Summary
Both canonical Wnt/β-catenin and TGFβ/Smad signaling pathways coordinately regulate pattern formation during embryogenesis as well as tumor progression. Evidence of cross-talk between these two pathways has been reported. Here we demonstrated that the Activin-like kinase 4 (Alk4)/Smad2 pathway facilitates the transcriptional activity of the oncogenic Wnt/β-catenin/Tcf4 pathway through a novel Smad4-independent mechanism. Upon activation, Smad2 physically interacted with Tcf4, β-catenin and the co-activator p300 to enhance transcriptional activity of β-catenin/Tcf4 through the histone acetyltransferase activity of p300. Transactivation by Smad2 was independent of a Smad-binding element (SBE) and Smad4. Indeed, the enhancement of β-catenin/Tcf4 transcriptional activity by activated Smad2 was negatively regulated by the presence of Smad4. Moreover, a tumor-derived missense mutant of Smad2, lacking the ability to bind to Smad4 was still able to enhance the Tcf4 transcriptional reporter in the presence of β-catenin and Tcf4. Our findings suggest that Smad2 may function as an activator of canonical Wnt/β-catenin/Tcf4 signaling through a SBE/Smad4-independent pathway.
Keywords: Activin, β-catenin, Smads, Tcf/Lef, TGFβ, Wnt
Introduction
Canonical Wnt/β-catenin/Tcf/Lef signaling is one of the major oncogenic pathways in various human cancers [1–4]. Deregulation of this pathway can initiate tumorigenesis in several organs including the intestine, skin and mammary gland [1, 2]. Approximately 90% of human colorectal cancers carry genetic alterations in either adenomatous polyposis coli (APC) or β-catenin genes resulting in constitutive activation of the canonical Wnt/β-catenin signaling pathway [2]. A subpopulation of human breast cancers overexpress Wnt ligands, this induces stabilization and nuclear accumulation of β-catenin through an autocrine mechanism without genetic alterations [5, 6]. Clinical evidences and animal models indicate that deregulation of the Wnt signaling pathway is important for initiation of tumorigenesis, especially in the adenoma-carcinoma sequence of colon [7, 8]. In addition, the Wnt/β-catenin pathway is involved in the induction of epithelial to mesenchymal transition (EMT), which correlates with metastasis of cancer cells [1–3, 9]. However, deregulation of the Wnt pathway is not sufficient to develop invasive tumors and sequential somatic mutations of at least four to five key genes are required before additional metastatic changes occur [10 11]. This suggests that aberrant molecular signaling, caused by a mutation in a tumor suppressor gene, may synergistically amplify Wnt target gene expression and thereby contribute to malignant progression. Several studies have indicated that the interaction between Wnt and other signaling pathways might also contribute to the expression of these Wnt target genes [11, 12]. Detailed mechanisms of such cross-talk during tumor initiation and metastasis remain largely unknown.
Genetic studies have shown that the accumulation of mutations in genes such as Smad2 and Smad4 contribute to the acceleration of colorectal tumorigenesis initiated and maintained by a deregulated Wnt/β-catenin signaling pathway [2, 7]. During early embryogenesis, the canonical Wnt/β-catenin signaling pathway coordinately regulates mesoderm induction and anterior-posterior patterning with the Activin/Nodal/Alk4/Smad signaling pathway [13, 14]. Binding of these TGFβ superfamily ligands to Activin type I receptor and type II receptor results in phosphorylation of intracellular Smad2 and Smad3 which can then hetero-oligomerize with Smad4. Smads function as transcriptional activators or repressors depending on the context [15]. In addition to exhibiting a tumor-suppressor role during early carcinogenesis, components of this pathway may be involved in the etiology and/or progression of human cancers in the later stages of carcinogenesis. In fact, phosphorylated Smad2 has been found in more than 90% of human breast cancers [16]. Induction of EMT which facilitates metastasis of epithelial cancers is induced by the TGFβ/Activin signaling pathway [15]. Nodal is overexpressed in several human breast cancer cell lines [17] and promotes tumor progression in melanoma [18]. The nodal co-receptor Cripto-1 is expressed in approximately 80% of human breast cancers and more than 70% of human primary and metastatic colorectal cancers [19, 20]. These findings suggest that both Wnt/β-catenin and TGFβ/Activin/Nodal/Smad signaling pathways are activated in at least some portion of human cancers.
Several studies have demonstrated a functional interaction between the canonical Wnt/β-catenin and TGFβ signaling pathways [12, 21–23]. For example, interaction between Smads and Lef-1 can enhance transcription of a subset of Wnt-target genes including Twin, Msx-2 or gastrin through the promoters that contain both a Tcf/Lef-binding element (TBE) and Smad-binding element (SBE) [21, 23, 24]. In mesenchymal cells, activation of TGFβ signaling synergistically induces the transcriptional activity of canonical Wnt/β-catenin signaling to control cell growth [22]. Global gene expression analysis of genetically manipulated mice revealed that TGFβ/Smad and Wnt/β-catenin signaling pathways are firmly intertwined [12]. However, details of the cross-talk between these two pathways remain to be elucidated. Here we demonstrate a novel transcriptional enhancer effect resulting from the physical interaction between Smad2, β-catenin, Tcf4 and p300. Smad2 synergistically enhanced the transcriptional activity of the Tcf/Lef-specific reporter (OT reporter) induced by activated β-catenin and Tcf4 through the histone acetyltransferase (HAT) activity of p300. Notably Smad4 negatively regulates the transactivation effect of Smad2 on the OT reporter. Our results demonstrate a previously unreported regulatory mechanism of transcriptional activity between the canonical Wnt/β-catenin pathway and the Activin/Nodal/Smad pathway.
Materials and Methods
Cell culture
Human breast cancer MCF-7 and human embryonic kidney 293T cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Human breast cancer cell lines MDA-MB-231 and MDA-MB-468, were cultured in Leibovitz's L-15 Medium supplemented with 10% fetal bovine serum. All cell lines were obtained from ATCC (Manassas, VA).
Expression vectors and transfection
Wild-type β-catenin vector [25], pcDEF3-FLAG(N)-Smads [26], p300 WT and p300 MutA2 vectors [27], OT/OF-pGL3 luciferase reporter vectors [28], Alk4-HA vector, (n2)7-Luc reporter and mFAST/pCS2 vector [29], have been previously described. A kinase-active form of Alk4, Alk4Thr206Asp (Alk4TD), a constitutive active form of β-catenin, Ser33Tyr β-catenin (S33Y β-catenin), and missense mutant Smad2, Smad2Asp450His (Smad2DH) were generated by PCR-based mutagenesis using appropriate vectors as templates. Glutathione S transferase (GST)-fused Smads vectors were constructed as follows. pcDEF3-FLAG(N) series of Smad2, 3 and 4 expression vectors were digested by XbaI, blunted and then digested by EcoRI. The inserts were then ligated into the EcoRI/SmaI site of pGEX-4T vector (Amersham Biosciences, Buckinghamshire, UK). GST-fused Smad2 deletions, GST-Smad2MH1 (3–185), GST-Smad2L (186–273) and GST-Smad2MH2 (271–467), were generated by PCR. Myc-tagged Tcf4 vector and HA-tagged p300 vector were purchased from Upstate (Chicago, IL). Tcf4 deletions were constructed by the following methods. To construct Tcf4ΔN (81–596), full length Tcf4 was digested by SmaI/XbaI and 2.3-kb fragment was purified and introduced into the EcoRV/XbaI site of pGEX-4T vector. Other deletions, Tcf4ΔC (1–412), Tcf4 ΔHMG+C (1–326) and Tcf4ΔHMG (1–326, 413–596) were generated by PCR. Transfections were performed with FuGENE6 (Roche Applied Science, Basel, Switzerland) at 70–80% confluency according to manufacturer’s instructions.
