Abstract
Sulfur mustard (SM) is an alkylating agent with a history of use as a chemical weapon. The chemical reactivity of sulfur mustard toward both proteins and nucleic acids coupled with the hours long delay between exposure and appearance of blisters has prevented the determination of the mechanism of blister formation. We have treated assembled keratin intermediate filaments with analogs of sulfur mustard to simulate exposure to SM. We find that treatment of intact filaments with chloroethyl ethyl sulfide (CEES) or mechlorethamine (MEC) produces aggregates of keratin filaments with little native appearing structure. Treatment of a mix of epidermal keratins 1/10 (keratin pair 1 and 10) and keratins 5/14 with a sulfhydryl-specific modification reagent also results in filament abnormalities. Our results are consistent with the hypothesis that modification of keratins by SM would result in keratin filament destruction, leading to lysis of epidermal basal cells and skin blistering.
Introduction
Sulfur mustard (SM, di(2-chloroethyl) sulfide) is probably most well known for its use as the chemical weapon mustard gas [1–4] although SM and related alkylating agents have been investigated as anti-tumor therapies. As a weapon, SM exposure may be lethal, but its main function is to cause incapacitating injury to the eyes, respiratory tract or skin [5]. Exposure to the liquid or aerosol (fine droplets) leads to the formation of blisters one to several hours after exposure [6]. While the vesicant activity of SM and related compounds is without dispute, the mechanism responsible for skin blistering is currently unknown, thus a rational approach to prevention/treatment is precluded.
Light microscope characterization of the blisters formed following exposure to SM reveals the location of the blister is between the dermis and epidermis [6]. One hypothesis for vesicant action is the activation of an endogenous protease by SM [7–10] and an endogenous inhibitor of this protease has been identified [11, 12]. Other investigators have analyzed the effects of SM on basal cell adhesion complex molecules [13–17], hypothesizing alkylation of basal cell adhesion complex molecules results in failure of the complex [16, 18], leading to blister formation, similar to inherited skin blistering diseases [19].
The effect of SM on keratin proteins within keratinocytes has also been studied using antibody-based methods. Dillman et al. report SM induced cross-linking of basal cell keratins k5 and k14 resulting in homodimer cross-link products k5-k5 and k14-k14 as well as the heterodimer, k5-k14 [17]. However, whether these cross-linked subunits were present in intact IFs or insoluble/nonfunctional aggregates was not resolved.
SM is a potent alkylating and crosslinking agent with reactivity toward numerous cellular constituents including both proteins and nucleic acids [20, 21]. Reactivity toward proteins is thought to occur preferentially at cysteines [20, 22], although reactivity at the alpha-amino group (ie, the amino terminus), the imino group of histidine, and carboxylic acid groups of aspartic and glutamic acid has been demonstrated [23–27].
We have investigated the effects of sulfur mustard analogs CEES (chloroethyl ethyl sulfide (also known as half mustard)) and MEC (mechlorethamine (also known as nitrogen mustard)) on keratin intermediate filaments (kIFs). In vitro, we find that both CEES and MEC are able to destroy keratin filament networks, whether composed of recombinant or natively isolated keratins. The sites of modification have not been identified, but our experiments show that cysteine (sulfhydryl) specific modification of epidermal keratins is sufficient to interfere with normal filament assembly and that such modified keratins act as dominant negatives, interfering with normal keratin assembly.
Results and Discussion
A pool of bovine keratins, containing k5/14 and k1/10, was isolated from bovine snout to ~90+% purity; recombinant human k8 and k18 were purified also to >90% purity as judged by SDS-PAGE (supplementary materials). After dialysis to remove urea [28], both the native bovine and recombinant human keratins formed long, smooth, native appearing 10 nm filaments as judged by electron microscopy (supplementary materials). Exposure of the filaments to either 1% ethanol or 1% DMSO (dimethyl sulfoxide), resulted in no significant changes (supplementary materials).
