate (to 25 mM), supplemented with required amino acids and, where indicated, NaCl. The medium was adjusted to either pH 7.0 or 8.3 and solidified with 0.25% Noble agar (Difco) as described earlier26. For experiments with strains in which mot genes were expressed from the amyE locus under control of the Pspac promoter, 0.6mM IPTG was included in the growth medium. Ampicillin at 100 μg/ml, chloramphenical at 5 μg/ml and spectinomycin at 150 μg/ml were added as needed for growing plasmid-bearing cells as well as for selecting transformants. Transformation of E. coli strains and all recombinant DNA manipulations were carried out by standard methods 63.
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5 αMCR | F−mcrAΔ 1 (mrr-hsd RMS-mcrBC) φ80dlacZΔ M15 Δ (lacZYA-argF)U169 deoR recA1 endA1 supE44 λ thi-1 gyr-496 relA1 | GIBCO-BRL |
| B. subtilis | ||
| BR151MA | lys3 trpC2 (wild type) | Grundy et al. (1994) |
| Δ PS | BR151MA Δ motPS::neo | Ito et al., 2004 |
| Δ ABΔ PS | BR151MA Δ motPS::neo Δ motAB::SpcR | This study |
| MotAB | Δ ABΔ PS amyE::PmotAB-motAB | This study |
| MotAB-Spac | Δ ABΔ PS amyE::Pspac-motAB | This study |
| MotPS | Δ ABΔ PS amyE::PmotAB-motPS | This study |
| MotPS-Spac | Δ ABΔ PS amyE::Pspac-motPS | This study |
| MotAS | Δ ABΔ PS amyE::PmotAB-motAS | This study |
| MotAS-Spac | Δ ABΔ PS amyE::PspacmotAS | This study |
| MotAS-M | Δ ABΔ PS amyE::PspacmotAS, up-motile variant | This study |
| MotPB | Δ ABΔ PS amyE::PmotAB-motPB | This study |
| MotPB-Spac | Δ ABΔ PS amyE::Pspac-motPB | This study |
| MotPB-M | Δ ABΔ PS amyE::Pspac-motPB, up-motile variant | This study |
| Plasmids | ||
| pMW118 | Cloning vector (Apr) | Nippon Gene |
| pDR67 | amyE integration vector with Cmr gene and Pspac promoter upstream of multiple cloning site | Ireton, K., et al. 1993. |
| pDG1730 | amyE integration vector with Ampr, Specr, and Ermr genes | Guerout-Fleury et al. 1996 |
| pDR-AB | pDR67 + motAB | This study |
| pDR-AS | pDR67 + motAS | This study |
| pDR-PS | pDR67 + motPS | This study |
| pDR-PB | pDR67 + motPB | This study |
| pDG-AB | pDG1730 + PmotAB-motAB | This study |
| pDG-AS | pDG1730 + PmotAB-motAS | This study |
| pDG-PS | pDG1730 + PmotAB-motPS | This study |
| pDG-PB | pDG1730 + PmotAB-motPB | This study |
Construction of a Δ motAB and Δ motPS double deletion mutant
For construction of a fragment carrying Emr gene cassette between upstream motA gene and downstream motB gene, a series of PCR reactions were performed with custom-made primers to create a product that could be directly transformed into the Δ PS strain51. Selection for Emr colonies yielded the strain designated Δ ABΔ PS, which was confirmed by PCR and Southern blot analyses to have a deletion of the motAB region. For this mutant and each type strain that was constructed using it, the phenotypes of several independent isolates was first assessed on soft agar plate assays to assure that the strain chosen for use in the studies was typical. Each strain was initially confirmed to have the expected PCR profile and was then directly shown to contain the expected sequence. The sequencing was performed by Hitachi System Sciences (Tokyo, Japan), using an ABI-100 model 377 sequencer.