Luciferase assay
MCF-7, 293T or MDA-MB468 cells were plated into 12-well culture plates and transfected with the indicated plasmids. Transfections contained 50 ng of S33Y β-catenin, 200 ng of Tcf4, 100 ng or 500 ng of Alk4TD, 500 ng of p300, 100 ng or 500 ng of Smad2, Smad3 or Smad2DH, 100 ng of Smad4, 50 ng of OT/OF reporter, 5 ng of pEF-Renilla reporter and pcDNA3.1V5His empty vector (Invitrogen, Carlsbad, CA) to adjust the total amount of DNA. Twenty four hours after transfection, cells were lysed and dual luciferase assays were carried out using a Dual Luciferase Reporter Assay Kit (Promega, Nadison, WI). All assays were performed in triplicates in at least two to four independent experiments and normalized by internal control (Renilla luciferase).
Western blot analysis
Cells were washed with PBS and lysed for 20 min on ice using a NP-40 buffer (20 mM Tris-HCl, pH7.5, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 0.5% NP-40, and protease inhibitors). For subcellular fractionation, MCF-7 cells were harvested using NE-PER Nuclear and Cytoplasmic extraction reagents (PIERCE, Rockford, IL). Proteins were resolved on 4–20% gradient SDS-PAGE gels (Invitrogen), transferred to PVDF membranes (Millipore, Billerica, MA) and detected using ECL reagent (Amersham). Antibodies were used at the following dilutions. Anti-total Smad2 (Invitrogen), anti-phospho-Smad2 (Ser465/467, Cell Signaling, Danvers, MA), anti-Tcf4 (clone 6H5-3, Upstate) and Anti-HA (Covance, Princeton, NJ) were used at 1:1000 dilutions, anti-β-catenin (R&D Systems, Minneapolis, MN), anti-Tubulin (Upstate), anti-Flag (Sigma-Aldrich, St. Louis MO) at 1:2000 dilutions, anti-lamin A/C (Santa Cruz, Santa Cruz, CA) at a 1:300 dilution, and anti-dephospho β-catenin antibody [30] at a 1:5 dilution.
Immunoprecipitation
293T cells were plated into 100-mm dishes and transfected with the indicated plasmids (2.5 µg of Tcf4, Tcf4ΔC or S33Y β-catenin, 5 µg of Alk4TD, 3 µg of Smad2, 2DH, 3 or 4 and/or 10 µg of p300). Seventy two hours after transfections, cells were lysed on ice for 40 min using a NP-40 buffer or TritonX-100 buffer (20 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.5% TritonX-100 and protease inhibitors). One milligram of proteins were incubated for 16 hours at 4°C with an anti-Myc agarose (Sigma-Aldrich) or anti-p300 antibody (Upstate) conjugated to protein G agarose (Roche). To detect endogenous interaction between Smad2 and Tcf4, MDA-MB-231 cells were seeded in 150-mm dishes and transfected with either 10 µg of empty vector or Alk4TD-HA vector. Seventy two hours after transfection, cells were lysed by mild sonication on ice in the buffer containing 20 mM Hepes, pH7.5, 50 mM NaCl, 0.1% Tween-20 and 20% glycerol and protease inhibitors. Four milligram of proteins was used for the immunoprecipitation analysis with an anti-Tcf4 antibody (Upstate) conjugated to protein G agarose. Immunoprecipitates were extensively washed 5 times by lysis buffer on ice and then analyzed by Western blotting.
DNA affinity precipitation
293T cells were transfected with indicated plasmids. Seventy two hours after transfection, cells were lysed by mild sonication on ice in HKMG buffer (10 mM Hepes, pH7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.1% NP-40 and proteinase inhibitors). After cell debris was removed by a centrifugation at 10000g for 20 min at 4°C, 400 µg of proteins were precleared at 4°C for 2 hours with streptavidin-agarose (Sigma-Aldrich) and then incubated at 4°C for 16 hours with 300 ng of biotinylated double-strand oligonucleotides corresponding to the canonical Tcf/Lef binding sequence (TBE) or mutant sequence [31] and 3 µg of poly[d(I-C)] (Roche). Precipitated DNA-protein complexes were washed 5 times with buffer and subsequently analyzed by Western blotting.
GST proteins and in vitro binding (GST-pull down) assay
GST-fused Smad2, 3, 4 or Smad2 deletion or GST alone were expressed in BL21 strain of Escherichia coli (Amersham). Bacteria were grown in 2 mL of 2xYT medium at 37°C for 16 hours. Two hundred microliter of the medium containing the bacteria were added into 1.8 mL of fresh 2xYT medium and incubated at 30°C for 4 hours. Isopropyl thio-β-D-galactoside (IPTG) was then added at a concentration of 0.3 mM and the bacteria were incubated at 30°C for another 20 hours. Harvested bacteria were then lysed on ice by mild sonication in the GST-pull down buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% TritonX-100 and 0.5% N-Lauroylsarcosine). After removal of cell debris by centrifugation at 10000g for 20 min at 4°C, proteins were incubated with Glutathione Sepharose 4B at 4°C for 2 hours. After washing 3 times with GST-pull down buffer, precipitates were incubated with in vitro translated full length Myc-tagged Tcf4 or Tcf4 deletions which were synthesized using TNT T7 Quick Master Mix (Promega) in the buffer at 4°C for 2 hours. Precipitates were washed 5 times with the buffer and analyzed by Western blotting.
Results
Kinase active form of Alk4 enhances transcriptional activity induced by oncogenic β-catenin/Tcf4 through Smad2
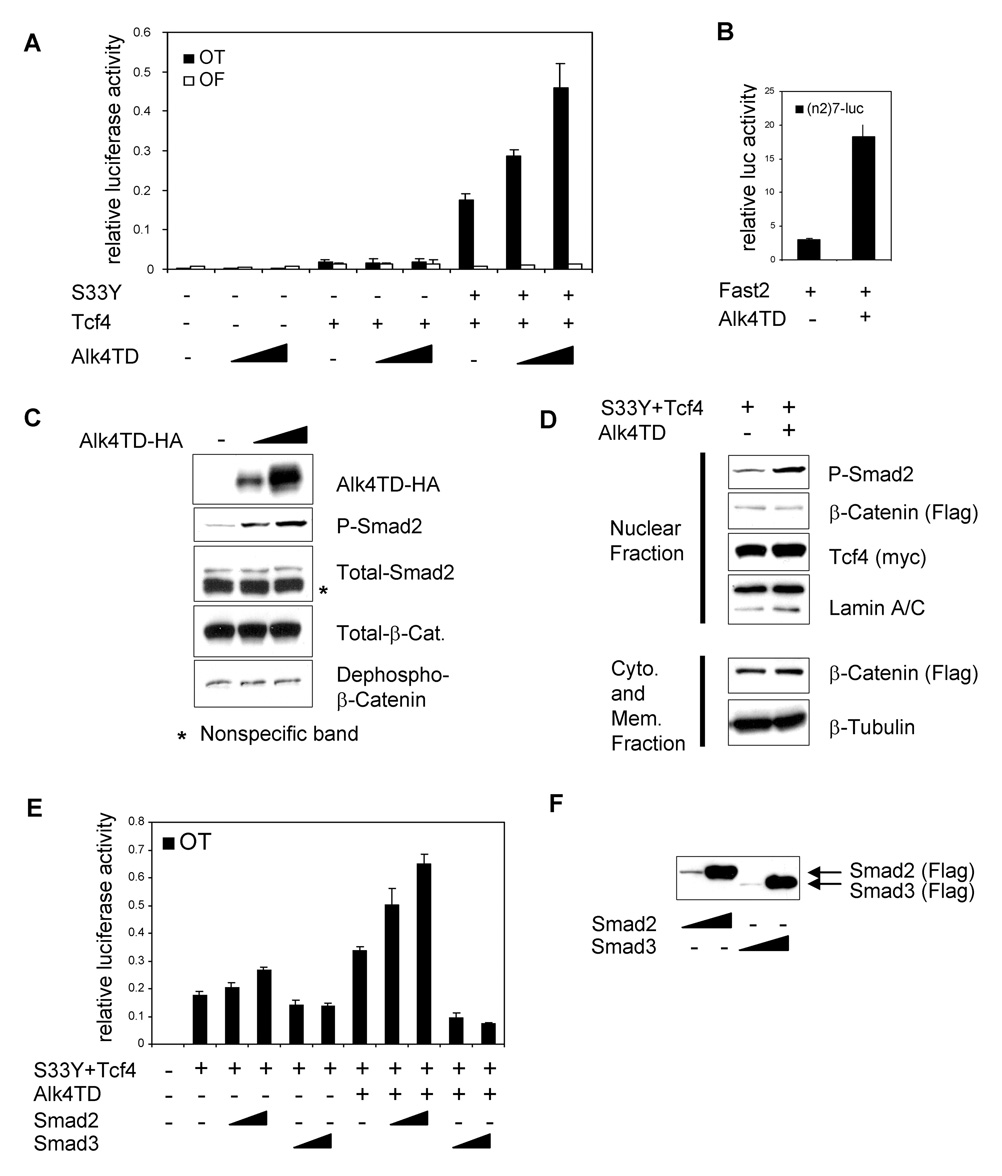
To ascertain if the Activin/Nodal/Smad pathway regulates the transcriptional activity of the canonical Wnt/β-catenin pathway in epithelial cancer cells, we utilized a constitutively active kinase form of the Acitivin type I receptor, Alk4T206D (Alk4TD) [32] and a specific β-catenin/Tcf/Lef reporter OT-pGL3 (OT) which contains consensus TBE or OF-pGL3 (OF) which contains mutated TBE [28]. When Alk4TD was co-transfected with OT or OF reporters into MCF-7 cells, Alk4TD itself did not show any significant effect on OT reporter activity (Fig. 1A). Tcf4, a member of the Tcf/Lef family of transcription factors, is expressed in mammary epithelial cells [33] and could induce a minimal level of OT reporter activity in MCF-7 cells. Alk4TD did not have any significant effect on OT reporter activity in the presence of Tcf4 alone (Fig. 1A). However, the combination of Tcf4 and a tumor-derived (constitutively active) mutant β-catenin (S33Y β-catenin) induced a high level of OT reporter activity. In addition, Alk4TD strongly enhanced the OT reporter activity induced by S33Y β-catenin and Tcf4 in a dose-dependent manner (Fig. 1A).