For the first experiments, bovine keratins were assembled into intermediate filaments [28] and treated with either 10 mM CEES or 10 mM MEC, for one hour. CEES, also known as half mustard, is similar to SM in reactivity, yet has only one reactive site and is unable to form crosslinks. MEC is similar to SM, but is a less reactive “nitrogen mustard”, with a nitrogen atom replacing the central sulfur atom. MEC is capable of forming cross-links. Samples of the treated filaments were removed after the hour incubation and applied to a formvar/carbon coated copper grid, stained with uranyl acetate and examined in the electron microscope [28]. Figure 1A and 1B, shows the type of aggregation and filament damage produced. Within each sample is a filamentous character, but long, smooth, IFs are absent. At 1 mM, CEES or MEC, IFs are disturbed, with slight aggregation but the over all IF network appears intact (supplementary materials).
Figure 1. Treatment of intact bovine kIFs with CEES or MEC.
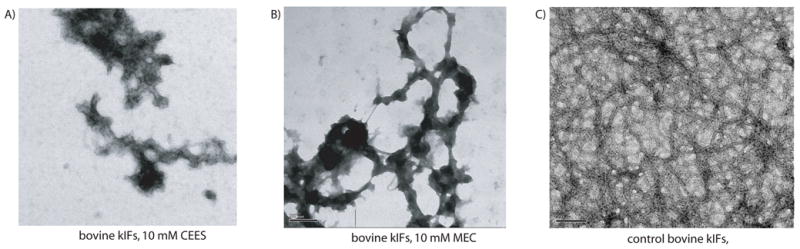
Bovine kIFs were incubated for one hour with either 10 mM CEES (panel A), 10 mM MEC (panel B) or untreated (panel C) and then prepared and viewed in the electron microscope. Scale bar represents 200 nm.
Similar results were seen with keratin 8/18 filaments assembled in vitro and treated with 10 mM CEES or 10 mM MEC. Figures 2A and 2B show representative views of k8/18 filaments treated with 10 mM CEES or 10 mM MEC. In each case, short regions of nearly normal filament diameter can be seen, but the majority of protein is found in large aggregates.
Figure 2. Modification of keratin proteins followed by IF assembly.
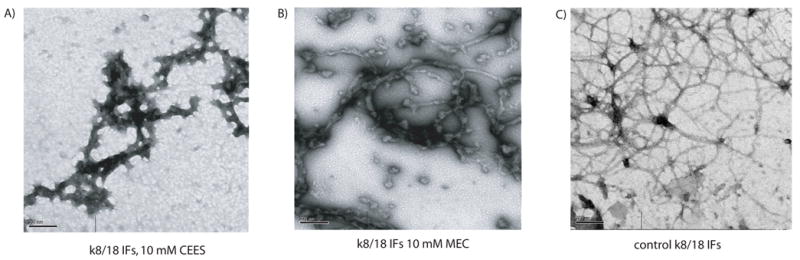
Purified keratin 8 and 18, assembled into kIFs were incubated for one hour with 10 mM CEES (panel A), 10 mM MEC (panel B) or untreated (Panel C). The scale bar represents 200 nm.
The use of different keratin pairs for chemical modification reveals the sensitivity of several different amino acid side chains to modification. A fundamental difference between the keratin pair k8/18 and either of the keratin pairs k5/14 and k1/10 is the presence of several cysteine residues in both k5/14 and 1/10, with none in either k8 or k18. Thus, the effect of CEES and MEC on the k8/18 pair requires that amino acids in addition to cysteine be alkylated at room temperature and near physiologic pH. Although we have not identified the modified amino acids or their locations within k8/18, previous characterization of SM reactivity indicates aspartic, glutamic [23, 24] or histidine [26, 27] side chains are modified by CEES and MEC, producing the effects shown in figures 2 and 3. Native keratins isolated from bovine snout are undoubtedly modified at the same positions, as well as endogenous cysteines.