Integration of selected mot genes into the amyE loci of particular mutant strains under a Pspac promoter and a PmotAB promoter
Four new derivatives of the Δ ABΔ PS strain, that had been prepared as described above, were constructed. Each strain contains a pair of mot genes encoding one of the four stators of interest. The genes were integrated into the chromosomal amyE locus under control of an IPTG-inducible Pspac or PmotAB promoter. Plasmid pDR67 was used for construction of strains using the IPTG-inducible Pspac promoter65, and pDG1730 was used for construction of strains using the PmotAB promoter66. Plasmid pDR67 contains fragments of the front and back ends of the amyE gene flanking a chloramphenicol resistance (Cmr) gene and also contains the Pspac promoter upstream of a multiple-cloning site. Plasmid pDG1730 contains fragments of the front and back ends of the amyE gene flanking a spectinomycin resistance (Spr) gene. (i) For construction of an IPTG-inducible Pspac promoter plasmid that was carrying intact motAB genes, PCR was performed on wild-type DNA with the appropriate primers. For construction of a plasmid that was carrying intact motPS genes under control of Pspac promoter, sets of primers were designed with nucleotides encoding a SphI site and an XmaI site. Each amplified fragment was cloned into SphI- and XmaI-digested pDR67, yielding pDR-AB and pDR-PS, respectively. (ii) For construction of a plasmid carrying the intact hybrid motAS genes under control of Pspac promoter, a series of PCR reactions were performed to create a product that, when digested with SphI and XmaI, could be cloned into SphI- and XmaI-digested pDR67, yielding pDR-AS. (iii) For construction of a plasmid carrying the intact hybrid motPB under control of Pspac promoter, a series of PCR reactions were performed to produce a product that, when digested with SphI and XmaI, could be cloned into SphI- and XmaI-digested pDR67, yielding pDR-PB. (iv) For construction of a plasmid that was carrying intact motAB genes under control of PmotAB promoter, a series of PCR reactions were performed to produce a product that, when digested with BamHI, could be cloned into BamHI-digested pDG1730, yielding pDG-AB. (v) For construction of a plasmid carrying the intact motPS genes under control of PmotAB promoter, a series of PCR reactions were performed to produce a product that, when digested with BamHI, could be cloned into BamHI-digested pDG1730, yielding pDG-PS. (vi) For construction of a plasmid carrying the intact hybrid motAS genes under control of PmotAB promoter, a series of PCR reactions were performed to produce a product that, when digested with BamHI, could be cloned into BamHI-digested pDG1730 yielding pDG-AS. (vii) For construction of a plasmid carrying the intact hybrid motPB genes under control of PmotAB promoter, a series of PCR reactions were performed to produce a product that could be cloned into EcoRV-digested pDG1730 yielding pDG-PB.
Each plasmid was used to transform particular mutants to a chloramphenicol-resistance, amylase-negative phenotype for pDR67 derivative, and to a spectinomycin resistance, amylase-negative phenotype for pDG1730 derivative. Recombinant transformants were selected by conventional techniques, and the presence of the insert was confirmed.
Visualization of flagella
The method described by Aono et al.67 as an adaptation of a standard flagella staining method was used. A loopful of several independent cultures of the double mutant and each single stator strain was transferred gently to a microscope slide, forming a thin film. The sample was air-dried and then heated briefly with a hair dryer. Staining was carried out for 30 minutes with a 2:1:0.1:0.15 (vol:vol) solution of: 10% w/v tannic acid, saturated potassium aluminum, saturated aniline and 5% w/v FeCl3, followed by ammoniac silver nitrate for 1 min. Observations of flagella were made using bright field microscopy (1000X). The number of flagella/cell and flagellar length were recorded for 50 cells. Negative staining of cells for transmission electron microscopy (JEM-1210 apparatus, JEOL) was carried out as described by Zillig et al. 68.
Motility on soft agar plates: isolation of up-motile strains and assays
Observations of motility of the newly isolated strains with hybrid stators were made in liquid. Observations of modest indications of motility were the basis for formulation of TY plating media solidified with 0.3% Noble agar for assessment of motility and isolation of up-motile strains. For MotAS and MotAS-M, the up-motile derivative of MotAS, the medium was at pH 8.5 and contained 200mM NaCl (with or without 0.6 mM IPTG). For MotPB and its up-motile derivative MotPB-M, the medium was at pH 6.0 (with or without 0.6 mM IPTG). A 1 μl drop from an overnight TY culture under the same conditions was spotted on to the centre of the plate. The plates were inoculated at 37 C for 36 h.
Assays of motility of B. subtilis strains were conducted exactly as described earlier26. Briefly, a colony from TBAB plates was used to inoculate overnight TY cultures; these overnight cultures served as the inoculation (1 μl in the centre of the modified Spizizen salt-glucose or malate plate) for the assays. The plates were incubated at 37 C, and the diameters of the colonies were measured at the indicated times.
Microscopic observation and recording of swimming speeds of strains in liquid cultures
Microscopic observation of swimming in liquid was carried out using a Leica DMLB100 microscope and Leica DC300F video camera system (Leica Geosystems, Tokyo), and recordings were made using a Leica IM50 version 1.20 system (Leica Geosystems, Tokyo) and analyzed using Display capture ARE software (http://www.vector.co.jp/soft/win95/art/se221399.html). For experimental assay of swimming speed, cultures were grown on TY medium, pH 7.0, overnight at 37 C. 20 μl of each culture was the inoculated into 1ml of a fresh TY medium and grown at 37 C for 6 hours; for the MotAS-M and Mot-M strains, 0.6 mM IPTG was added to the growth medium during this period. Experiments were initiated by diluting the 6-hour culture (which was at a A600 of 0.45-0.50) 50-fold into TY medium at the pH and with additions that are specified for each experiment. Swimming was immediately observed by dark field microscopy, recordings were made from which the swimming speed of individual cells was calculated. All results shown are the averages of three independent experiments in which the speed of 20 different cells was measured. The patterns of responses to all effectors by each strain and relative to one another were highly reproducible, with standard deviations of the means for each condition, i.e. the values shown in the figures, being within 15% of the mean.