Fig. 1.
Enhancement of transcriptional activity induced by oncogenic β-catenin/Tcf4 by the kinase active form of Alk4 (Alk4TD). (A) OT/OF reporter assay was performed in MCF-7 cells transiently transfected with the indicated expression vectors. Different amounts of Alk4TD vector (100, 500 ng/well) were used. (B) (n2)7 reporter luciferase assay was performed in MCF-7 cells transiently transfected with the indicated expression vectors. (C) Western blot analysis was performed in MCF-7 cells transiently transfected with different amounts of Alk4TD-HA vector (100, 500 ng DNA/well). (D) Western blot analysis was performed for subcellular fractionated proteins from MCF-7 cells. (E) OT reporter assay performed in MCF-7 cells transiently transfected with the indicated expression vectors. Different amounts of Smad2 or Smad3 expression vector (100, 500 ng DNA/well, respectively) were used. (F) Western blot analysis was performed in MCF-7 cells transiently transfected with Smad2 or Smad3 at the same concentration as panel E. Error bars indicate standard deviations of triplicate experiments.
Smad2, Smad3 and Smad4 are known to be expressed in MCF-7 cells [34]. Smad2 can also be phosphorylated after stimulation by TGFβ [16]. When Alk4TD was transfected into MCF-7 cells, an Activin/Nodal reporter (n2)7-Luc that contains mouse Fast binding sites from the nodal left side-specific enhancer (nodal ASE) [35] showed enhanced activity (Fig. 1B). Western blot analysis of MCF-7 cells transfected with either HA-tagged Alk4TD (Alk4TD-HA) or empty vector revealed that Smad2 phosphorylation was strongly induced by Alk4TD expression in a dose-dependent manner (Fig. 1C). These results demonstrated that the Smad signaling pathway was functionally active following stimulation by an activated Alk4 in MCF-7 cells. However, the amount of total β-catenin and also dephospho-β-catenin, which is the active form of β-catenin, was not affected by Alk4TD expression (Fig. 1C). To confirm this, a nuclear fraction from MCF-7 cells was analyzed following transfection with Flag-tagged S33Y β-catenin (Flag-S33Y β-catenin) and Myc-tagged Tcf4 (Myc-Tcf4) and/or Alk4TD-HA. The amount of nuclear β-catenin was not affected by Alk4TD expression (Fig. 1D). In contrast, the amount of nuclear phospho-Smad2 was increased by expression of Alk4TD (Fig. 1D). These results demonstrate that constitutively active Alk4 could enhance the transcriptional activity of β-catenin and Tcf4 without affecting the amounts of active, nuclear level of β-catenin.
To identify downstream components of the Alk4/Smad pathway responsible for enhancement of transcriptional activity of β-catenin/Tcf4, either Smad2 or Smad3 was co-transfected with or without Alk4TD into MCF-7 cells (Fig. 1E). Smad2 itself could minimally enhance the OT reporter activity induced by S33Y β-catenin and Tcf4. However, combination of Smad2 and Alk4TD markedly enhance the OT reporter activity induced by S33Y β-catenin and Tcf4. In contrast to Smad2, Smad3 repressed the OT reporter activity that was enhanced by Alk4TD in the presence of S33Y β-catenin and Tcf4 (Fig. 1E). The level of expression of both Flag-tagged Smad2 and Smad3 (Flag-Smad2 and Flag-Smad3) was comparable (Fig. 1F). These results indicate that Smad2, but not Smad3, function as a transcriptional activator in the presence of Alk4 by enhancing the transcription of the oncogenic β-catenin/Tcf4 activity.
Tcf4 interacts with Smad2
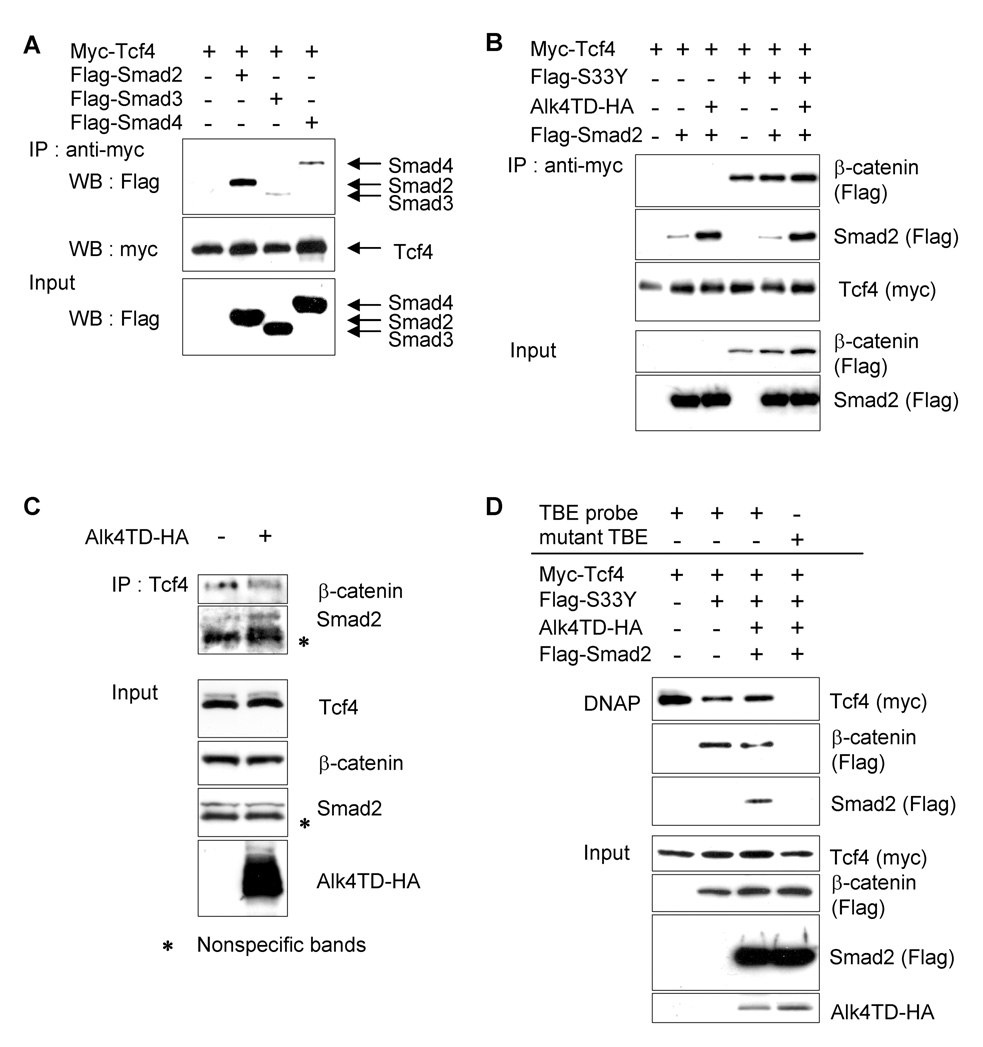