Figure 3. Sulfhydryl modification of bovine keratins.
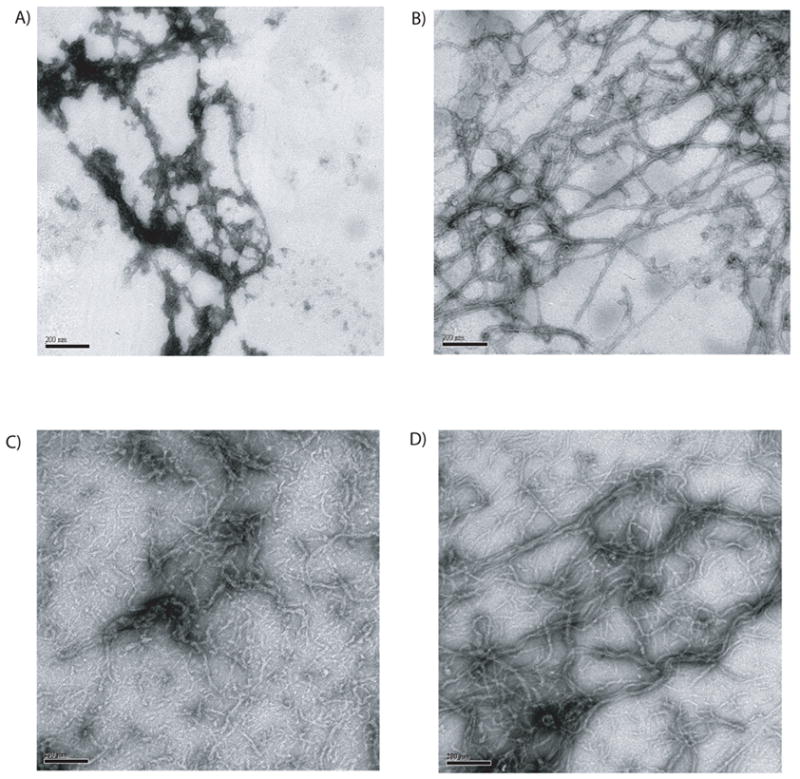
A pool of k1/10 and 5/14, isolated from bovine snout, were treated with the sulfhydryl specific reagent CE-MTS. Panel A shows a representative view of kIFs assembled from the modified keratin proteins. Panel B shows CE-MTS treated proteins kIFs assembled in the presence of DTT. Panel C shows kIFs assembled from a 1:1 mixture of CE-MTS treated and untreated bovine keratins. Panel D is a 90:10 mixture of unmodified keratins to modified keratins, respectively. The scale bar located in the bottom of each panel represents 200 nm.
To investigate the sensitivity of cysteine side chains to modification and subsequent interference with kIF assembly, we used the sulfhydryl reactive reagent 2-chloroethyl-MTS to modify k5/k14 and k1/k10 cysteines. Treatment of cysteine containing proteins with this reagent results in the modification of the cysteine with a CE (chloro-ethyl) group attached. Figure 3 shows a series of electron micrographs depicting the changes to k5/14 and k1/10 keratin filaments produced by modification with CE-MTS. Panel A shows keratin filaments assembled from CE-MTS modified keratin proteins assembled in the absence of DTT; protein aggregates decorate a filamentous backbone.
The modification of proteins with the sulfhydryl specific reagent CE-MTS is easily reversible; simple dialysis of modified proteins in the presence of DTT results in removal of the CE group. In the presence of DTT, and therefore with all modifications removed, CE-MTS treated keratins formed long filaments after dialysis (panel B). Thus, CE-MTS modification of keratins introduces sufficient change to interfere with normal assembly, but removal of the modification restores the ability of the keratins to assemble.