Acknowledgments
We thank David Blair for critically reading this manuscript and Tina Henkin for early encouragement to examine hybrid stators of B. subtilis MotPS and MotAB. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Area “Genomic Biology” and the 21rst Century Center of Excellence program and high-technology research centers organized by the Ministry of Education, Culture, Sports, Sciences and Technology of Japan (to M.I.) and research grant GM28454 from the National Institute of General Medical Sciences (to TAK).
Abbreviations used
- EIPA
5-(N-ethyl-N-isopropyl)-amiloride
- IPTG
isopropyl-1-thio-β-D-galactopyranoside
- LB
Luria Bertani broth
- pmf
proton motive force
- PVP
polyvinylpyrrolidone
- smf
sodium motive force
- TMS
transmembrane segment of a membrane protein
- TY
tryptone-yeast extract medium
References
- 1.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 2.Larsen SH, Adler J, Gargus JJ, Hogg RW. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 4.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 5.Berry RM. Theories of rotary motors. Philos Trans R Soc Lond B Biol Sci. 2000;355:503–509. doi: 10.1098/rstb.2000.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yorimitsu T, Mimaki A, Yakushi T, Homma M. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J Mol Biol. 2003;334:567–583. doi: 10.1016/j.jmb.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 7.Yorimitsu T, Homma M. Na+-driven flagellar motor of Vibrio. Biochim Biophys Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 8.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platzer J, Sterr W, Hausmann M, Schmitt R. Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J Bacteriol. 1997;179:6391–6399. doi: 10.1128/jb.179.20.6391-6399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S, Dapice M, Reese TS. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 11.Ridgway HG, Silverman M, Simon MI. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J Bacteriol. 1977;132:657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato K, Homma M. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 15.Chun SY, Parkinson JS. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 16.Khan S, Khan IH, Reese TS. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J Bacteriol. 1991;173:2888–2896. doi: 10.1128/jb.173.9.2888-2896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan S, Ivey DM, Krulwich TA. Membrane ultrastructure of alkaliphilic Bacillus species studied by rapid-freeze electron microscopy. J Bacteriol. 1992;174:5123–5126. doi: 10.1128/jb.174.15.5123-5126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun TF, Blair DF. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry. 2001;40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 19.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry. 2004;43:35–45. doi: 10.1021/bi035406d. [DOI] [PubMed] [Google Scholar]
- 20.Shioi JI, Imae Y, Oosawa F. Protonmotive force and motility of Bacillus subtilis. J Bacteriol. 1978;133:1083–1038. doi: 10.1128/jb.133.3.1083-1088.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura S, Shioi J, Imae Y. Motility in Bacillus subtilis driven by an artificial protonmotive force. FEBS Lett. 1977;82:187–190. doi: 10.1016/0014-5793(77)80581-4. [DOI] [PubMed] [Google Scholar]
- 23.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 24.Hirota N, Imae Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 25.Hirota N, Kitada M, Imae Y. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+ FEBS Lett. 1981;132:278–280. [Google Scholar]
- 26.Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers B, Zvi L, Uematsu K, Krulwich TA. MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis. Mol Microbiol. 2004;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. [DOI] [PubMed] [Google Scholar]
- 27.Hase CC, Barquera B. Role of sodium bioenergetics in Vibrio cholerae. Biochim Biophys Acta. 2001;1505:169–178. doi: 10.1016/s0005-2728(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 28.Takami H, Takaki Y, Uchiyama I. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 2002;30:3927–3935. doi: 10.1093/nar/gkf526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarter LL. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 30.McCarter LL. Multiple modes of motility: a second flagellar system in Escherichia coli. J Bacteriol. 2005;187:1207–1209. doi: 10.1128/JB.187.4.1207-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuura S, Shioi JI, Imae Y, Iida S. Characterization of the Bacillus subtilis motile system driven by an artificially created proton motive force. J Bacteriol. 1979;140:28–36. doi: 10.1128/jb.140.1.28-36.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aizawa S, Zhulin IB, Marquez-Magana L, Ordal GW. Chemotaxis and motility. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its Closest Relatives from Genes to Cells. ASM Press; Washington, DC: 2002. [Google Scholar]
- 33.Ordal GW, Marquez-Magana, Chamberlin MJ. Motility and chemotaxis. In: Sonenshein AL, Hock JA, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press; Washington, D.C: 1993. pp. 765–784. [Google Scholar]