To investigate the mechanism by which this synergism between two signaling pathways may occur, we examined the ability of components in these two pathways to bind each other by performing immunoprecipitation assays using epitope-tagged constructs. First, we examined the interaction between the transcription factor Tcf4 and Smad2, 3, and/or 4. Myc-Tcf4 alone or together with either Flag-Smad2, Flag-Smad3 or Flag-tagged Smad4 (Flag-Smad4) were transfected into 293T cells and protein lysates were immunoprecipitated using anti-Myc agarose. High amounts of Flag-Smad2 were co-immunoprecipitated with Myc-Tcf4, whereas precipitated Flag-Smad3 or 4 was faint (Fig. 2A). We then ascertained potential interaction between Tcf4, β-catenin and Smad2 with or without Alk4TD expression. Immunoprecipitation analysis revealed that binding between Smad2 and Tcf4 was higher when Smad2 was co-transfected with Alk4TD, compared to Smad2 alone. The presence of activated S33Y β-catenin did not affect the binding of Smad2 with Tcf4 (Fig. 2B). We next investigated the endogenous interaction between Smad2, β-catenin and Tcf4 in MDA-MB-231 breast cancer cells. In these cells, the Wnt/β-catenin pathway is constitutively active by an autocrine mechanism [36] and the components in the TGFβ/Smad pathway are intact [16]. Immunoprecipitation analysis was performed using an anti-Tcf4 antibody in transiently Alk4TD- or empty vector (EV)-transfected MDA-MB231 cells. Western blot analysis using anti-Smad2 or anti-β-catenin specific antibodies revealed an endogenous interaction of Tcf4 with Smad2 or β-catenin (Fig. 2C). The interaction between Smad2 and Tcf4 was enhanced when endogenous Smad2 was activated by exogenous expression of Alk4TD (Fig. 4C). Furthermore, to determine interaction between Tcf4, β-catenin and Smad2 with DNA containing the TBE, a DNA affinity precipitation (DNAP) assay was performed. Myc-Tcf4, Flag-S33Y β-catenin, Alk4TD-HA and Flag-Smad2 were co-transfected and immunprecipitation was carried out using a biotin-labeled double-stranded DNA probe that contained a consensus or mutated TBE [31]. Subsequent Western blot analysis revealed that Tcf4, β-catenin and Smad2, in the presence of Alk4TD, formed a complex with the consensus TBE, but not with the oligonucleotide containing the mutated TBE (Fig. 2D).
Fig. 2.
Interactions among β-catenin, Tcf4 and Smads. (A) Indicated plasmids were co-transfected into 293T cells and lysates were immnoprecipitated using anti-Myc agarose. Precipitates were analyzed by Western blotting with the indicated antibodies. (B) Interactions between Flag-Smad2, Flag-S33Y β-catenin and Myc-Tcf4 after co-expression of Alk4TD-HA were examined by co-immunoprecipitation analysis using anti-Myc agarose in 293T cells. (C) Endogenous interactions between Tcf4, Smad2 and β-catenin in MDA-MB-231 cells transiently transfected with or without Alk4TD-HA were analyzed by co-immunoprecipitation using anti-Tcf4 specific antibody. (D) Interactions among Flag-Smad2, S33Y β-catenin and Myc-Tcf4 on DNA were analyzed by DNAP analysis using a consensus or mutant TBE probe.
Fig. 4.
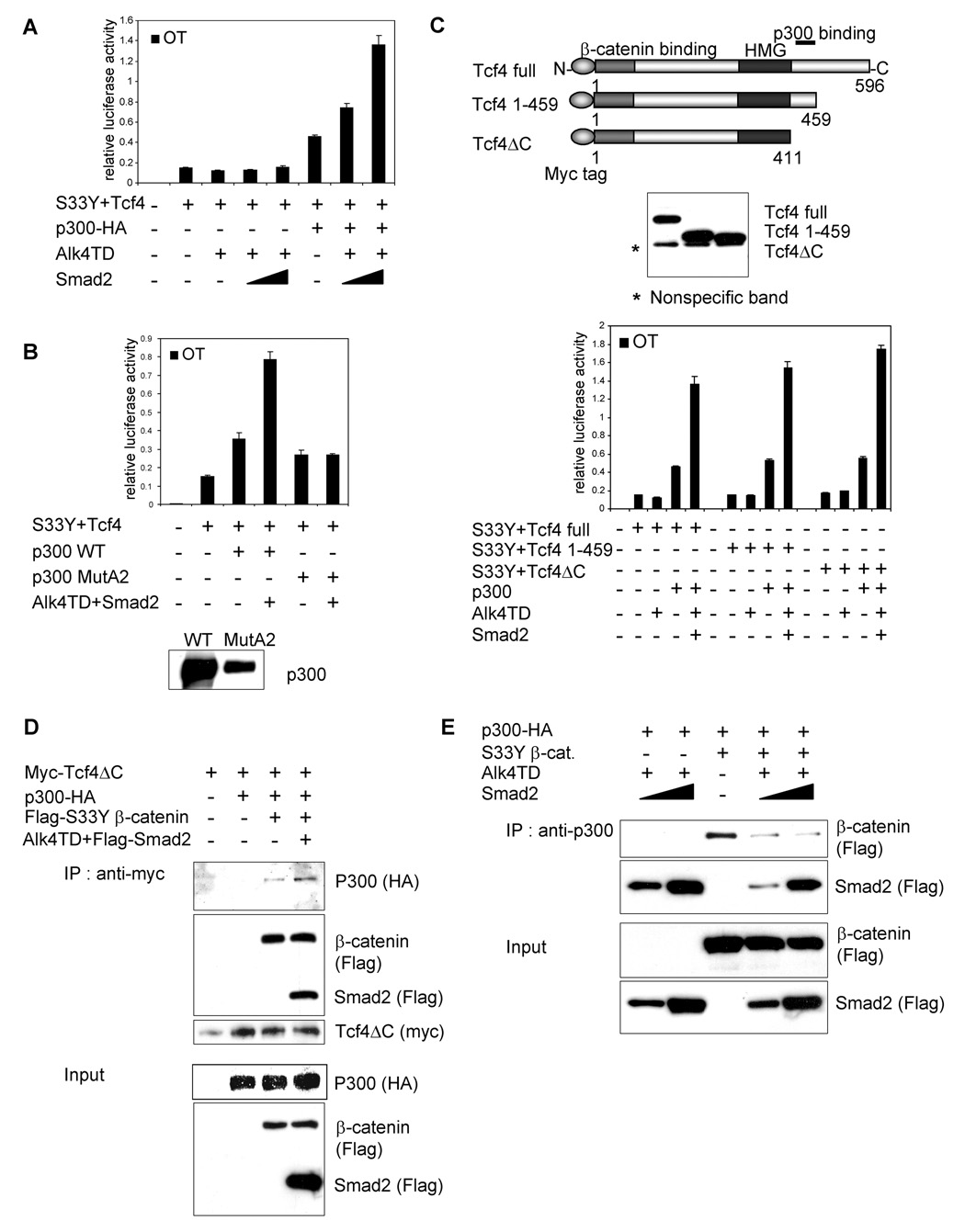
Requirement of HAT activity of p300 for the transactivation effect of Smad2 on β-catenin/Tcf4 activity. (A) OT reporter assays were performed in 293T cells transiently transfected with the indicated expression vectors. Different amount of Smad2 expression vector (100, 500 ng/well) were used. (B) OT reporter assays were performed in 293T cells transiently transfected with the indicated expression vectors. Western blot analysis was performed using an anti-p300 antibody (bottom). (C) Schematic of the Myc-Tcf4 deletion constructs (upper panel). Binding site of p300 is represented. Center panel shows the expression of the Myc-tagged Tcf4 deletion constructs in 293T cells by Western blot analysis using anti-Myc tag antibody. OT reporter assay was performed in 293T cells transiently transfected with the indicated expression vectors (lower panel). (D) Protein-protein interaction among p300-HA, Flag-Smad2, Flag-S33Y β-catenin and Myc-Tcf4ΔC in transiently transfected 293T cells. (E) Protein lysates were immunoprecipitated using anti-p300 antibody conjugated protein G agarose. Proteins interacting with p300 were visualized by Western blot analysis using indicated antibodies. Error bars indicate standard deviations of triplicate experiments.