An additional experiment was performed to determine whether modified keratins acted as dominant negatives when mixed with unmodified proteins. Unmodified keratins produce a thick mass of long keratin filaments. Inclusion of an equal mass of CE-MTS modified keratins produces very few long filaments and yields mainly incomplete filaments with few identifiable normal looking filaments (figure 3C). Mixing unmodified keratins with CE-MTS modified keratins at a 9:1 ratio, also produces filament abnormalities (figure 3D). Extrapolating our results to the in vivo situation, the presence of SM modified keratins would be predicted to interfere with normal IF biogenesis until the almost all of the modified keratin protein has been removed.
Our results show that CEES, MEC and CE-MTS are each capable of damaging filaments. Extrapolating from our in vitro results to proteins modified in vivo by exposure to SM, we believe that protein modification by vesicants would produce IF aggregation, as demonstrated by Dillman et al., resulting in filament network collapse with subsequent cell lysis and skin blistering. Our results leave open the possibility that SM destruction of the kIF network is the mechanism behind the vesicant action of SM. The concentration of CEES and MEC we have used is higher than used in some tissue culture models [11, 16, 17] but it is similar to the concentrations used in live animal studies [13, 14, 24, 25, 29–31].
The specific, limited and reversible modification of keratins using CE-MTS reveals that a relatively small number of modifications interfers with proper filament assembly. Keratin 5 and k14 each contain 4 cysteine residues (<1%), 2 each in the head and tail domains. Keratin proteins modified by this reagent are incapable of assembling into native looking filaments. These results are consistent with experiments published by Steinert and Parry who found that modification of keratin 1 or 10 by iodoacetate prevented filament assembly [32]. Extrapolation of our results to in vivo conditions leads to the conclusion that a minor level of SM modified proteins could have a very deleterious effect on the keratinocyte IF network, results that are implied by the data of Dillman et al [17].
Dillman et al have characterized keratin proteins isolated from keratinocytes treated with SM and report that basal cell keratins k5 and k14 are cross-linked into both homodimers and heterodimers. However, whether the cross-linking produces any changes to the filament network, or filaments themselves was not examined. In light of the previous work performed by IF researchers, demonstrating the ability of cross-linked proteins to form bona-fide IFs (see figure 9 of Coulombe and Fuchs, figure 1 of Geisler et al or figure 10 of Quinlan et al, [33–36], the simple observation of protein cross-linking does not justify the conclusion that the keratinocyte network must be collapsed.
Our experiments demonstrate that IFs are resistant, but not immune, to destruction by chemical modification. Both generalized modification by SM analogs and limited numbers of modifications produced by cysteine specific probes can have a profound effect on IFs. By comparison, in vivo, a single amino acid change within k5 or k14 is responsible for severe skin blistering. Thus, our experiments leave open the possibility that modification of keratin filaments and protein subunits are is responsible for SM induced blister formation as a result of kIF aggregation and IF network collapse.
Materials and Methods
CEES and MEC were purchased from Aldrich and stored at room temperature. Prior to use, each was diluted in DMSO (Aldrich) or 100% ethanol. cDNA clones for human k8 and k18 were provided by Bishr Omary (Stanford University, Palo Alto, CA). K8 and k18 cDNA sequences were inserted into pT7-7 for bacterial expression [37]. Bacterially expressed k8 or k18 readily formed inclusion bodies and these were purified from bacterial cell pellets [38, 39] Briefly, inclusion bodies were dissolved in buffer A (10 mM Tris, pH 8.0, 1 mM EDTA) plus 8 M urea and purified by gel filtration using a Pharmacia FPLC system. Native bovine keratins 5/14 and 1/10 were isolated from bovine snout using a protocol provided by Roy Quinlan (University of Durham, UK) followed by gel filtration chromatography. Proteins were quantified by BCA assay (Pierce, Rockford, IL) using a BSA standard and stored at −80°C.
Treatment of filaments was performed by adding 1 microliter of the stock CEES or MEC (1M or 100mM) reagent solution to 99 microliters of protein solution. Solvent controls were performed analogously. Treatments were performed for one hour in a laboratory fume hood. Following treatment, samples were spotted onto EM grids and negatively stained with 10% aqueous uranyl acetate; following drying, the grids were removed from the hood. Electron microscopy was performed as described by Quinlan and coworkers [28].