- 34.West JT, Estacio W, Marquez-Magana L. Relative roles of the fla/che PA, PD-3, and PsigD promoters in regulating motility and sigD expression in Bacillus subtilis. J Bacteriol. 2000;182:4841–4848. doi: 10.1128/jb.182.17.4841-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugiyama S, Cragoe EJ, Jr, Imae Y. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J Biol Chem. 1988;263:8215–8219. [PubMed] [Google Scholar]
- 36.Atsumi T, Maekawa Y, Tokuda H, Imae Y. Amiloride at pH 7.0 inhibits the Na+-driven flagellar motors of Vibrio alginolyticus but allows cell growth. FEBS Lett. 1992;314:114–116. doi: 10.1016/0014-5793(92)80954-f. [DOI] [PubMed] [Google Scholar]
- 37.Atsumi T, Sugiyama S, Cragoe EJ, Jr, Imae Y. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J Bacteriol. 1990;172:1634–1639. doi: 10.1128/jb.172.3.1634-1639.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiyama S, Magariyama Y, Kudo S. Forced rotation of Na+-driven flagellar motor in a coupling ion-free environment. Biochim Biophys Acta. 2004;1656:32–36. doi: 10.1016/j.bbabio.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant GF, Simon MI. Synthesis of bacterial flagella. II PBS1 transduction of flagella-specific markers in Bacillus subtilis. J Bacteriol. 1969;99:116–124. doi: 10.1128/jb.99.1.116-124.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senesi S, Ghelardi E, Celandroni F, Salvetti S, Parisio E, Galizzi A. Surface-associated flagellum formation and swarming differentiation in Bacillus subtilis are controlled by the ifm locus. J Bacteriol. 2004;186:1158–1164. doi: 10.1128/JB.186.4.1158-1164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minamino T, Imae Y, Oosawa F, Kobayashi Y, Oosawa K. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J Bacteriol. 2003;185:1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida S, Sugiyama S, Hojo Y, Tokuda H, Imae Y. Intracellular Na+ kinetically interferes with the rotation of the Na+-driven flagellar motors of Vibrio alginolyticus. J Biol Chem. 1990;265:20346–20350. [PubMed] [Google Scholar]
- 44.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 45.Kudo S, Imai N, Nishitoba M, Sugiyama S, Magariyama Y. Asymmetric swimming pattern of Vibrio alginolyticus cells with single polar flagella. FEMS Microbiol Lett. 2005;242:221–225. doi: 10.1016/j.femsle.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Muramoto K, Kawagishi I, Kudo S, Magariyama Y, Imae Y, Homma M. High-speed rotation and speed stability of the sodium-driven flagellar motor in Vibrio alginolyticus. J Mol Biol. 1995;251:50–58. doi: 10.1006/jmbi.1995.0415. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 2005;24:2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toutain CM, Zegans ME, O'Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosink KK, Hase CC. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol. 2000;182:4234–4240. doi: 10.1128/jb.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henkin TM. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 51.Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 52.Asai Y, Kawagishi I, Sockett RE, Homma M. Hybrid motor with H+- and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J Bacteriol. 1999;181:6332–6338. doi: 10.1128/jb.181.20.6332-6338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asai Y, Kawagishi I, Sockett RE, Homma M. Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 2000;19:3639–3648. doi: 10.1093/emboj/19.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asai Y, Yakushi T, Kawagishi I, Homma M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J Mol Biol. 2003;327:453–463. doi: 10.1016/s0022-2836(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 55.Jaques S, Kim YK, McCarter LL. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci USA. 1999;96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations in putative channel components. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 57.Kojima S, Kuroda M, Kawagishi I, Homma M. Random mutagenesis of the pomA gene encoding a putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology. 1999;145:1759–1767. doi: 10.1099/13500872-145-7-1759. [DOI] [PubMed] [Google Scholar]
- 58.Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J Bacteriol. 1996;178:5024–5026. doi: 10.1128/jb.178.16.5024-5026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 60.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 61.Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 62.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook J, Frisch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 64.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 66.Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 67.Aono R, Ogino H, Horikoshi K. pH-dependent flagella formation by facultative alkaliphilic Bacillus sp C-125 . Biosci Biotechnol Biochem. 1992;56:48–53. doi: 10.1271/bbb.56.48. [DOI] [PubMed] [Google Scholar]
- 68.Zillig W, Holz I, Janekovic D, Klenk HP, Imsel E, Trent J, Wunderl S, Forjaz VH, Coutinho R, Ferreira T. Hyperthermus butylicus, a hyperthermophilic sulfur-reducing archaebacterium that ferments peptides. J Bacteriol. 1990;172:3959–3965. doi: 10.1128/jb.172.7.3959-3965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]