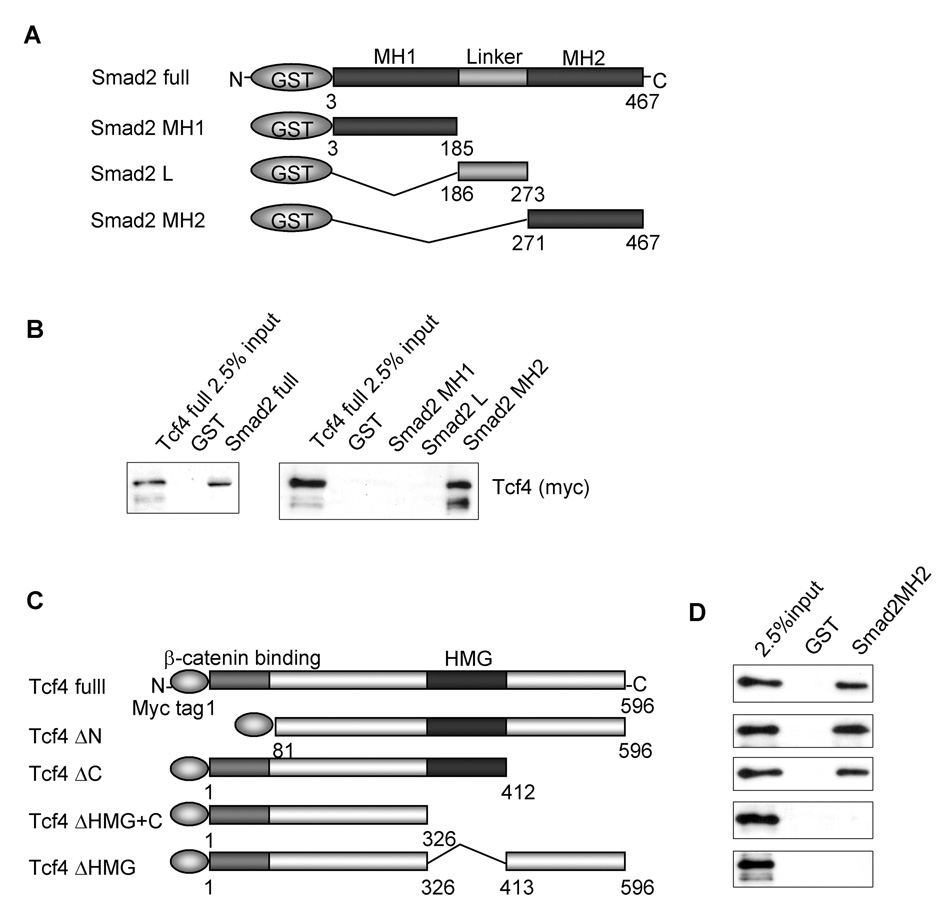
To confirm the physical binding between Tcf4 and Smad2, in vitro binding assays were performed (Fig. 3A). As shown in Fig. 3B, GST-fused full length Smad2 physically bound to Myc-Tcf4 protein. To ascertain the domain within Smad2 that is capable of binding to Tcf4, a series of GST-fused Smad2 deletion constructs were tested (Fig. 3A). The results clearly demonstrated that the MH2 domain of Smad2 was responsible for the binding of Smad2 to Tcf4 (Fig. 3B). Reciprocally, to determine the Smad2 binding region in Tcf4 a series of Myc-Tcf4 deletion constructs were tested (Fig. 3C). The results demonstrated that the HMG domain of Tcf4 was responsible for the binding to the Smad2 MH2 domain (Fig. 3D). However, Smad2 could not directly bind to in vitro translated Myc-tagged β-catenin (data not shown).
Fig. 3.
Physical interaction between Smad2 and Tcf4. (A) GST-fused Smad2 deletion constructs. (B) In vitro binding (GST-pull down) assay was performed between bacterially expressed GST-fused full-length Smad2 protein and in vitro translated Myc-tagged full-length Tcf4 (Myc-Tcf4) (left panel) and between GST-fused Smad2MH2 (271–467) domain and in vitro translated Myc-Tcf4 (right panel). Protein interaction was visualized by Western blot analysis with anti-Myc tag antibody. (C) Myc-tagged Tcf4 deletion constructs. (D) GST-pull down assays were performed using GST-fused Smad2MH2 domain (Smad2MH2) and Myc-Tcf4 or indicated Tcf4 deletion constructs.
HAT activity of co-activator p300 is essential for the transactivation effect of Smad2 on oncogenic β-catenin/Tcf4
To elucidate the mechanism by which the synergistic transcriptional activation of the Wnt/β-catenin/Tcf4 pathway by the Alk4/Smad2 pathway might occur, we examined the involvement of p300 which has been shown to interact with both of these pathways, as a transcriptional co-activator. p300 also directly binds to Smad2/3 [37], β-catenin [38] and Tcf4 [39]. We used 293T cells for the OT reporter assays since the endogenous p300 function in 293T cells is largely suppressed by the adenoviral E1A protein [40]. When Alk4TD and Smad2 were co-transfected with S33Y β-catenin and Tcf4 in 293T cells, OT reporter activity induced by S33Y β-catenin and Tcf4 was not enhanced by Alk4TD and/or Smad2 (Fig. 4A). However, expression of exogenous p300 enhanced OT reporter activity in the presence of S33Y β-catenin and Tcf4. In addition, Alk4TD and Smad2 markedly enhanced OT reporter activity only when p300 was simultaneously transfected (Fig. 4A). These results clearly demonstrate an obligatory role of p300 in regulating the synergistic transactivation of the OT reporter through Alk4/Smad2 and β-catenin/Tcf4 signaling pathways.
Previous studies have shown that the HAT activity of p300 is not required for its co-activator function in regulating the transcriptional activity of β-catenin/Tcf [40]. We therefore examined whether HAT activity was required for the transactivation effect of Alk4/Smad2 on β-catenin/Tcf4. Wild-type p300 or p300 MutA2 which lacks HAT activity [27] were used for assessment in the OT reporter assay. Both wild-type p300 and p300 MutA2 had similar activity on the OT reporter induced by S33Y β-catenin and Tcf4 (Fig. 4B). However, Alk4TD and Smad2 enhanced the OT reporter activity induced by S33Y β-catenin and Tcf4 only in the presence of wild-type p300 but not in the presence of p300 MutA2 (Fig. 4B). This result indicated that the transactivation effect of the Alk4/Smad2 pathway upon β-catenin/Tcf4 pathway was dependent on the HAT activity of p300.
A previous study demonstrated that Tcf4 physically interacts with p300 via a region in the COOH terminal domain of Tcf4 [39]. We therefore assessed whether binding between p300 and Tcf4 was required for the transactivation effect of the Alk4TD/Smad2 pathway on the Wnt/β-catenin/Tcf4 pathway. We analyzed three different deletion constructs of Tcf4 presented in Fig. 4C. Full length Tcf4 and Tcf4 1–459 constructs include the p300 binding region while the p300 binding region was deleted in the Tcf4ΔC construct. A series of OT reporter assays demonstrated that Alk4/Smad2 pathway retained the same transactivation effect with the Tcf4ΔC construct on enhancing OT reporter activity in the presence of p300 (Fig.4C). This result suggests that a physical interaction between p300 and Tcf4 was not required for the transactivation effect of Alk4/Smad2.