Chloroethyl-MTS (chlororethyl-methanethiosulfonate, Toronto Research Chemicals, Toronto, Canada) treatment of k5/14 and 1/10 was performed by treating proteins with 100 micromolar TCEP to reduce cystines followed by incubation with CE-MTS (1mM CE-MTS final concentration). These conditions have been found to result in ~100% spin labeling of intermediate filament proteins in similar reactions [38, 40–42]. Following an 1 hour incubation, samples were dialyzed against filament assembly buffers either with or without DTT.
Filaments were assembled using a multistep dialysis procedure. Purified keratin subunits in 8 M urea plus buffer A (10mM tris, pH 8.0, 1mM EDTA) were mixed, placed into dialysis tubing (SpectraPor, regenerated cellulose, 10K cutoff) and dialyzed against fresh buffer A plus 8 M urea (1 hour), followed by buffer A plus 4M urea (2 hours), then 10 mM Tris pH 8.0 (2 hours), 10 mM tris pH 7.0, 1 mM MgCl2 (2 hours), and finally, 10 mM Tris pH 7.0, 1 mM MgCl2, 50 mM NaCl (overnight).
Supplementary Material
Figure 1. Protein purification and filament assembly controls.
A. Keratin 8 and 18 proteins were purified using a bacterial expression system followed by chromatography. Bovine keratins were purified from bovine snout by gel filtration chromatography. Approximately 10 micrograms of each protein pool was electrophoresed on a SDS-PAGE and proteins visualized by staining with Coomassie blue. Lane 1, recombinant k8; lane 2, recombinant k18; lane 3, purified bovine keratins, lane 4, protein standards (Invitrogen Benchmark ladder).
B. Recombinant k8 and k18 IFs assembled and viewed in the EM as described in materials and methods.
C. Bovine keratins assembled into kIFs, then prepared and viewed in the EM (electron microscope). Scale bar is 200 nm.
D. Recombinant kIFs treated with 1% ethanol, viewed in EM.
E. Recombinant kIFs treated with 10% ethanol, viewed in EM.
Figure 2. Treatment of keratin IFs with 1 mM CEES or 1mM MEC.
A. Keratin IFs treated with 1mM CEES for 1 hour followed by visualization in the EM.
B. Keratin IFs treated with 1mM MEC for 1 hour followed by visualization in the EM. Asterisks mark sites of protein disassembly.
Acknowledgments
This work was made possible by funding from the US Army, Grant DAMD 17-02-1-0664 to JFH and the NIH R01 NEI EY08747 to PGF. Thanks to Joshua Pittenger and Gordon Lee for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dacre JC, Goldman M. Toxicology and pharmacology of the chemical warfare agent sulfur mustard. Pharmacol Rev. 1996;48:289–326. [PubMed] [Google Scholar]
- 2.Momeni AZ, Enshaeih S, Meghdadi M, Amindjavaheri M. Skin manifestations of mustard gas. A clinical study of 535 patients exposed to mustard gas. Arch Dermatol. 1992;128:775–780. see comments. [PubMed] [Google Scholar]
- 3.Smith KJ, Hurst CG, Moeller RB, Skelton HG, Sidell FR. Sulfur mustard: its continuing threat as a chemical warfare agent, the cutaneous lesions induced, progress in understanding its mechanism of action, its long-term health effects, and new developments for protection and therapy. J Am Acad Dermatol. 1995;32:765–776. doi: 10.1016/0190-9622(95)91457-9. [DOI] [PubMed] [Google Scholar]
- 4.Requena L, Requena C, Sanchez M, Jaqueti G, Aguilar A, Sanchez-Yus E, Hernandez-Moro B. Chemical warfare. Cutaneous lesions from mustard gas. J Am Acad Dermatol. 1988;19:529–536. doi: 10.1016/s0190-9622(88)70208-x. [DOI] [PubMed] [Google Scholar]