We next determined the protein binding between S33Y β-catenin, Tcf4, Smad2 and p300. Myc-tagged Tcf4ΔC (Myc-Tcf4ΔC) was co-transfected with HA-tagged p300 (p300-HA). As expected, we failed to detect a direct interaction between p300 and Tcf4ΔC (Fig. 4D). However, when S33Y β-catenin was co-transfected with Tcf4ΔC and p300, p300 was co-immunoprecipitated with myc-Tcf4ΔC (Fig. 4D). Moreover, when Smad2 and Alk4TD were transfected with S33Y β-catenin, p300 and Tcf4βC, more p300 was bound to Tcf4βC (Fig. 4D).
In contrast, when p300 was immunoprecipitated using an anti-p300 specific antibody in transiently transfected 293T cells, the association between S33Y β-catenin and p300 was decreased by introduction of increasing amounts of Smad2 with Alk4TD (Fig. 4E). This suggests that S33Y β-catenin and activated Smad2 bind to regions in close proximity within the p300 protein.
Transactivation effect of activated Smad2 on the Wnt/β-catenin/Tcf4 pathway was negatively regulated by Smad4
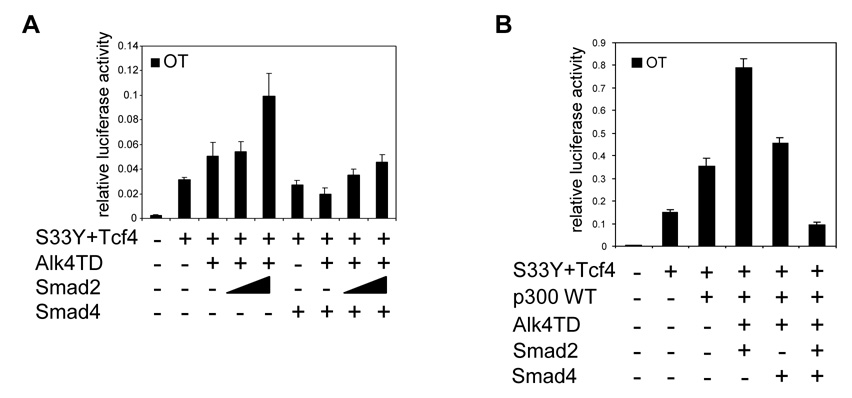
Following activation of Alk4, Smad2 is phosphorylated and activated leading to the formation of a complex of Smad2 with Smad4 [14, 15]. Smad4 is an essential component in all canonical TGFβ superfamily pathways that function through an SBE [15]. Smad4 has been reported as a tumor suppressor, since this gene is frequently mutated in pancreatic cancer and metastatic colon cancer [7, 41]. To determine the effect of Smad4 on the transactivation effect of Smad2 in the β-catenin/Tcf4 pathway, we performed OT reporter assays using the human breast cancer cell line MDA-MB-468 which has a Smad4 deletion [16]. Alk4TD was capable of producing a slight enhancement of OT reporter activity induced by S33Y β-catenin and Tcf4. This effect on the OT reporter activity could be further enhanced when Smad2 was co-transfected with Alk4TD (Fig. 5A), suggesting that the effect of Smad2 on the OT reporter occurs through a Smad4-independent mechanism. Furthermore, when Smad4 was co-transfected with Smad2 and Alk4TD, the stimulatory effect of activated Smad2 on OT reporter activity was abolished (Fig. 5A). In 293T cells which express endogenous Smad4, this repressive effect of Smad4 on the synergistic activation of OT reporter by Smad2 was greater when Smad4 was co-transfected together with Alk4TD and Smad2 than with Alk4TD alone (Fig. 5B). This suggests that Smad2 can function as a repressor of the Wnt/β-catenin/Tcf4 signaling pathway when Smad4 is highly expressed.
Fig. 5.
Smad4-independent activity of Smad2 to enhance canonical Wnt/β-catenin/Tcf4 signaling pathway. (A) OT reporter assays were performed in MDA-MB468 cells transiently transfected with the indicated expression vectors. Different amount of Smad2 expression vector (100, 500 ng/well) were used. (B) OT reporter assay was performed in 293T cells transiently transfected with the indicated expression vectors. Error bars indicate standard deviations of triplicate experiments.
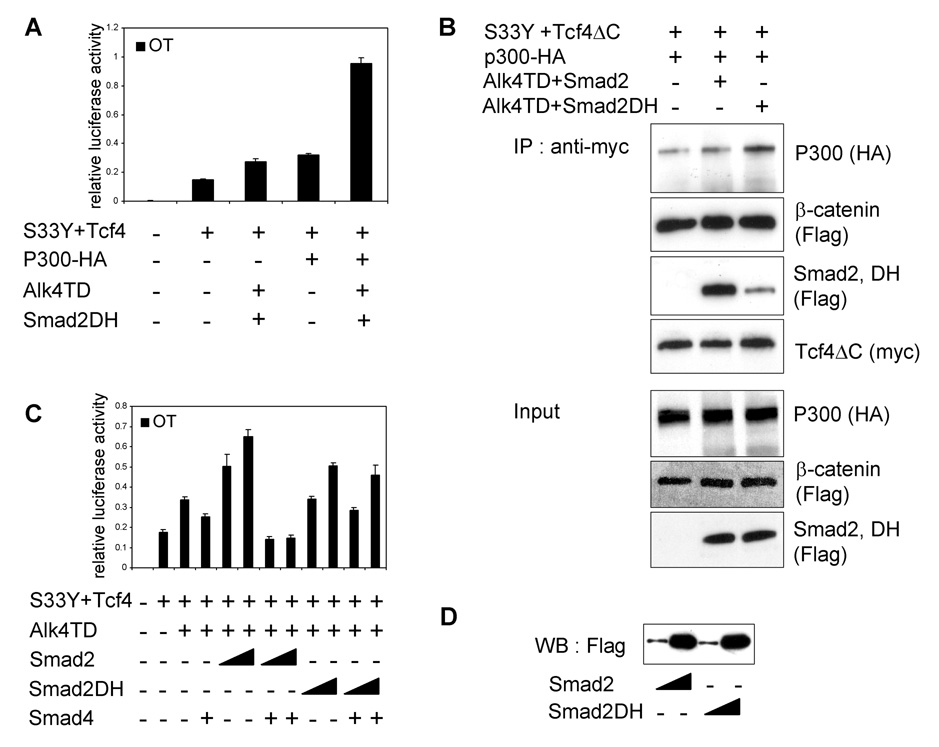
In order to confirm the Smad4-independent activity on transcriptional activity of β-catenin/Tcf4, we utilized a tumor-derived missense mutant of Smad2, Smad2DH, which lacks the ability to bind and to form a complex with Smad4 [42, 43]. OT reporter assay in 293T cells revealed that Smad2DH retained the capacity to synergistically activate the OT reporter activity (Fig. 6A). We then performed immunoprecipitation assays using transfected 293 cells to assess the protein-protein interactions of Smad2DH with S33Y β-catenin, Tcf4ΔC, and p300. Smad2DH weakly co-immunoprecipitated with myc-Tcf4ΔC (Fig. 6B). p300 and S33Y β-catenin were simultaneously co-immunoprecipitated with myc-Tcf4ΔC. Furthermore, the inhibitory effect of Smad4 on the enhanced OT reporter activity was not observed on OT reporter activity enhanced by Smad2DH (Fig. 6C–D). These results confirmed that the synergistic activation of canonical Wnt/β-catenin/Tcf pathway by Smad2 is independent of both Smad4 and SBE.
Fig. 6.
Inhibitory activity of Smad4 on the oncogenic β-catenin/Tcf4 pathway is blocked by a tumor-derived missense mutant Smad2DH. (A) OT reporter assay was performed in 293T cells transiently transfected with the indicated expression vectors. (B) Interactions among Flag-Smad2DH, p300-HA, Flag-S33Y β-catenin and Myc-Tcf4ΔC in transiently transfected 293T cells. (C) OT reporter assay was performed in MCF-7 cells transiently transfected with the indicated expression vectors. Different amounts of Smad2 or Smad2DH expression vector (100, 500 ng DNA/well) were used. (D) Western blot analysis was performed in MCF-7 cells transiently transfected with Smad2 or Smad2DH at the same concentration as panel C.
Discussion
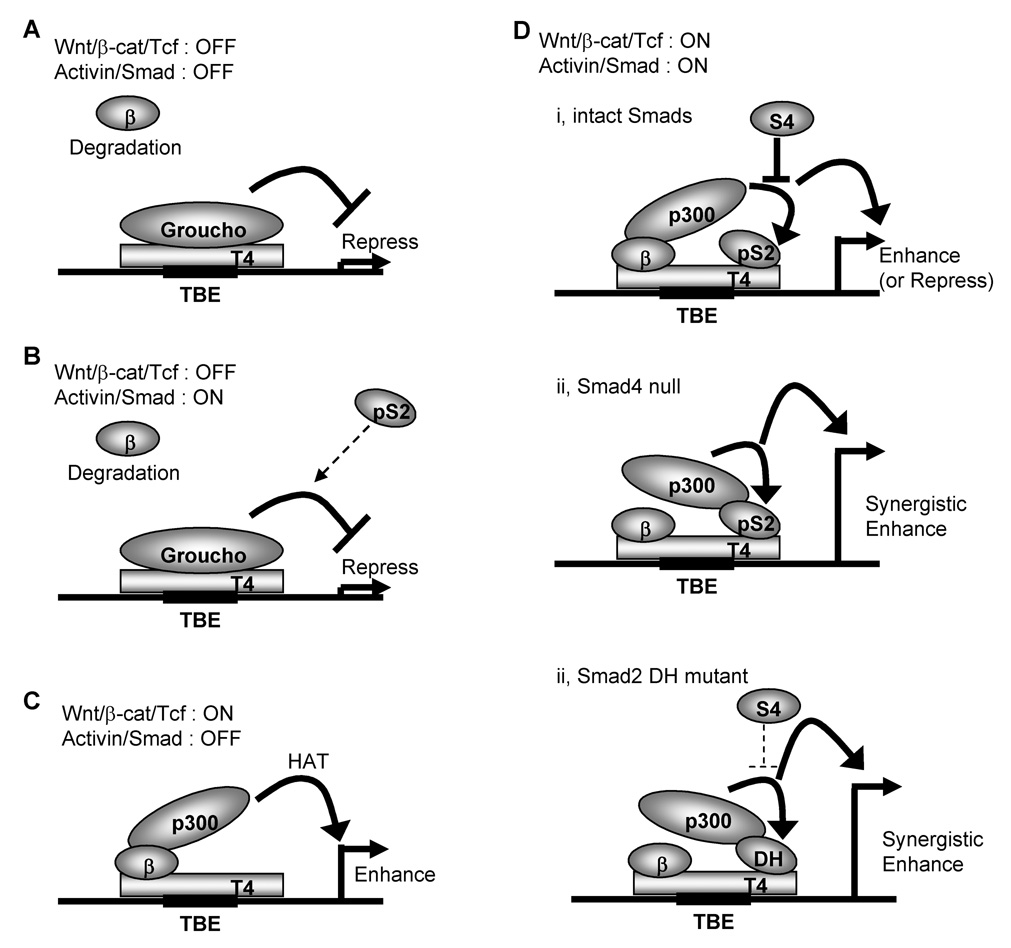
Our present study demonstrated that receptor-activated Smad2 synergistically enhances the canonical Wnt/β-catenin pathway in epithelial cancer cells through an SBE-independent mechanism. Activation of the Wnt/β-catenin pathway requires an inhibition of degradation complexes and the stabilization of β-catenin to regulate the down stream target genes in cooperation with Tcf/Lef transcriptional factors [1–4]. Without the presence of nuclear β-catenin, Tcf/Lef forms a repressive complex with the transcriptional repressor Groucho which recruits histone deacetylases to block transcription of Wnt target genes [4] (Fig. 7A). Stimulation with Wnt ligands or mutations of components in the Wnt signaling pathway, such as APC or β-catenin results in stabilization and accumulation of nuclear β-catenin. This β-catenin binds with Tcf/Lef to enhance the transcriptional activity of Wnt target genes [1, 4] (Fig. 7C). Our results suggest that receptor-activated Smad2 has no effect on the canonical β-catenin/Tcf transcriptional activity when nuclear β-catenin is absent probably because Tcf/Lef forms a repressive complex with Groucho (Fig. 7B). However, when the canonical Wnt/β-catenin pathway is activated and nuclear β-catenin and Tcf form an activation complex, receptor-activated Smad2 synergistically enhances the transcriptional activity of Wnt/β-catenin target genes through the HAT activity of p300 (Fig. 7D). Our results suggest that a common Smad, Smad4, which forms a heterogeneous complex with phosphorylated Smad2 or Smad3 and is required for the canonical TGFβ/Smad signaling activity [15], has a negative effect on the synergistic enhancement of β-catenin/Tcf transcriptional activity by Smad2 in Smad4-intact cells (Fig. 7Di). However, in cells with deletions or mutations of Smad4 or mutations of Smad2 in the MH2 domain, which occur frequently in several types of human cancers such as pancreatic or colorectal cancers [7, 41, 42, 44], this negative effect of Smad4 is absent and the transactivation effect of Smad2 on the canonical Wnt/β-catenin pathway is accelerated (Fig. 7Dii, iii). These findings provide a novel signaling mechanism in which Smad2 may act as a potential tumor-enhancer by facilitating oncogenic Wnt/β-catenin pathway and may explain the synergism that has been observed between these two pathways [12].
Fig. 7.
A proposed model for transcriptional regulatory mechanism of canonical Wnt/β-catenin/Tcf4 signaling pathway by Smad2 and Smad4.
Our results suggest that the transactivation effect of the canonical Wnt/β-catenin pathway is specific for Smad2, not Smad3. The different binding affinities with Tcf4 or p300 between Smad2 and Smad3 may be responsible for their different effects on the Wnt/β-catenin pathway. This difference in the transactivation effect on the canonical Wnt/β-catenin pathway might be a possible explanation for the differential roles of Smad2 and Smad3 in development and carcinogenesis [45]. In fact, there is a correlation between the deregulation of Wnt/β-catenin pathway and Smad2 expression in colorectal and breast cancers [16, 46]. In contrast, Smad3 expression is reduced in high grade breast cancers [47].
This model may also provide an explanation for the mechanism by which Smad4 can function as a tumor suppressor [7, 41]. Several animal models have demonstrated a synergistic effect between the presence of Smad4 mutations and the canonical Wnt/β-catenin signaling on tumor invasion [8, 48, 49]. For example, compound heterozygous mice that have both Apc and Smad4 genes knocked out rapidly develop colorectal cancers with invasive characteristics not found in colon tumors of the littermates carrying only the Apc gene deletion [49]. The present study provides a possible molecular mechanism for the tumor suppressor functions of Smad4 as it relates to the oncogenic Wnt/β-catenin signaling pathway. Mutations in Smad4 that compromise the ability of Smad4 to bind to Smad2 may lead to an augmentation in the ability of Smad2 to transactivate the β-catenin/Tcf4 activity through the HAT activity of p300.
Likewise, missence mutations in Smad2 which disrupt the binding to Smad4 may have a similar effect. Most Smad2 mutations found in colorectal and lung cancers are in the MH2 domain which can lead to a loss of stability or disruption of homo- or hetero-oligomerization [42, 44, 50]. Mutation of D450 residue is reported to abrogate the oligomerization between Smad2 and Smad4 [50]. The present data demonstrate that the missense mutant Smad2DH still retains the ability to form a complex with p300, β-catenin and Tcf4 and can still function as a co-activator of β-catenin/Tcf4 (Fig. 7Diii). Importantly, Smad2DH is resistant to the tumor suppressor effect of Smad4 with respect to the ability of Smad4 to function as a repressor of β-catenin/Tcf4 signaling.