- 5.Wormser U. Toxicology of mustard gas. Trends Pharmacol Sci. 1991;12:164–167. doi: 10.1016/0165-6147(91)90534-y. [DOI] [PubMed] [Google Scholar]
- 6.Smith WJ, Dunn MA. Medical defense against blistering chemical warfare agents. Arch Dermatol. 1991;127:1207–1213. [PubMed] [Google Scholar]
- 7.Smith WJ, Cowan FM, Broomfield CA. increased proteolytic activity in human epithelial cells following exposure to sulfur mustard. Faseb J. 1991;5:828. [Google Scholar]
- 8.Cowan FM, Broomfield CA, Smith WJ. Effect of sulfur exposure on protease activity in human peripheral blood lymphocytes. Cell Biol Toxicol. 1991;7:239–248. doi: 10.1007/BF00250978. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti AK, Ray P, Broomfield CA, Ray R. Purification and characterization of protease activated by sulfur mustard in normal human epidermal keratinocytes. Biochem Pharmacol. 1998;56:467–472. doi: 10.1016/s0006-2952(98)00160-9. [DOI] [PubMed] [Google Scholar]
- 10.Shakarjian MP, Bhatt P, Gordon MK, Chang YC, Casbohm SL, Rudge TL, Kiser RC, Sabourin CL, Casillas RP, Ohman-Strickland P, Riley DJ, Gerecke DR. Preferential expression of matrix metalloproteinase-9 in mouse skin after sulfur mustard exposure. J Appl Toxicol. 2006 doi: 10.1002/jat.1134. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti AK, Ray P. Novel endogenous inhibitor of sulfur mustard-stimulated protease in cultured human epidermal keratinocytes: possible application in vesicant intervention. J Appl Toxicol. 2000;20(Suppl 1):S59–61. doi: 10.1002/1099-1263(200012)20:1+<::aid-jat688>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Ray P, Chakrabarti AK, Broomfield CA, Ray R. Sulfur mustard-stimulated protease: a target for antivesicant drugs. J Appl Toxicol. 2002;22:139–140. doi: 10.1002/jat.829. [DOI] [PubMed] [Google Scholar]
- 13.Monteiro-Riviere NA, Inman AO. Ultrastructural characterization of sulfur mustard-induced vesication in isolated perfused porcine skin. Microsc Res Tech. 1997;37:229–241. doi: 10.1002/(SICI)1097-0029(19970501)37:3<229::AID-JEMT8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Monteiro-Riviere NA, Inman AO, Babin MC, Casillas RP. Immunohistochemical characterization of the basement membrane epitopes in bis(2-chloroethyl) sulfide-induced toxicity in mouse ear skin. J Appl Toxicol. 1999;19:313–328. doi: 10.1002/(sici)1099-1263(199909/10)19:5<313::aid-jat582>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Werrlein RJ, Madren-Whalley JS. Multiphoton microscopy: an optical approach to understanding and resolving sulfur mustard lesions. J Biomed Opt. 2003;8:396–409. doi: 10.1117/1.1584687. [DOI] [PubMed] [Google Scholar]
- 16.Werrlein RJ, Madren-Whalley JS. Effects of sulfur mustard on the basal cell adhesion complex. J Appl Toxicol. 2000;20(Suppl 1):S115–123. doi: 10.1002/1099-1263(200012)20:1+<::aid-jat682>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 17.Dillman JF, 3rd, McGary KL, Schlager JJ. Sulfur mustard induces the formation of keratin aggregates in human epidermal keratinocytes. Toxicol Appl Pharmacol. 2003;193:228–236. doi: 10.1016/j.taap.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Peters BP, Monteiro-Riviere NA. Assessment of sulfur mustard interaction with basement membrane components. Cell Biol Toxicol. 1995;11:89–101. doi: 10.1007/BF00767494. [DOI] [PubMed] [Google Scholar]
- 19.Irvine AD, McLean WHI. Human keratin diseases: The increasing spectrum of disease and subtlety of the phenotype-genotype correlation. British Journal of Dermatology. 1999;140:815–828. doi: 10.1046/j.1365-2133.1999.02810.x. [DOI] [PubMed] [Google Scholar]
- 20.Ross WCJ. Biological Alkylating Agents. Butterworths; London: 1962. [Google Scholar]
- 21.Wheeler GP. Studies related to the mechanisms of action of cytotoxic alkylating agents: a review. Cancer Res. 1962;22:651–688. [PubMed] [Google Scholar]
- 22.Byrne MP, Broomfield CA, Stites WE. Mustard gas crosslinking of proteins through preferential alkylation of cysteines. J Protein Chem. 1996;15:131–136. doi: 10.1007/BF01887394. [DOI] [PubMed] [Google Scholar]
- 23.Goodlad GA. Esterification of protein and amino acid carboxyl groups by mustard gas and related compounds. Biochim Biophys Acta. 1957;24:645–646. doi: 10.1016/0006-3002(57)90264-0. [DOI] [PubMed] [Google Scholar]
- 24.Moore S, Stein WH, Fruton JS. Chemical reactions of mustard gas and related counds. II. The reaction of mustard gas with carboxy groups and with the amino groups of amino acis and peptides. J Org Chem. 1946;11:675–680. doi: 10.1021/jo01176a008. [DOI] [PubMed] [Google Scholar]
- 25.Herriott RM, Anson ML, Northrup JH. Reaction of enzymes and proteins with mustard gas. J Gen Physio. 1946;30:185–210. doi: 10.1085/jgp.30.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis SB, Ross WF. The reaction of mustard gas with proteins. J Am Chem Soc. 1947;69:1177–1185. doi: 10.1021/ja01197a056. [DOI] [PubMed] [Google Scholar]
- 27.Noort D, Verheij ER, Hulst AG, de Jong LP, Benschop HP. Characterization of sulfur mustard induced structural modifications in human hemoglobin by liquid chromatography--tandem mass spectrometry. Chem Res Toxicol. 1996;9:781–787. doi: 10.1021/tx9502148. [DOI] [PubMed] [Google Scholar]
- 28.Carter JM, Hutcheson AM, Quinlan RA. In vitro studies on the assembly properties of the lens proteins CP49, CP115: coassembly with alpha-crystallin but not with vimentin. Experimental Eye Research. 1995;60:181–192. doi: 10.1016/s0014-4835(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 29.Baxter MA, Chahwala SB, Hickman JA, Spurgin GE. The effects of nitrogen mustard (HN2) on activities of the plasma membrane of PC6A mouse plasmacytoma cells. Biochem Pharmacol. 1982;31:1773–1778. doi: 10.1016/0006-2952(82)90683-9. [DOI] [PubMed] [Google Scholar]
- 30.Kim YB, Lee YS, Choi DS, Cha SH, Sok DE. Inactivation of microsomal Ca(2+)-ATPase by 2-chloroethylethyl sulfide. Chem Biol Interact. 1995;97:239–246. doi: 10.1016/0009-2797(95)03619-w. [DOI] [PubMed] [Google Scholar]
- 31.van der Schans GP, Noort D, Mars-Groenendijk RH, Fidder A, Chau LF, de Jong LP, Benschop HP. Immunochemical detection of sulfur mustard adducts with keratins in the stratum corneum of human skin. Chem Res Toxicol. 2002;15:21–25. doi: 10.1021/tx0100136. [DOI] [PubMed] [Google Scholar]
- 32.Steinert PM, Parry DA. The conserved H1 domain of the type II keratin 1 chain plays an essential role in the alignment of nearest neighbor molecules in mouse and human keratin 1/keratin 10 intermediate filaments at the two- to four-molecule level of structure. J Biol Chem. 1993;268:2878–2887. [PubMed] [Google Scholar]
- 33.Coulombe PA, Fuchs E. Elucidating the early stages of keratin filament assembly. J Cell Biol. 1990;111:153–169. doi: 10.1083/jcb.111.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers KR, Herrmann H, Franke WW. Characterization of disulfide crosslink formation of human vimentin at the dimer, tetramer, and intermediate filament levels. Journal of Structural Biology. 1996;117:55–69. doi: 10.1006/jsbi.1996.0069. [DOI] [PubMed] [Google Scholar]