The present data suggest a mechanism by which the TGFβ/Activin activity can switch from functioning as a tumor suppressor during the early stages of transformation to promoting tumorigenesis and metastasis at later stages [15, 51]. Moreover, our results provide a mechanism for a Smad4-independent function of Smad2. In this regard, previous gene targeting studies have indicated that Smad4 is non-essential for mesoderm induction since Smad4 knockout embryos are able to undergo early mesodermal development in the presence of wild-type visceral endoderm [52], while Nodal or Smad2 null mice fail to form the mesoderm induction [14, 53, 54]. The proposed model may explain one possible molecular mechanism involving co-activation of the Wnt/β-catenin signaling pathway by which Smad2 can function independent of Smad4.
Acknowledgments
We thank Ms. Brenda W. Jones for excellent technical assistance. We are grateful to Dr. Roger Herrell, Dr. Kohei Miyazono, Dr. W. Lee Kraus, Dr. Bert Vogelstein, Dr. Hiroshi Hamada and Dr. Hans Clevers for kindly providing the appropriate expression vectors or antibodies. We also thank NIH Fellows Editorial Board for assistance with editing. This work was supported by NIH Intramural Funding.
Abbreviations used are
- Alk
activin-like kinase
- EV
empty vector
- EMT
epithelial-mesenchymal transition
- GST
glutathione S transferase
- HAT
histone acetyltransferase
- SBE
Smad-binding element
- TBE
Tcf/Lef-binding element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Polakis P. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Giles RH, van Es JH, Clevers H. Biochim Biophys Acta. 2003;1653(1):1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 3.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 4.Gordon MD, Nusse R. J Biol Chem. 2006;281(32):22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 5.Mohinta S, Wu H, Chaurasia P, Watabe K. Front Biosci. 2007;12:4020–4033. doi: 10.2741/2368. [DOI] [PubMed] [Google Scholar]
- 6.Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. Cancer Cell. 2004;6(5):497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler KW, Vogelstein B. Cell. 1996;87(2):159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 8.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, Kanamaru R, Kanegae Y, Saito I, Nakamura Y, Shiba K, Noda T. Science. 1997;278(5335):120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 9.Neth P, Ries C, Karow M, Egea V, Ilmer M, Jochum M. Stem Cell Rev. 2007;3(1):18–29. doi: 10.1007/s12015-007-0001-y. [DOI] [PubMed] [Google Scholar]
- 10.Fearon ER, Vogelstein B. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 11.Katoh M. Stem Cell Rev. 2007;3(1):30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- 12.Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Cancer Res. 2007;67(1):75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 13.Marikawa Y. Semin Cell Dev Biol. 2006;17(2):175–184. doi: 10.1016/j.semcdb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Shen MM. Development. 2007;134(6):1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Massague J. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 16.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Cancer Res. 2002;62(2):497–505. [PubMed] [Google Scholar]
- 17.Normanno N, De Luca A, Bianco C, Maiello MR, Carriero MV, Rehman A, Wechselberger C, Arra C, Strizzi L, Sanicola M, Salomon DS. J Cell Physiol. 2004;198(1):31–39. doi: 10.1002/jcp.10375. [DOI] [PubMed] [Google Scholar]
- 18.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Nat Med. 2006;12(8):925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 19.Saloman DS, Bianco C, Ebert AD, Khan NI, De Santis M, Normanno N, Wechselberger C, Seno M, Williams K, Sanicola M, Foley S, Gullick WJ, Persico G. Endocr Relat Cancer. 2000;7(4):199–226. doi: 10.1677/erc.0.0070199. [DOI] [PubMed] [Google Scholar]
- 20.Bianco C, Strizzi L, Normanno N, Khan N, Salomon DS. Curr Top Dev Biol. 2005;67:85–133. doi: 10.1016/S0070-2153(05)67003-2. [DOI] [PubMed] [Google Scholar]
- 21.Labbe E, Letamendia A, Attisano L. Proc Natl Acad Sci U S A. 2000;97(15):8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner DR, Greene RM, Pisano MM. FEBS Lett. 2005;579(17):3539–3546. doi: 10.1016/j.febslet.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 23.Hussein SM, Duff EK, Sirard C. J Biol Chem. 2003;278(49):48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- 24.Lei S, Dubeykovskiy A, Chakladar A, Wojtukiewicz L, Wang TC. J Biol Chem. 2004;279(41):42492–42502. doi: 10.1074/jbc.M404025200. [DOI] [PubMed] [Google Scholar]
- 25.Song LN, Herrell R, Byers S, Shah S, Wilson EM, Gelmann EP. Mol Cell Biol. 2003;23(5):1674–1687. doi: 10.1128/MCB.23.5.1674-1687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Embo J. 1998;17(14):4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus WL, Manning ET, Kadonaga JT. Mol Cell Biol. 1999;19(12):8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih IM, Yu J, He TC, Vogelstein B, Kinzler KW. Cancer Res. 2000;60(6):1671–1676. [PubMed] [Google Scholar]
- 29.Watanabe K, Hamada S, Bianco C, Mancino M, Nagaoka T, Gonzales M, Bailly V, Strizzi L, Salomon DS. J Biol Chem. 2007;282(49):35772–35786. doi: 10.1074/jbc.M707351200. [DOI] [PubMed] [Google Scholar]
- 30.van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. J Biol Chem. 2002;277(20):17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- 31.Valenta T, Lukas J, Korinek V. Nucleic Acids Res. 2003;31(9):2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attisano L, Wrana JL, Montalvo E, Massague J. Mol Cell Biol. 1996;16(3):1066–1073. doi: 10.1128/mcb.16.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker N, Huls G, Korinek V, Clevers H. Am J Pathol. 1999;154(1):29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamichi Y, Waidmann O, Hein R, Eleftheriou P, Giehl K, Menke A. Biol Chem. 2005;386(3):225–236. doi: 10.1515/BC.2005.028. [DOI] [PubMed] [Google Scholar]
- 35.Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, Miyazono K, Whitman M, Hamada H. Mol Cell. 2000;5(1):35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- 36.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Breast Cancer Res. 2007;9(5):R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng XH, Zhang Y, Wu RY, Derynck R. Genes Dev. 1998;12(14):2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. Mol Cell Biol. 2004;24(24):10689–10702. doi: 10.1128/MCB.24.24.10689-10702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hecht A, Stemmler MP. J Biol Chem. 2003;278(6):3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 40.Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. Embo J. 2000;19(8):1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Massague J, Blain SW, Lo RS. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 42.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. Cell. 1996;86(4):543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 43.Prunier C, Mazars A, Noe V, Bruyneel E, Mareel M, Gespach C, Atfi A. J Biol Chem. 1999;274(33):22919–22922. doi: 10.1074/jbc.274.33.22919. [DOI] [PubMed] [Google Scholar]
- 44.Uchida K, Nagatake M, Osada H, Yatabe Y, Kondo M, Mitsudomi T, Masuda A, Takahashi T, Takahashi T. Cancer Res. 1996;56(24):5583–5585. [PubMed] [Google Scholar]
- 45.Brown KA, Pietenpol JA, Moses HL. J Cell Biochem. 2007;101(1):9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 46.Korchynskyi O, Landstrom M, Stoika R, Funa K, Heldin CH, ten Dijke P, Souchelnytskyi S. Int J Cancer. 1999;82(2):197–202. doi: 10.1002/(sici)1097-0215(19990719)82:2<197::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 47.Jeruss JS, Sturgis CD, Rademaker AW, Woodruff TK. Cancer Res. 2003;63(13):3783–3790. [PubMed] [Google Scholar]
- 48.Xu X, Brodie SG, Yang X, Im YH, Parks WT, Chen L, Zhou YX, Weinstein M, Kim SJ, Deng CX. Oncogene. 2000;19(15):1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]
- 49.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Cell. 1998;92(5):645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 50.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massague J, Shi Y. Mol Cell. 2001;8(6):1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- 51.Bachman KE, Park BH. Curr Opin Oncol. 2005;17(1):49–54. doi: 10.1097/01.cco.0000143682.45316.ae. [DOI] [PubMed] [Google Scholar]
- 52.Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern SE, Rossant J, Mak TW. Genes Dev. 1998;12(1):107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. Development. 1994;120(7):1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 54.Nomura M, Li E. Nature. 1998;393(6687):786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]