- 35.Geisler N, Schunemann J, Weber K. Chemical crosslinking indicates a staggered and antiparellel protofilament of desmin intermediate filaments and characterizes one higher level complex between protofilaments. Eur J Biochemistry. 1992;206:841–852. doi: 10.1111/j.1432-1033.1992.tb16992.x. [DOI] [PubMed] [Google Scholar]
- 36.Quinlan RA, Cohlberg JA, Schiller DL, Hatzfeld M, Franke WW. Heterotypic tetramer (A2D2) complexes of non-epidermal keratins isolated from cytoskeletons of rat hepatocytes and hepatoma cells. J Mol Biol. 1984;178:365–388. doi: 10.1016/0022-2836(84)90149-9. [DOI] [PubMed] [Google Scholar]
- 37.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess JF, Voss JC, FitzGerald PG. Real-time observation of coiled-coil domains and subunit assembly in intermediate filaments. J Biol Chem. 2002;277:35516–35522. doi: 10.1074/jbc.M206500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai K, Thøgersen HC. Synthesis and sequence-specific proteolysis of hybrid proteins produced in Escherichia coli. Methods in Enzymology. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 40.Hess JF, Budamagunta MS, Shipman RL, FitzGerald PG, Voss JC. Characterization of the linker 2 region in human vimentin using site-directed spin labeling and electron paramagnetic resonance. Biochemistry. 2006;45:11737–11743. doi: 10.1021/bi060741y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess JF, Budamagunta MS, Fitzgerald PG, Voss JC. Characterization of Structural Changes in Vimentin Bearing an Epidermolysis Bullosa Simplex-like Mutation Using Site-directed Spin Labeling and Electron Paramagnetic Resonance. J Biol Chem. 2005;280:2141–2146. doi: 10.1074/jbc.M412254200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hess JF, Budamagunta MS, Voss JC, Fitzgerald PG. Structural Characterization of Human Vimentin Rod 1 and the Sequencing of Assembly Steps in Intermediate Filament Formation in Vitro Using Site-directed Spin Labeling and Electron Paramagnetic Resonance. J Biol Chem. 2004;279:44841–44846. doi: 10.1074/jbc.M406257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. Protein purification and filament assembly controls.
A. Keratin 8 and 18 proteins were purified using a bacterial expression system followed by chromatography. Bovine keratins were purified from bovine snout by gel filtration chromatography. Approximately 10 micrograms of each protein pool was electrophoresed on a SDS-PAGE and proteins visualized by staining with Coomassie blue. Lane 1, recombinant k8; lane 2, recombinant k18; lane 3, purified bovine keratins, lane 4, protein standards (Invitrogen Benchmark ladder).
B. Recombinant k8 and k18 IFs assembled and viewed in the EM as described in materials and methods.
C. Bovine keratins assembled into kIFs, then prepared and viewed in the EM (electron microscope). Scale bar is 200 nm.
D. Recombinant kIFs treated with 1% ethanol, viewed in EM.
E. Recombinant kIFs treated with 10% ethanol, viewed in EM.
Figure 2. Treatment of keratin IFs with 1 mM CEES or 1mM MEC.
A. Keratin IFs treated with 1mM CEES for 1 hour followed by visualization in the EM.
B. Keratin IFs treated with 1mM MEC for 1 hour followed by visualization in the EM. Asterisks mark sites of protein disassembly.


